Abstract
Plant defense responses against pathogens are mediated by activation and repression of a large array of genes. Host endogenous small RNAs are essential in this gene expression reprogramming process. Here, we discuss recent findings on pathogen-regulated host microRNAs (miRNAs) and small interfering RNAs (siRNAs) and their roles in plant-microbe interaction. We further introduce small RNA pathway components, including Dicer-like proteins (DCLs), double-stranded RNA (dsRNA) binding protein, RNA-dependent RNA polymerases (RDRs), small RNA methyltransferase HEN1, and Argonaute (AGO) proteins, that contribute to plant immune responses. The strategies that pathogens have evolved to suppress host small RNA pathways are also discussed. Collectively, host small RNAs and RNA silencing machinery constitute a critical layer of defense in regulating the interaction of pathogens with plants.
Keywords: miRNAs, siRNAs, dicer, argonaute, RNA silencing suppressors
INTRODUCTION
Small RNAs are 20 to 40 nucleotide (nt)-long noncoding RNA molecules present in most eukaryotic organisms that regulate gene expression in a sequence-specific manner either transcriptionally or posttranscriptionally (5, 11, 98, 99). On the basis of their biogenesis and precursor structure, small RNAs are placed in two distinct groups: microRNAs (miRNAs) and small interfering RNAs (siRNAs). Small RNAs regulate a multitude of biological processes in plants, including development, metabolism, maintenance of genome integrity, immunity against pathogens, and abiotic stress responses. Increasing evidence suggests that small RNAs play a critical role in regulating the interaction of pathogens with plants.
SMALL RNA BIOGENESIS PATHWAYS IN PLANTS
Small RNA pathways in plants have been best characterized in the model plant Arabidopsis, and seminal pieces of work involving both forward and reverse genetic screens have delineated the cellular proteins that are involved in biogenesis and function of miRNAs and siRNAs. In this section, we present a brief summary of different kinds of small RNA pathways known in Arabidopsis (Figure 1). For detailed information, please refer to other reviews (11, 44, 47, 53, 66, 92, 98, 99, 107, 115).
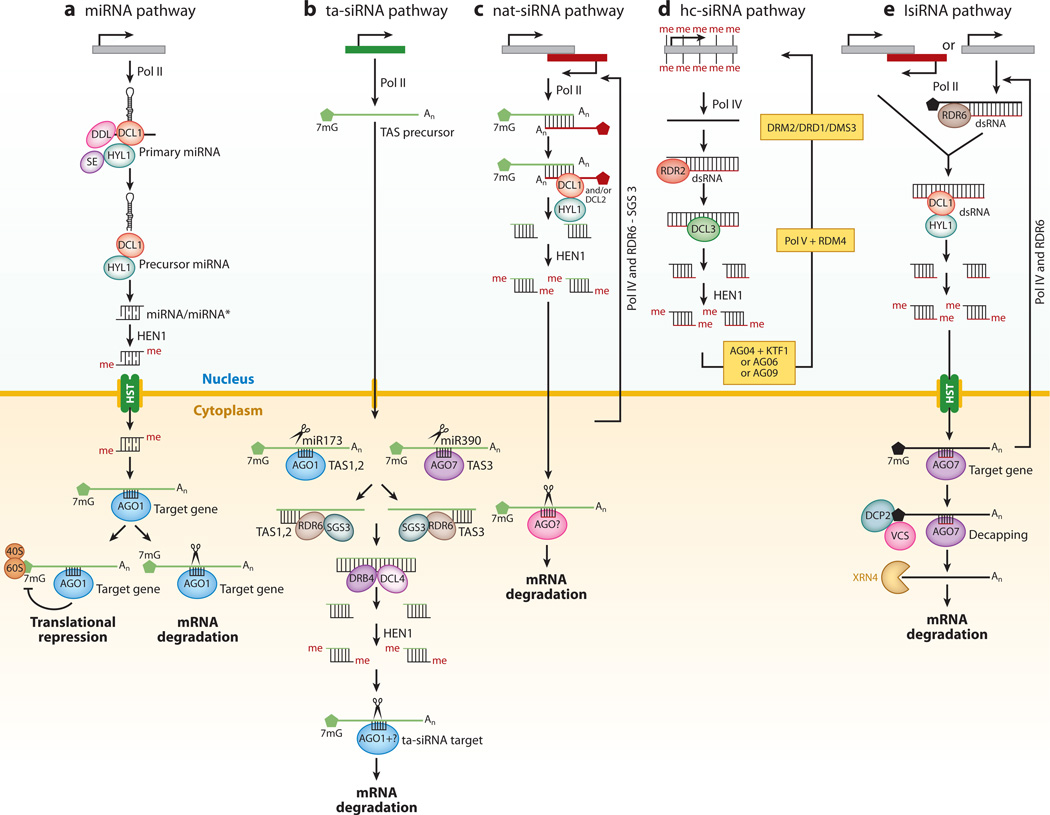
Figure 1.
