Abstract
In Schizosaccharomyces pombe, heterochromatin spread, which is marked by histone H3 lysine 9 methylation (H3K9me), requires the chromodomains (CD) of the H3K9 methylase Suv39/Clr4 and the HP1/Swi6 protein. It is unclear how the actions of these two H3K9 methylation recognizing CDs are coordinated. We find that the intrinsic preference of Suv39/Clr4 is to generate dimethylated H3K9 product. Recognition of pre-existing H3K9me marks by the CD of Suv39/Clr4 stimulates overall catalysis, enabling accumulation of small amounts of trimethylated product in vivo. Coincidentally, the Suv39/Clr4 CD, unlike the HP1/Swi6 CD, has been shown to prefer the trimethyl state over dimethyl. We show that this preference enables efficient heterochromatin spread in vivo by reducing competition with HP1 proteins for the more prevalent dimethyl state. Our results reveal a strategy by which writers and readers of a chromatin mark exploit different methylation states on the same residue to facilitate collaboration and avoid competition.
INTRODUCTION
Constitutive heterochromatin is required for long-term silencing of large stretches of the genome as well as for its structural integrity. This type of heterochromatin is signaled by methylation at lysine 9 of histone 3 (H3K9) and is conserved from Schizosaccharomyces pombe (S. pombe) to humans (Elgin and Grewal, 2003). The capacity of H3K9 methylation (H3K9me) and its associated silencing functions to initiate at specific DNA sequences and then spread beyond initiation sites in a sequence-indifferent manner is key to the normal function of constitutive heterochromatin (Grewal and Jia, 2007). Moreover, such spreading behavior can lead to silencing of tumor suppressor genes and contribute to malignancy when heterochromatin is ectopically initiated in disease states (Bhalla, 2005; Carbone et al., 2006; Ceol et al., 2011; Reed-Inderbitzin et al., 2006).
The regulation of heterochromatin formation can be divided into initiation and spread steps. Initiation requires the recruitment of the SET-domain H3K9 methylase (“writer”) enzyme to an initiation site. The initiation process has been extensively studied in S. pombe, where the H3K9me writer Suv39/Clr4 (Rea et al., 2000) exists in a protein complex called CLRC (Hong et al., 2005; Horn et al., 2005; Jia et al., 2005). Qualitatively distinct initiation mechanisms recruit either the Suv39/Clr4 methylase or the non-catalytic subunits of CLRC to trigger methylation (Buhler et al., 2007; Jia et al., 2004; Zofall et al., 2012). Following initiation, H3K9 methylation spreads via the combined action of the methylase and the two HP1 proteins, Swi6 and Chp2, and its extent is contained in part by the action of boundary elements (Noma et al., 2001).
Despite the central biological roles played by heterochromatin, the molecular mechanisms underlying its assembly and spread have remained opaque. One reason for the lack of mechanistic understanding is the combinatorial complexity of the process. H3K9 methylation adopts three distinct states: H3K9 monomethylation (H3K9me1), dimethylation (H3K9me2), and trimethylation (H3K9me3), which can have distinct biological outputs. In S. pombe, Suv39/Clr4 is the sole H3K9me writer, yet all three methylation states of H3K9 have been documented (Yamada et al., 2005). Therefore, the observed distribution of the H3K9 methyl states either reflects the intrinsic activity of Suv39/Clr4 or is additionally regulated by other factors acting on this enzyme or its products. In metazoans, in contrast, different methylation states are achieved with different enzymes that can co-operate to generate specific H3K9 methylation states (Cheutin et al., 2004; Fritsch et al., 2010; Peters et al., 2003; Rice et al., 2003; Tachibana et al., 2005). Adding to the combinatorial complexity is the participation of several heterochromatin proteins that recognize the H3K9me mark via a chromodomain (CD). The CDs of the HP1 proteins, Swi6 and Chp2, recognize the H3K9me mark and coat heterochromatin to generate a platform that recruits diverse silencing regulators. Additionally, Swi6 is required for heterochromatin spread. At the same time, the N-terminal chromodomain (CD) of Suv39/Clr4 is also required for efficient spread of H3K9 methylation and concomitant spread of CLRC (Noma et al., 2004; Zhang et al., 2008). This requirement has given rise to a feedback hypothesis whereby recognition of the Suv39/Clr4 methylation product by its CD promotes further deposition of H3K9 methylation.
These observations raise the following questions: 1) What is the mechanism of the positive feedback from the reaction product and 2) How are the activities of the writer and reader CDs coordinated to allow efficient spread rather than non-productive competition?
Using a combination of biochemical dissection, a new method to compare relative methylation levels at specific loci in vivo (Q-ChIP), and genetic approaches, we have uncovered a basic regulatory circuit that is coded within the intrinsic properties of Suv39/Clr4 and HP1/Swi6. We find that positive feedback from H3K9 methylation enhances catalysis by Suv39/Clr4 in vitro and is required for accumulation of the H3K9 trimethyl state in vivo. We further find that the previously documented selectivity of the Suv39/Clr4 CD for this modification state allows efficient heterochromatin spread by avoiding competition with HP1 proteins, which do not significantly distinguish between H3K9 trimethyl and the more prevalent dimethyl states.
Our data uncover a division of labor strategy in heterochromatin formation wherein the recognition modules of “writer” and “reader” proteins are tuned to recognize distinct modification states of the heterochromatic mark, enabling productive collaboration between the two proteins.
RESULTS
To test proposals for the role of Suv39/Clr4 in the spread of H3K9 methylation, we first biochemically reconstituted an elemental reaction: H3K9 methylation across an asymmetric dinucleosome with one nucleosome containing pre-existing H3K9 methylation. Then, to study the cellular implications of the biochemical parameters identified by this approach, we developed a novel quantitative chromatin immunoprecipitation (ChIP) procedure, termed Q-ChIP, that utilizes external calibration with recombinant modified nucleosome standards to directly compare the amounts of H3K9me1, me2 and me3 formed in vivo. Using these approaches we then investigated the impact of CD specificity on spreading in vivo.
An H3K9me3 mark stimulates Suv39/Clr4 catalysis, but not binding, on an adjacent nucleosome
We first characterized the activity of Suv39/Clr4 on a mononucleosome compared to H31-20 peptide substrates using a fully recombinant system in which all components were bacterially expressed and purified. All reactions were performed under saturating concentrations of S-adenosyl methionine (Supplementary Figure 2A). We found that Suv39/Clr4 displays increased catalytic specificity for nucleosomal substrates over tail peptides. This specificity likely derives from a decreased KM in the context of the nucleosome (Supplementary Figure 1 and Supplementary experimental procedures). We then directly tested the prevalent hypothesis that pre-existing H3K9me marks enhance the activity of Suv39/Clr4 on chromatin by using a di-nucleosome template (Figure 1A). The dinucleosome comprises one substrate nucleosome (N1) and one effector nucleosome (N2), which is either methylated at H3K9 using Methyl-Lysine Analog (MLA) technology (Simon et al., 2007) or contains a non-methylatable arginine at this position (K9R, Supplementary Figure 2B). MLA substrates differ from unmodified substrates in harboring sulfur rather than carbon in the γ position of the lysine side chain (denoted as H3Kc9). This alteration can significantly affect the affinity of some CD-containing proteins (Munari et al., 2012; Seeliger et al., 2012). To assess whether the Suv39/Clr4 CD can efficiently recognize an MLA at H3K9, we compared the ability of H3Kc9me3 and H3K9me3 peptides to compete off fluorescently labeled H3K9me3 peptide bound to the Suv39/Clr4-CD. We find that the competitive abilities of H3Kc9me3 and H3K9me3 peptides are very similar, with their apparent KI differing only by ~1.6 fold (Supplementary Figure 3A). These results implied that the CD of Suv39/Clr4 recognizes an MLA at H3K9 with similar efficacy as a bona-fide methyl mark.
