Abstract
Small RNAs regulate a multitude of cellular processes, including development, stress responses, metabolism, and maintenance of genome integrity, in a sequence-specific manner. Accumulating evidence reveals that host endogenous small RNAs and small RNA pathway components play important roles in plant immune responses against various pathogens, including bacteria, fungi, oomycetes, and viruses. Small-RNA-mediated defense responses are regulated through diverse pathways and the components of these pathways, including Dicer-like proteins, RNA-dependent RNA polymerases, Argonaute proteins, and RNA polymerase IV and V, exhibit functional specificities as well as redundancy. In this review, we summarize the recent insights revealed mainly through the examination of two model plants, Arabidopsis and rice, with a primary focus on our emerging understanding of how these small RNA pathway components contribute to plant immunity.
Plants have evolved multiple layers of immune responses to detect and defend themselves against various invading pathogens such as bacteria, fungi, oomycetes, and viruses. The first layer involves the detection of conserved pathogen-associated molecular patterns (PAMPs), which triggers PAMP-triggered immunity (PTI) that limits pathogen spread (Chisholm et al. 2006; Jones and Dangl 2006). In order to survive and propagate, some pathogens have evolved effector proteins, which can suppress PTI. To prevent infection, plants have evolved resistance (R) proteins that recognize the effectors and activate effector-triggered immunity (ETI), a more rapid and robust immune defense response (Chisholm et al. 2006; Jones and Dangl 2006). Increasing evidence has shown the important roles of small RNAs (sRNAs) in the regulation of these intricate defense responses against pathogens (Katiyar-Agarwal and Jin 2010; Padmanabhan et al. 2009; Ruiz-Ferrer and Voinnet 2009).
sRNAs are short, noncoding RNA molecules that guide silencing of genes either through transcriptional gene silencing (TGS) or post-transcriptional gene silencing (PTGS) (Baulcombe 2004). They are divided into two subgroups, micro-RNAs (miRNAs) and short interfering RNAs (siRNAs), based on their origin and biogenesis. In plants, siRNAs can be further categorized as trans-acting siRNAs (ta-siRNAs), heterochromatic siRNAs (hc-siRNAs), natural antisense transcript-derived siRNAs (nat-siRNAs), or long siRNAs (lsiRNAs) (Chapman and Carrington 2007; Katiyar-Agarwal and Jin 2010). Over the last decade, extensive studies have unveiled the diverse pathways, including their various cellular components, which regulate sRNA biogenesis and mode of action (Baulcombe 2004; Chapman and Carrington 2007; Katiyar-Agarwal and Jin 2010; Kim 2005). Although there are differences among the pathways, they generally involve three critical reactions: i) generation of double-stranded RNA (dsRNA), ii) processing of dsRNA into sRNAs, and iii) sequence-specific targeting and silencing by sRNA-incorporated effector complexes.
Although sRNAs have regulatory roles in a multitude of basic biological processes, including immune responses, accumulating evidence indicates that the cellular components associated in sRNA pathways are also directly involved in defense responses to pathogens, as schematically summarized in Figure 1. Dicer-like proteins (DCL), RNA-dependent RNA polymerases (RDR), and Argonaute (AGO) proteins are key components in sRNA pathways (Baulcombe 2004; Chapman and Carrington 2007; Vaucheret 2006). In addition, plant-specific RNA polymerase (Pol) IV and Pol V are essential for the biogenesis and function of hc-siRNAs, which mediate TGS by RNA-directed DNA methylation (RdDM) or histone modification (Haag and Pikaard 2011). Plant sRNA components, in particular, exhibit considerable variation and partial functional redundancy because they are encoded by multi-protein families (e.g., Arabidopsis has four DCL, six RDR, and 10 AGO proteins; and rice has eight DCL, five RDR, and 19 AGO proteins). Some of these components are involved in sRNA-mediated plant defense responses (Table 1). This review will focus on the functions of sRNA pathway components in plant immunity.
Fig. 1.
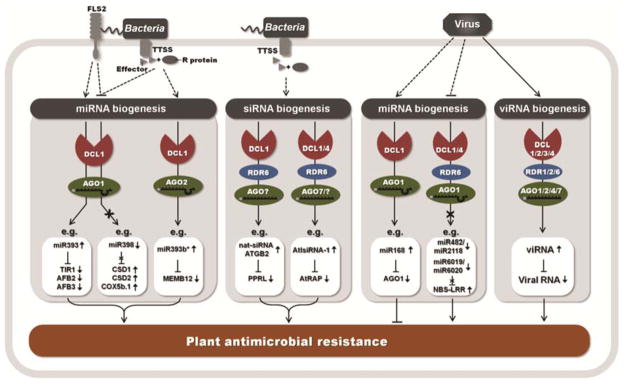
Plant small RNAs (sRNAs) and RNA interference pathway components contribute to plant immunity. Plants modulate sRNA pathways upon recognition of pathogen-associated molecular patterns or effectors of pathogens. Each pathway involves a distinct context of sRNA pathway components. FLS2, a flagellin-sensitive receptor kinase; TTSS, type III secretion system; miRNA, microRNA; siRNA, short interfering RNA; viRNA, virus-derived siRNA; DCL, Dicer-like protein; AGO, Argonaute; RDR, RNA-dependent RNA polymerase; nat-siRNA, natural antisense transcript-derived siRNA.
Table 1.
