Summary
HP1 proteins are central to the assembly and spread of heterochromatin containing histone H3K9 methylation. The chromodomain (CD) of HP1 proteins specifically recognizes the methyl mark on H3 peptides, but the same extent of specificity is not observed within chromatin. The chromoshadow domain of HP1 proteins promotes homodimerization, but this alone cannot explain heterochromatin spread. Using the S. pombe HP1 protein, Swi6, we show that recognition of H3K9 methylated chromatin in vitro relies on a newly identified interface between two CDs. This interaction causes Swi6 to tetramerize on a nucleosome, generating two vacant CD sticky ends. On nucleosomal arrays, methyl-mark recognition is highly sensitive to inter-nucleosomal distance, suggesting that the CD sticky ends bridge nearby methylated nucleosomes. Strengthening the CD-CD interaction enhances silencing and heterochromatin spread in vivo. Our findings suggest that recognition of methylated nucleosomes and HP1 spread on chromatin are structurally coupled, and imply that methylation and nucleosome arrangement synergistically regulate HP1 function.
Introduction
Histone H3 lysine 9 methylated (H3K9me3) heterochromatin, conserved from yeast to humans, is a highly versatile nuclear structure. It is required for centromere formation, heritable gene silencing, repression of recombination, sister chromatid cohesion, and maintenance of telomere stability (Grewal and Jia, 2007). A hallmark of this type of heterochromatin is the formation of macromolecular assemblies that can spread along chromatin from specific nucleation sites (Hall et al., 2002). The structural features that allow H3K9me3 based heterochromatin to spread and fulfill its various nuclear functions, however, are not well understood.
At the core of heterochromatic macromolecular assemblies lies the HP1-H3K9me3 chromatin complex, which is thought to mediate the many functions of heterochromatin through the recruitment of diverse sets of regulators (Grewal and Jia, 2007; Smothers and Henikoff, 2000). In gene silencing, HP1 proteins are thought to reduce RNA polymerase occupancy by both recruiting accessory silencing factors (Fischer et al., 2009) and by forming less accessible chromatin structures (Danzer and Wallrath, 2004). HP1 proteins have been proposed to enable post-transcriptional gene silencing by recruiting RNA processing machinery (Iida et al., 2008).Understanding how HP1 proteins recognize and bind H3K9me3 chromatin is thus central to understanding both the molecular mechanisms of heterochromatin assembly and how this type of heterochromatin fulfills its wide range of functions.
Previous work has described individual aspects of the HP1/H3K9me3 nucleosome complex. HP1 proteins contain three recognized protein domains: 1) a chromodomain (CD), 2) an evolutionarily related chromoshadow domain (CSD), and 3) a poorly defined hinge (H) region between the CD and CSD. The CD is part of a family of proteins that contain a specialized hydrophobic cage, formed by aromatic residues, that bind methyl marks on histones with high specificity but low affinity (Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002). The CSD is involved in dimerization of HP1 proteins (Cowieson et al., 2000) and is important for the silencing function of HP1 proteins (Sadaie et al., 2008). The H region is thought to be required for sequence-independent DNA binding of HP1 proteins, as observed in vitro (Meehan et al., 2003; Zhao et al., 2000). Despite these key findings, several questions remain about how the functions of these individual domains are integrated to allow stable recognition of the physiological template, H3K9 methylated chromatin. For example, it is not clear whether the weak binding of the CD for methylated tail peptides observed in vitro is sufficient to guide heterochromatin assembly to the correct sites in vivo. In particular, the strong non-specific binding of HP1 proteins to inter-nucleosomal DNA (Meehan et al., 2003; Yamada et al., 1999) raises the question of how specificity for the methyl mark is attained in the context of chromatin. Finally, while HP1 proteins can dimerize via the CSD, such homodimerization alone appears insufficient to explain the ability of these proteins to spread along chromatin.
To address these questions, we used the S. pombe HP1 protein, Swi6, as a model system. S. pombe contains only a single H3K9 methyltransferase, Clr4, along with two HP1 proteins, Chp2 and Swi6, of which Swi6 is more abundant (Grewal and Jia, 2007; Sadaie et al., 2008). We reconstituted the core Swi6-H3K9me3 chromatin complex using recombinant Swi6 and chromatin templates that are homogeneously methylated at H3K9 using methyl lysine analog (MLA) technology (Simon et al., 2007). We analyzed the biochemical properties of this complex and tested our key conclusions in vivo. Our results suggest a mechanism of heterochromatin formation in which HP1 proteins utilize a process of step-wise higher order oligomerization. This process is mediated by interactions between CDs to interpret information encoded in both the methylation state and the underlying nucleosomal arrangement of chromatin.
Results
Swi6 recognizes the H3K9 methyl mark within mononucleosomes and forms oligomers on mononucleosomes and in solution
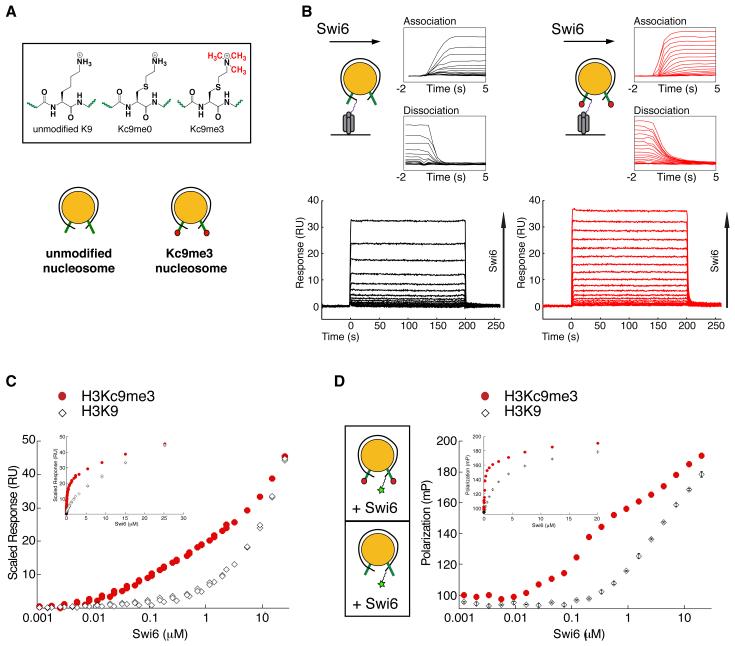
Previous studies have reported on the ability of Swi6 to preferentially bind the H3K9me3 mark in the context of H3 tail peptides (Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002; Yamada et al., 2005). However, the magnitude of discrimination observed within H3 tail peptides has not been recapitulated in the context of chromatin, largely due to the challenge of generating homogeneously methylated chromatin. We produced homogenously methylated nucleosomes using methyl lysine analogs (MLAs), then investigated the ability of recombinant Swi6 to specifically recognize methylated nucleosomes using two different equilibrium approaches. For both approaches, unmodified (H3K9) and methylated (H3Kc9me3) nucleosomes were assembled on 147 base pairs of the nucleosome positioning sequence 601 (Figure 1a).
Figure 1. Swi6 recognizes the H3K9 methyl mark within mononucleosomes and forms oligomers on mononucleosomes.
(A) Schematics of the unmodified K9 and methyl lysine analog (MLA) Kc9me0 and Kc9me3 substrates (top panel) and of the unmodified (H3K9) and MLA methylated (H3Kc9me3) mononucleosomes assembled on the 147 bp 601 sequence (bottom).
(B) Bottom Panels: Representative dose responses for H3K9 (black) and H3Kc9me3 (red) mononucleosomes. Schematic: H3K9 and H3Kc9me3 mononucleosomes captured on a streptavidin derivatized SPR chip. Top panels: Close up of the kinetics of association and dissociation.
(C) Scaled isotherms for three independent dose responses of Swi6 against H3K9 (open diamonds) and H3Kc9me3 (filled circles) mononucleosome surfaces plotted on a semi-log scale. Plotted points represent the response at equilibrium, determined by averaging the signal over the final ten seconds of the sample injection. Inset: isotherms plotted on a linear scale.
(D) Schematics: Mononucleosomes with fluorescein (green star) attached by a flexible linker at one end of the 147 bp DNA template. Average of three independent fluorescent polarization experiments for H3K9 (open diamonds) and H3Kc9me3 (filled circles) mononucleosomes are shown. Error bars represent s.e.m. All Swi6 concentrations represent monomer concentrations.