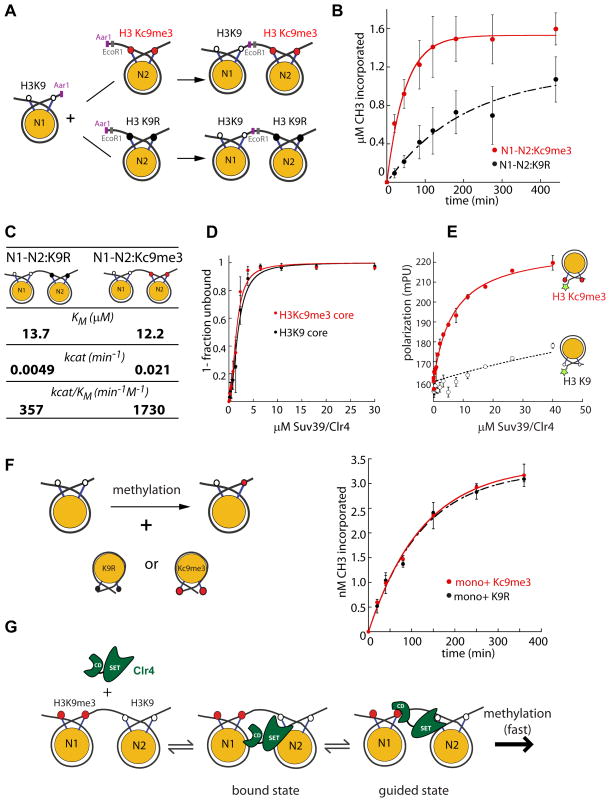
Figure 1. H3K9me3 raises the kcat of Suv39/Clr4 only when present in cis in a dinucleosome context.
A. Scheme for the production of asymmetric dinucleosomes.
B. Dinucleosome methylation time courses at saturating (20μM) Suv39/Clr4. 100nM N1-N2:K9R (black) or N1-N2:Kc9me3 dinucleosomes (red) were used as substrates. The kcat values for Suv39/Clr4 for N1-N2:K9R and N1-N2:Kc9me3 were 0.0049min−1 and 0.021min−1, respectively. The kcat was derived from normalized data points from 4 separate experiments. Error bars denote standard error of five repeats.
C. Enzymological parameters for Suv39/Clr4 activity on N1-N2:K9R or N1-N2:Kc9me3 dinucleosomes.
D. Electrophoretic mobility shift experiments with H3K9 (black circles) and H3Kc9me3 (red circles) core mononucleosomes to determine the overall binding K1/2. The K1/2 of Suv39/Clr4 for H3K9 and H3Kc9me3 core mononucleosomes was 1.8μM and 1.5μM, respectively. Error bars denote standard error of three repeats.
E. Fluorescence polarization (FP) on nucleosomes. H3K9 (open black circles) and H3Kc9me3 (red circles) core mononucleosomes were fluorescently labeled at the 5′ end of the 147-bp positioning sequence. FP detected by this label is sensitive to local binding events, such as at the nearby H3 tail, as previously described (Canzio et al.,2011). The Suv39/Clr4 dependent FP increase at this position is at least 25- fold more sensitive to the presence of the H3Kc9me3 mark over the H3K9 control.
In this panel the bars shown do not denote standard error, but instead show the variation between two independent repeats.
F. Methylation of mononucleosomes in presence of effector nucleosomes in trans. LEFT: Experimental design.
RIGHT: Methylation time courses at 2μM Suv39/Clr4 (below the KM for di- and mononucleosomes). 300nM wild-type core mononucleosomes were used as substrates. K9R (black) or Kc9me3 core mononucleosomes (red) were added as effector in trans at 10μM. In this panel the bars shown do not denote standard error, but instead show the variation between two independent repeats.
G. Model for role of the Suv39/Clr4 CD in catalytic enhancement of H3K9 methylation on chromatin. Formation of the enzyme-substrate ground state complex (bound state) is independent of pre-existing H3K9 methyl marks. Catalysis requires formation of a guided state, which optimally positions the active site relative to the H3K9 substrate. A pre-existing H3K9 methyl mark stabilizes the guided state. The absence of a pre-existing H3K9 methyl mark, slows catalysis as formation of the guided state is destabilized.
We followed overall methyl incorporation using a tritium (3H) labeled S-adenosyl methionine (SAM) methyl-donor tracer with saturating total SAM (Supplementary Figure 2A). By measuring methylation at subsaturating concentrations of Suv39/Clr4, we obtained values for the specificity constant, kcat/KM. The presence of a methyl mark on the adjacent nucleosome increased kcat/KM by ~ 5-fold (Figure 1C). This result validates the basic premise of the positive feedback model proposed for Suv39/Clr4 action (Elgin and Grewal, 2003; Hall et al., 2002; Zhang et al., 2008).
The increase in kcat/KM could reflect increased binding to the template or increased catalysis. If the increase is due to stronger binding of Suv39/Clr4 to the pre-methylated template, then kcat would be the same for both substrates and the KM value would be lower for the pre-methylated substrate. We found that kcat is ~4.3-fold faster for N1-N2MLA (0.021min−1), as compared to N1-N2K9R (0.0049min−1) (Figure 1B and C) and recapitulates most of the stimulatory effect seen for kcat/KM. This suggests that the pre-methylated template increases catalysis, rather than binding, by the methylase. This conclusion was supported by our observation of similar KM values for the two dinucleosome substrates (Figure 1C and data not shown). Consistent with this conclusion, we found that Suv39/Clr4 binds H3Kc9me3 and H3K9 mononucleosomes with similar affinities when measured by electrophoretic mobility shift assays (Figure 1D; K1/2 = 1.8μM &1.5μM for H3K9 and H3Kc9me3 nucleosomes, respectively). When binding is detected specifically proximal to the H3 tail using a fluorescence polarization assay, however, we find that the Suv39/Clr4-CD is capable of recognizing H3K9 methylation on the mononucleosome (Figure 1E). We interpret these results to suggest that, similar to HP1/Swi6 (Canzio et al.,2011), Suv39/Clr4 interacts with the nucleosome in binding modes specific for H3K9 methylation as well as alternative modes where the CD does not engage the methyl mark. The lack for obvious binding preference for methylated versus unmethylated nucleosomes when all binding events are measured indicates that alternative binding modes must dominate ground state interactions of Suv39/Clr4 and the nucleosome. Together, these results indicate that the increased specificity for N1-N2MLA dinucleosomes derives primarily from a post-binding catalytic effect. To determine whether this effect in fact derives from CD engagement with MLA-installed H3K9 methylation, we tested the impact of mutating the Suv39/Clr4 CD on catalysis. We find that the catalytic effect of H3Kc9me3 is indeed dependent on a functional Suv39/Clr4 CD (Supplementary Figure 4C).
In principle, two models could account for the observed H3K9me3-dependent increase of the kcat. 1) An allosteric model. In this model, when the Suv39/Clr4 CD is engaged at an H3K9me3 mark, it induces an allosteric change in the active site, directly increasing catalysis. 2) A “guided state” model. In this model, following initial substrate binding, the CD-H3K9me3 interaction guides the Suv39/Clr4 active site so that it is properly oriented with respect to the substrate H3 tail. The allosteric model most simply predicts that the active site will be stimulated when the H3K9me3 mark is both in cis as well as in trans (not linked on the same substrate). In contrast, a guided state model would require the methyl mark to be present in cis. To test these predictions, we asked whether an H3Kc9me3 modified effector mononucleosome can stimulate methylation of a substrate mononucleosome in trans in a methylation sensitive manner. We performed these experiments at sub-saturating concentrations of Suv39/Clr4 (Supplementary Figure 1C, D) and with effector nucleosome concentrations above their K1/2 values for binding Suv39/Clr4 (Figure 1D). In contrast to the behavior in cis, H3Kc9me3 effector mononucleosomes added in trans do not show greater stimulation than H3K9R effector nucleosomes (Figure 1F). We obtained similar results using peptide substrates or peptide effectors (Supplementary Figure 2C and D).