Diversity of the essential components involved in small-RNA-mediated plant immunity
| Componenta | Function in antimicrobial resistanceb | References |
|---|---|---|
| DCL | ||
| DCL1 | Biogenesis of miRNA, nat-siRNA, and lsiRNA involved in bacterial resistance and viRNA production (from DNA viruses) | Baulcombe 2004; Blevins et al. 2006; Katiyar-Agarwal et al. 2006, 2007; Moissiard and Voinnet 2006; Voinnet 2009 |
| DCL2 | viRNA biogenesis | Bouche et al. 2006; Deleris et al. 2006; Diaz-Pendon et al. 2007 |
| DCL3 | hc-siRNA and viRNA biogenesis | Baulcombe 2004; Bouche et al. 2006; Deleris et al. 2006; Diaz-Pendon et al. 2007 |
| DCL4 | Biogenesis of lsiRNA involved in bacterial resistance and miRNA and viRNA involved in viral resistance | Bouche et al. 2006; Deleris et al. 2006; Diaz-Pendon et al. 2007; Katiyar-Agarwal et al. 2007; Li et al. 2012 |
| dsRNA | ||
| HYL1 | Biogenesis of miRNA, nat-siRNA and lsiRNA involved in bacterial resistance | Katiyar-Agarwal et al. 2006, 2007; Voinnet 2009 |
| DRB4 | viRNA biogenesis | Ding 2010; Jakubiec et al. 2012 |
| RDR | ||
| RDR1 | viRNA biogenesis | Ding 2010; Ding and Voinnet 2007; Qi et al. 2009; Qu 2010 |
| RDR2 | viRNA biogenesis | Donaire et al. 2008; Garcia-Ruiz et al. 2010 |
| RDR6 | Biogenesis of ta-siRNA, nat-siRNA, lsiRNA and secondary siRNA involved in bacterial resistance and viRNA production | Ding 2010; Ding and Voinnet 2007; Katiyar-Agarwal et al. 2007; Li et al. 2012; Qi et al. 2009; Qu 2010; Shivaprasad et al. 2012 |
| AGO | ||
| AGO1 | miRNA-directed gene silencing and viRNA-directed viral RNA silencing | Li et al. 2010; Mallory and Vaucheret 2010; Morel et al. 2002; Navarro et al. 2006; Qu et al. 2008 |
| AGO2 | miRNA-directed gene silencing and viRNA-directed viral RNA silencing | Harvey et al. 2011; Jaubert et al. 2011; Wang et al. 2011; Zhang et al. 2011b, 2012 |
| AGO4 | viRNA-directed inactivation of DNA viruses | Buchmann et al. 2009; Ding 2010; Raja et al. 2008 |
| AGO7 | Biogenesis of lsiRNA involved in antibacterial resistance, ta-siRNA-directed gene silencing, and viRNA production | Fahlgren et al. 2006; Katiyar-Agarwal et al. 2007; Montgomery et al. 2008; Morel et al. 2002; Qu et al. 2008 |
Protein family and component. DCL, Dicer-like protein; dsRNA, double-stranded RNA binding protein; RDR, RNA-dependent RNA polymerase; and AGO, Argonaute protein.
miRNA, microRNA; siRNA, short interfering RNA; nat-siRNA, natural antisense transcript-derived siRNA; lsiRNA, long siRNA; viRNA, virus-derived siRNA; hc-siRNA, heterochromatic siRNAs; ta-siRNA, trans-acting siRNAs.
DCL and dsRNA-binding proteins
DCL belong to the RNase III family of endoribonucleases that specifically process dsRNAs into sRNA duplexes. DCL typically contain a helicase domain, a DUF283 domain, a PAZ domain, two tandem RNase III domains, and two tandem dsRNA-binding domains (Carmell and Hannon 2004). Arabidopsis encodes four DCL (DCL1 to DCL4) (Baulcombe 2004; Chapman and Carrington 2007; Katiyar-Agarwal and Jin 2010). DCL1 predominantly generates miRNAs but is also involved in producing some endogenous siRNAs, such as nat-siRNAs and lsiRNAs (Borsani et al. 2005; Katiyar-Agarwal and Jin 2010; Katiyar-Agarwal et al. 2006, 2007; Ron et al. 2010; Voinnet 2009; Zhang et al. 2012). DCL2, DCL3, and DCL4 mainly process long dsRNA precursors produced by RDR, natural antisense transcription, or inverted repeats (Baulcombe 2004; Chapman and Carrington 2007; Katiyar-Agarwal and Jin 2010). DCL3 is responsible mainly for the processing of Pol IV- and RDR2-generated dsRNAs and gives rise to hc-siRNAs that guide RdDM at target genomic loci (Baulcombe 2004; Law et al. 2010). DCL4 takes part in the generation of predominantly 21-nucleotide (nt) ta-siRNAs in an RDR6-dependent manner (Chapman and Carrington 2007). A recent study revealed that DCL4 also has a siRNA-independent function in cleaving nascent transcripts and promoting transcription termination of the FCA gene (Liu et al. 2012).
DCL play an important role in conferring bacterial resistance. The dcl1 mutant is more susceptible to bacterial pathogen Pseudomonas syringae pv. tomato DC3000 hrcC, a nonpathogenic strain that lacks the functional type III secretion system but is capable of eliciting PTI (Navarro et al. 2008). This result indicates that DCL1 is a positive regulator of PTI, which may be due to the fact that DCL1 is responsible for the biogenesis of almost all miRNAs. High-throughput sRNA profiling analyses have shown that the expression of some sRNAs is differentially regulated upon bacterial infection in both model plant species and economically important crops (Fahlgren et al. 2007; Katiyar-Agarwal and Jin 2010; Li et al. 2010; Subramanian et al. 2008; Wang et al. 2009; Zhang et al. 2011a). Some miRNAs, including miR393, miR393*, and miR160, positively regulate plant defense responses, whereas other miRNAs, including miR773 and miR398, negatively regulate antibacterial immunity (Jagadeeswaran et al. 2009; Navarro et al. 2006; Zhang et al. 2011b). Identification of pathogen-regulated sRNAs will help elucidate the molecular mechanisms of host defense responses or disease pathogenicity. For example, recently, we found that miR399 was induced specifically in citrus plants that have succumbed to huanglongbing (HLB), a devastating citrus disease, but not in the citrus plants with citrus stubborn disease that causes similar disease symptoms (Zhao et al. 2013). HLB is associated with bacteria of the genus ‘Candidatus Liberibacter’ whereas citrus stubborn is caused by Spiroplasma citri. Because miR399 is induced by phosphorus starvation in other plant species, we examined the phosphorus content and found a 35% reduction of phosphorus in HLB-positive trees compared with healthy trees (Zhao et al. 2013). Applying phosphorus oxyanion solutions reduced HLB symptom severity and significantly improved fruit production in a field trial. Thus, sRNA studies can help elucidate the regulatory mechanisms of plant–microbe interactions and help develop effective means for disease detection and management.
DCL1 is also involved in the generation of several endogenous siRNAs regulating R-gene-mediated ETI. For example, nat-siRNAATGB2 and AtlsiRNA-1, which are both processed by DCL1 and are induced specifically by avirulent P. syringae pv. tomato carrying an effector gene avrRpt2, contribute to resistance gene RPS2-mediated bacterial resistance (Fig. 1) (Katiyar-Agarwal et al. 2006, 2007).
DCL also contribute to defense responses to various fungal and oomycete pathogens. The dcl4 but not dcl2 mutant displays enhanced susceptibility to Verticillium dahliae and exhibits more severe stunting and necrosis than wild-type plants, which suggests that DCL4 plays a positive role in defense response against Verticillium wilt disease (Ellendorff et al. 2009). However, the responsible sRNAs that are dependent on DCL4 have yet to be discovered. Genome-wide studies in several crops have revealed an array of miRNAs that are differentially expressed in response to infection by various fungal and oomycete pathogens, such as Fusarium virguliforme (Radwan et al. 2011), Erysiphe graminis (Xin et al. 2010), V. dahliae (Yin et al. 2012), Cronartium quercuum (Lu et al. 2007), and Phytophthora sojae (Guo et al. 2011). These miRNAs, most of which are processed by DCL1, are likely to regulate the expression of their target genes and, thereby, contribute to the host resistance responses against these pathogens.