In the first approach, surface plasmon resonance (SPR) was used to assay binding of Swi6 to H3K9 and H3Kc9me3 nucleosomes (Figure 1b). Analysis of the binding kinetics (traces in Figure 1b, inset) revealed no large differences in the association rates, but comparison of the dissociation traces reveals that Swi6 dissociates more rapidly from H3K9 nucleosomes compared to H3Kc9me3 nucleosomes, consistent with specific binding of Swi6 to methylated nucleosomes (Figure 1b). Because kinetic analysis of SPR data can be problematic and at times unreliable, we further optimized the assay for equilibrium measurements. The equilibrium binding isotherms clearly reveal two features (Figure 1c; see also Figure S1b&c). At low concentrations (10 nM – 1 μM), there is a methylation specific interaction that approaches but does not reach saturation. At high concentrations (>1 μM), there is apparently a weak, non-saturable interaction, and the concentration dependence of this interaction is similar for the H3K9 and the H3Kc9me3 nucleosome surfaces. We were, however, unable to fit a physically meaningful model to the data because (i) the data do not reveal saturation and therefore cannot be used to determine a final stoichiometry and (ii) HP1 proteins are known to oligomerize in solution, so the concentration will change as function of the oligomeric state of Swi6 (See Figure S1e&f for more detailed discussion).
Despite the inability to fit a quantitative model to the data, the Swi6 concentration dependence reveals interesting features of the interaction of Swi6 with nucleosomes. The results imply the presence of at least two types of Swi6 binding events: one that occurs at concentrations below 1 μM and involves recognition of the methyl mark, and a second that occurs primarily at higher concentrations, is less sensitive to the presence of the methyl mark and is suggestive of step-wise Swi6 oligomerization.
To further investigate the Swi6 behavior observed by SPR, we measured Swi6 binding to core nucleosomes using a fluorescence polarization based approach. Using nucleosomal DNA labeled at one end with fluorescein, we monitored the gain in fluorescence polarization as a function of Swi6 concentration (Figure 1d, schematic, also see Extended Experimental Procedures). Analogous to the SPR data, we observe a binding profile that contains a methylation specific concentration regime and a non-saturable concentration regime.
The above results raised the question of what physical processes underlie the different types of binding events implied by the unusual concentration dependence. We hypothesized that the binding events in the methyl mark specific concentration regime reflect direct binding of Swi6 to the nucleosome and the H3K9 residue, while the binding events in the non-saturable concentration regime reflect mainly Swi6-Swi6 interactions that are scaffolded by the initial Swi6-nucleosome complex. The non-saturable behavior would then arise because addition of each Swi6 molecule would generate a new binding site for another Swi6 molecule, reflecting an intrinsic property of Swi6 to self-associate. To test this hypothesis, we investigated the oligomeric states adopted by Swi6 in solution under the two concentration regimes.
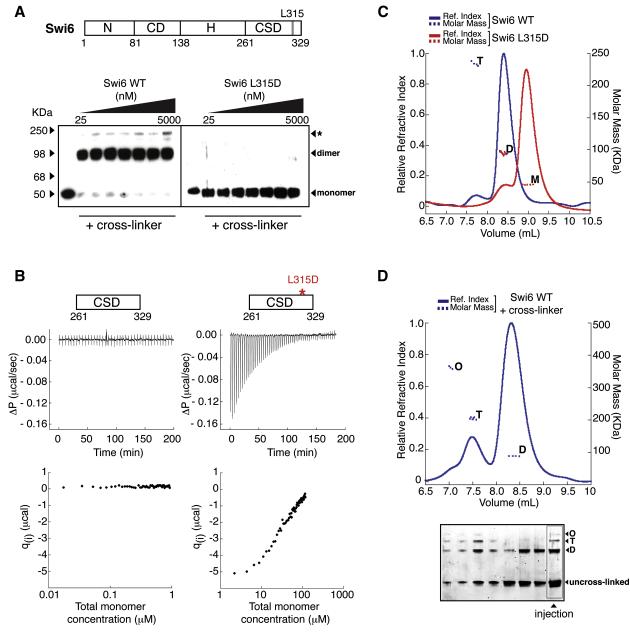
To determine the oligomeric state of Swi6 in the methylation specific concentration regime, we used two complementary approaches: (i) a cross-linking based approach and (ii) isothermal titration calorimetry (ITC). Over concentrations ranging from 25-5000 nM, cross-linker treated wild-type Swi6 migrates on SDS-PAGE gels at a mass consistent with a dimer, while the previously described dimer-disrupting CSD mutant, L315D, migrates at a mass consistent with a monomer (Cowieson et al., 2000) (Figure 2a). We then used ITC to obtain a more quantitative estimate of the Kd of the known dimerization domain of Swi6, the CSD (Figure 2b). Consistent with the cross-linking data, titrations of the WT Swi6 CSD into buffer produced no detectable heat release even at 17 nM indicating that Kd for CSD self-association is below 17 nM (Figure 2b, left panel). In contrast, titrations for the CSD domain containing the L315D mutation produce significant heat release and suggest a Kd for self-association of this mutant CSD in the high micromolar range (Figure 2b, right panel). Together, these two approaches indicate that at low nanomolar concentrations, Swi6 mainly exists as a dimer in the absence of nucleosomes.
Figure 2. Swi6 forms distinct oligomeric states in the absence of chromatin.
(A) Wild-type Swi6 (schematic on top) is largely a pre-formed dimer at low nM concentrations. Swi6 WT (left) and L315D (right) were treated at indicated concentrations with EDC and NHS cross-linkers. Treated proteins were separated by SDS-PAGE and detected by anti-FLAG western. Swi6 concentrations: uncross-linked 50 nM, cross-linked 25, 50, 100, 200, 500, 1000 and 5000 nM.
(B) The CSD-CSD dimerization Kd < 17 nM. Top: Representative ITC thermogram profiles for the dissociation of WT CSD dimer (left) and L315D CSD dimer (right) at 15°C. Bottom: Graphs represent the respective binding isotherms plotted as heat changes per injection (qi) vs. total monomer concentration.
(C) Multiangle light scattering (MALS) measurements for 20 μM WT Swi6 (blue) and 20 μM L315D mutant (red). Relative refractive index signals (solid lines, left y-axis) and derived molar masses (dotted lines, right y-axis) shown as a function of the elution volume. M: monomer, D: dimer, T: tetramer. (D) Top panel: Higher order oligomeric species of Swi6 stabilized by cross-linking. MALS measurements conducted as in (A). M: monomer, D: dimer, T: tetramer, O: octamer. Bottom panel: Aliquots of fractions collected from chromatography in (Top) were separated by denaturing SDS-PAGE and visualized by Sypro Red staining. The distribution of distinct oligomeric states thus visualized directly correlates with the oligomeric masses observed by MALS, while the presence of un-cross-linked Swi6 demonstrates Swi6 is not over cross-linked. All Swi6 concentrations represent monomer concentrations.
We next determined the oligomeric states that can be adopted by Swi6 in the non-saturable concentration regime. We had noticed that under cross-linking conditions, Swi6 can form oligomers larger than a dimer (Figure 2a, indicated by asterisk), consistent with previous studies on HP1 (Yamada et al., 1999; Zhao et al., 2000). To investigate the formation of defined higher-order oligomers and obtain true masses independent of oligomer shape we used a multi-angle light scattering (MALS) approach (Extended Experimental Procedures). The WT Swi6 protein forms mainly dimers at 20 μM (Figure 2c). Interestingly, approximately 5% of the protein is tetrameric, suggesting that Swi6 is capable of forming oligomers beyond a dimer. In contrast, the L315D mutation drastically reduces the ability of Swi6 to dimerize: more than 90% of the L315D is monomeric at 20 μM, in agreement with the ITC data (Figure 2b). The inter-molecular cross-linking approach described above enabled further stabilization of the higher order oligomeric states for analysis by MALS. Using this approach we found that WT Swi6 can form discrete complexes corresponding to dimeric, tetrameric, and octameric states (Figure 2d), whereas the L315D mutant is strongly impaired in forming such oligomeric states (Figure S2a). These data indicate that Swi6 can form well-defined higher order complexes in solution. Further, the Swi6 concentration regime in which states beyond dimer become populated correlates with the non-saturable concentration regimes of Figures 1c and d, suggesting that the non-saturable concentration regime mainly reflects Swi6-Swi6 interactions.
The above characterization of the oligomeric states of Swi6 indicates that Swi6 exists as a preformed dimer in the concentration regime in which we observed discrimination between H3Kc9me3 and H3K9 mononucleosomes. Further, the intrinsic property of Swi6 to form higher order oligomers suggests a potential for such oligomerization in binding across multiple nucleosomes within a nucleosomal array. To examine this possibility, we isolated the steps involved in direct recognition of the H3 tail within a mononucleosome, then used the information derived from these studies to better understand how Swi6 functions in the context of multiple nucleosomes.