Endogenous small RNA pathways in Arabidopsis. (a) microRNA (miRNA) pathway. miRNAs are generated by transcription of noncoding miRNA genes by RNA Pol II. The primary miRNAs possess stem-loop structures that are acted upon by DCL1-HYL1-SE protein complex. DDL protein is known to be involved in the formation of precursor miRNA (pre-miRNA). The DCL1-HYL1 complex further processes pre-miRNA into 21-nucleotide (nt) miRNAs. The miRNA (miRNA:miRNA*) duplexes are then methylated at their 3’ ends by HEN1. These methylated miRNAs are transported into cytoplasm by an exportin homolog, HASTY (HST). The mature miRNA is incorporated into the RNA-induced silencing complex (RISC) containing AGO1 protein. The RISC is recruited to the target gene on the basis of sequence complementarity, and AGO1 represses gene expression by either mRNA degradation or translational repression. (b) trans-acting small interfering RNA (ta-siRNA) pathway. The process of TAS precursor is triggered by an miRNA-mediated cleavage. The resulting 5′ fragment (in case of TAS1a–c and TAS2) and 3′ fragment (in case of TAS3) act as templates for the formation of long double-stranded RNA (dsRNA) by concerted action of RDR6 and SGS3. These long dsRNAs are then recognized by the DCL4-DRB4 complex and cut into phased 21-nt small RNAs that undergo further methylation by HEN1. The ta-siRNAs are incorporated into a RISC containing AGO7 (in the case ofTAS3) or AGO1 (in the case of TAS1 and 2), which results in target cleavage. (c) natural antisense transcript-derived siRNA (nat-siRNA) pathway. Natural antisense transcripts produced by Pol II form dsRNA within their overlapping regions. The dsRNAs are processed by DCL1 and/or DCL2 into siRNAs that target antisense transcripts through an unidentified AGO protein containing RISC complex. RDR6-SGS3, together with Pol IV, forms an amplification loop to generate more nat-siRNAs, which reinforce the cleavage of antisense transcript. (d) heterochromatic siRNA (hc-siRNA) pathway. Transcription of heterochromatic regions, repeat regions or transposons by Pol II and/or Pol IV results in the formation of single-stranded RNA(ssRNA), which is converted into dsRNA by the action of RDR2. This dsRNA is processed into predominantly 24-nt long hc-siRNAs by DCL3. These 24-nt siRNAs associate with AGO4 (or AGO6, or AGO9) through an adaptor protein, KTF1, to form an RNA-directed DNA methylation (RdDM) effector complex that directly or indirectly recruits proteins involved in heterochromatin formation, including DRM2, DRD1, and DMS3, to the hc-siRNA target loci. (e) long siRNA (lsiRNA) pathway. lsiRNAs are generated by DCL1 from coding or noncoding genes, or overlapping regions of antisense transcription, or dsRNAs from the action of Pol IV and RDRs. lsiRNAs are methylated by HEN1 and repress the expression of target genes by guiding mRNA decapping mediated by DCP2 (decapping 2) and VCS (Varicose) and 5’–3’ RNA decay mediated by exoribonuclease XRN4.
miRNA Pathway
miRNAs are derived from the transcripts of miRNA genes generated by RNA polymerase II. The primary miRNA (pri-miRNA) transcript forms a fold-back structure, which is processed into a stem-loop precursor known as precursor miRNA (pre-miRNA). A protein named DAWDLE (DDL) has been proposed to play an important role in miRNA biogenesis by recruiting predominantly DICER-like protein 1 (DCL1) to pri-miRNA for downstream processing (110). The pre-miRNA is acted upon by DCL1 together with HYL1 (HYPONASTIC LEAVES 1) and SE (SERRATE) to form the small RNA duplex. The small RNA duplex is then methylated at the 3′ ends by HEN1 (HUA ENHANCER 1) and is exported to the cytoplasm by an exportin homolog, HST (HASTY). Mature miRNA is preferentially incorporated into AGO1 (or AGO10) and guides the complex to the target mRNA for cleavage or translational inhibition on the basis of sequence complementarity.
siRNA Pathways
In contrast to miRNAs that are derived from imperfectly base-paired hairpin loop structures, siRNAs are derived from perfectly paired double-stranded RNA (dsRNA) precursors. These dsRNA precursors are derived either from antisense transcription or by the action of a cellular RNA-dependent RNA polymerase (RDR). Four different types of siRNAs are known in plants: trans-acting siRNAs (ta-siRNAs), natural antisense transcripts (NATs)-derived siRNAs (nat-siRNAs), heterochromatic siRNAs (hc-siRNAs) or repeat-associated siRNAs (ra-siRNAs), and long siRNAs (lsiRNAs). RNA Pol II transcribes noncoding TAS genes, and the long primary transcript products are initially cleaved by miRNAs loaded with RNA-induced silencing complexes (RISCs), resulting in a 5′ fragment or a 3′ fragment. These fragments then act as a template for synthesis of a complementary strand by the concerted action of RDR6 and SGS3 (46, 99). The resulting dsRNA molecule is acted upon by DCL4 and DRB4 to trigger the subsequent production of ta-siRNAs (37, 46). nat-siRNAs are produced from the overlap regions of sense and antisense transcripts of cis-NATs. A significant proportion of most eukaryotic genomes encode overlapping cis-NAT genes, which have the potential to generate siRNAs when base pairing between sense and antisense transcripts occurs. Though nat-siRNAs have been shown to play an important function in both abiotic and biotic stresses (8, 55), their roles in other plant processes remains to be investigated. The cellular components involved in production of nat-siRNAs are DCL1 and/or DCL2, HYL1, and HEN1 (8, 55). The nat-siRNAs studied also partially depend on RDR6, SGS3, and Pol IV (8, 55). hc-siRNAs or ra-siRNAs are usually 24 nt in length and are primarily derived from transposons, repeat elements, and heterochromatin regions. Their biogenesis is dependent on the DCL3-RDR2-Pol IV pathway (65, 66, 78). hc-siRNAs or ra-siRNAs function in the RNA-dependent DNA methylation (RdDM) pathway by mediating DNA methylation and/or histone modification at the target sites. In addition to 21 to 24 nt siRNAs, a class of lsiRNAs in the size range of 30 to 40 nt was discovered (54). The biogenesis of lsiRNAs is dependent on DCL1, HYL1, HEN1, AGO7, and HST and partially dependent on RDR6 and Pol IV (54). AtlsiRNA-1 is induced by bacterial pathogen Pseudomonas syringae and triggers silencing of the target by destabilizing the target mRNA through decapping and 5′–3′degradation (54).
HOST ENDOGENOUS SMALL RNAS IN PLANT-MICROBE INTERACTIONS
Plants have evolved multiple layers of defense in response to pathogen attacks, and bacterial pathogens provide useful examples of how pathogens are encountered at various levels. The preliminary interaction between the pathogen and its host is responsible for pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) in plants (15, 52). Bacteria counteract PTI by secreting and injecting effector proteins into plant cells, which leads to suppression of PTI. Host plants, in turn, have evolved resistance components such as resistance (R) proteins that can recognize effectors and elicit effector-triggered immunity (ETI) (15, 52).
An important role of small RNAs was first demonstrated in plant growth and development (63, 74). There is increasing evidence, however, that small RNAs are involved in regulating plant responses to adverse conditions, including biotic stresses (13, 51). Antiviral defense involving virus-derived small RNAs is an important example of an interaction between plant and pathogen that is mediated by small RNAs. However, these small RNAs are derived from viruses and are therefore exogenous in origin. Unlike viruses that replicate inside the host cell, bacteria, fungi, and other microbes interact with plants without undergoing DNA or RNA replication and transcription inside the plant cell. In these interactions, host endogenous small RNAs play an important role in counteracting these pathogens. Recent reports have shown that plant endogenous small RNAs, including miRNAs and siRNAs, are integral regulatory components of plant defense machinery against bacteria and fungi.
miRNAs
A number of miRNAs have been linked to biotic stress respsonses in plants. In this section we discuss the role of these miRNAs in plants when infected by different types of pathogens such as bacteria, viruses, and fungi. Moreover, it is now known that these small RNAs are also important in regulating plant-microbe interaction during nitrogen fixation by Rhizobium and tumor formation by Agrobacterium.