In antiviral immunity, all four DCL are involved in producing virus-derived siRNAs (viRNAs). DCL4 has the predominant role in antiviral defense and targets dsRNAs derived from virus replication to produce 21-nt viRNAs, whereas DCL2 acts as an alternative enzyme in a hierarchical manner to produce 22-nt viRNAs (Bouche et al. 2006; Deleris et al. 2006; Diaz-Pendon et al. 2007; Zhang et al. 2012). DCL4 is sufficient for antiviral silencing in inoculated leaves but both DCL2 and DCL4 are involved in antiviral silencing in systemic tissues (Garcia-Ruiz et al. 2010). DCL3 produces 24-nt viRNAs in plants infected with DNA viruses (Bouche et al. 2006; Deleris et al. 2006; Diaz-Pendon et al. 2007). Although the role of these 24-nt viRNAs in antiviral resistance is controversial, recent studies have shown that geminiviruses encode suppressors that specifically target the DNA methylation process (Buchmann et al. 2009; Yang et al. 2011). The function of these DCL3-produced 24-nt viRNAs may not become apparent unless DNA viruses with defective silencing suppressors are used. A detectable level of 21-nt viRNAs accumulated in the dcl2/dcl3/dcl4 mutant plants infected with both RNA and DNA viruses, suggesting a role for DCL1 in viRNA generation (Blevins et al. 2006; Deleris et al. 2006; Moissiard and Voinnet 2006). Specifically, DCL1 processes 21-nt siRNAs from the leader region of Cauliflower mosaic virus (CaMV) (Blevins et al. 2006). Furthermore, the accumulation of all three 21-, 22-, and 24-nt viRNA species decreased significantly in dcl1 mutant plants infected with CaMV, suggesting that DCL1 is likely to be involved in sRNA-mediated defense (Blevins et al. 2006; Moissiard and Voinnet 2006). On the other hand, DCL1 is also implicated in negative regulation of DCL4 and DCL3, resulting in repression of antiviral RNA silencing (Azevedo et al. 2010; Qu et al. 2008).
The rice genome encodes eight DCL genes: OsDCL1a, OsDCL1b, OsDCL1c, OsDCL2a, OsDCL2b, OsDCL3a, OsDCL3b, and OsDCL4. Among them, OsDCL1b, OsDCL1c, OsDCL2a, and OsDCL2b do not have all the functional domains (Kapoor et al. 2008). OsDCL1a, OsDCL2, OsDCL3a, and OsDCL4 are ubiquitously expressed, whereas the others are expressed differentially in various developmental stages. Like its Arabidopsis orthologous gene AtDCL1, OsDCL1a is responsible for OsmiRNA processing (Liu et al. 2005). OsDCL4 and OsDCL3b have been found to be responsible for the biogenesis of 21- and 24-nt phased sRNAs in rice, respectively (Song et al. 2012).
In addition to the well-established role of viRNA-directed RNA silencing in plant immunity against viruses, emerging evidence implicates a role for host miRNAs in plant–virus interactions in rice. We have showed that two distinct viruses, Rice dwarf virus (RDV) and Rice stripe virus (RSV), have distinct impacts on rice sRNA profiles (Du et al. 2011). In RSV-infected rice plants, many miRNA*s accumulated to higher levels than in noninfected plants, whereas their corresponding miRNA sequences did not show obvious changes (Du et al. 2011). These include miRNA*s of miR160, miR166, miR167, miR171, and miR396 families and miRNA*s of miR1318, miR1425, miR159a, miR168, miR172d, miR390, miR444b.2, and miR528 (Du et al. 2011). In addition, Os11g15060, a predicted target of miR1425*, was downregulated in RSV-infected plants (Du et al. 2011). Furthermore, RSV infection also induced the expression of novel phased miRNAs from several conserved miRNA precursors (Du et al. 2011). However, these changes were not observed in RDV-infected rice.
dsRNA-binding proteins (DRB) are one of the essential cofactors of DCL (Katiyar-Agarwal and Jin 2010; Voinnet 2009). HYL1 (also known as DRB1) and DRB4 are associated with DCL1 and DCL4, respectively. Evidence shows that HYL1 is involved in antibacterial defense. The hyl1 mutant not only is susceptible to Pseudomonas syringae pv. tomato (avrRpt2) but also is deficient in the accumulation of nat-siRNAATGB2 and AtlsiRNA-1, which positively regulate bacterial resistance (Katiyar-Agarwal et al. 2006, 2007). DRB4 contributes to antiviral defense as a cofactor of DCL4 in sRNA metabolism (Haas et al. 2008). Furthermore, a recent study suggested a potential function of DRB4 in the regulation of viral RNA translation by directly interacting with viral RNAs (Jakubiec et al. 2012). The function of rice DRB in disease resistance pathways has yet to be determined.
Host RDR
Arabidopsis encodes six RDR proteins (RDR1 to RDR6). They play important roles in siRNA biogenesis by converting single-stranded RNAs (ssRNAs) to dsRNAs, which are then processed by DCL to generate siRNAs (Wassenegger and Krczal 2006).
RDR6 is required for the biogenesis of bacterial-induced nat-siRNAATGB2 and AtlsiRNA-1 (Katiyar-Agarwal et al. 2006, 2007). The observation that the rdr6 mutant is more susceptible to P. syringae pv. tomato (avrRpt2) suggests that the RDR6-dependent mechanism is important for antibacterial defense (Katiyar-Agarwal et al. 2006). Further evidence supporting the importance of RDR6 in plant immunity has come from several recent studies on secondary siRNAs that target R genes. In certain plant species, such as those in the Solanaceous and Leguminosa families, several miRNAs can target nucleotide-binding site (NBS) leucine-rich repeat (LRR) R genes and trigger the generation of secondary siRNAs in an RDR6-dependent manner. These siRNAs regulate the expression of R genes (Li et al. 2012; Shivaprasad et al. 2012; Zhai et al. 2011). Tomato miR482 and miR2118 families can target the P-loop motif of the coiled-coil (CC)-NBS-LRR type of R genes and trigger secondary siRNA formation through RDR6 (Shivaprasad et al. 2012). Viral and bacterial infection suppresses miR482-mediated silencing of R genes. In tobacco, miR6019 (22 nt) and miR6020 (21 nt) target Toll and interleukin-1 receptor (TIR)-NBS-LRR gene N that confers resistance to Tobacco mosaic virus (TMV) (Li et al. 2012). Cleavage of N transcripts triggers phased secondary siRNAs that are DCL4 and RDR6 dependent. Overexpression of nta-miR6019/miR6020 attenuated N-mediated resistance to TMV. Eight additional miRNA families were identified from tobacco, tomato, and potato using a bioinformatics approach in this study. In addition to the ones that target several antiviral R genes, some also target R genes that confer resistance to fungal and oomycete pathogens. For example, miR6021, miR6022, and miR6023 target a tomato homolog of the Cladosporium fulvum resistance 9 gene, which encodes a transmembrane protein and confers resistance to leaf mold fungus C. fulvum. miR482 family members could target potato R2 and RB genes, which confer resistance to late blight oomycete Phytophthora infestans. Thus, various types of R genes, including both TIR-NBS-LRR and CC-NBS-LRR types and receptor-like transmembrane type, could be targeted by miRNAs and initiate secondary siRNA formation to silence the expression of these resistance genes.