Swi6 displays lower specificity for the H3K9me3 mark in mononucleosomes compared to H3 tail peptides
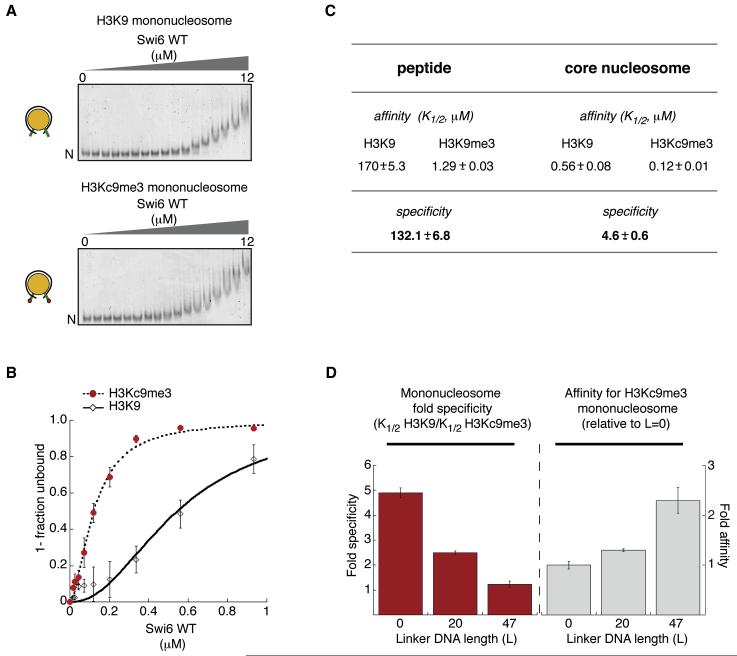
We reasoned that, by following the disappearance of the unbound nucleosomes in a gel mobility shift assay, we could better separate direct binding of Swi6 to the nucleosome from subsequent binding events that might entail mainly Swi6-Swi6 contacts. We measured the Swi6 concentration dependence for disappearance of unbound nucleosomes and obtained a value for K1/2, which represents the concentration of Swi6 at which half of the nucleosomes remain unshifted. Most of the unbound MLA nucleosomes completely disappear by 1 μM Swi6 (Figure 3a). At higher concentrations we observe further, apparently continuous upshifting of the complexes, consistent with the nucleosome-scaffolded oligomerization behavior inferred from Figures 1c and d.
Figure 3. Swi6 displays lower specificity for the H3K9me3 mark in mononucleosomes compared to that in H3 tail peptides.
(A) Representative gel shift using H3K9 or H3Kc9me3 mononucleosomes. Swi6 concentrations vary from 0 to 12 μM (0.6 fold dilutions). Unbound nucleosomes (N).
(B) Quantification of three gel shift experiments using H3K9 (open diamonds) and H3Kc9me3 (filled circles) to determine K1/2 and specificity (K1/2 H3K9 / K1/2 H3Kc9me3). Hill coefficient = 1.7 (H3Kc9me3) and = 2 (H3K9).
(C) Swi6 specificities for H3Kc9me3 mononucleosome and H3K9me3 peptide. K1/2 values (μM) for peptides were measured by fluorescence anisotropy and K1/2 (μM) for nucleosomes are from (B) with n=3.
(D) Increasing linker DNA length (L, in “bp”) decreases Swi6’s ability to discriminate the methyl mark on mononucleosomes. Left graph: Swi6 discrimination for H3Kc9me3 over unmodified mononucleosomes. Right graph: Swi6 affinity for H3Kc9me3 mononucleosomes, normalized to core (L=0) nucleosomes. All error bars represent s.e.m. All Swi6 concentrations represent monomer concentrations.
Using the above approach of quantifying K1/2 values, we found that Swi6 prefers H3Kc9me3 over H3K9 nucleosomes by 5-fold (Figure 3a, right panel; specificity is expressed as a ratio of K1/2 for H3K9 to that for H3Kc9me3 nucleosomes). Swi6 binds H3Kc9me0 nucleosomes with the same affinity as H3K9 nucleosomes (Figure S3a). We obtained the same 5-fold specificity for H3Kc9me3 over H3K9 nucleosomes using an equilibrium binding assay, in which the two types of nucleosomes compete with a fluorescently labeled DNA molecule for binding to Swi6 (Figure S3e).
Both the above assays indicate that the specificity for the methyl mark on core nucleosomes is substantially lower than that observed for the methyl mark on H3 tail peptides (Figures 3b&c and S3b). The results suggest a model in which Swi6 can bind to a core nucleosome in alternative orientations that lack interactions between the H3K9 residue and the CD, in addition to orientations that recognize the H3K9 residue. The binding orientations that lack interactions between the CD and H3K9 could arise from the previously described abilities of the hinge and the CSD domains to interact with other regions of the nucleosome, such as the DNA and a globular region of H3, respectively (Dawson et al., 2009; Lavigne et al., 2009; Meehan et al., 2003). The above model predicts that increasing alternative binding interactions between Swi6 and the nucleosome will decrease the observed specificity for the methyl mark, as a smaller proportion of Swi6 molecules would bind in H3K9 recognizing orientations. At the same time we expect that the overall affinity will increase, as increasing the number of alternative binding orientations will increase the binding options of Swi6. Given that the affinity of HP1 proteins for free DNA increases with DNA length (Zhao et al., 2000) and given our similar observations for Swi6 (Figure S3d), increasing the flanking DNA could be one way to increase the number of alternative binding modes. Consistent with these predictions, we find that increasing the flanking DNA length on one or both sides of a nucleosome results in a reduction in specificity but a gain in overall affinity (Figure 3d and Figure S3c).
Application of a simple quantitative model suggests that, for Swi6-H3Kc9me3 core nucleosome complexes, 94% of the Swi6 molecules are bound in H3K9-specific orientations and 6% are bound in alternative orientations (Extended Experimental Procedures). In contrast, for Swi6-H3K9 core nucleosome complexes, only 0.1% of the Swi6 molecules are bound in H3K9-specific orientations and >99% are bound in alternative orientations. Thus, in the context of H3K9 nucleosomes, the large fraction of Swi6 molecules bound in alternative orientations is expected to mask the binding contributions from molecules bound in H3K9-specific orientations. Together, the above observations raise the possibility that the specificity of HP1 proteins for the H3K9me3 mark could be controlled in part by regulating alternative binding orientations. The experiments that follow provide a structural and energetic framework to understand how such regulation might occur.
The core unit of Swi6 binding to a mononucleosome is a tetramer
Quantification of the gel mobility shifts results suggests that binding of Swi6 to either H3Kc9me3 or H3K9 core nucleosomes occurs cooperatively with Hill coefficients of ~1.7 and 2.0, respectively (Figure 3b) suggesting that at least two molecules of Swi6 bind to one nucleosome. Further, the analysis in figures 2a and b indicates that, at the concentrations used in the native gel-shift assay, Swi6 is a dimer in solution. The cooperative binding could then reflect an additional interaction between two or more Swi6 dimers on the nucleosome. Indeed, the MALS data from Figure 2 indicate that Swi6 can form tetramers and octamers in the absence of nucleosomes at high concentrations. Alternatively, the two Swi6 dimers may not directly interact, but binding by two or more dimers may be required to stably upshift the nucleosomes on a native gel.
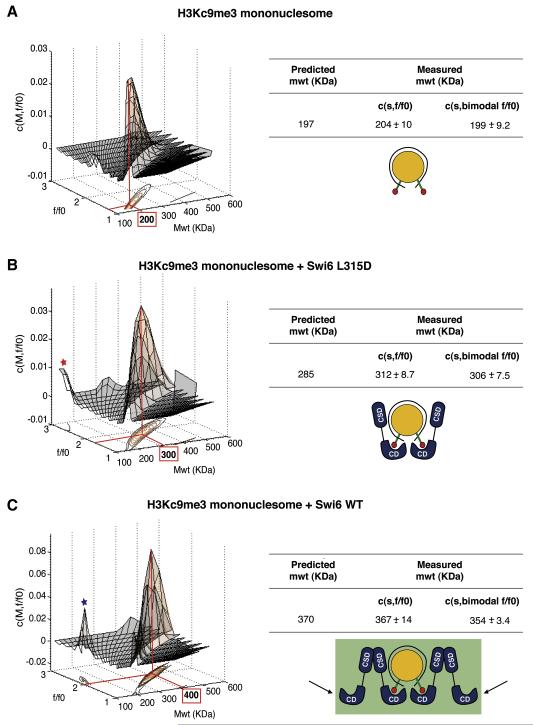
To determine how many Swi6 molecules directly interact with the nucleosome, we used sedimentation velocity analytical ultracentrifugation (SV-AUC). SV-AUC allows the differentiation of multiple species present in the sample based on their mass dependent migration. Recent improvements in the analysis tools for SV-AUC data allow the determination of masses of multi-protein complexes while directly accounting for differences in shapes (Brown and Schuck, 2006). We performed three independent experiments each for samples containing H3Kc9me3 core nucleosomes alone (Figure 4a), H3Kc9me3 core nucleosomes bound by L315D Swi6 (Figure 4b), or H3Kc9me3 core nucleosomes bound by WT Swi6 (Figure 4c). We used Swi6 and nucleosome concentrations based on titration experiments (See Experimental Procedures). Each experiment was analyzed using two independent models for data fitting: (i) a continuous two-dimensional function c(s,f/f0) for sedimentation coefficient s and hydrodynamic translational frictional ratio f/f0, (Figure S4a), and (ii) a continuous function c(s) for sedimentation coefficient s with a bi-modal f/f0 distribution (Figure S4b) (f/f0 is a measure of the shape of the complex, see Experimental Procedures).