Bacterial infection
In Arabidopsis, the first miRNA discovered to play a role in defense against pathogens was miR393 (70). miR393 is induced by a bacterial flagellin-derived PAMP, Flg22. miR393 negatively regulates auxin signaling by targeting auxin receptors TIR1 (transport inhibitor response 1), AFB2 (auxin signaling F-box protein 2), and AFB3. However, a TIR1 paralog, AFB1, was found to be partially resistant to miR393-mediated cleavage because of extra mismatches in the miRNA target site. Transgenic lines expressing Myc-AFB1 in tir1-1 background were more susceptible to virulent Pseudomonas syringae pv. tomato strain DC3000 (Pst DC3000) and displayed enhanced disease symptoms. To determine whether miR393 is involved in race-specific resistance, these transgenic plants were inoculated with avirulent Pst DC3000 carrying an effector gene, avrRpt2. Bacterial growth in transgenic lines expressing Myc-AFB1 did not differ from non-transformed plants even at 4 days postinoculation (dpi). These data suggest that miR393 has a role in imparting basal resistance but not race-specific resistance. Induction of miR393 was further confirmed by Fahlgren et al. (31) when they carried out deep sequencing of Arabidopsis leaves at 1 h and 3 h postinoculation (hpi) with a nonpathogenic strain, Pst DC3000 hrcC−, which has a mutation in the type III secretion system (TTSS). Relative to uninfected leaves, miR393 was induced tenfold in the infected leaves at 3 hpi. Overexpression of miR393a from a strong constitutive promoter resulted in lower levels of TIR1 mRNA in transgenic lines, and these lines exhibited restricted bacterial growth. Besides miR393, two other miRNA families, miR160 and miR167, were also upregulated at 3 hpi. These miRNAs target members of the auxin-response factor (ARF) family of transcription factors that are also involved in auxin signaling (85). Thus, in response to bacterial infection, plants suppress multiple components of the auxin signaling pathways. Another miRNA, miR825, which is predicted to target remorin, zinc finger homeobox family protein, and frataxin-related protein, was also elevated during Pst hrcC− infection (31).
The interaction between the plant and another bacterium, Agrobacterium tumefaciens, is of general interest because of the widespread use of A. tumefaciens for transferring genes into plant genomes. Pruss et al. (79) showed that an oncogenic strain of A. tumefaciens induced miR393 at the infiltrated zones, whereas the disarmed strain (i.e., a strain lacking tumor-inducing properties) did not induce miR393. Interestingly, the levels of miR393 and miR167, which repress the auxin signaling pathway, were greatly reduced in tumors induced by A. tumefaciens infection. Derepression of the auxin signaling pathway promotes tumor growth. Moreover, roots and stems of miRNA-deficient mutants, dcl1 and hen1, were immune to A. tumefaciens infection (28). All of these results indicate a possible role of miR393 and miR167 in regulating auxin responses in tumor tissue. However, the levels of all other miRNAs studied were moderately altered in tumor tissue. All these results demonstrate the importance of miR393 and miR167 in coordinating the interaction between plants and A. tumefaciens.
Recognition of pathogens by plants can initiate an oxidative burst resulting in enhanced abundance of reactive oxygen species (ROS) (25, 58, 72, 94, 104). Whereas miR398 is down-regulated in response to oxidative stress imposed by high Cu2+ or high Fe3+ in Arabidopsis, the levels of the target mRNAs, Cu/Zn superoxide dismutatses 1 and 2 (CSD1 and CSD2), increase significantly (91). To investigate effects of microbe infection on the levels of miR398 and its targets, plants were infected with both virulent Pst DC3000 and avirulent Pst DC3000 carrying effector avrRpt2 or avr-Rpm1 (48). miR398 was down-regulated by infection with Pst (avrRpm1) and Pst (avrRpt2) but was unaffected by Pst DC3000 infiltration. miR398 cleaves its target mRNA, thereby decreasing the levels of CSD1 and CSD2. During Pst (avrRpm1) infection, CSD1 but not CSD2 exhibited strong upregulation in transcript levels. The differential response of CSD2 mRNA to diverse stress conditions suggests the existence of multiple mechanisms that are either dependent or independent of miR398-guided regulation in plants.
Fungal infection
RNA silencing is a robust strategy developed by plants to defend against pathogens, including fungi. Posttranscriptional gene silencing (PTGS) was shown to affect fungal resistance in Arabidopsis in studies employing the RNA silencing mutants sgs2, sgs3, ago7, dcl4, nrpd1a, and rdr2, which exhibited enhanced susceptibility to Verticillium strains (30).
Lu et al. (61) tested whether small RNAs are involved in the infection of loblolly pine by the endemic rust fungus, Cronartium quercuum f. sp. fusiforme. Infection with this fungus causes fusiform rust disease, which is characterized by stem and/or branch galls. Small RNAs were cloned from the developing xylem of pine, and 26 miRNAs belonging to four conserved and seven loblolly pine-specific miRNA families were identified. Using small RNA expression profiling, miRNAs involved in disease development were delineated and compared in uninfected pine trees and trees infected with the fusiform rust fungus. The transcript levels for these 11 families of miRNAs were unchanged in roots and in stems above the galls, but transcript levels for 10 of these miRNA families were significantly suppressed in galled stem. These reduced levels of miRNAs in galled stems relative to healthy stems were correlated with increased levels of their target transcripts relative to healthy stems. Interestingly, although the expression of these miRNAs was unchanged in the stem above the gall, their target transcripts were significantly upregulated in stem above the gall as compared with healthy stems. This result suggests that fungal infection at the gall probably immunizes the uninfected stem and may provide protection ahead of the spreading infection. Taken together, these data highlight the complexity of plant-microbe interactions mediated by small RNAs in the galled tissue and the tissue surrounding the gall. The signals responsible for the upregulation of defense responsive genes in the uninfected tissue around the gall remain to be identified.