Extensive studies have shown that RDR1 and RDR6 participate in antiviral defense and viRNA biogenesis (Ding 2010; Ding and Voinnet 2007; Jiang et al. 2012; Qi et al. 2009; Qu 2010; Wang et al. 2010). RDR-dependent amplification of viRNAs is required to confer effective RNA-based antiviral defense in Arabidopsis (Wang et al. 2010). RDR1 is primarily responsible for the accumulation of the majority of viRNAs whereas RDR6 acts as a surrogate for RDR1 (Ding 2010; Qu 2010). However, it is more likely that RDR1 and RDR6 have specific roles in RNA-based antiviral defense because a recent sRNA profiling study revealed that viRNAs associated with RDR1 mostly mapped to the 5′ terminus of viral RNAs, whereas those associated with RDR6 mapped mainly to 3′ regions (Wang et al. 2010). Knocking out both RDR1 and RDR6 results in a dramatic decrease in viRNA accumulation, indicating that RDR1 and RDR6 synergistically contribute to the production of viRNAs and antiviral defense. Notably, RDR2 was also shown to be involved in the biogenesis of Tobacco rattle virus-derived siRNAs and antiviral resistance (Donaire et al. 2008). In addition, it has been shown that RDR2 acts in coordination with RDR1 and RDR6 to restrict Turnip mosaic virus infection in inoculated leaves (Garcia-Ruiz et al. 2010). RDR2 functions mainly with DCL3 and generates hc-siRNAs that guide DNA methylation and histone modification in the RdDM pathway. Whether the involvement of RDR2 in antiviral defense against RNA viruses is dependent on RdDM pathway has yet to be determined.
RDR also play a key role in antiviral immunity in rice. OsRDR1 is required for RNA silencing triggered by an ssRNA virus. In OsRDR1 mutant lines, the antiviral silencing pathway was impaired by the infection with Brome mosaic virus (an ssRNA virus) but not Wheat dwarf geminivirus (an ssDNA virus). Recently, OsRDR6 was shown to play an antiviral role against RSV, a negative-strand RNA virus. Transgenic OsRDR6IR RNA interference lines are more susceptible to RSV infection and accumulate more RSV genomic RNA (Jiang et al. 2012). Furthermore, deep sequencing showed reduced accumulation of RSV-derived viRNA in OsRDR6IR plants (Jiang et al. 2012).
Studies of the roles of plant RDR in fungus–plant interaction are rather limited. RDR2 and RDR6 were shown to positively regulate defense responses against Verticillium wilt disease (Ellendorff et al. 2009). The rdr2 and rdr6 mutants displayed enhanced susceptibility to Verticillium wilt disease and accumulated more fungal biomass compared with wild-type plants.
AGO proteins
AGO proteins are the core molecules of the effector complex in sRNA-directed gene silencing. sRNAs are loaded into AGO proteins to guide sequence-specific regulation of gene expression. AGO proteins comprise four major domains: N-terminal domain, PAZ domain, MID domain, and PIWI domain. Plant AGO proteins all belong to the AGO subfamily. Arabidopsis encodes 10 AGO proteins, which can be divided into three major clades based on their sequence similarity: the AGO1/AGO5/AGO10 clade, AGO2/AGO3/AGO7 clade, and AGO4/AGO6/AGO8/AGO9 clade (Vaucheret 2008).
In Arabidopsis, AGO1, AGO2, and AGO7 primarily bind 21- to 22-nt sRNAs (either miRNAs or endogenous and exogenous siRNAs) and direct PTGS (Mi et al. 2008; Montgomery et al. 2008), whereas AGO4, AGO6, and AGO9 predominately bind 24- to 26-nt sRNAs and are involved in TGS (Havecker et al. 2010; Qi et al. 2006; Zheng et al. 2007; Zilberman et al. 2003).
AGO1 contributes to plant innate immunity, which is supported by the evidence that the ago1 mutant plants displayed attenuated PTI responses (Li et al. 2010). AGO1 functions through the action of its associated miRNAs. Some miRNAs positively regulate PTI. For instance, miR393, which is induced by a bacterial elicitor, flg22, contributes to bacterial resistance by targeting mRNAs encoding the auxin receptor, transport inhibitor response1 (TIR1), and related proteins, thereby negatively regulating auxin signaling and shifting energy from growth to basal defense (Fig. 1) (Navarro et al. 2006). Other miRNAs, on the other hand, negatively regulate plant innate immunity. For example, flg22 and avirulent Pseudomonas syringae pv. tomato strains downregulate miR398, resulting in upregulation of its target genes, such as copper/zinc superoxide dismutase, thereby positively regulating bacterial resistance (Jagadeeswaran et al. 2009; Li et al. 2010; Zhang et al. 2011b).