Figure 4. The core unit of Swi6 binding to a mononucleosome is a tetramer.
(A) Sedimentation Velocity Analytical Ultracentrifugation (SV-AUC) on H3Kc9me3 core nucleosomes. Left: The c(M, f/f0) distribution generated from SV–AUC experiments shown as a two-dimensional distribution. x-axis: molecular weight (Mwt); y-axis: hydrodynamic translational frictional ratio (f/f0). Below, the c(M, f/f0) surface is shown as a contour plot of the distribution projected onto the M-f/f0 plane, where the magnitude of c(M, f/f0) is indicated by contour lines at constant c(M, f/f0) for equidistant intervals of c. Right: Table showing measured average masses (versus theoretically predicted masses) from three independent experiments using either a continuous two-dimensional function c(s,f/f0) for sedimentation coefficient s and hydrodynamic translational frictional ratio f/f0, or a continuous function c(s) with a bi-modal f/f0 distribution c(s, bimodal f/f0). Errors represent s.e.m.
(B) SV-AUC on H3Kc9me3 core nucleosome with L315D Swi6. Representation and table as in (A). Red asterisk: free L315D Swi6.
(C) SV-AUC on H3Kc9me3 core nucleosome with WT Swi6. Representation and table as in (A). Blue asterisk: free WT Swi6. Black arrows represent sticky ends. The measured masses are used to derive structural models for the stoichiometry of the complexes as shown.
Both analysis methods indicate that the majority of the complexes have a stoichiometry of four WT Swi6 proteins to one core nucleosome (Figure 4c). These findings suggest that two WT Swi6 dimers bind to the nucleosome to form a tetramer. Further, the molar mass obtained for the core nucleosome-L315D Swi6 complex reveals a stoichiometry of two Swi6 proteins to one core nucleosome (Figure 4b).
These observations suggest a model in which the two unoccupied Swi6 chromodomains in the Swi6 tetramer can serve as sticky ends (Figure 4c, black arrows) that can bind methyl marks on nearby nucleosomes. Binding of proximal nucleosomes via this specific type of sticky ends architecture would be predicted to energetically favor H3K9-specific binding orientations of the Swi6 tetramer over alternative binding orientations, resulting in greater specificity for the methyl mark. To test this hypothesis, we determined whether Swi6 binds with greater specificity to methylated di- and polynucleosome constructs compared to mononucleosomes.
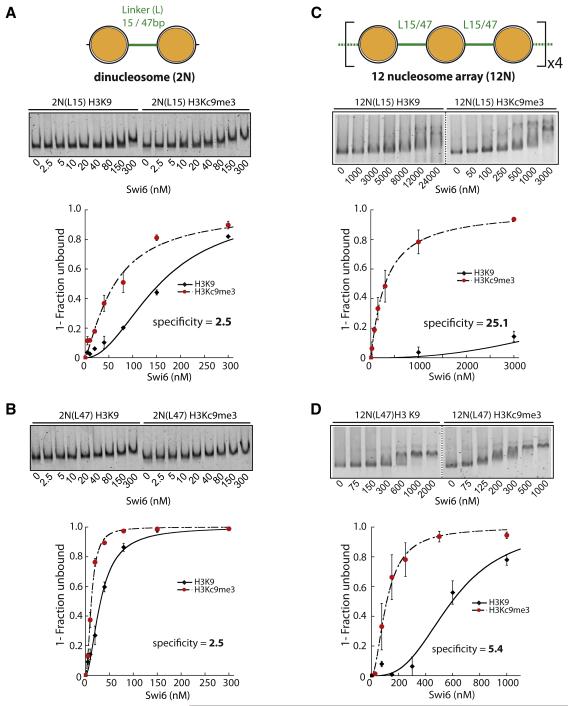
Swi6 binds with similar specificity to mono and dinucleosomes
Dinucleosomes were first assembled on a DNA construct containing 15 base pairs of linker DNA (L15) between two 601 positioning sequences (Figure 5a, diagram). The relatively short linker length is designed to mimic inter-nucleosomal distances prevalent in S. pombe (Godde and Widom, 1992; Lantermann et al., 2010). Native gel mobility shift assays show that Swi6 binds to methylated 2N(L15) with approximately 2.5-fold higher affinity than to the unmethylated control (Figure 5a). This specificity is comparable to that observed for mononucleosomes containing 20 bp of flanking DNA and is likely due to nonspecific binding of Swi6 to the linker DNA (Figure 3d and Fig. S3c). These results suggest that L15 dinucleosomes do not increase specific binding by Swi6. To test if these closely spaced nucleosomes sterically interfere with Swi6 binding to the H3 tails of both nucleosomes, we also measured Swi6 binding to a 2N(L47) dinucleosome, linked by 47 base pairs of DNA. Swi6 binds to methylated 2N(L47) with 2.5-fold higher affinity than unmethylated 2N(L47), ruling out a simple steric interference model (Figure 5b).
Figure 5. Amplification of Swi6 specificity towards H3Kc9me3 occurs on nucleosome arrays and is sensitive to nucleosomal placement.
Dinucleosome (2N) or 12-nucleosome arrays (12N) constructs contain either 15 bp (L15) or 47 bp (L47) internucleosomal linkers.
(A) Swi6 displays 2.5 fold specificity towards 2N(L15) H3Kc9me3 dinucleosomes. Representative gel shift shown. K1/2 for H3Kc9me3 and H3K9 2N(L15) substrates are 62 and 156 nM, respectively. Specificity = K1/2 H3Kc9me3 / K1/2 H3K9.
(B) Swi6 displays similar specificity towards H3Kc9me3 2N(L47) as for 2N(L15) dinucleosomes. Gel shift and analysis as in (A). K1/2 for H3Kc9me3 and H3K9 2N(L47) substrates were 12 and 32 nM, respectively.
(C) Swi6 displays ~10× amplified specificity towards H3Kc9me3 12N(L15) arrays vs. H3Kc9me3 2N(L15) dinucleosomes. Swi6-bound and unbound arrays were separated by agarose gel electrophoresis. Representative gel shift shown. K1/2 and specificity were determined as above.
(D) Amplification of Swi6 specificity on 12N(L15) arrays is reduced on 12N(L47) arrays. Gel shift and analysis as in (C).
K1/2 for array substrates: see Figure S6d. All error bars represent s.e.m. Swi6 concentrations represent monomer concentrations.
The above results suggest that, in the context of Swi6 binding, a dinucleosome substrate behaves like two unlinked mononucleosomes with flanking DNA and does not show any amplification of specificity. In vivo, however, Swi6 binds along many nucleosomes (Noma et al., 2001), leaving the possibility that the sticky ends mechanism may have evolved to have a larger effect in the context of a long stretch of nucleosomes.
Nucleosome arrays provide a highly specific substrate for Swi6
We next measured Swi6 binding to a 12 nucleosome array containing the same 15 base pair linker length as used in the dinucleosome construct (Figure 5c, diagram and Figure S5a). Native gel mobility shifts show that this 12N(L15) array substrate substantially increases Swi6 specificity for the methyl mark, to ~25-fold (Figure 5c & S5d,e). This represents a ~10-fold amplification in specificity compared to that measured for the corresponding dinucleosome construct. The large gain in specificity on nucleosomal arrays is consistent with our model (Figure 4c), in which bridging interactions between nucleosomes, mediated by vacant CD sticky ends, favor binding of Swi6 in H3K9-specific orientations over alternative orientations. The observation that H3K9me specificity is amplified only in the context of 12N arrays but not dinucleosome substrates, suggests that Swi6 bridging requires nucleosome conformations that cannot be accessed by dinucleosomes.
If bridging nearby nucleosomes is required for correctly orienting Swi6 complexes, then increasing the distance between nucleosomes on a 12N array is expected to reduce such bridging and result in lower specificity. We therefore measured the specificity of Swi6 for the methyl mark in the context of arrays with more widely spaced nucleosomes, containing 47 base pair linker DNA (Figure S5b). As predicted, Swi6 binds to the methylated 12N(L47) substrate with lower specificity (5.4-fold) than to methylated 12N(L15) arrays (Figure 5d). Moreover, we observe a greater reduction of specificity with increasing flanking DNA in the context of an array compared to a mononucleosome (Figure 3c & S3d). Together these results are consistent with a model in which appropriate nucleosome placement is important for bridging of proximal nucleosomes and for energetically favoring H3K9-specific binding orientations (see Supplemental Discussion).