Viral infection
As discussed earlier, host miRNAs respond to attack by pathogenic fungi and bacteria. This prompts the question of whether host miRNAs respond to viral infection. Two miRNAs, bra-miR158 and bra-miR1885, were greatly upregulated when Brassica rapa was infected by Turnip mosaic virus (TuMV) (42). The induction of bra-miR158 and bra-miR1885 is highly specific to TuMV infection because infection of B. rapa and B. napus with Cucumber mosaic virus, Tobacco mosaic virus (TMV), or the fungal pathogen Sclerotinia sclerotiorum had no such change. The putative target for bra-miR1885 is predicted to be a member of the TIR-NBS-LRR class of disease-resistant proteins. It is suggested that bra-miR1885 is a novel miRNA generated from gene-duplication events from the TIR-NBS-LRR class of proteins. Understanding the mechanism of plant defense responses against viruses will require the identification of additional miRNAs that are regulated by viral infection.
Symbiotic nitrogen fixation
Nitrogen fixation in soybean and other legumes is the result of a symbiotic association between the leguminous plant and rhizobial bacteria. This mutually beneficial association involves the exchange of chemical signals leading to the formation of specialized nitrogen-fixing structures known as root nodules (17). Understanding and elucidating this symbiotic association will require the identification of the molecular determinants and their regulators at different stages of the interaction leading to nodule development. Two strategies have been employed for identifying small RNAs that could participate in this interaction: One involves the identification of conserved miRNAs in soybean based on the homology of known miRNAs in other plant species, and the other involves the use of high-throughput sequencing and cloning of small RNAs differentially expressed in soybean roots inoculated with the bacterium Bradyrhizobium japonicum compared to mock-inoculated roots (90). Approximately 55 families of miRNAs were identified, of which 35 were found to be novel. Further expression analysis of B. japonicum–responsive miRNAs revealed that miR168 and miR172 were transiently upregulated up to 3 hpi but were downregulated to basal levels by 12 hpi. Although miR159 and miR393 exhibited significant upregulation at 3 hpi, miR160 and miR169 showed downregulation. Rhizobial infection changes the levels of miR160, miR393, miR164, and miR168, which target ARFs (ARF10, ARF16, and ARF17), TIR1, NAC1, and AGO1, respectively. These results strongly support the role of auxin homeostasis/signaling in nodulation (Figure 2).
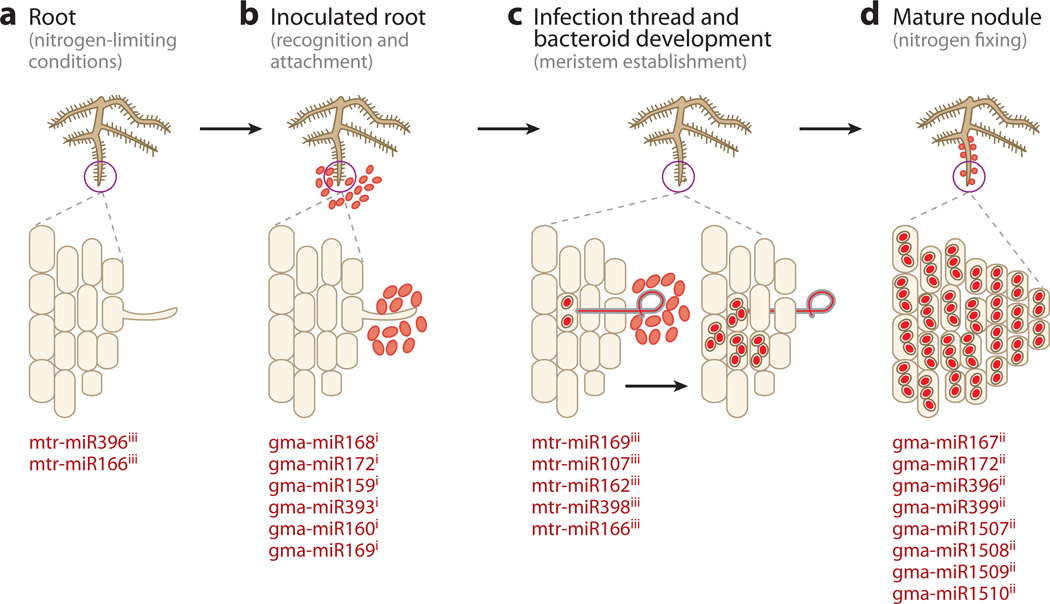
Figure 2.
Small RNAs regulate rhizobia-legume symbioses, resulting in nodule development for nitrogen fixation. Different steps in nodule formation are shown along with the miRNAs predicted to be involved at specific steps. (a) The interaction of nitrogen-starved plants with rhizobial bacteria results in the exchange of chemical signals as plants secrete flavonoids and bacteria produce lipochitooligosaccharides. (b) The recognition of signals results in the attachment of bacterial cells to root hairs. (c) Changes in ionic equilibrium lead to the deformation of root hairs and transcription of nodulation-specific genes. Curling of root hairs to engulf bacteria results in the formation of infection thread that transports bacteria deep into the root tissue followed by bacteroid development. (d) Within 2 to 3 weeks postinoculation, mature nitrogen-fixing nodules are formed. Superscripts i, ii, and iii represent the miRNAs identified by Subramanian et al. (90), Wang et al. (101), and Lelandais-Briére et al. (59), respectively. mtr, Medicago truncatula; gma, Glycine max.
To understand the additional components involved in root nodulation in legumes, researchers have been unraveling the regulatory network at the later phase of the soybean-rhizobial interaction. Cloning and sequencing of miRNAs from functional nodules of soybean identified small RNAs belonging to 11 miRNA families (101). Four of these belonged to conserved miR167, miR172, miR396, and miR399 families, whereas another four families had sequences homologous to gma-miR1507, gma-miR1508, gma-miR1509, and gma-miR1510, which were also reported previously by Subramanian et al. (90). Three novel miRNA families (gma-miR222, gma-miR383, and gma-miR235) that possibly play a role in nitrogen fixation were reported. These miRNAs exhibited differential expression in root nodules. High levels of gma-miR172 and gma-miR222 but low levels of gma-miR1508 and gma-miR1510 were detected in root nodules, prompting the authors to speculate that these miRNAs play critical roles in nodule maturation and nitrogen fixation (Figure 2). Further investigations on functions of these miRNAs and their putative targets would provide insights on legume-rhizobium interactions during symbiosis and ultimately about the mechanism of nitrogen fixation.