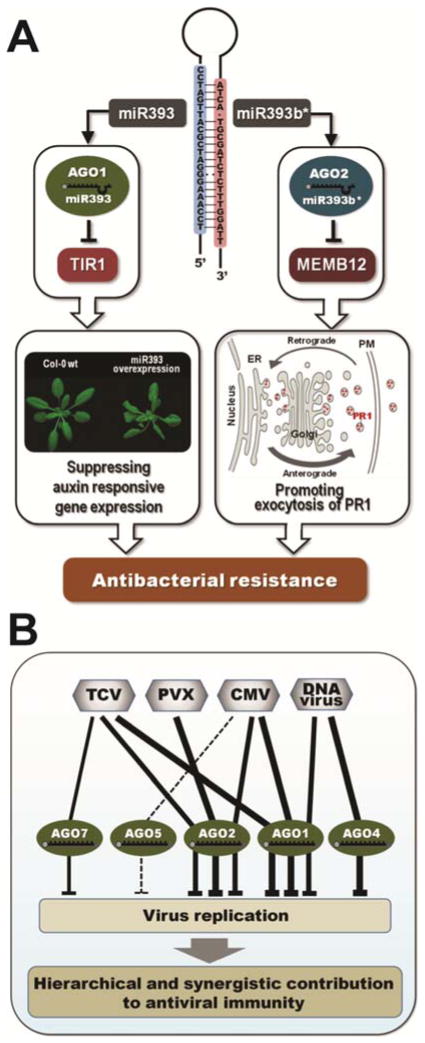
Our recent study has showed that Arabidopsis AGO2 plays an important role in antibacterial immune responses. Among the 10 Arabidopsis AGO proteins, only AGO2 is highly induced by infection with both virulent P. syringae pv. tomato and avirulent P. syringae pv. tomato (avrRpt2) strains, and ago2 mutant plants displayed enhanced susceptibility to both strains (Zhang et al. 2011b). Interestingly, sequencing of AGO2-associated sRNAs after immunoprecipitation revealed that one of the most abundant miRNAs associated with AGO2 is miR393b*, whereas its corresponding miR393, which is involved in PTI, is associated with AGO1. The miR393b*-AGO2 complex targets a Golgi-localized gene, MEMB12, which encodes a SNARE protein that controls retrograde trafficking. Induction of AGO2 allows increased miR393* loading and induces silencing of MEMB12, which suppresses protein endocytosis and recycling, thereby resulting in increased exocytosis of antimicrobial pathogenesis-related protein PR1. Secretion of antimicrobial peptides directly confers resistance against bacteria. This study provided an example of a miRNA-miRNA* pair that both contribute to plant immunity through two distinct AGO proteins: miR393 associates with AGO1 whereas miR393* associates with AGO2 (Zhang et al. 2011b) (Fig. 2A). Not only miRNA-miRNA* pairs generate more than one functional miRNA entity; some miRNA precursors with long stem structures could be sequentially processed by DCL1 and give rise to multiple miRNA duplexes, and some of these miRNA-like RNAs are most likely functional (Zhang et al. 2010). We have also observed that some miRNA genes could give rise to both DCL1-dependent 21-nt miRNAs and DCL3- and RDR2-dependent 24-nt siRNAs at the same loci. The miRNAs silence the targets through PTGS while some of the siRNAs could silence the targets through TGS (Chellappan et al. 2010). Some of the miRNA gene-derived miRNA-like RNAs and siRNAs are differentially regulated by pathogen-infection, suggesting that they have a potential function in regulating target expression during plant–microbe interaction.
Fig. 2.
Role of Argonaute (AGO) proteins in plant immunity. A, MicroRNA (miRNA)/miRNA* pair contributes to plant antibacterial immunity through two distinct AGO proteins, miR393 and miR393b*, which are induced by Pseudomonas syringae pv. tomato infection, specifically associate with AGO1 and AGO2, respectively. The miR393/AGO1 complex contributes to pathogen-associated molecular-pattern-triggered immunity by targeting TRANSPORT INHIBITOR RESPONSE1 (TIR1) and related proteins, thereby suppressing auxin signaling; the miR393*-AGO2 complex targets a Golgi-localized SNARE gene, MEMB12, resulting in increased exocytosis of antimicrobial proteins which directly confer resistance against bacteria. PR1, pathogenesis-related protein 1. B, Hierarchical and synergistic contribution of AGO proteins to plant antiviral immunity. AGO1 acts as a major antiviral RNA slicer in antiviral defense against Cucumber mosaic virus (CMV) and Turnip crinkle virus (TCV), while AGO2 functions as a surrogate of AGO1. AGO5 is known to bind to CMV-derived virus-derived siRNAs (viRNAs), although its function in antiviral defense is not clear. AGO7 has a different mode of action from AGO1 in antiviral defense against TCV. AGO2 mediates viRNA-directed defenses against Potato virus X (PVX) in Arabidopsis. Some DNA viruses are targeted by AGO4 for the methylation of viral DNAs. AGO1-associated 21- and 22-nucleotide viRNAs are also considered to be involved in antiviral defense against DNA viruses. Thickness of the lines indicates relative hierarchical contribution of each AGO protein to antiviral immunity.
AGO7 belongs to the same subfamily as AGO2 and AGO3. Although a mutation in AGO3 has no obvious effect on plant innate immunity, a mutation in AGO7 weakly attenuates ETI triggered by P. syringae pv. tomato (avrRpt2), whereas no obvious effect was observed in PTI responses (Katiyar-Agarwal et al. 2007; Li et al. 2010; Zhang et al. 2011b). AGO7 participates in the biogenesis of AtlsiRNA-1, an endogenous siRNA regulating RPS2-mediated ETI (Katiyar-Agarwal et al. 2007). Furthermore, AGO7 predominantly associates with miR390, which triggers the generation of TAS3 ta-siRNAs that subsequently regulate the expression of auxin-signaling components; namely, ARF genes (Fahlgren et al. 2006; Montgomery et al. 2008). Although different ARF could be either positive or negative regulators of auxin signaling, downregulation of the auxin signaling pathway could indirectly contribute to plant defense. Furthermore, the ago7 mutant exhibited enhanced susceptibility to fungal pathogen Verticillium (Ellendorff et al. 2009), suggesting that AGO7 also plays a positive role in anti-fungal defense against Verticillium spp.
A genetic screen identified an ago4 mutant that exhibits reduced resistance to both virulent P. syringae pv. tomato and avirulent P. syringae pv. tomato (avrRpm1), indicating that AGO4 is required for bacterial resistance in Arabidopsis (Agorio and Vera 2007). However, AGO4 and AGO6 are down-regulated by bacterial infection and flg22 treatment, implying a negative role of AGO4 and AGO6 in plant antibacterial immunity (Yu et al. 2013). Although AGO4 is involved in the RdDM pathway, single mutants of other components within the RdDM pathway did not affect bacterial resistance (Agorio and Vera 2007). It is possible that these components have partial functional redundancy and to assess the effect of double or triple mutants would be more reliable.
AGO proteins also play critical roles in antiviral defense (Fig. 2B). Upregulation of AGO1 is a general response in plants infected with viruses (Csorba et al. 2007; Havelda et al. 2008; Zhang et al. 2006). Genetic studies showed that RNA-based antiviral defense is attenuated in single ago1 and ago7 mutants (Morel et al. 2002; Qu et al. 2008). AGO1 is the major AGO protein that associates with viRNAs (Zhang et al. 2006). Cucumber mosaic virus (CMV) 2b suppressor protein specifically inhibits AGO1 slicer activity to counter plant defense (Zhang et al. 2006). Several plant viruses, including CMV, Turnip crinkle virus (TCV), and Cymbidium ringspot virus, can compromise host RNA silencing by inducing the expression of miR168, which targets AGO1 mRNA to repress translation (Varallyay et al. 2010). AGO7 has a mode of action different from AGO1 in antiviral defense against TCV (Qu et al. 2008); AGO7 targets less-structured RNAs and functions together with DRB4 and RDR6 (Qu et al. 2008) in the amplification of secondary viRNAs, similar to its function in the ta-siRNA pathway (Montgomery et al. 2008). Similar to AGO1, AGO2 and AGO5 have also been shown to bind viRNAs (Harvey et al. 2011; Ruiz-Ferrer and Voinnet 2009; Takeda et al. 2008). AGO2 functions in defense against RNA viruses, including TCV, Potato virus X, CMV, and Tomato bushy stunt virus (Harvey et al. 2011; Jaubert et al. 2011; Scholthof et al. 2011; Wang et al. 2011; Zhang et al. 2012). Accumulating evidence suggests that AGO1 and AGO2 have nonredundant and cooperative functions in antiviral defense, and the other AGO proteins, in addition to AGO1, may play a greater role in antiviral defense than previously thought (Alvarado and Scholthof 2011). Antiviral defense against some DNA viruses involves AGO4-mediated mechanisms (Buchmann et al. 2009; Raja et al. 2008), which is directly linked with the RdDM pathway (details below).