The results from above experiments argue against a simple allosteric mechanism for stimulation by the product (model 1). Instead, our results suggest that the H3K9me3 product acts in cis in a step subsequent to enzyme–substrate complex formation to stabilize a “guided state” that orients the Suv39/Clr4 active site with respect to the adjacent H3K9 substrate tail (model 2, Figure 1G).
Suv39/Clr4 is a preferential H3K9 dimethylase of nucleosomal substrates
Unlike in metazoans, in S. pombe, all three H3K9 methylation states are produced by one enzyme (Yamada et al., 2005). The ability of Suv39/Clr4 to produce all three methylation states is in part predicted from sequence homology of key active site residues to those of other H3K9 methylation writers (Couture et al., 2008; Wu et al., 2010) and has been qualitatively demonstrated on peptide substrates (Dirk et al., 2007). However, the methylation state that is preferentially generated by Suv39/Clr4 on chromatin substrates is not known.
To determine the product distribution intrinsic to Suv39/Clr4, we measured the kinetics of H3K9 mono-, di-, and trimethylation on core mononucleosomes using saturating and excess Suv39/Clr4. We detected the H3K9me1, me2, and me3 states by Western blot using antibodies raised to recognize the specific methylation state. Traditional Western blot assays are limited in their quantitative resolution, hampering kinetic analysis. We therefore adapted a fluorescence-based system that allows measurement of methylation signals over a large dynamic range. The two fluorescence channels in this system allowed us to detect and quantify the H3K9me1, me2, or me3 signals and separately measure H4 levels for internally controlled normalization. As a standard for calibrating the Western signals, we used histone octamers uniformly modified to be mono-, di-, or trimethylated with Methyl-Lysine Analog (MLA) technology (Simon et al., 2007). The vendor-specified selectivity of antisera does not always match the observed specificity. We therefore experimentally quantified the selectivity of the three antisera used here (Supplementary Figure 3B). We found the anti-sera to be largely highly selective for the modification state they were raised against. In the case of the anti-H3K9me2 antisera, we quantify about 10% cross-reactivity with H3K9me3.
The rate constant for the transition of unmethylated to monomethylated nucleosomes is defined as k1, that from the mono- to dimethylated state is defined as k2, and that from the di- to trimethylated state is defined as k3 (Figure 2A, top panel). Qualitatively, we observe a rapid rise and fall of H3K9me1, a gradual rise of H3K9me2, and a very slow rise of H3K9me3 (Figure 2A, bottom panel). The lags evident in H3K9me2 and me3 are expected from the consecutive nature of the methylation reactions. It is technically difficult to extract k1, k2, and k3 from this type of time course data, as the parameters are underdetermined in this setting. To overcome this limitation, we used MLA technology to produce intermediates of the overall reaction and used them as substrates to extract k1, k2, and k3 for the sub-reactions in Figure 2A. To test the validity of using MLA substrates to mimic Suv39/Clr4 reaction intermediates, we compared the time-courses for conversion of H3Kc9 nucleosomes to H3Kc9me2 nucleosomes with those for conversion of H3K9 nucleosomes to H3K9me2 nucleosomes, under the same conditions. We find that the two time-courses essentially overlap (Supplementary Figure 3C), arguing that H3K9 and H3Kc9 nucleosomes are equally good methylation substrates for Suv39/Clr4 and validating the use of these substrates for extracting individual rate constants.
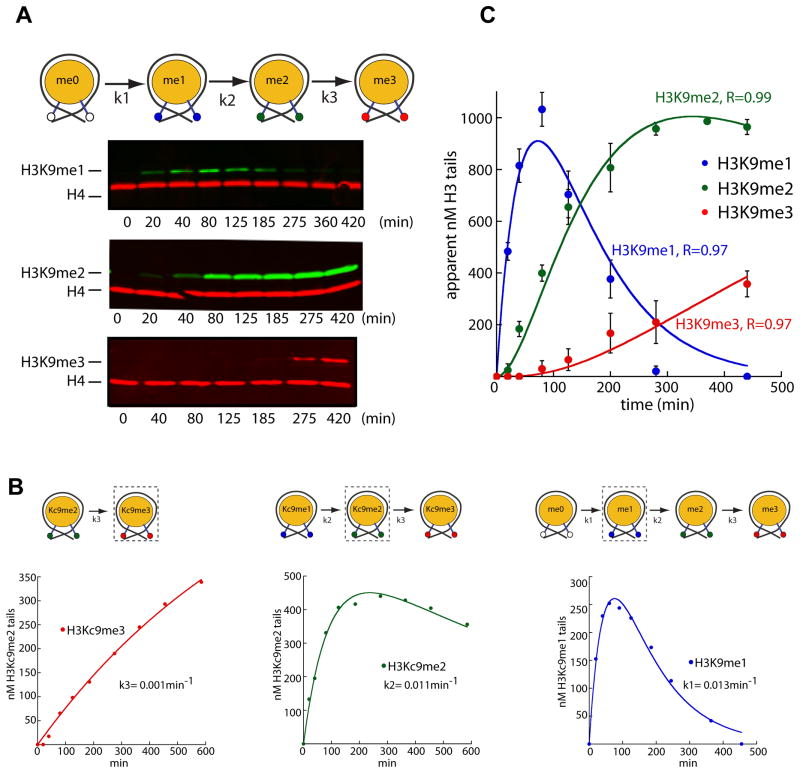
Figure 2. Suv39/Clr4 is primarily a H3K9 dimethylase on mononucleosomes.
A. TOP: H3K9 methylation by Suv39/Clr4 proceeds in a series of consecutive steps: monomethylation (k1), dimethylation (k2) and trimethylation (k3). BOTTOM: Quantitative western blots of H3K9me1, me2 and me3. Methylation reactions were performed at 20μM Suv39/Clr4 and 125nM core mononucleosomes. Reactions were stopped and separated by SDS-PAGE and probed for H4 (red) and H3K9me1 (green, top panel), H3K9me2 (green, middle panel) or H3K9me3 (red, bottom panel).
B. Determination of k1, k2 and k3. Methyl-Lysine Analog (MLA) core mononucleosomes representing intermediate steps of the consecutive reaction depicted in (A) were used as methylation substrates as in (A).
The concentration of methylated tails in nanomolar (nM) is derived from the H4-adjusted amount of H3K9me1, H3Kc9me2 or H3Kc9me3 at each time point as determined by H3Kc9me1, me2 or me3 standards.
C. Fitting the consecutive methylation reaction. k1, k2 and k3 as determined from (B) were used to model the formation of H3K9me1, me2 and me3 starting from unmethylated core mononucleosomes. The lines describing the model fit well to the data obtained from unmethylated H3K9 nucleosomes. H3K9me1, me2 and me3 quantities determined as in (B). Error bars denote standard error of three timecourses.
We first determined k3, measuring the rate of conversion of H3Kc9me2 to H3Kc9me3 nucleosomes using anti-H3K9me3 antisera. As this is a single conversion step, no lag is evident (Figure 2B, left panel) and the data can be fit to a single exponential, giving a k3 of 0.0011 min−1. This value is ~ 10-fold lower than the overall kcat. To extract k2, we measured the rate of H3Kc9me2 formation starting from H3Kc9me1 mononucleosomes. This allowed us to fit the data with a model containing the directly determined k3 value, yielding a k2 value of 0.011 min−1 (Figure 2B, middle panel). Finally, we determined k1 by following H3K9me1 formation using unmodified nucleosomes as the substrate. Fitting this data by a model containing the k2 value determined as described above, yielded a k1 value of 0.013 min−1 (Figure 2B, right panel). In an additional validation of the above approach, a model with the k1, k2, and k3 values fixed to the experimentally determined values recapitulates well the methylation time-course on unmodified nucleosomes (Figure 2C; R = 0.97,0.99 and 0.97 for mono-, di-, and trimethylation, respectively).