The levels of MtHAP2-1, a transcription factor of Medicago truncatula strongly upregulated during nodule development, are controlled by miR169 (16). MtHAP2-1 encodes a HAP2-type transcription factor (29) and is abundant in and limited to cells of the nodule meristematic zone (16), which further suggests its role in nitrogen fixation. MtHAP2-1 RNAi transgenic lines exhibited delayed nodule maturation because of arrested growth and internment of the bacterium Sinorhizobium melitoti within the developing nodule. miR169 limits the expression of MtHAP2-1 to the nodule meristematic zone, and the root growth of miR169-resistant MtHAP2-1 transgenic plants was reduced. All of these results strongly suggest that, during rhizobial infection of M. truncatula, MtHAP2-1 is critical in differentiation of nodule cells and that its expression levels are temporally and spatially tuned by miR169. The involvement of miR169 in regulating MtHAP2-1 levels further emphasizes the role of small RNAs in symbiotic interaction between bacteria and plants.
Genome-wide small RNA profiling of root apices and nodules of M. truncatula identified 100 novel candidate miRNAs in addition to the miRNAs that are homologous to known miRNAs from other species (59). Northern blot analysis and in situ localization studies revealed several miRNAs that were differentially expressed between roots and nodules. miR167 is highly expressed in the nodule peripheral vascular tissues, whereas miR172 and miR398 are specifically localized in the infection zone, which suggests that they have roles in cell differentiation or infection by the symbiotic bacteria. These studies in soybean and Medicago indicate that miRNAs indeed contribute to gene regulation of nodulation.
siRNAs
Although plants contain only several hundred miRNAs, they contain huge numbers of endogenous siRNAs. As discussed earlier, these siRNAs have been classified into groups based mainly on their biogenesis: the ta-siRNAs, ra-siRNAs, nat-siRNAs, and lsiRNAs. Their biological roles, however, are not well understood. Our laboratory has identified a nat-siRNA and a lsiRNA specifically induced by the bacterial pathogen Pst (avrRpt2), and these siRNAs contribute to plant antibacterial immunity (54, 55).
nat-siRNAs
The first plant-endogenous nat-siRNA identified as involved in plant immunity is nat-siRNAATGB2, which regulates R-gene mediated ETI (55). This small RNA is generated from the overlapping region of a NAT pair, which encodes a Rab2-like GTP-binding protein gene ATGB2 and a pentatricopepetide protein (PPR)-like gene PPRL. The endogenous RNA is specifically and strongly induced by Pst (avrRpt2) and cleaves antisense PPRL mRNA for silencing. To define the components involved in its biogenesis, we examined the accumulation of ATGB2 nat-siRNAs in Pst (avrRpt2)-challenged small RNA-biogenesis mutants. This small RNA is processed by the DCL1-HYL1 complex, stabilized by HEN1-mediated methylation, and amplified by RDR6, SGS3, and RNA Pol IV. We further demonstrated that the induction of nat-siRNAATGB2 by Pst (avrRpt2) also requires the cognate host R gene RPS2 and its resistance signaling components, including NDR1. Constitutive overexpression of the target gene PPRL without the nat-siRNA target site in transgenic Arabidopsis plants resulted in a delayed hypersensitive response (HR) and attenuated RPS2-mediated disease resistance. These results strongly suggest that PPRL acts as a negative regulator of RPS2-mediated resistance. PPRL is an atypical tandem PPR and is likely to be localized in mitochondria (43, 62), which may contribute to Pst (avrRpt2)-triggered oxidative burst, HR, and programmed cell death (PCD). Proteins containing the PPR domain are believed to be involved mainly in posttranscriptional processes in organelles, including RNA editing (87), mRNA silencing by cleavage (103), and translational regulation (86).
lsiRNAs
During the search for pathogen-induced small RNAs by Northern blot analysis, we identified a novel class of endogenous siRNA, the lsiRNAs (54). As the name suggests, lsiRNAs are longer than the normal 21-to 24-nt siRNAs and are in the size range of 30 to 40 nt. We found several lsiRNAs that are mainly induced by bacterial infection or specific growth conditions. The biogenesis of AtlsiRNA-1 involves components of distinct small RNA pathways, including DCL1, HYL1, HEN1, HST, AGO7, RDR6, NRPD1a, and NRPD1b. Pst (avrRpt2)-mediated induction of AtlsiRNA-1 specifically targets the AtRAP gene, which encodes a RNA-binding protein containing a putative RNA-binding RAP domain (RNA binding domain abundant in Api-complexans). AtlsiRNA-1 employs a unique mechanism to degrade target mRNA by DCP2-VCS (Decapping 2 and Varicose)-mediated decapping followed by an exoribonuclease XRN4-mediated 5′–3′ decay. AtRAP is a negative regulator of PTI and ETI because the knock-out mutant of this gene resulted in enhanced resistance to both avirulent Pst (avrRpt2) and a virulent strain Pst DC3000.
To be cost effective, plants have developed a sophisticated regulatory mechanism to suppress the defense response systems under normal conditions, possibly by employing negative regulators of plant defense, such as PPRL and AtRAP. As the defense systems must be switched on rapidly upon pathogen attack, small RNAs such as miRNA393, nat-siRNAATGB2, and AtlsiRNA-1, are induced in the early phases of infection and cleave or degrade the mRNA of these negative regulators of plant immunity, thereby mounting rapid counterdefense mechanisms.
siRNAs generated from R gene loci (RPP5 locus and N gene MITEs)
Plant resistance (R) proteins recognize specific pathogens by directly and indirectly interacting with corresponding avirulence (avr) effectors and triggering a cascade of events leading to disease resistance (21, 64). These R genes are generally clustered in the genome and encode proteins with common motifs. To keep pace with the continuous and rapid evolution of pathogens, R genes undergo coevolution resulting in variable gene clusters. The Arabidopsis thaliana ecotype Columbia RPP4 locus (known as RPP5 in Landsberg erecta for recognition of the oomycetes Hyaloperonospora parasitica 5) is composed of seven TIR-NBS-LRR class-R genes along with three related and two unrelated genes (73, 109). Two R genes in this locus, named RPP4 and SNC1 (for suppressor of npr1-1, constitutive 1), impart resistance to both P. syringe pv. maculicola and H. parasitica (89, 97, 108, 114). These two R genes are coordinately regulated by transcriptional activation and siRNA-mediated RNA silencing. Endogenous siRNAs generated at the RPP4 locus apparently target SNC1 because enhanced transcript levels of SNC1 were observed in small RNA biogenesis-deficient mutants such as dcl4, upf1, and ago1 as well as in transgenic plants expressing P1/HC-Pro suppressor. This fine-tuning of R-gene expression is important because it substantially reduces the fitness cost for constitutive activation of defense pathways. In other words, the siRNAs generated from the RPP4 locus may control the resistance responses, thereby enhancing plant health and fitness.