Our finding that specificity for the methyl mark is amplified in a manner that is sensitive to internucleosomal distance is consistent with model in which the tetrameric Swi6 architecture depicted in Figure 4c enables bridging across nucleosomes. In this context, the intrinsic ability of Swi6 to form a tetrameric state (Figure 2c,d) suggests that, in addition to the CSD-CSD interface, there are other Swi6-Swi6 interfaces that promote tetramerization.
Swi6 tetramerization is mediated by the chromodomain
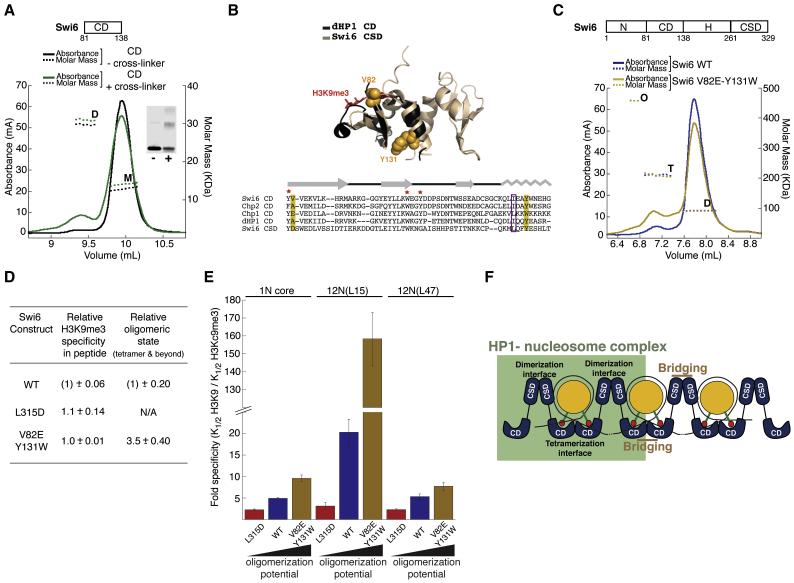
Because the CSD domain alone shows no oligomer formation beyond a dimer, even at concentrations where the intact Swi6 protein forms tetrameric species (Figure 2c and S2b), we used a domain deletion approach using Swi6 constructs lacking the CSD domain to identify the domain responsible for Swi6 tetramer formation. We used gel filtration to measure the extent of protein self-association for these proteins. At high concentrations, the CD alone (aa 81-137) is able to dimerize in solution, to the same extent as a Swi6 protein lacking only the CSD (NCDH) (Figure S6a). This suggested that the CD is the major component of the additional protein-protein interface. The weak CD self-association could be further stabilized by cross-linking (Figure 6a). The CD-CD interaction also helps explain the non-saturable addition of Swi6 to nucleosomes observed by SPR and fluorescence anisotropy.
Figure 6. The chromodomain contains the Swi6 tetramerization interface and couples tetramerization on the nucleosome surface to H3K9me3 recognition.
(A) The chromodomain of Swi6 can homodimerize. MALS measurements for uncross-linked (black) and cross-linked (green) Swi6 chromodomain (CD) showing UV absorbance signal in mA (solid lines, left y-axis) and derived molar masses (dotted lines, right y-axis) as a function of the elution volume. The CD was injected at ~50 μM. M: monomer, D:dimer. Cross-linked CD shows increased D. Inset: SDS-PAGE analysis for the uncross-linked (−) and cross-linked (+) samples used in the MALS measurements.
(B) Top: Superimposition of the structure of monomeric dHP1 CD (black, pdb 1KNE) with dimeric Swi6 CSD (light brown, pdb 1E0B) shows structural similarity between the two evolutionarily related domains. Bottom: Alignment of the CD of the three HP1-like proteins in S. pombe with dHP1 CD and Swi6 CSD. Yellow boxes: conserved residues V82 and Y131. Purple box: hydrophobic residue (L or I) central to CSD dimerization. Red star: CD hydrophobic cage residues required for H3K9me3 recognition. Gray: secondary structure schematic for dHP1 CD and Swi6 CSD.
(C) MALS measurements for WT Swi6 (blue) and V82E Y131W Swi6 (yellow), shows UV absorbance signal (solid lines, left y-axis) and derived molar masses (dotted lines, right y-axis) as a function of the elution volume. WT and V82E-Y131W Swi6 were injected at ~20 μM. The V82E-Y131W protein shows a higher proportion of species in tetrameric (T) and octameric (O) oligomeric states.
(D) Relationship between peptide specificity and oligomeric states (tetramer and beyond) for WT, CSD mutant (L315D) and the CD double mutant V82E-Y131W. H3K9me3 specificity for each protein is calculated as K1/2H3K9/K1/2H3K9me3. All data are reported as fold differences relative to the WT protein. Errors represent s.e.m.
(E) H3K9me3 specificity is regulated by the oligomeric state of Swi6. y-axis: Fold specificity for methylated mononucleosome (1N) and indicated 12N array substrates.
(F) A model to depict how the CSD-CSD and CD-CD interactions enable orientation of Swi6 to correctly recognize the methyl mark in a nucleosome and generate sticky ends that bridge nearby nucleosomes and further enhance specific orientations.Swi6 concentrations represent monomer concentrations.
Our data agree with previous reports that the CD of human HP1 forms higher-order oligomers when cross-linked (Yamada et al., 1999). However, the interface through which such CD-CD interactions occur has not yet been identified. Given the high level of structural similarity between the CD and the evolutionary related CSD (Figure 6b), we hypothesized that the region of the CD corresponding to the sole alpha helix in the CSD that is primarily responsible for CSD dimerization might play a similar role in CD self-association. This hypothesis was further supported by analysis of the previously determined crystal structure of the dHP1 CD (Jacobs and Khorasanizadeh, 2002). The crystallographic unit of this structure contains two CD monomers that appear to engage in contacts via the alpha helix (Figure S6b). Over 30 different point mutants were made in an attempt to reduce CD dimerization, but all of these also resulted in a loss of H3K9me3 peptide binding function (data not shown). We were, however, able to obtain two gain-of-function mutants that increase CD dimerization without significantly, or not all, disrupting peptide binding: the single mutant Y131W and the double mutant V82E-Y131W (Figures 6b). The single mutant V82E replaces a Swi6 residue with a residue normally found at this location in Chp1, another chromodomain containing protein in S. pombe (Schalch et al., 2009).
When introduced in the full-length protein, the Y131W single and the V82E-Y131W double mutant respectively displayed ~ 1.6-fold and ~3.5-fold increased tetramer formation over WT as determined by MALS, suggesting that this region of the CD is involved in Swi6 tetramerization (Figure 6c&d and S6c). The V82E single mutant by itself does not significantly increase tetramer formation (Figure S6c). We find that the V82E single substitution increases binding to H3K9me3 tail peptides by ~ 3-fold, consistent with previous work (Schalch et al., 2009). The double mutant V82E-Y131W however displays similar affinity for the H3K9me3 tail peptide as WT (Figure 6d).
Specificity for the methyl mark is dependent on both the CD-CD and the CSD-CSD interactions
The observation that CD-CD self association helps form Swi6 tetramers was particularly intriguing because our model, in which both H3K9 methyl marks are bound by CDs of different Swi6 dimers, places those two CDs in close proximity to self-associate (Figure 4c). We therefore hypothesized that binding in the specific orientation would strongly favor Swi6 tetramerization via CD-CD self-association and conversely, Swi6 tetramerization via self-association of two CDs would strongly favor binding in the specific orientation. If so, any disruption of the tetramer architecture depicted in Figure 4c would reduce specific recognition of the H3K9me3 mark on the nucleosome, while any strengthening of the specific architecture would increase specificity for the H3K9me3 mark.
To test these predictions, we measured specificity towards H3Kc9me3 core nucleosomes for WT Swi6 and for the L315D and V82E-Y131W mutants (Figures 6e & S6c). The L315D mutation, which significantly decreases higher-order oligomerization by disrupting CSD self-association (Figures 2c and S2a), displays 2.5-fold reduced specificity for methylated core nucleosomes relative to WT Swi6 (Figure 6e). Conversely, the V82E-Y131W double mutant, which increases tetramer formation 3.5-fold in solution by increasing CD self-association (Figure 6c&d), displays 2-fold increased specificity for methylated core nucleosomes (Figure 6e). Interestingly, both the L315D and the V82E-Y131W mutant proteins bind the H3K9me3 tail peptide with specificities comparable to the WT protein (Figure 6d). The observation that the mutations alter methyl mark discrimination only in the nucleosomal context suggests that the effects are a result of altered oligomerization states. These results indicate that specific recognition of the nucleosomal H3K9me3 mark by Swi6 is dependent on both CSD-mediated dimerization and CD-mediated tetramerization on the nucleosome surface.