Suv39/Clr4 displays almost identical k1 and k2 values, indicating that it does not distinguish unmethylated and monomethylated chromatin substrates. In contrast, k3 is 10-fold lower than k1 and k2, indicating that dimethylated mononucleosomes are poor substrates for Suv39/Clr4. These results imply that the preferred product of Suv39/Clr4 is dimethylated H3K9.
H3K9me2 is the predominant methylation state in vivo
We next asked if the in vitro preference for dimethylation is reflected in vivo. TheH3K9 methylation state distribution can additionally be regulated by multiple factors in vivo, including, but not limited to, H3K9 demethylases. However, to date, the distribution of H3K9 methylation states at specific heterochromatin loci has not been quantified. The primary tool to detect different histone marks at specific loci is chromatin immunoprecipitation (ChIP) using highly specific ChIP antibodies (Peters et al., 2003). Since the immunoprecipitation efficiency of antibodies varies widely, the amounts of the different histone marks cannot be directly compared. To enable a direct quantitative comparison between different methylation states in vivo, we developed a new method to standardize the ChIP signals deriving from H3K9me1, me2, and me3 antibodies (Figure 3A, “Q-ChIP”). We calibrated the ChIP externally by adding defined concentrations of H3Kc9me1, me2, and me3 core mononucleosomes assembled on the “601” sequence to the individual ChIP reactions. These concentrations spanned a 100-fold range. Calculating the amount of “601” DNA in the ChIP reactions allowed us to extract the relative efficiency of each antibody and normalize the ChIP signals for the three H3K9 methylation states (Figure 3A).
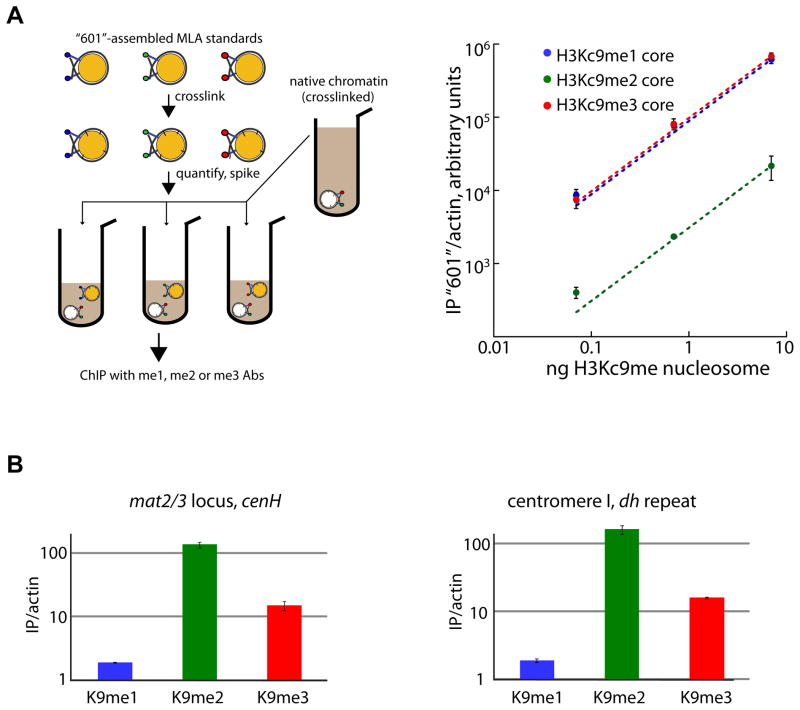
Figure 3. The dimethyl state is the predominant H3K9 methylation state in vivo alongside a small trimethyl pool.
A. The Q-ChIP method. LEFT: overview of the H3Kc9me nucleosome ChIP spiking scheme. RIGHT: Double log plot of amount of crosslinked H3Kc9me1, me2, or me3 core mononucleosomes spiked into ChIP reactions (x-axis), vs. amount of precipitated, actin-normalized 601 nucleosome DNA (y-axis) as quantified by RT-qPCR. Error bars denote standard error of three immunoprecipitations (IPs).
B. ChIP with anti-H3K9me1 (blue), anti-H3K9me2 (green) or anti-H3K9me3 (red) antisera at the mating type locus cenH element (LEFT) and the centromeric dh repeat (RIGHT). Enrichment is represented as the ratio of the actin-normalized signal in Suv39/Clr4 WT over the clr4Δ (no methylation) mutant. Relative H3K9me1, me2 and me3 signals are normalized by a parallel Q-ChIP reaction and shown on a log scale. Error bars denote standard error of three IPs.
Should the intrinsic catalytic capability of Suv39/Clr4 dominate the in vivo methylation profile, we would predict more H3K9me2 than H3K9me3 if the slower trimethylation reaction does not go all the way to completion. Using Q-ChIP at two primary heterochromatic loci, the centromeric dh repeat, and the cenH initiation site of the silent mating type (mat2/3) region, this is indeed what we observe: 1–2% H3K9me1, ~84–88% H3K9me2, and 10–15% H3K9me3 (Figure 3B and data not shown).
Product guidance is required for maintenance of H3K9me3 levels in vivo
We next asked how product guidance impacts the distribution of H3K9 methylation states produced by Suv39/Clr4. We adopted the quantitative Western blot approach shown in Figure 2 using saturating concentrations of enzyme. To avoid interference from the H3Kc9me3 mark in the westerns, we biotin-tagged the dinucleosome at the N1 end of the DNA, captured the N1-N2 substrate, and removed N2MLA or N2K9R by EcoRI cleavage prior to analysis by Western (Scheme Figure 4A). We observed that k1, k2, and k3 are all elevated on N1-N2MLA versus N1-N2K9R (Figure 4B). The comparable increase of all three rates results in more H3K9me3 being produced on N1-N2MLA versus N1-N2K9R at the later time points. Since H3K9 methylation is a series of consecutive, irreversible reactions, and given that k3 is the slowest rate, H3K9me3 levels are expected to be the most sensitive to disrupting product guidance.
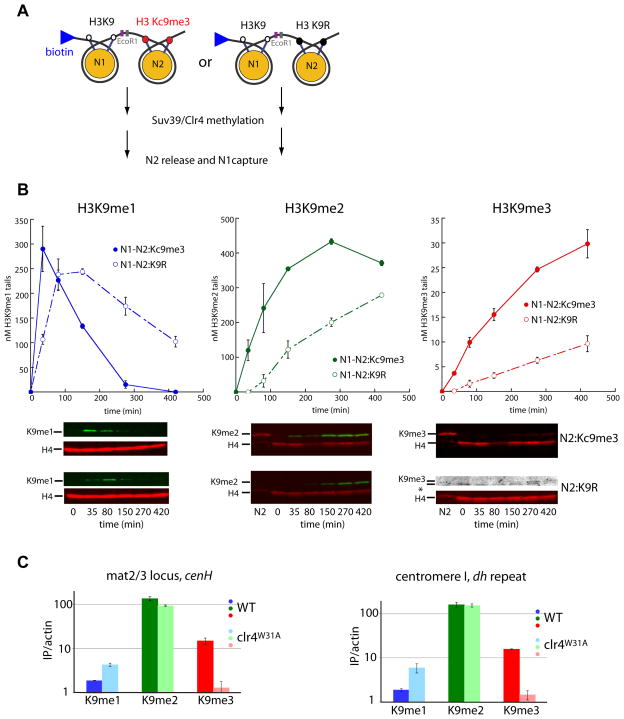
Figure 4. Impact of product guidance on H3K9me3 accumulation in vitro and in vivo.
A. Scheme for measuring mono-, di- and trimethylation on Biotin-N1-N2K9R and Biotin-N1-N2MLA dinucleosomes. Methylation reactions were performed as in Figure 2A, reactions were stopped with 2mM SAH. Following reactions, N2 effector nucleosomes were released by EcoRI restriction digestion and N1 nucleosomes separated by SDS-PAGE and probed with indicated antisera.