MITEs (miniature inverted repeat transposable elements) are truncated DNA transposons that are generally fewer than 600 base pairs (bp) long, lack open reading frames, and depend on the activity of transposons for their mobility (49, 50). MITEs are present in high copy numbers in several plant genomes and are predicted to be regulatory elements in plant gene expression (33, 50). They are also thought to serve as a major evolutionary element in transposon-mediated gene regulation in plants by generating small RNAs. The complexity of the TMV R gene N is maintained by MITE-mediated creation of new gene structures (56). Virus infection may lead to temporary inhibition of PTGS for N expression and siRNA-guided cleavage. This may induce premature translation termination, provide a polyadenylation signal, or introduce deletions/insertions leading to gene diversity. Kuang et al. (56) suggested that biogenesis of most MITE-derived small RNAs (MiS) depends on the NR-PDla/RDR2/DCL3/AGO4 pathway. DCL4 is also implicated in the accumulation of MiS small RNAs (56). It will help our understanding of R gene evolution to elucidate the mechanisms of generating small RNAs from MiS and their functionality in gene regulation in plants.
COMPONENTS OF THE SMALL RNA BIOGENESIS PATHWAY PLAY AN IMPORTANT ROLE IN PLANT DEFENSE
Many plant genomes encode multiple DCLs, RDRs, and AGOs in the RNAi silencing machinery. The components within the same family have distinct or sometimes partially overlapping functions in different small RNA pathways. Arabidopsis has four DCLs, six RDRs, and ten AGOs, many of which are involved in plant-defense signaling pathways.
Dicer-Like Proteins and Their Associated Proteins
Arabidopsis contains four DCLs that process dsRNA or fold-back RNA precursors to generate siRNAs and miRNAs, respectively. Genetic experiments using single, double, or triple mutants of DCLs have dissected the roles of individual DCLs and their compensatory functions in the production of virus-derived small RNAs (viRNAs). Recent studies demonstrate that loss-of-function mutations in both DCL4 and DCL2 are necessary and sufficient to make plants highly susceptible to several (+)ssRNA viruses (9, 22, 24, 35).
Deleris et al. (22) employed virus-induced gene silencing (VIGS) to knock out endogenous phytoene desaturase (PDS) by inoculating Arabidopsis plants with modified tobacco rattle virus (TRV). The virus was modified such that the plant PDS gene replaced the RNA2-encoded 2b and 2c sequences in the viral genome. After infection, the modified virus did not cause disease because of the siRNA-mediated antiviral response. That is, the siRNAs targeted TRV-PDS and generated PDS-targeting siRNAs. These PDS siRNAs initiated degradation of endogenous PDS mRNA, which results in extensive photo-bleaching of the plants. The inoculation of the modified TRV-PDS in wild-type and dcl mutants showed that DCL4 is the primary sensor and produces 21-nt siRNAs that program a RISC effector complex against viruses. In the absence of DCL4, DCL2 acts as a subordinate antiviral defense protein by producing 22-nt siRNAs. Moreover, double mutants of dcl2 and dcl4 exhibited hypersusceptibility to the viral infection, which suggests the combined action of these two specific proteins in antiviral defense. Unlike DCL2 and DCL4, DCL1 and DCL3 were not found to be involved in antiviral immunity in this study. A similar study by Qu et al. (82), which aimed to determine the contribution of key RNA-silencing pathway components in antiviral silencing, found that all four DCL proteins are involved in mounting an antiviral defense in plants. This study confirmed that DCL2 and DCL4 are primary proteins, whereas DCL3 has a minor role in this process. Interestingly, DCL1 was also implicated in antiviral silencing but apparently plays a negative role, as it down-regulates the expression of DCL4 and DCL3. These studies corroborate that there is a functional hierarchy of different plant DCL proteins (DCL4>DCL2>DCL3>DCL1) in processing viral RNAs into viRNAs (22). DCL3 also has clear antiviral roles in natural DNA virus infections (7, 67). Arabidopsis plants infected with CaMV had an increased viral load in dcl2-dcl3-dcl4 mutants. However, viral load determined by immunoblot analysis of coat protein was not significantly different between wild-type plants and single mutants (dcl2, dcl3, dcl4) or double mutants (dcl2-dcl3, dcl2-dcl4, dcl3-dcl4). DCL1 plays a positive role in antiviral defense by facilitating synthesis of viRNAs derived from the intramolecular hairpins formed in the 35S-leader sequence of CaMV. These hairpins resemble miRNA precursors, and DCL1 probably identifies this structure, excises it, and then presents it to other Dicers for further processing.
Small dsRNA-binding proteins (DRBs) are essential cofactors of DCL proteins (45, 69). These DRBs, however, do not exhibit hierarchical redundancy as do DCLs (19). DCL1 and DCL4 interact with DRB1/HYL1 and DRB4, respectively (45, 69). DRB4 contributes to antiviral defense, possibly by interacting with DCL4 (82). In contrast, DCL2 and DCL3 do not require any DRB for production of viRNAs (19). Another protein that contains a dsRNA binding domain is HEN1, which plays an important role in small RNA metabolism (75). The Arabidopsis hen1 mutant exhibits hyper-susceptibility to CMV by a fivefold increase in the accumulation of the viral RNA when compared with that in wild type, which suggests that HEN1 contributes to resistance against the virus (10).
DCL proteins are also involved in the generation of small RNAs that contribute to antibacterial immunity in plants. The dcl1 mutant showed enhanced susceptibility to Pst DC3000 hrcC−, a nonpathogenic strain that can elicit PTI (71). DCL1 is required for generating miR393. The accumulation of the target transcripts of miR393 (TIR1, AFB2, and AFB3) is increased in the dcl1–9 mutant (70) when treated with flg22 peptide. DCL1 is also involved in the generation of nat-siRNAATGB2 (55) and AtlsiRNA-1 (54).
HYL1, the dsRNA-binding protein associated with DCL1, is also involved in resistance against bacterial infection, as the hyl1 mutant was susceptible to Pst (avrRpt2) (55). Moreover, the hyl1 mutant exhibited compromised accumulation of nat-siRNAATGB2 and AtlsiRNA-1 (54, 55).