A specific CD-CD interface implies that the sticky ends that bridge nearby nucleosomes would entail CD-CD interactions in addition to interactions between the unoccupied CD and a nearby methyl mark. Therefore, in the context of nucleosomal arrays, the CD-CD interaction would further promote the H3K9 specific orientations via bridging interactions with nearby nucleosomes (see also Supplementary discussion). Such a model then makes two key predictions: (i) strengthening the CD-CD interaction would increase the specificity on nucleosomal arrays to a greater extent than on mononucleosomes, and (ii) any amplification of specificity would be very sensitive to the inter-nucleosomal distance. To test these predictions, we compared the specificity of the V82E-Y131W mutant to that of WT Swi6 on the 12N(L15) and 12N(L47) nucleosomal arrays.
As predicted by the model, we found that the V82E-Y131W mutant shows a large increase in specificity (~7-fold) on the 12N(L15) arrays compared to WT Swi6 (Figure 6e). Interestingly, this raises the specificity for the H3K9me3 mark to ~130 fold. Further, most of the observed gain in specificity arises from a large decrease in binding to the H3K9 array and a small increase in binding to the H3Kc9me3 array (Figure S6d). These results suggest that the combination of strengthening the CD-CD interface and binding across multiple nucleosomes eliminates most of the alternative binding modes adopted by Swi6. No significant amplification of specificity is observed in the context of the 12N(L47), confirming that the CD-CD nucleosome bridging interaction is sensitive to inter-nucleosomal distance (Figure 6e).
To further investigate the role of Swi6 oligomerization in the context of nucleosomal arrays, we tested the effects of disrupting the CSD-CSD interface, which is also expected to disrupt the ability of Swi6 dimers to bridge across nucleosomes (Figure 6e). The L315D mutant shows greatly reduced specificity on the 12N(L15) template. Intriguingly, the L315D mutant discriminates between methylated and unmethylated 12N(L15) arrays to a similar degree (2-3 fold) as in the context of 12N(L47) arrays and mononucleosomes (Figure 6e). Therefore, the L315D Swi6 mutant is insensitive to the distance between nucleosomes. The L315D mutant thus uncovers the baseline ability of HP1 proteins to recognize a nucleosomal H3K9 methyl mark in the absence of significant oligomerization and nucleosome bridging-dependent effects.
Increased tetramerization of Swi6 results in increased silencing at an artificial heterochromatic locus and higher recruitment to centromeres
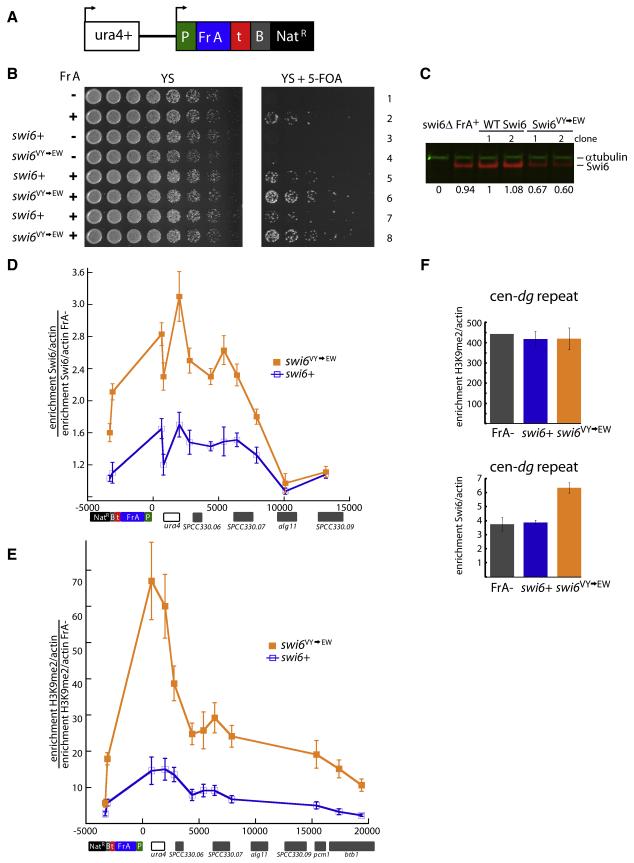
To test whether these biochemically derived mechanistic conclusions are relevant to the ability of Swi6 to form functional heterochromatin in vivo, we investigated whether strengthening the CD-CD interface via the V82E-Y131W double mutant causes enhanced silencing and Swi6 occupancy in vivo.
To test for such an effect, we utilized a reporter system that measures silencing of the ura4+ gene at its endogenous location on Chromosome 3 (S.S., K. Finn, H.D.M., unpublished). In this reporter construct, a centromeric fragment, under control of a promoter, is inserted 1.8 kb downstream of the ura4+ gene (Figure 7a). We chose a 1.7 kb fragment (Fragment A; Fr A) from a library of fragments derived from the centromeric dh repeats. Fr A shows very weak silencing of the ura4+ gene, leading to minimal growth of cells on 5-FOA, which provides a sensitive assay for mutants that enhance silencing (Figure 7b). We introduced the swi6V82E, Y131W allele by chromosomal integration into strains containing Fr A. As a control, we constructed isogenic swi6+ strains. To control for strain-to-strain variability, we isolated and characterized 6 independent genetic isolates for both swi6V82E, Y131W and swi6+ alleles in the Fr A background. As shown in Figure 7b for two independent strains, swi6V82E, Y131W increased Fr A-dependent silencing of ura4+ (compare rows 6 and 8 to rows 5 and 7). A side by side comparison of all 6 independent isolates of the swi6+ and swi6V82E, Y131W alleles further confirms stronger growth on 5-FOA for all the swi6V82E, Y131W strains (Fig. S7). The mutant Swi6 protein is not expressed at a higher level than the WT Swi6 protein, ruling out a trivial explanation for the gain of silencing effects (Figure 7c).
Figure 7. Increased tetramerization of Swi6 translates into increased silencing and heterochromatin spreading at an artificial heterochromatic locus.
(A) Schematic of the reporter cassette integrated downstream of endogenous ura4+ gene. Cassette contains a promoter (P) driving the expression of a centromeric dh fragment (Fr A), an intergenic region from two convergent regions (t), a boundary element (B) that contains synthetic TFIIIC binding sites (known to limit the spread of heterochromatin in S. pombe) and a Nat drug resistance marker (NatR).
(B) The V82E-Y131W mutant shows increased silencing of the fragment A cassette (Fr A). Serial dilutions of indicated S. pombe strains. Strains containing Fr A show silencing of ura4+ and are able to grow on media containing 5-FOA. swi6+ or swi6VY→EW alleles were introduced into strains containing the whole cassette with or without Fr A (Fr A− ); 2 independent clones are shown for each swi6 allele. Fr A− strains contain the entire cassette as shown in (A) but lack the centromeric fragment.
(C) The V82E-Y131W mutant expresses slightly lower levels of Swi6 than WT. Extracts from respective strains were separated on SDS-PAGE gels and probed for α-tubulin (green) or Swi6 (red). Quantification of the Swi6 band normalized for the α-tubulin control is shown relative to the value obtained for swi6+ clone 1.
(D) The Swi6 V82E-Y131W mutation induces increased Swi6 recruitment to the Fr A locus. Chromatin immunoprecipitation (ChIP) with anti-Swi6 antisera was performed in swi6+, swi6 VY→EW or Fr A− backgrounds. Fr A specific Swi6 enrichment is represented as the ratio of the actin-normalized signal at indicated amplicons in swi6+ or swi6VY→EW strains divided by the actin-normalized signal in the Fr A− strain. Error bars represent s.e.m. of unicate ChIP experiments from the 6 genetic isolates of swi6+ or swi6VY→EW alleles. X-axis: distance in base pairs relative to the Fr A cassette promoter (P). Genomic features near the Fr A cassette insertion site are aligned below the graph.
(E) The Swi6 V82E-Y131W mutation increases H3K9 methylation at and beyond the Fr A cassette. ChIP experiments were performed with anti-H3K9me2 antisera. The Fr A specific H3K9me2 enrichment is calculated as in (D). Error bars as in (D).
(F) The Swi6 V82E-Y131W mutation leads to increased Swi6 recruitment at endogenous heterochromatin. Top: H3K9me2 ChIP. H3K9me2 fold enrichment over actin at the centromeric dg repeat for the Fr A- strain and swi6+ or swi6VY→EW alleles. Bottom: Swi6 ChIP. Swi6 fold enrichment over actin at the centromeric dg repeat for the same strains as (Top). Error bars for swi6+ and swi6VY→EW as in (D). Error bar for Fr A− (bottom) represents s.e.m. for three independent IPs from the Fr A− strain.