B. TOP: Quantification of Western blot data as in Figure 2B. BOTTOM: Representative western blots. N2 represents effector nucleosomes from t=0 min time point. The N1-N2:K9R anti-H3K9me3 blot is enhanced to show the very small pool of detectable H3K9me3. Error bars denote standard error of three timecourses.
C. The Suv39/Clr4 W31A mutation leads to loss of H3K9me3 at the mat2/3 locus cenH element or centromere dh repeat. Q-ChIP was performed as in Figure 3B. The Suv39/Clr4W31A is denoted in light colors. Wild-type data are the same as in Figure 3B and shown for comparison. Error bars denote standard error of three IPs.
To examine the role of product guidance in vivo, we mutated the critical W31 residue in the hydrophobic cage of the Suv39/Clr4 CD (W31A in this study). Such W31 mutants are not capable of recognizing H3K9me3 (Zhang et al., 2008) (Supplementary Figure 4A), but are still capable of H3K9 methylation on peptides (Nakayama et al., 2001) (Supplementary Figure 2C). Further, we find that Suv39/Clr4W31A has similar H3K9 mono-, di-, and trimethylation rate constants on mononucleosomes as wild-type Suv39/Clr4 (Supplementary Figure 4B), demonstrating that this CD mutant does not impact intrinsic methyltransferase activity. Consistent with previous observations (Noma et al., 2004), our Suv39/Clr4W31A mutant showed significantly reduced spread of H3K9 methylation outside the cenH initiation region of the mat2/3 locus (data not shown). We then used Q-ChIP to investigate the relative distributions of H3K9me1, me2, and me3 at the centromeric dh repeat and the cenH initiation element in the Suv39/Clr4W31A mutant. We find that compromising product guidance results in a near-complete loss of H3K9me3 at both those regions (Figure 4C), with much smaller effects on the other two methylation states. The H3K9me1 levels are increased 2–3-fold at both loci, while H3K9me2 is almost unaffected. These results are consistent with the predictions from the biochemical analysis in Figure 4B.
The H3K9me3 selectivity of the Suv39/Clr4 CD is required for spread and prevention of nonproductive competition
In trying to understand the significance of the H3K9 methyl state distribution in vivo and its relation to spreading, we recalled prior studies, which have shown that the CD of Suv39/Clr4 is unique among the CDs of heterochromatin factors in S. pombe in its H3K9 methyl state selectivity. The HP1 proteins Swi6 and Chp2 and the RITS-associated Chp1 protein have CDs that discriminate only 1.5–2- fold between H3K9me3 over H3K9me2 (Fischle et al., 2003; Sadaie et al., 2008; Schalch et al., 2009; Yamada et al., 2005), while the CD of Suv39/Clr4 has a 5–6-fold preference for H3K9me3 over H3K9me2 (Schalch et al., 2009; Zhang et al., 2008). This selectivity of the Clr4 CD is comparable to the selectivities of H3K4me3-specific PHD finger factors (Li et al., 2006; Pena et al., 2006). We tested whether this difference had a functional impact on the spread of H3K9 methylation.
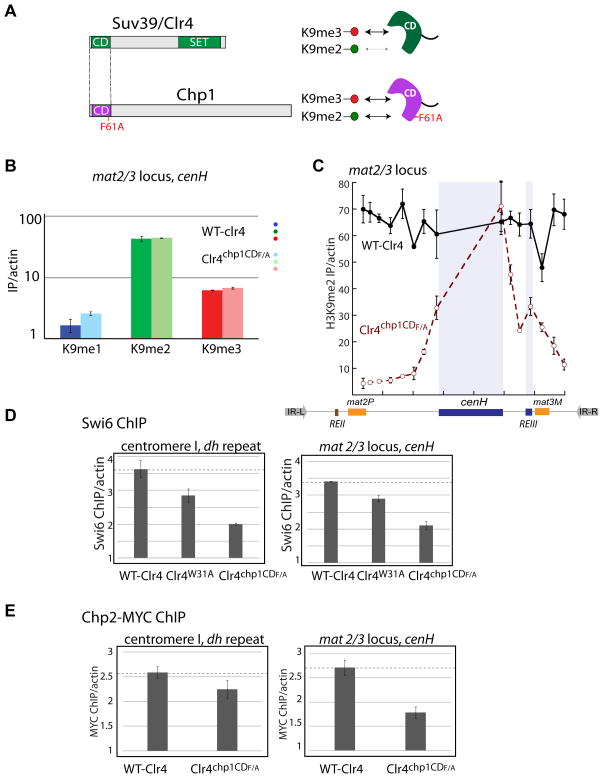
We swapped the CD of Suv39/Clr4 with that of Chp1 mutated at F61 to alanine (Chp1CDF61A). This mutation yields a similar affinity for H3K9me3 as the wild-type CD of Suv39/Clr4 but exhibits 3-fold tighter affinity for H3K9me2 (Schalch et al., 2009), effectively reducing the selectivity of Suv39/Clr4 for H3K9me3. We find that Clr4chp1CDF61A rescues the silencing function of Suv39/Clr4 at an ectopic locus, unlike the CD functional null Suv39/Clr4W31A (Supplementary Figure 5B). However, while the distribution of H3K9 methylation states in Clr4chp1CDF61A at endogenous initiation sites is similar to wild-type Suv39/Clr4 (Figure 5B and Supplementary Figure 5C), the spread of methylation beyond presumptive initiation sites (cenH and REIII) in the mat2/3 silenced region is dramatically reduced (Figure 5C), albeit not abolished, as in Suv39/Clr4W31A.
Figure 5. Impact of changing H3K9me3 selectivity of the Suv39/Clr4 CD on methylation spread HP1 assembly in vivo.
A. The CD of Suv39/Clr4 (residue 1–64) was swapped for the F61A mutant CD of Chp1 (1–77). The F61A Chp1 CD has a similar preference for H3K9me2 and H3K9me3, while the Clr4 CD is more H3K9me3 specific.
B. H3K9me1, me2 or me3 signals in Suv39/Clr4 WT or Suv39/Clr4chp1CDF61A at the cenH initiation region probe of the mat2/3 locus were quantified as in Figure 3B. Error bars represent standard error of three IPs.
C. H3K9me2 signals across the mat2/3 locus in Suv39/Clr4 WT or Suv39/Clr4chp1CDF61A backgrounds. Error bars represent standard error of three IPs.
D. The Suv39/Clr4chp1CDF61A CD domain swap decreases HP1/Swi6 at initiation sites. Swi6 enrichment over a clr4Δ mutant, normalized to an actin control is shown. Error bars represent standard error of three of three IPs.
E. The Suv39/Clr4chp1CDF61A CD domain swap decreases Chp2:7xMYC at the mat2/3 locus and to a lesser degree at the centromere. Chp2:7xMYC enrichment over a clr4Δ mutant normalized to an actin control is shown. Error bars represent standard error of three IPs.
Given that we converted the CD of Suv39/Clr4 into one that could in principle compete better with the CDs of HP1 proteins for H3K9me2, and given that HP1 proteins are themselves required for the spread of heterochromatin (Hall et al., 2002; Kanoh et al., 2005), we hypothesized that displacement of HP1 proteins by the engineered CD might account for part of the defect in heterochromatin spread. Consistent with this notion, we observed that HP1/Swi6 levels at the centromeric dh repeat or the cenH initiation element are reduced ~2 fold in the mutant (Figure 5D), even though overall H3K9 methylation is not significantly altered at these sites (Figure 5B and Supplementary Figure 5C). Additionally, the HP1 protein Chp2 is reduced to a similar extent at the mating-type locus cenH initiation site, while it is only mildly decreased at the centromeric dh element (Figure 5E). These results demonstrate that the binding preference of the CD of Suv39/Clr4 for the trimethylated state of H3K9 is critical for the lateral spread of heterochromatin in vivo, we hypothesize, in part because it reduces non-productive competition with HP1 proteins (see discussion).