The rice genome encodes 19 AGO proteins which were first classified into six main clades: AGO2/AGO3/AGO7 (SHL4) clade, AGO4/AGO15/AGO16 clade, AGO5 (MEL1)/AGO11/ AGO12/AGO13/AGO14 clade, AGO1/AGO10 (PHN1) clade, AGO17 clade, and AGO18 clade (Kapoor et al. 2008; Mallory and Vaucheret 2010). Among them, only five OsAGO (AGO1, AGO4, AGO5, AGO7, and AGO16) have been studied. OsAGO1a, OsAGO1b, and OsAGO1c have slicer activity and mediate PTGS by primarily associating with 21-nt sRNAs with a 5′ U (Wu et al. 2009). OsAGO4a, OsAGO4b, OsAGO16, and their associated 24-nt sRNAs are involved in TGS by guiding DNA methylation (Wu et al. 2010). Studies on rice AGO proteins in plant defense have started to emerge. Our recent study showed that RSV infection significantly elevated the expression of AGO18 and AGO2 but RDV infection only affected the expression of certain OsRDR, not OsAGO (Du et al. 2011). RSV also induces the expression of many miRNA*s (Du et al. 2011). We speculate that these miRNA*s are likely to be associated with OsAGO2 during bacterial infection, similar to the case with Arabidopsis AGO2.
RNA Pol IV and V and the RdDM pathway
A newly emerging, important aspect of plant immune responses is the modulations that occur at the epigenetic level, including the dynamic changes in DNA methylation and histone modification. Infection by P. syringae pv. tomato induces DNA hypomethylation at centromeric regions and alters hetero-chromatin patterns (Pavet et al. 2006). Recently, a genome-wide single-base resolution DNA methylome study using whole-genome bisulfite sequencing revealed dynamic DNA methylation changes in numerous genomic regions in response to infection with bacterial pathogens—virulent P. syringae pv. tomato or avirulent P. syringae pv. tomato (avrPphB)—and treatment with the hormone salicylic acid (SA) (Dowen et al. 2012). Arabidopsis DNA methyltransferase mutants met1 (deective in maintenance of CG methylation) and ddc (drm1 drm2cmt3 triple mutant, defective in de novo non-CG methylation) displayed markedly enhanced resistance to P. syringae pv. tomato (Dowen et al. 2012). Similarly, a modest increase in resistance was also observed in the RdDM pathway mutants dcl2/3/4, rdr2, nrpd1a (carrying a mutation in the largest subunit of RNA Pol IV), and drd1, indicating that the RdDM pathway contributes to the regulation of plant immunity. It is likely that the RdDM pathway mediated by hc-siRNAs is important for silencing plant immunity genes such as R genes and signaling components (e.g., SA pathway genes) under normal conditions and, upon infection by biotrophic or hemibiotrophic pathogens, this silencing is suppressed and the R genes and immunity signaling genes are upregulated. In this study, no alteration in disease resistance was observed in the nrpe1 mutant, which carries a mutation in the largest subunit of RNA Pol V; however, interestingly, another study by Lopez and associates (2011) observed enhanced resistance to P. syringae pv. tomato in nrpe1. This discrepancy is likely due to the different infection conditions used in these two studies (Dowen et al. 2012; Lopez et al. 2011). More recently, Yu and associates (2013) have shown that some transposable elements, which are targets of RdDM, are demethylated and transcriptionally reactivated during antibacterial defense. They also demonstrated that DNA demethylation is associated with an activation of some disease resistance genes carrying repeats or siRNA clusters in their vicinity (Yu et al. 2013).
Host resistance against necrotrophic fungal pathogens, such as Botrytis cinerea and Plectosphaerella cucumerina, is dependent on the jasmonic acid (JA) signaling pathway, whereas resistance against biotrophic or hemibiotrophic pathogens, such as P. syringae pv. tomato, largely relies on the SA signaling pathway. The JA signaling pathway is generally antagonistic to the SA pathway (Glazebrook 2005). As predicted, enhanced susceptibility to B. cinerea and P. cucumerina was observed in ago4 and nrpe1 (Lopez et al. 2011).
Arabidopsis plants suppress geminiviruses at the transcriptional level by inducing both cytosine and histone methylation of the viral genomes (Raja et al. 2008). Methylation-deficient Arabidopsis mutants carrying mutations in cytosine or histone H3 lysine 9 (H3K9) methyltransferases (drm1, drm2, cmt3, met1, and kyp2), RdDM pathway components (ago4, ddm1, and nrpd2a), or methyl cycle enzymes (adk1 and adk2) are hypersensitive to geminivirus infection (Raja et al. 2008). Finally, the most abundant viRNAs in plants infected with geminiviruses are 24 nt long (Rodriguez-Negrete et al. 2009), providing a link between the methylation of viral DNA and the AGO4-mediated RdDM pathway.
Conclusions and future perspectives
High-throughput strategies have revealed many endogenous sRNAs that regulate gene expression reprogramming upon bacterial, fungal, or viral infection. This scheme is supported by genetic studies showing that sRNA pathway components are required for plant immune responses. In summary, DCL, which are implicated in the generation of various classes of sRNAs, are important factors for defense against various pathogens. Although DCL1 is an important contributor to antibacterial defense, hierarchical and synergistic cooperation between DCL4 and DCL2 proteins are necessary to establish antiviral immunity. DCL4 positively regulates defense responses against Verticillium wilt disease. RDR contribute to the generation of secondary siRNAs, some of which are involved in plant immunity. Arabidopsis RDR6 is important for the biogenesis of secondary siRNAs that target genes involved in antimicrobial defense, such as NBS-LRR R genes. Recent researches have shown that RDR1 and RDR6 are important for antiviral immunity in both Arabidopsis and rice. sRNAs are loaded onto AGO proteins to direct PTGS or TGS of viral RNAs and DNAs. Characterization of various Arabidopsis ago mutants has revealed that AGO1 and AGO2 are the major regulators for antiviral and antibacterial immunity. We expect to see more studies on sRNA pathway components and their role in regulating host–microbe interactions in various plant species and crops.