We next probed the molecular features of the silenced region using ChIP. We first examined Swi6 localization across the Fr A cassette locus, and found a reproducible 2-3 fold increase in Swi6 enrichment in the swi6V82E, Y131W alleles versus the swi6+ alleles (Figure 7d), consistent with the increased specific binding observed on nucleosomal arrays in vitro. However, the overall enrichment was low, probably reflecting the low degree of silencing at this artificial locus. Next, we examined H3K9me2 levels at and around the Fr A locus. Since Fr A-dependent Swi6 localization spreads beyond Fr A into adjacent euchromatic regions at the unbounded 5′ end (Figure 7d), H3K9 methylation may also exhibit some Swi6-dependent spread (Hall et al., 2002). Indeed, we found that H3K9me2 levels are robustly increased in the swi6V82E, Y131W alleles, and remain elevated at regions well outside (~20 kb) the Fr A initiating element (Figure 7e). The fact that H3K9me2 enrichment can be observed outside the zone of detectable Swi6 enrichment is likely due to the differential sensitivity of the two ChIP experiments. Increased localization of Swi6 in the context of the V82E-Y131W mutation, concomitant with robustly increased H3K9me2 levels and elevated ura4+ silencing, suggests that increasing the oligomerization capacity of the Swi6 protein enhances the ability of Swi6 to establish and spread heterochromatin at the artificial locus.
Next we asked whether the V82E-Y131W mutation has an effect on Swi6 activity at endogenous heterochromatin loci. We examined recruitment of Swi6 by ChIP at the dg repeat of centromere 1. Since H3K9 methylation at the centromere is Swi6-independent (Nakayama et al., 2001) examining centromeric heterochromatin should allow us to uncouple Swi6 recruitment from deposition of H3K9 methylation. In such a situation, changes in Swi6 recruitment should directly report on the ability of the protein to recognize H3K9 methylated chromatin in vivo. Indeed, when we examined H3K9me2 methylation at the dg repeat in the no Fr A control, swi6+ and swi6V82E, Y131W Fr A-containing alleles, we found no change in the enrichment level of H3K9me2 at the dg repeat (Figure 7f, top panel). In contrast, when we tested for Swi6 recruitment, we found a small but reproducible increase of Swi6 residence at the dg repeat only in the context of the Swi6 V82E-Y131W mutant (Figure 7f, bottom panel). This result suggests that when Swi6 oligomerization is increased, Swi6 recruitment is increased at endogenous heterochromatin loci where H3K9 methylation is Swi6-independent. These data help strengthen our model that CD-mediated oligomerization is critical for Swi6-dependent heterochromatin formation.
Discussion
To understand how HP1 proteins assemble on physiological chromatin templates, we reconstituted and characterized the core HP1-H3K9me3 chromatin complex. Our data suggest that recognition of H3K9me3 by HP1 proteins is coupled to its oligomerization on the nucleosome through a chromodomain-chromodomain interface that promotes silencing in vivo. The mechanistic implications of these observations are discussed below.
A new role for the HP1 chromodomain in heterochromatin formation
Swi6 mutants that have increased tetramerization mediated via the CD-CD interaction exhibit increased specificity for H3K9me3 nucleosomes, suggesting that interactions between two CDs on a nucleosome restricts the number of non-H3K9me3 specific binding modes. Mutants that increase tetramerization, and thus H3K9me3 specificity in vitro, also exhibit increased heterochromatin spread and silencing at an artificially induced heterochromatic locus in vivo. The CD of HP1 proteins was previously known to recognize peptides containing methylated H3K9 (Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002). Our work suggests that the CD has an additional critical role in the context of chromatin: orienting HP1 proteins via CD-CD interactions to ensure that HP1 proteins can distinguish the methyl mark from other overlapping binding surfaces presented by a nucleosome.
Sticky end chromodomains in the Swi6 tetramer-nucleosome complex present polymerizable surfaces for higher order oligomerization across chromatin
Our data imply that H3K9me3 recognition and chromatin coating by Swi6 are mechanistically coupled and intrinsic to the fundamental architecture of the tetrameric HP1/Swi6 complex on the nucleosome (Figure 6f) as follows: i) Dimerization via the strong CSD-CSD interaction and tetramerization via the weaker CD-CD interaction couples recognition of the two methyl marks in a nucleosome to the generation of two unoccupied CDs. These unoccupied CDs can serve as sticky ends that bridge and recruit neighboring methylated nucleosomes, which might be either adjacent or located on different chromatin fibers. ii) Interactions between the CD of a chromatin bound HP1 dimer and that of an incoming HP1 dimer can promote deposition of the incoming HP1 dimer in an H3K9me3-recognizing orientation.
This ability to bridge nucleosomes via polymerizable CDs may represent the primary underlying mechanism that allows HP1 proteins to spread (Hall et al., 2002) along the chromatin fiber and establish the extent of the heterochromatic domain. Further, since this mechanism is dependent on a high density of H3K9me3 methylation on chromatin (Nakayama et al., 2001; Noma et al., 2001), it may enable Swi6 to sense regions of high local Clr4 methylase activity, preventing ectopic heterochromatin formation. Coupling between oligomerization and recognition of H3K9methyl marks has also been proposed in the context of the vertebrate chromodomain containing protein CDYL1b (Franz et al., 2009).
Chromatin architecture and implications for heterochromatin spread
The bridging architecture depicted in Figure 6f places specific steric and distance constraints on any Swi6-mediated heterochromatin assembly and spread, restricting the number of chromatin architectures accessible to heterochromatin assembly by Swi6. In fact, we find that Swi6 gains specificity on nucleosome arrays over unlinked nucleosomes only in the context of short DNA linkers (Figures 5c&d and 6e). We therefore hypothesize that HP1 proteins assess both the nucleosome arrangement in addition to the H3K9me3 mark, thereby integrating two signals for heterochromatin assembly.
If only a subset of chromatin architectures is permissive to template the assembly of HP1 proteins on H3K9me3 chromatin, such architectures might be regulated in vivo to allow specification of HP1 protein binding sites. In fact, in metazoans, the nucleosome architecture of heterochromatic loci shows significant differences compared to euchromatin sites. For example, in Drosophila melanogaster, constitutive heterochromatin is characterized by more evenly spaced nucleosomes (Danzer and Wallrath, 2004; Wallrath and Elgin, 1995) compared to euchromatin. It has been suggested that the ACF chromatin remodeling complex is involved in generating such chromatin architectures in Drosophila (Fyodorov et al., 2004). In S. pombe, there is some evidence that local nucleosome arrangement in heterochromatin impacts Swi6 association. Several protein complexes collaborate in S. pombe to maintain heterochromatin regions. A key such effector is a bi-functional enzyme complex called SHREC, containing both the histone deacetylase (HDAC) Clr3 and the SNF2 chromatin-remodeling factor homolog Mit1 (Sugiyama et al., 2007). The Clr3 subunit of SHREC is required for Swi6 localization, in a manner that appears uncoupled from its effects on H3K9 methylation (Nakayama et al., 2001; Yamada et al., 2005); S.I.S. Grewal, personal communication). This effect may result either from 1) the absence of Clr3’s HDAC activity, resulting in an increase in acetylated and phosphorylated histones that may affect Swi6’s ability to associate with those nucleosomes (Yamada et al., 2005), or 2) effects of the SHREC complex on nucleosome arrangement (Sugiyama et al., 2007). We speculate that SHREC and/or other chromatin regulators may promote a nucleosome arrangement that enables Swi6 to bridge H3K9me3-marked nucleosomes and therefore to spread. Further work will be needed to identify what exact chromatin architectures are compatible with Swi6 bridging and how such structures may be generated and maintained in vivo.
Experimental Procedures
(Detailed Methods are described in “Extended Experimental Procedures” in Supplementary Information)
Protein cloning and purification
Full length Swi6 was cloned into pET30a (Novagen), mutants made using site directed mutagenesis and proteins purified from E. coli. Tagged Swi6 containing N-terminal 6xHis and C-terminal FLAG tags was used for the MALS, AUC, cross-linking, native gel-shift, nucleosome competition and peptide binding assays. Untagged Swi6 was used for the SPR-based and polarization-based nucleosome binding measurements. Removing the tags slightly increases overall affinity for nucleosomes and arrays (~2.5 fold) but does not affect specificity (data not shown).
Mononucleosome, dinucleosome, and array reconstitution
Gradient salt dialysis was used to assemble mononucleosomes on DNA templates containing the 147 bp long 601 positioning sequence, dinucleosomes on DNA templates containing two 601 sequences linked by 15 or 47 bp of DNA and arrays on DNA templates containing 12 copies of the 601 nucleosome positioning sequence separated by either 15 or 47 bp linkers. H3Kc9me3, H3Kc9me0 and H3K9 histones were prepared as described (Luger et al., 1999; Simon et al., 2007). Arrays with >95% assembly were used for gel shift assays.