RNA-Dependent RNA Polymerases
In plants, RDRs are important for siRNA formation because they synthesize dsRNAs for downstream processing by DCL. Initial studies showed that virus infection enhances RDR activity, which led to the hypothesis that RDRs are one of the many host factors that assist virus replication (27). Subsequently, extensive studies have implicated RDRs in antiviral defense in plants (4, 106, 112). Xie et al. (106) found that RDR1 activity is induced not only by virus infection but also by defense signaling compounds such as salicylic acid (SA). Reducing the expression levels of RDR1 in transgenic antisense Arabidopsis plants resulted in enhanced accumulation of viral RNAs and increased susceptibility to TMV and potato virus X (PVX) infection. AtRDR1, an ortholog of NtRDR1, is also known to impart defense against tobamovirus and tobravirus because Arabidopsis rdr1 mutant plants had enhanced levels of viral RNAs (112). NtRDR1 is also involved in combating potato virus Y (PVY) infection; knocking down expression of RDR1 in transgenic tobacco plants resulted in reduced expression of other defense-related genes such as AOX1 and ERF5 (84). These studies raise the question of whether different RDR proteins (for instance, the six members in Arabidopsis thaliana) have distinct or overlapping functions.
Arabidopsis RDR6 (SDE1/SGS2) was initially considered to be important for transgene-induced PTGS and apparently had no role in plant antiviral response because a RDR6 mutant allele sde1 did not exhibit enhanced viral susceptibility (20, 68). Extensive studies by Qu et al. (81), however, demonstrated that NbRDR6 (a functional homolog of AtRDR6) plays an important role in antiviral defense. The transgenic tobacco plants wherein NbRDR6 was downregulated showed hypersusceptibility to many different viruses, and this response was more pronounced at elevated temperatures. This was consistent with earlier reports, which demonstrated that there is increased siRNA generation at higher temperatures (93). This study provided strong evidence that NbRDR6 contributes to general antiviral defense by RNA silencing and that environmental conditions influence the plant-virus interactions. In a recent elegant study, Wang et al. (102) examined single, double, and triple mutants of RDR1, RDR2, and RDR6 using a mutated CMV with no viral suppressor (VSR) 2b, and demonstrated the role of both RDR1 and RDR6 in secondary viRNA formation. Small RNA profiling revealed that RDR1 preferentially amplifies viRNAs that mapped at the 5′-terminal viral RNAs, whereas RDR6-dependent viRNAs mapped to the 3′-terminal half of viral RNAs. RDR6 interacts with a coiled-coil protein, SGS3, to produce secondary viRNAs (57, 99). An sgs3 mutant shows enhanced susceptibility to cucumovirus (68), which indicates that SGS3 also contributes to antiviral resistance in plants. The generation of nat-siRNAATGB2 (55) and AtlsiRNA-1 (54) requires RDR6. Pst DC3000 (avrRpt2) displayed enhanced growth in the rdr6 mutant (55), which provided direct evidence for the function of RDR6 in plant immunity.
Argonautes
AGOs are associated with small RNAs and form RISC complexes to induce silencing of target genes (41). Arabidopsis contains 10 AGOs, and their role in plant immunity is yet to be determined. There is emerging evidence that the methylation status of plant genomes is altered in response to attack by pathogens, including viruses, bacteria, and fungi (34, 39, 76, 88). hc-siRNAs trigger transcriptional gene silencing (TGS) by guiding RNA-directed DNA methylation (RdDM) and histone modification in plants (47, 66, 96). AGO4 is a major nuclear RNAi effector associated with hc-siRNAs or ra-siRNAs that direct DNA methylation (60, 80). Involvement of AGO4 in the disease-resistance response links DNA methylation and plant defense. When attacked by viruses, plants employ DNA methylation to repress viral transcription and/or replication. Upon infection by either of two geminiviruses, cabbage leaf curl virus (CaLCuV) or beet curly top virus (BCTV), Arabidopsis plants silence viral chromatin by both cytosine and histone methyltransferases (83). This is evident from the hypersusceptibility of the methylation-deficient mutants to geminiviruses, including mutants of cytosine methyltransferases (drm1, drm2, cmt3, and met1), histone H3K9 methyltransferase (kyp2), RdDM pathway components (ago4, ddm1, and nrpd2A), or methyl cycle enzymes (adk1 and adk2) (83). Viral suppressors AL2 and L2 inhibit the activity of adenosine kinase (ADK), a cellular enzyme involved in the generation of S-adenosylmethionine (a methyltransferase cofactor). Therefore, plants infected with virus lacking L2 had hypermethylation of viral DNA. Additionally, recovery of the virus-infected plants from the disease symptoms required AGO4 (83). Chromatin methylation may be a generalized process adopted by plants to evade infection by DNA. VIGS of two Nicotiana benthamiana homologs of Arabidopsis AGO4 blocked the R gene–mediated antiviral responses through translational suppression of viral RNAs (6). This result suggests that AGO4 may have additional functions besides its role in the RdDM pathway. It is also possible that the NbAGO4 may not function the same way as Arabidopsis AGO4, or there might be another unidentified N. benthamiana AGO that is more closely related to Arabidopsis AGO4.
AGO4 is also involved in antibacterial defenses. Mutant ago4–2 was identified from a genetic screening using a H2O2-responsive Ep5C promoter-driven GUS reporter (2). Assessment of disease susceptibility revealed that ago4-2 exhibits reduced resistance to virulent Pst DC3000 as well as to avirulent Pst (avrRpm1). However, mutants of DCL3 and RDR2, the up-stream components of AGO4, and mutants of chromomethylase 3 (CMT3) and defective in RNA-directed DNA methylation (DRD1) and domains rearranged methyltransferase 1 and 2 (DRM1 and DRM2), the downstream components of AGO4 in the RdDM pathway, showed no change in Pst growth. These results suggest that either these components are functionally redundant with their close homologous genes or AGO4 simply has an unidentified RdDM-unrelated function in plant defense. These possibilities should be investigated by testing the disease resistance responses of double and triple mutants of these components in the RNAi and RdDM pathways.
In addition to AGO4, Qu et al. (82) conclusively showed that AGO1 and AGO7 have an important role in slicing viral RNAs. AGO1 is the primary slicer because it targets viral RNAs with more compact structures, but AGO7 is a surrogate slicer whose targets are less structured (82). The biogenesis of AtlsiRNA-1 is known to involve AGO7, as ago7 mutant does not accumulate AtlsiRNA-1 (54). However, other ago mutant plants, including ago3, ago4, and ago9, showed no significant change in the level of AtlsiRNA-1 as compared with wild type. AGO7 is also associated with TAS3 ta-siRNA (1, 32, 36). The accumulation of bacteria-induced AtlsiRNA-1 is dependent on AGO7, suggesting a role of AGO7 in antibacterial defense.