DISCUSSION
In S. pombe, the spread of heterochromatin relies on the recognition of H3K9me marks by both “readers” as well as the enzymatic “writers” that put on the mark. Two central questions emerge from this arrangement: 1) How does recognition of the “writer” reaction product, H3K9me, regulate Suv39/Clr4 to promote spread and 2) How do “readers” and “writers” co-ordinate H3K9me recognition through their respective CDs to avoid non-productive competition for this mark? We find that a key to answering these questions lies in the central finding presented here that different modification states of the same chromatin mark, in this case H3K9me2 and H3K9me3, take on distinct roles.
Catalytic stimulation underlies positive feedback by the Suv39/Clr4 product
The finding that H3K9me recognition modules in “writer” and “reader” proteins are required for heterochromatin spread has led to a positive feedback hypothesis, where it is postulated that pre-existing methylation events help stimulate subsequent rounds of H3K9 methylation. The mechanistic basis of such feedback is not fully understood. Two broadly recognized classes of mechanisms have been proposed for how a mark might promote its own formation via a reader-writer protein: enhanced binding or allosteric activation. We find that H3K9me3, present on both tails in cis on a neighboring nucleosome, stimulates catalysis by Suv39/Clr4, rather than substrate binding. We further exclude a straightforward allosteric mechanism (Figure 1F and Supplemental Figure 2C and D). The stimulatory effect is most simply accounted for by a model in which the CD:H3K9me3 interaction guides the correct alignment of the substrate reactive groups relative to the active site residues. In such a model, the CD:H3K9me3 interaction occurs after binding to stabilize a high energy, guided state in which the H3 tails of the adjacent nucleosome are optimally positioned within the SET domain active site (Figure 1G). In principle, the guiding nucleosome can be methylated on one, or both (as in our experiments) tails. If the formation of a guided state were highly sensitive to the steric arrangement of methylated tail and substrate, then a nucleosome methylated on one tail would exercise guidance only if such constraints were to be satisfied. Alternatively, should guidance not be sensitive to which of the two effector nucleosome tails is methylated, singly-methylated nucleosomes would be at least 2-fold less efficient at catalytic stimulation, and full stimulation would require methylation of both tails.
Formation of the guided state will most likely rely on a conformational change within Suv39/Clr4 following initial engagement with the nucleosome. Narrowly interpreted, this model would predict that binding of Suv39/Clr4 to chromatin in vivo should not depend on its CD. However, Chromatin immunoprecipitation experiments, a proxy for binding in vivo, indicate that CLRC components localize specifically to heterochromatic regions in a manner dependent on the Suv39/Clr4 CD (Zhang et al., 2008). Therefore, it is likely that other subunits of the CLRC complex, or other regulators, recognize features of heterochromatin (e.g. bound HP1/Swi6) to further stabilize the high energy guided state of Suv39/Clr4, resulting in CD-dependent ChIP of CLRC components.
Regulation from the reaction product is common in chromatin biology (Kirmizis et al., 2007; Lan et al., 2007; Margueron et al., 2009), and has been extensively analyzed in the context of H3K27 methylation by the PRC2 complex (Margueron et al., 2009). In this case there is strong evidence for canonical allosteric activation as the product peptide provided in trans stimulates methylation by the catalytic EZH2 subunit (Margueron et al., 2009). In contrast to this type of mechanism, guided state based mechanisms require activators in cis and may allow enzymes like Suv39/Clr4 to integrate specificity determinants distributed over a complex substrate such as a dinucleosome. Analogous post-binding catalytic effects of enzymatic residues that are distant from the active site have been documented in other enzymatic systems (Narlikar and Herschlag, 1998, reviewed in Jencks, 1987).
Product guidance mechanism is required for the accumulation of H3K9me3 in vivo
In vitro, the absence of a product guidance mark lowers the rates for generating each methylation state. Since H3K9me3 is the terminal product, there is a large cumulative impact on H3K9me3 levels at any given time (Figure 4B). Consistent with these in vitro observations, compromising product guidance in vivo leads to a severe reduction in H3K9me3 at presumptive initiation sites (Figure 4C). Previous work has shown that compromising the ability of the Suv39/Clr4 CD to recognize the H3K9 methyl mark greatly diminishes H3K9 methylation spread beyond the initiation sites (Noma et al., 2004). Our data raise the possibility that the lack of spread observed previously is mechanistically related to the reduction in H3K9 trimethylation levels at the initiation sites. We discuss this possibility further in the next section (Figure 6).
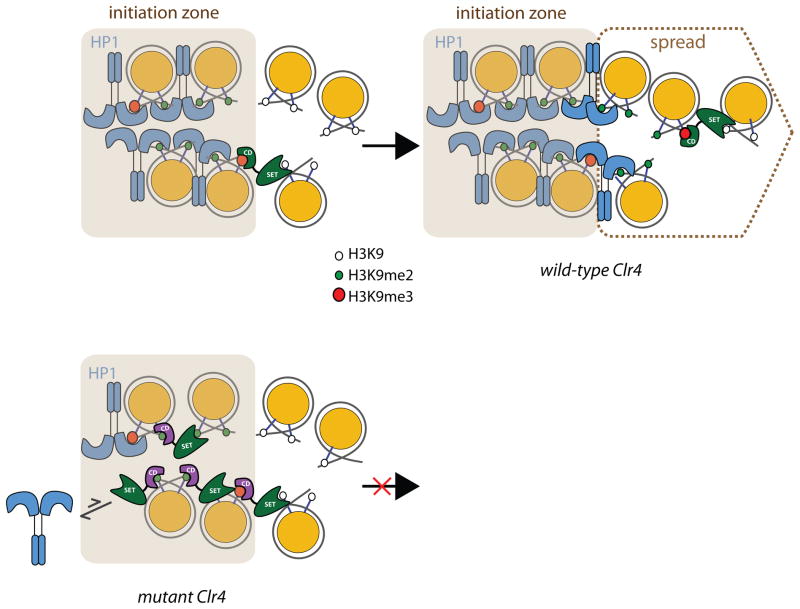
Figure 6. Model for co-ordination of Suv39/Clr4 and HP1 protein chromodomains in H3K9 methylation spread across nucleosomes.
In the initiation zone, a pool of H3K9me2 and H3K9me3 marks is formed by directly recruited Suv39/Clr4 molecules. The CDs of HP1 proteins such as Swi6 and Chp2 cover most of the central H3K9me2 and H3K9me3 marks. Specific H3K9me2/3 recognition of some HP1 proteins like Swi6 requires oligomerization mediated by the chromatin template (Canzio et al., 2011), exposing H3K9me2/3 at the edge of the heterochromatic domain. In the context of wild-type enzyme, exposed H3K9me3 marks act as a guidance mark for Suv39/Clr4. Any nearby unmethylated nucleosome transiently available for the guided state of Suv39/Clr4 will be methylated, resulting mostly in H3K9 dimethylation, and a small H3K9 trimethylated pool. These new sites are covered by the HP1 proteins, preferentially exposing H3K9 methylated tails at the edge of the growing heterochromatin structure. Suv39/Clr4 does not significantly compete for HP1 proteins in the central domain due its high preference for H3K9me3, lacking in HP1 proteins, which efficiently recognize H3K9me2 marks and are also present in higher concentration. In the context of Suv39/Clr4chp1CDF61A (“mutant Clr4”), the increased affinity for H3K9me2 now allows Suv39/Clr4 to engage the guided state at a majority of methylated nucleosomes, competing non-productively with HP1 proteins.