Recently, the function of hc-siRNA-mediated transcriptional regulation in plant immunity has come to light. The use of Arabidopsis RdDM pathway mutants and methylome sequencing has revealed that pathogen infection induces changes in DNA methylation. Mutations in MET1, DRM1/DRM2/CMT3, DCL 2/3/4, RDR2, POL IV, POL V, and DRD1 render the plants more resistant to bacterial infection, suggesting that many resistance-related genes are under the epigenetic regulation by the RdDM pathway; demethylation of these loci in response to pathogen attack activates plant immunity pathways. On the other hand, AGO4 and POL V mutations render plants more susceptible to necrotrophic fungal pathogens, which activate an antagonistic pathway. As genome-wide high-throughput technologies and bioinformatics approaches continue to advance, we expect new discoveries of the roles of sRNAs, the associated pathway components, and epigenetic regulation in plant immunity. On the side of the pathogen, we also expect to discover more effector molecules, not limited to only proteins, that suppress host immunity.
Acknowledgments
Research in H. Jin’s laboratory is supported by NSF Career Award MCB-0642843, NIH grant R01GM093008, California Department of Food & Agriculture (CDFA) grant 23741, and California Citrus Board grant 5210-131. We apologize for not citing some publications owing to space limitations. Research in the Li laboratory is supported by grants from the Natural Science Foundation of China (30910103904 and 31030005) and 973 program (2012CB114004).
LITERATURE CITED
- Agorio A, Vera P. ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell. 2007;19:3778–3790. doi: 10.1105/tpc.107.054494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado VY, Scholthof HB. AGO2: a new Argonaute compromising plant virus accumulation. Front Plant Sci. 2011;2:112. doi: 10.3389/fpls.2011.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo J, Garcia D, Pontier D, Ohnesorge S, Yu A, Garcia S, Braun L, Bergdoll M, Hakimi MA, Lagrange T, Voinnet O. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 2010;24:904–915. doi: 10.1101/gad.1908710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F, Jr, Hohn T, Pooggin MM. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO (Eur Mol Biol Organ) J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmann RC, Asad S, Wolf JN, Mohannath G, Bisaro DM. Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J Virol. 2009;83:5005–5013. doi: 10.1128/JVI.01771-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Chellappan P, Xia J, Zhou X, Gao S, Zhang X, Coutino G, Vazquez F, Zhang W, Jin H. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res. 2010;38:6883–6894. doi: 10.1093/nar/gkq590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Csorba T, Bovi A, Dalmay T, Burgyan J. The p122 subunit of Tobacco mosaic virus replicase is a potent silencing suppressor and compromises both small interfering RNA- and microRNA-mediated pathways. J Virol. 2007;81:11768–11780. doi: 10.1128/JVI.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- Diaz-Pendon JA, Li F, Li WX, Ding SW. Suppression of antiviral silencing by Cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell. 2007;19:2053–2063. doi: 10.1105/tpc.106.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire L, Barajas D, Martinez-Garcia B, Martinez-Priego L, Pagan I, Llave C. Structural and genetic requirements for the biogenesis of Tobacco rattle virus-derived small interfering RNAs. J Virol. 2008;82:5167–5177. doi: 10.1128/JVI.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA. 2012;109:E2183–2191. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Wu J, Zhang J, Zhao S, Zheng H, Gao G, Wei L, Li Y. Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog. 2011;7:e1002176. doi: 10.1371/journal.ppat.1002176. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellendorff U, Fradin EF, de Jonge R, Thomma BP. RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J Exp Bot. 2009;60:591–602. doi: 10.1093/jxb/ern306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One. 2007;2:e219. doi: 10.1371/journal.pone.0000219. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell. 2010;22:481–496. doi: 10.1105/tpc.109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against bio-trophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Guo N, Ye WW, Wu XL, Shen DY, Wang YC, Xing H, Dou DL. Microarray profiling reveals microRNAs involving soybean resistance to Phytophthora sojae. Genome. 2011;54:954–958. doi: 10.1139/g11-050. [DOI] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- Haas G, Azevedo J, Moissiard G, Geldreich A, Himber C, Bureau M, Fukuhara T, Keller M, Voinnet O. Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO (Eur Mol Biol Organ) J. 2008;27:2102–2112. doi: 10.1038/emboj.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey JJ, Lewsey MG, Patel K, Westwood J, Heimstadt S, Carr JP, Baulcombe DC. An antiviral defense role of AGO2 in plants. PLoS One. 2011;6:e14639. doi: 10.1371/journal.pone.0014639. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. The Arabidopsis RNA-directed DNA methylation Argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell. 2010;22:321–334. doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelda Z, Varallyay E, Valoczi A, Burgyan J. Plant virus infection-induced persistent host gene downregulation in systemically infected leaves. Plant J. 2008;55:278–288. doi: 10.1111/j.1365-313X.2008.03501.x. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G, Saini A, Sunkar R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta. 2009;229:1009–1014. doi: 10.1007/s00425-009-0889-3. [DOI] [PubMed] [Google Scholar]
- Jakubiec A, Yang SW, Chua NH. Arabidopsis DRB4 protein in antiviral defense against Turnip yellow mosaic virus infection. Plant J. 2012;69:14–25. doi: 10.1111/j.1365-313X.2011.04765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaubert M, Bhattacharjee S, Mello AF, Perry KL, Moffett P. ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 2011;156:1556–1564. doi: 10.1104/pp.111.178012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Qian D, Zheng H, Meng LY, Chen J, Le WJ, Zhou T, Zhou YJ, Wei CH, Li Y. RNA-dependent RNA polymerase 6 of rice (Oryza sativa) plays role in host defense against negative-strand RNA virus, Rice stripe virus. Virus Res. 2012;163:512–519. doi: 10.1016/j.virusres.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Arora R, Lama T, Nijhawan A, Khurana JP, Tyagi AK, Kapoor S. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics. 2008;9:451. doi: 10.1186/1471-2164-9-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Jin H. Role of small RNAs in host–microbe interactions. Annu Rev Phytopathol. 2010;48:225–246. doi: 10.1146/annurev-phyto-073009-114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr, Zhu JK, Staskawicz BJ, Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 2007;21:3123–3134. doi: 10.1101/gad.1595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010;152:2222–2231. doi: 10.1104/pp.109.151803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li P, Li X, Liu C, Cao S, Chu C, Cao X. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 2005;139:296–305. doi: 10.1104/pp.105.063420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Bakht S, Dean C. Cotranscriptional role for Arabidopsis DICER-LIKE 4 in transcription termination. Science. 2012;335:1621–1623. doi: 10.1126/science.1214402. [DOI] [PubMed] [Google Scholar]