Surface plasmon resonance
A BiaCore T100 instrument was used for SPR analysis of Swi6 interaction with the mononucleosome substrates. H3K9 and H3Kc9me3 substrates assembled on a 5′ biotinylated 601 sequence were immobilized to 25 to 60 RU on active flow cells. Immobilization levels of mononucleosomes ranged from 25 to 60 RU. Surface stability and assay quality were judged by the reproducibility of a 10 μL control sample (100 nM Swi6) injection that followed each sample concentration (Figure S1d).
Fluorescence polarization binding measurements
All H3 tail peptides were produced as described (Simon et al., 2007). Nucleosomes for polarization-based binding measurements were assembled on a 6-carboxyfluorescein-labeled 601 positioning sequence.
Native gel mobility shift assays
Different concentrations of Swi6 protein were incubated with 5-10 nM mononucleosome or 1.25 nM dinucleosome. Samples were run on native polyacrylamide gels, stained with SyBR Gold (Invitrogen), visualized on a Molecular Dynamics Typhoon scanner and quantified using Image Quant software. The K1/2 for each binding curve and Hill coefficient were calculated with Kaleidograph software using a simple equilibrium model. Swi6 gel shifts with 12N arrays were performed using agarose gels with 1 nM array (12 nM nucleosomes) and analyzed as described for mononucleosomes.
Protein cross-linking
Cross-linking assays were performed using EDC/NHS cross-linking (Pierce) (see Extended Experimental Procedures). The samples were boiled and analyzed on 4-12% NuPAGE gradient gels (Invitrogen) under denaturing conditions, then visualized on a Typhoon scanner by Sypro Red staining or by anti-FLAG western blotting.
Isothermal titration calorimetry
The heat released by dissociation of CSD dimers into monomers was measured with a Microcal, Inc., Omega microcalorimeter. Dilution ITC experiments involved sequential injections from a concentrated protein stock (5 μM for WT Swi6 and 625 μM for L315D Swi6) in 5 μl increments into the 1.4 ml calorimeter cell initially containing only buffer.
Size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS/UV/RI)
Protein samples were injected into an analytical size exclusion silica gel KW804 chromatography column (Shodex). The chromatography system was coupled to an 18-angle light scattering detector (DAWN HELEOS II, Wyatt Technology) and a differential refractometer (Optilab rEX, Wyatt Technology).
Sedimentation velocity analytical ultracentrifugation
Sedimentation velocity experiments were conducted using an analytical ultracentrifuge (Beckman Coulter) equipped with an absorption optical scanner. The binding reaction was set up such that a) both nucleosome and Swi6 concentrations were above the K1/2 value measured by native gel and b) the Swi6 concentration was sufficient to titrate all the nucleosomes as assayed by native gel shift.
Data were analyzed using the Sedfit software (Schuck, 2004). Three independent analyses: c(s), c(s,fr), and c(s,bi-modal fr) were used to study the sedimentation properties and the molar mass of each sample. Solution density (ρ), solution viscosity (η) were calculated in SEDNTERP (Schuck, 2004).
In vivo silencing assays
A fragment of the dh centromeric repeat was placed 1.8 kb downstream of the ura4+ gene on chromosome III using homologous recombination. Transcription of this fragment is driven by the padh1+ promoter and is sufficient to induce silencing of ura4+ in a manner that requires clr4+ and dcr1+ (S.S., K. Finn and H.D.M., unpublished). Silencing of ura4+ gene was assayed by growth on 2 mg/mL 5-fluoroorotic acid (5-FOA). The endogenous swi6+ was replaced with either swi6V82E,Y131W marked with a 5′ G418R selectable marker, or the wild-type allele and the same marker.
Chromatin immunoprecipitation (ChIP)
ChIP experiments with Fr A−, swi6+ and swi6V82E,Y131W strains were performed using anti-H3K9me2 (Abcam) or anti-Swi6 (Nakayama et al., 2000) antisera. Details see Extended Experimental procedures.
Supplementary Material
Acknowledgements
We thank Shiv I.S. Grewal for a generous gift of anti-Swi6 antisera, Christine Rumpf for help with ITC measurements, Dan Southworth and Janet Yang for assistance with MALS set-up, Kalyan Sinha and Peter Schuck for critical advice and comments on the AUC experiments and data interpretation and Robert Fletterick for critical advice and comments on the SPR experiments and data interpretation. We thank Barbara Panning, Jonathan Weissman, Karim-Jean Armache and members of the Narlikar Lab for helpful discussion and comments on the manuscript. This work was supported by grants from the National Institutes of Health (5R01GM071801 to H.D.M. and 1R01GM073767 to G.J.N.), the UCSF Program for Breakthrough Biomedical Research (PBBR) Award to G.J.N, and the Beckman Foundation (to G.J.N.). G.J.N. and H.D.M. are Scholars of the Leukemia and Lymphoma Society. D.C. is a Genentech, Inc. Predoctoral Fellow. E.C. is a ARCS Foundation, Inc. Fellow. S.S. is a fellow of the Leukemia and Lymphoma Society. K.M.K is supported by a training grant to the Biophysics Graduate Group (NIH T32GM008284).M.D.S. is a fellow of the Helen Hay Whitney foundation. B.A. is a fellow of the Jane Coffin Childs Memorial Fund for Medical Research. D.C. wishes to dedicate this work to the memory of Dr. D.O. Harris for his guidance throughout his undergraduate carrier.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr Biol. 2000;10:517–525. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- Danzer JR, Wallrath LL. Mechanisms of HP1-mediated gene silencing in Drosophila. Development. 2004;131:3571–3580. doi: 10.1242/dev.01223. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1 alpha from chromatin. Nature. 2009;461:819–U879. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, Grewal SIS. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci USA. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz H, Mosch K, Soeroes S, Urlaub H, Fischle W. Multimerization and H3K9me3 binding are required for CDYL1b heterochromatin association. J Biol Chem. 2009;284:35049–35059. doi: 10.1074/jbc.M109.052332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 2004;18:170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godde JS, Widom J. CHROMATIN STRUCTURE OF SCHIZOSACCHAROMYCES-POMBE - A NUCLEOSOME REPEAT LENGTH THAT IS SHORTER THAN THE CHROMATOSOMAL DNA LENGTH. Journal of Molecular Biology. 1992;226:1009–1025. doi: 10.1016/0022-2836(92)91049-u. [DOI] [PubMed] [Google Scholar]
- Grewal SIS, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma KI, Ayoub N, Cohen A, Grewal SIS. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Iida T, Nakayama J.-i., Moazed D. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol Cell. 2008;31:178–189. doi: 10.1016/j.molcel.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nature Structural & Molecular Biology. 2010;17:251–U215. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- Lavigne M, Eskeland R, Azebi S, Saint-Andre V, Jang SM, Batsche E, Fan HY, Kingston RE, Imhof A, Muchardt C. Interaction of HP1 and Brg1/Brm with the globular domain of histone H3 is required for HP1-mediated repression. PLoS Genet. 2009;5:e1000769. doi: 10.1371/journal.pgen.1000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Chromatin. Academic Press Inc; San Diego: 1999. Preparation of nucleosome core particle from recombinant histones; pp. 3–19. [DOI] [PubMed] [Google Scholar]
- Meehan RR, Kao CF, Pennings S. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. Embo Journal. 2003;22:3164–3174. doi: 10.1093/emboj/cdg306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Klar AJ, Grewal SI. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SIS. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Sadaie M, Kawaguchi R, Ohtani Y, Arisaka F, Tanaka K, Shirahige K, Nakayama J. Balance between Distinct HP1 Family Proteins Controls Heterochromatin Assembly in Fission Yeast. Molecular and Cellular Biology. 2008;28:6973–6988. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Job G, Noffsinger VJ, Shanker S, Kuscu C, Joshua-Tor L, Partridge JF. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol Cell. 2009;34:36–46. doi: 10.1016/j.molcel.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. A model for sedimentation in inhomogeneous media. I. Dynamic density gradients from sedimenting co-solutes. Biophysical Chemistry. 2004;108:187–200. doi: 10.1016/j.bpc.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers JF, Henikoff S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K.-i., Zofall M, Kobayashi R, Grewal SIS. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Wallrath LL, Elgin SCR. Position Effect Variegation in Drosophila Is Associated with an Altered Chromatin Structure. Genes & Development. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SIS. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell. 2005;20:173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Yamada T, Fukuda R, Himeno M, Sugimoto K. Functional domain structure of human heterochromatin protein HP1 (Hsalpha): involvement of internal DNA-binding and C-terminal self-association domains in the formation of discrete dots in interphase nuclei. J Biochem. 1999;125:832–837. doi: 10.1093/oxfordjournals.jbchem.a022356. [DOI] [PubMed] [Google Scholar]
- Zhao T, Heyduk T, Allis CD, Eissenberg JC. Heterochromatin protein 1 binds to nucleosomes and DNA in vitro. J Biol Chem. 2000;275:28332–28338. doi: 10.1074/jbc.M003493200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.