Division of labor between the chromodomains of HP1 and Suv39/Clr4 is required for productive heterochromatin assembly
We find that the distribution of H3K9 methylation states catalyzed by Suv39/Clr4 is critical for allowing H3K9me spread and for avoiding non-productive competition with the two central S. pombe HP1 proteins, Swi6 and Chp2. Our in vitro measurements show that formation of H3K9me3 from H3K9me2 is 10-fold slower than formation of either H3K9me2 or H3K9me1 (Figure 2). Correspondingly in vivo, using a novel quantitative method to measure individual methylation states, we find that cellular levels of H3K9me3 are ~10-fold lower than H3K9me2 levels (Figure 3). At the same time, our results with the Suv39/Clr4 CD mutant indicate that the spread of methylation is correlated with the presence of the trimethyl state in the initiation sites (Figure 4B&C). Why then are the H3K9me3 levels maintained at a low level in vivo? We believe the different levels of H3K9me3 and H3K9me2 reflect the need to separate the role of H3K9me3 in regulating Suv39/Clr4 spread from the role of H3K9me2 in directing structural assembly by HP1 proteins.
In support of the above hypothesis we find that selective recognition of the Suv39/Clr4 CD for the trimethylated state of H3K9 is required for the spread of heterochromatin in vivo (Figure 5C). Suv39/Clr4 has a higher preference for the H3K9me3 mark than the S. pombe HP1 proteins (Nakayama et al., 2000; Schalch et al., 2009; Zhang et al., 2008). Engineering the Suv39/Clr4 CD to increase its affinity for the dimethyl state, while maintaining the same affinity for the trimethyl state (Figure 5A), results in significant loss of H3K9 methylation spread (Figure 5B). Importantly, this increase in selectivity for the dimethyl state results in a reduction in the recruitment of the HP1 proteins, Swi6 and Chp2, at initiation loci (Figure 5D and E). The simplest explanation for this reduced recruitment is that the engineered Suv39/Clr4 protein is now better at competing with HP1 proteins for binding to H3K9me2. Such a competitive scenario requires H3K9me “reader” proteins to be present in excess of H3K9me binding sites so that H3K9me tails are, in fact, limiting. This is broadly consistent with published data, which indicate that HP1/Swi6 accumulates to about 20,000 copies per cell (Sadaie et al., 2008). This represents about a 10-fold excess over H3K9me tails that participate in heterochromatin initiating regions (~ 120kb). Together, these results provide a biological rationale for why Suv39/Clr4 prefers to generate dimethyl over trimethyl H3K9.
Together, our data suggest a model in which the small cellular H3K9me3 pool plays roles in spreading while the dominant H3K9me2 pool plays roles in assembly (Figure 6). At initiation sites, the recruitment of Suv39/Clr4 leads mostly to local H3K9 dimethylation and small amounts of H3K9me3. We hypothesize that the CD of Suv39/Clr4 primarily engages the H3K9me3 mark. This leaves the predominant H3K9me2 species free to be bound by the CDs of HP1 proteins, which do not distinguish significantly between the H3K9me2 and H3K9me3 methylation states. When Suv39/Clr4 encounters an available H3K9me3 nucleosome, the methylation of adjacent unmethylated nucleosomes is stimulated. We speculate that the HP1 protein Swi6 is non-redundantly required for spread at the mat2/3 locus by masking previously methylated regions and preventing Suv39/Clr4 from getting trapped in these methylated regions.
Overall, our data reveals strategies that allow “writer” enzyme and “reader” proteins to productively engage in the formation of a heterochromatic domain. This is accomplished in two ways; First, the affinities of the chromatin mark binding modules are tuned differentially such that “writers” and “readers” are sensitive to different modification states. Second, the “writer” enzyme has access to principally one of the modification states, and this state stimulates the “writer” enzyme on the chromatin template to achieve enhanced catalysis dependent on proximity to previously deposited reaction products. Together, these two strategies allow for productive collaboration between “reader” and “writer” proteins in the spread of a chromatin mark.
EXPERIMENTAL PROCEDURES
Mononucleosome and asymmetric dinucleosome reconstitution
Core mononucleosomes were reconstituted on the 147bp “601” positioning sequence (Thastrom et al., 1999), with or without a 5′ fluorescein label, as described (Canzio et al., 2011). To produce asymmetric dinucleosomes (N1-N2K9R/MLA), “601” DNA fragments containing a site for the type I restriction endonuclease AarI (designed cut site ‘CCCT’) were PCR amplified, AarI digested, assembled separately with WT, H3K9R (K9R) or H3Kc9me3 (MLA) octamers to produce N1 or N2 nucleosomes, which were ligated to produce N1-N2K9R or N1-N2MLA dinucleosomes. For detailed methods, see Supplementary Experimental Procedures.
3HSAM incorporation assays with biotinylated H31-20 peptides and nucleosomes
For peptide reactions, biotinylated H31-20 peptide, or H31-15 peptide effector were produced as described (Canzio et al., 2011). For nucleosome reactions, 70–300nM of core mononucleosomes or dinucleosomes were used. Reactions contained 70–100 μM cold SAM (as either Chloride or Iodide salt, Sigma) and 0.3–0.4μM 3HSAM tracer (55–75Ci/mmole, Perkin-Elmer) and were incubated with varying amounts of Suv39/Clr4. Reactions were performed at 25°C, in reaction buffer (RB) 100mM Tris pH8.5, 100mM KCl, 10% glycerol, 1mM MgCl2, 20μM ZnSO4, 10mM β-mercaptoethanol. To degrade the inhibitory reaction product S-adenosyl-homocysteine, the reaction further contained 20μM SAHH enzyme in the cae of peptide reactions. Methylation reactions were stopped by the addition of 5x SDS-loading buffer and brief incubation at 100°C. Reactions with biotinylated peptides were spotted on SAM2 Biotin capture membrane (Promega) and washed 3x with 1M NaCl and 2x with ddH2O. The spots were dried and counted in a scintillation counter. Nucleosome methylation reactions were separated on 15% SDS-PAGE gels, dried and imaged on Tritium screens. Signals were visualized on a Molecular Dynamics Typhoon imager and quantified using ImageQuant software.
Quantitative western blots
Reactions were performed as above, except for biotin tagged N1-N2K9R/MLA dinucleosomes (bN1-N2K9R/MLA): Reactions were stopped by the addition of 2mM S-adenosyl- homocysteine, and bN1-N2K9R/MLA captures on magnetic streptavidin beads (Dynal). Beads were washed twice in RB and bN1-N2K9R/MLA digested on beads with 200 units EcoR1 for 90min at 37°C to remove N2K9R/MLA. Beads were washed again twice with RB and resuspended in 1x SDS-loading buffer.
Reactions were separated on 4–20% SDS-PAGE gels and transferred to PVDF membrane, blocked in 1:1 PBS:Li-Cor Odssey blocking buffer (Li-Cor), and probed with either anti –H3K9me1 (monoclonal, Active Motif), –H3K9me2 (monoclonal, Abcam), – H3K9me3 (polyclonal, Millipore), and -H4 (polyclonal, Active Motif) antisera. Signals were detected by LiCor secondary antibodies IRDye680CW (H3K9me3 and H4) or IRDye800CW (H3K9me1 and H3K9me2). Methods employed to determine H3K9me1, me2 and me3 concentrations and extract the underlying k1, k2 and k3 rate constants is described in Supplementary Experimental Procedures.
Chromatin immunoprecipitation
Q-ChIP method: H3Kc9me1, me2 and me3 core mononucleosomes were reconstituted as above and dialyzed into 50mM HEPES pH7.5, 2mM DTT, 0.5mM EDTA. Nucleosomes were cross-linked 15′ with 1% formaldehyde and the reaction quenched with 250mM glycine. Cross-linked nucleosomes, long with a DNA standard, were separated on a native PAGE gels, stained with SyBR Gold, visualized on a Molecular Dynamics Typhoon scanner and quantified using Image Quant software as described (Canzio et al., 2011). Strain growth and chromatin preparation were as described (Canzio et al., 2011) and ChIP performed with H3K9 methylation state specific antisera (further details see Supplementary Experimental Procedures). For H3K9me1, me2 or me3 and Swi6 ChIP experiments, enrichments were plotted as the enrichment over a clr4Δ mutant, normalized to an actin control. For Swi6 ChIP, a polyclonal antiserum was used as described (Canzio et al., 2011), while for Chp2:7xMYC a commercial monoclonal anti-MYC antiserum was used (Abcam, Ab32).