- Lopez A, Ramirez V, Garcia-Andrade J, Flors V, Vera P. The RNA silencing enzyme RNA polymerase v is required for plant immunity. PLoS Genet. 2011;7:e1002434. doi: 10.1371/journal.pgen.1002434. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Sun YH, Amerson H, Chiang VL. MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J. 2007;51:1077–1098. doi: 10.1111/j.1365-313X.2007.03208.x. [DOI] [PubMed] [Google Scholar]
- Mallory A, Vaucheret H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell. 2010;22:3879–3889. doi: 10.1105/tpc.110.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, Chen S, Hannon GJ, Qi Y. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard G, Voinnet O. RNA silencing of host transcripts by Cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc Natl Acad Sci USA. 2006;103:19593–19598. doi: 10.1073/pnas.0604627103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. Fertile hypomorphic ARGO-NAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Navarro L, Jay F, Nomura K, He SY, Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan C, Zhang X, Jin H. Host small RNAs are big contributors to plant innate immunity. Curr Opin Plant Biol. 2009;12:465–472. doi: 10.1016/j.pbi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Pavet V, Quintero C, Cecchini NM, Rosa AL, Alvarez ME. Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by Pseudomonas syringae. Mol Plant-Microbe Interact. 2006;19:577–587. doi: 10.1094/MPMI-19-0577. [DOI] [PubMed] [Google Scholar]
- Qi X, Bao FS, Xie Z. Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS One. 2009;4:e4971. doi: 10.1371/journal.pone.0004971. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi YJ, He XY, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Qu F. Antiviral role of plant-encoded RNA-dependent RNA polymerases revisited with deep sequencing of small interfering RNAs of virus origin. Mol Plant-Microbe Interact. 2010;23:1248–1252. doi: 10.1094/MPMI-06-10-0124. [DOI] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci USA. 2008;105:14732–14737. doi: 10.1073/pnas.0805760105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwan O, Liu Y, Clough SJ. Transcriptional analysis of soybean root response to Fusarium virguliforme, the causal agent of sudden death syndrome. Mol Plant-Microbe Interact. 2011;24:958–972. doi: 10.1094/MPMI-11-10-0271. [DOI] [PubMed] [Google Scholar]
- Raja P, Sanville BC, Buchmann RC, Bisaro DM. Viral genome methylation as an epigenetic defense against geminiviruses. J Virol. 2008;82:8997–9007. doi: 10.1128/JVI.00719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Negrete EA, Carrillo-Tripp J, Rivera-Bustamante RF. RNA silencing against geminivirus: complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J Virol. 2009;83:1332–1340. doi: 10.1128/JVI.01474-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Alandete Saez M, Eshed Williams L, Fletcher JC, McCormick S. Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 2010;24:1010–1021. doi: 10.1101/gad.1882810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- Scholthof HB, Alvarado VY, Vega-Arreguin JC, Ciomperlik J, Odokonyero D, Brosseau C, Jaubert M, Zamora A, Moffett P. Identification of an ARGONAUTE for antiviral RNA silencing in Nicotiana benthamiana. Plant Physiol. 2011;156:1548–1555. doi: 10.1104/pp.111.178764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Li P, Zhai J, Zhou M, Ma L, Liu B, Jeong DH, Nakano M, Cao S, Liu C, Chu C, Wang XJ, Green PJ, Meyers BC, Cao X. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 2012;69:462–474. doi: 10.1111/j.1365-313X.2011.04805.x. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O. Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics. 2008;9:160. doi: 10.1186/1471-2164-9-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins. Plant Cell Physiol. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- Varallyay E, Valoczi A, Agyi A, Burgyan J, Havelda Z. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO (Eur Mol Biol Organ) J. 2010;29:3507–3519. doi: 10.1038/emboj.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Wang XB, Wu Q, Ito T, Cillo F, Li WX, Chen X, Yu JL, Ding SW. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:484–489. doi: 10.1073/pnas.0904086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX, Gasciolli V, Vaucheret H, Ding SW. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative Argonautes in Arabidopsis thaliana. Plant Cell. 2011;23:1625–1638. doi: 10.1105/tpc.110.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li P, Cao X, Wang X, Zhang A, Li X. Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochem Biophys Res Commun. 2009;378:799–803. doi: 10.1016/j.bbrc.2008.11.140. [DOI] [PubMed] [Google Scholar]
- Wassenegger M, Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Wu L, Zhang Q, Zhou H, Ni F, Wu X, Qi Y. Rice MicroRNA effector complexes and targets. Plant Cell. 2009;21:3421–3435. doi: 10.1105/tpc.109.070938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y. DNA methylation mediated by a microRNA pathway. Mol Cell. 2010;38:465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z, Sun Q. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.) BMC Plant Biol. 2010;10:123. doi: 10.1186/1471-2229-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Xie Y, Raja P, Li S, Wolf JN, Shen Q, Bisaro DM, Zhou X. Suppression of methylation-mediated transcriptional gene silencing by betaC1-SAHH protein interaction during gemini-virus-betasatellite infection. PLoS Pathog. 2011;7:e1002329. doi: 10.1371/journal.ppat.1002329. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Li Y, Han X, Shen F. Genome-wide profiling of miRNAs and other small non-coding RNAs in the Verticillium dahliae-inoculated cotton roots. PLoS One. 2012;7:e35765. doi: 10.1371/journal.pone.0035765. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Lepere G, Jay F, Wang J, Bapaume L, Wang Y, Abraham AL, Penterman J, Fischer RL, Voinnet O, Navarro L. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci USA. 2013;110:2389–2394. doi: 10.1073/pnas.1211757110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, Gonzalez AJ, Yan Z, Kitto SL, Grusak MA, Jackson SA, Stacey G, Cook DR, Green PJ, Sherrier DJ, Meyers BC. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Gao S, Zhou X, Xia J, Chellappan P, Zhang X, Jin H. Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol. 2010;11:R81. doi: 10.1186/gb-2010-11-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhang X, Fromuth N, Coutino G, Coffey M, Jin H. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol. 2011a;75:93–105. doi: 10.1007/s11103-010-9710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhao H, Gao S, Wang WC, Katiyar-Agarwal S, Huang HD, Raikhel N, Jin H. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(*)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell. 2011b;42:356–366. doi: 10.1016/j.molcel.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Singh J, Li D, Qu F. Temperature-dependent survival of Turnip crinkle virus-infected Arabidopsis plants relies on an RNA silencing-based defense that requires dcl2, AGO2, and HEN1. J Virol. 2012;86:6847–6854. doi: 10.1128/JVI.00497-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sun R, Albrecht U, Padmanabhan C, Wang A, Coffey MD, Girke T, Wang Z, Close TJ, Roose M, Yokomi RK, Folimonova S, Vidalakis G, Rouse R, Bowman KD, Jin H. Small RNA profiling reveals phosphorus deficiency as a contributing factor in symptom expression for citrus huanglongbing disease. Mol Plant. 2013;6:301–310. doi: 10.1093/mp/sst002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO (Eur Mol Biol Organ) J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]