Native Gel Mobility Shift Assays
Indicated concentrations of full length wild-type Suv39/Clr4 were incubated with 20nM 147bp “601” DNA assembled wild-type or H3Kc9me3 mononucleosomes. Mobility shift assays and quantifications were performed as described (Canzio et al., 2011).
Strain construction
To swap the CD of Chp1 of the CD of Suv39/Clr4, we replaced the N-terminal 64 residues of Suv39/Clr4 with the N-terminal 77 residues of Chp1. The endogenous clr4+ was replaced with either Suv39/Clr4W31A or Suv39/Clr4chp1CDF61A or a wild-type sequence and marked with a 5′G418 R selectable marker flanked 5′ with 500bp of the clr4+ promoter. For chp2 ChIP, the endogenous chp2+ gene was replaced with a C-terminally 7x MYC tagged chp2 construct marked with a 3′Hygromycin R selectable marker.
Supplementary Material
Highlights.
H3K9 methylation on an adjacent nucleosome stimulates catalysis by Suv39/Clr4
H3K9me2 is the predominant Suv39/Clr4 product produced in vitro and in vivo
Production of a small H3K9me3 pool is required for heterochromatin spread
Distinct intrinsic binding preferences of HP1 and Suv39/Clr4 CDs avoids competition
Acknowledgments
We would like to thank Shiv IS Grewal for the generous gift of anti-Swi6 anitsera, Raymond C Trievel for his generous gift of the S-adenosyl-homocysteine hydrolyase clone, Carrie Shiau for production of H3Kc9me1 and me2 histones, Adam Larson for providing fluorescein-labeled nucleosomes and Lindsey Pack for providing H3K9me31-15 and H3Kc9me31-15 peptides. We thank Daniele Canzio for helping establish initial parameters of Suv39/Clr4 methylation on peptide substrates and Jennifer Garcia for helpful comments on the Q-ChIP method. Further, we thank Carol A Gross, Jesse Zalatan, Richard S. Isaac and Phillip Dumesic for critical reading of the manuscript and members of the Narlikar and Madhani labs for helpful comments and discussion. Research in the Narlikar laboratory was supported by grants from the American Cancer Society, the Leukemia and Lymphoma Society and the National Institutes of Health. Chromatin research in the Madhani laboratory is supported by a grant from the National Institutes of Health (GM071801). B.A-S. was a fellow of the Jane Coffin Childs Memorial Fund for Medical research.
Footnotes
Author contributions
B.A.-S., H.D.M. and G.J.N. designed experiments, B.A.-S. performed all experiments, B.A.-S., H.D.M. and G.J.N. analyzed and discussed the data. B.A.-S., H.D.M. and G.J.N. prepared the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bhalla KN. Epigenetic and Chromatin Modifiers As Targeted Therapy of Hematologic Malignancies. J Clin Oncol. 2005;23:3971–3993. doi: 10.1200/JCO.2005.16.600. [DOI] [PubMed] [Google Scholar]
- Buhler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone R, Botrugno OA, Ronzoni S, Insinga A, Di Croce L, Pelicci PG, Minucci S. Recruitment of the Histone Methyltransferase SUV39H1 and Its Role in the Oncogenic Properties of the Leukemia-Associated PML-Retinoic Acid Receptor Fusion Protein. Mol Cell Biol. 2006;26:1288–1296. doi: 10.1128/MCB.26.4.1288-1296.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, Gorski SA, May KM, Singh PB, Misteli T. In Vivo Dynamics of Swi6 in Yeast: Evidence for a Stochastic Model of Heterochromatin. Mol Cell Biol. 2004;24:3157–3167. doi: 10.1128/MCB.24.8.3157-3167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases. Anal Biochem. 2005;342:86–92. doi: 10.1016/j.ab.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Couture JF, Dirk LM, Brunzelle JS, Houtz RL, Trievel RC. Structural origins for the product specificity of SET domain protein methyltransferases. P Natl Acad Sci USA. 2008;105:20659–20664. doi: 10.1073/pnas.0806712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirk LM, Flynn EM, Dietzel K, Couture JF, Trievel RC, Houtz RL. Kinetic manifestation of processivity during multiple methylations catalyzed by SET domain protein methyltransferases. Biochemistry. 2007;46:3905–3915. doi: 10.1021/bi6023644. [DOI] [PubMed] [Google Scholar]
- Elgin SCR, Grewal SIS. Heterochromatin: silence is golden. Current Biology. 2003;13:R895–R898. doi: 10.1016/j.cub.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang YM, Jacobs SA, Kim YC, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 bv Polvcomb and HP1 chromodomains. Genes & Development. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, Harel-Bellan A, Ameyar-Zazoua M, Ait-Si-Ali S. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Molecular cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Grewal SIS, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma KI, Ayoub N, Cohen A, Grewal SIS. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Hong EJ, Villen J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2005;2:106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- Horn PJ, Bastie JN, Peterson CL. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 2005;19:1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jencks WP. Catalysis in Chemistry and Enzymology. New York: Dover Publications; 1987. Binding Energy, Specificity, and Enzymic Catalysis: The Circe Effect; pp. 615–797. [DOI] [PubMed] [Google Scholar]
- Jia S, Kobayashi R, Grewal SIS. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol. 2005;7:1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- Jia ST, Noma K, Grewal SIS. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, Ishikawa F. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol. 2005;15:1808–1819. doi: 10.1016/j.cub.2005.09.041. [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munari F, Soeroes S, Zenn HM, Schomburg A, Kost N, Schroeder S, Klingberg R, Rezaei-Ghaleh N, Stuetzer A, Gelato KA, et al. Methylation of K9 in histone H3 directs alternative modes of highly dynamic interaction of heterochromatin protein hHP1beta with the nucleosome. The Journal of biological chemistry. 2012 doi: 10.1074/jbc.M112.390849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Klar AJ, Grewal SI. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Herschlag D. Direct demonstration of the catalytic role of binding interactions in an enzymatic reaction. Biochemistry. 1998;37:9902–9911. doi: 10.1021/bi980495t. [DOI] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SIS. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AHFM, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AAHA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. Partitioning and Plasticity of Repressive Histone Methylation States in Mammalian Chromatin. Molecular Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Reed-Inderbitzin E, Moreno-Miralles I, Vanden-Eynden SK, Xie J, Lutterbach B, Durst-Goodwin KL, Luce KS, Irvin BJ, Cleary ML, Brandt SJ, et al. RUNX1 associates with histone deacetylases and SUV39H1 to repress transcription. Oncogene. 2006;25:5777–5786. doi: 10.1038/sj.onc.1209591. [DOI] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Molecular cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- Sadaie M, Kawaguchi R, Ohtani Y, Arisaka F, Tanaka K, Shirahige K, Nakayama J. Balance between Distinct HP1 Family Proteins Controls Heterochromatin Assembly in Fission Yeast. Molecular and Cellular Biology. 2008;28:6973–6988. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Job G, Noffsinger VJ, Shanker S, Kuscu C, Joshua-Tor L, Partridge JF. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol Cell. 2009;34:36–46. doi: 10.1016/j.molcel.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger D, Soeroes S, Klingberg R, Schwarzer D, Grubmuller H, Fischle W. Quantitative assessment of protein interaction with methyl-lysine analogues by hybrid computational and experimental approaches. ACS Chem Biol. 2012;7:150–154. doi: 10.1021/cb200363r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes & development. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thastrom A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. Journal of Molecular Biology. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, Allali-Hassani A, Campagna-Slater V, Vedadi M, Arrowsmith CH, et al. Structural biology of human H3K9 methyltransferases. PLoS One. 2010;5:e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SIS. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell. 2005;20:173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SIS. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SI. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.