RNA SILENCING SUPPRESSORS OF PATHOGENS
Viral Suppressors of RNA Silencing
Many viruses encode specific proteins that suppress the host antiviral silencing response and thereby benefit viral infection. These viral suppressors of RNA silencing (VSRs) can act at three levels, i.e., they can (a) inhibit generation of viRNAs, (b) inhibit loading of viRNAs in RISC by binding to the viRNA, and (c) inhibit components of RISC. Ectopically expressed VSRs, in conjugation with a sensor, have been used to decipher functions of many VSRs. The use of viruses with disabled or modified VSRs, however, has recently proven to be a very effective approach for determining VSR function. Two recent reviews provide comprehensive coverage on this topic (23, 26). Table 1 summarizes what is known about the mode of action of VSRs in plants.
Table 1.
Mode of action of viral silencing suppressors in plants
| Suppressor | Source | Mode of action | Reference |
|---|---|---|---|
| AC4 | Geminivirus | Competes with AGOs by binding to single-stranded siRNA and thereby preventing RISC assembly | 12 |
| AC2 | Begomovirus | Transcriptional activator. Induces expression of any gene, which might be a silencing suppressor. | 95 |
| HcPro | Potyvirus | Mimics hen1 mutations. viRNAs are oligo-uridylated and partially degraded due to lack of 2′-O-methylation. Interacts with a calmodulin-related protein, overexpression of which suppresses silencing. Amino acids 180, 205, and 396 of HcPro are critical for suppression of miRNA, ta-siRNA, and VIGS pathway but not for sense PTGS. |
111, 3, 105 |
| P6 | Cauliflower mosaic virus | Is imported in the nucleus and binds to DRB4 protein. Suppresses RNA silencing pathway, possibly by inactivating DRB4, which is an essential component required for DCL4 action. | 40 |
| 2b | Cucumber mosaic virus | Interacts physically with siRNA-loaded RISC and inhibits its slicing action. In vitro assays suggest that 2b binds to siRNAs to a lesser extent than to long dsRNAs. 2b inhibits the production of RDR1-dependent viral siRNAs. |
113, 38, 24 |
| P0 | Polerovirus | Promotes ubiquitin-dependent proteolysis of AGO1. | 77 |
| P69 | Tymovirus | Inhibits viRNA amplification. | 14 |
| AL2 | Curtovirus | Interacts with adenosine kinase, whose inhibition possibly prevent methylation of viral DNA. | 100 |
| p126 | TMV | Encodes methyltransferase and helicase. Binds duplex siRNA and inhibits HEN1-dependent methylation and degradation. | 7 |
| RNAse III | Closterviridae | In vitro assays suggest that RNAse III suppresses siRNA silencing by cleaving 21, 22, and 24 bp siRNAs into 14-bp fragments. | 18 |
Bacteria-Encoded Suppressors of RNA Silencing
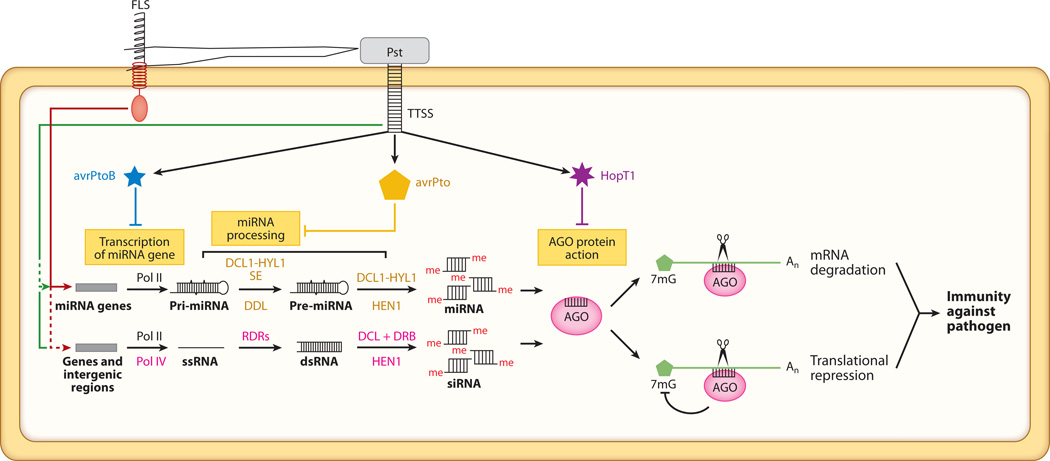
As noted in the previous paragraph, viruses encode VSRs that suppress host antiviral silencing machinery and thereby promote their pathogenesis. Because small RNAs and RNA-silencing machinery are also important for antibacterial defense, the question arises as to whether bacterial pathogens also developed similar silencing suppressors to counter antibacterial defense responses in plants. Recently, Navarro et al. (71) identified several Pst type III secretion effectors that suppress host RNA silencing machinery and therefore increase disease susceptibility (Figure 3). AvrPtoB represses transcription of miRNA genes and results in a low level of pri-miR393. Some pri-miRNAs were unaffected, however, and it is therefore unlikely that AvrPtoB is a general transcriptional suppressor of the miRNA pathway. AvrPtoB might suppress a specific component involved in plant defense that is required for transcription of miR393 genes. Another effector, AvrPto, interferes with processing of some miRNA precursors and downregulates the level of mature miR393. It remains to be determined whether AvrPto directly suppresses miRNA-processing components, such as DCL1 and HYL1, or the components required for miRNA stability, such as HEN1. In addition, HopT1 inhibits the action of the AGO1 protein in the RISC complex. Thus, as with viruses, bacteria have evolved an array of effectors that target different steps of the miRNA pathway. We speculate that other pathogens, such as fungi and oomycetes, have also developed RNAi suppressors to counteract host antipathogen RNA-silencing mechanisms.
Figure 3.
Mechanism of action of bacterial suppressors of RNA silencing (BSRs) in plants. Different steps in small RNA pathways are suppressed by different effectors encoded by Pseudomonas syringae. FLS: flagellin receptor; TTSS: type III secretion system of bacterial pathogen; Pst: Pseudomonas syringae pv. tomato DC3000.
CONCLUDING REMARKS
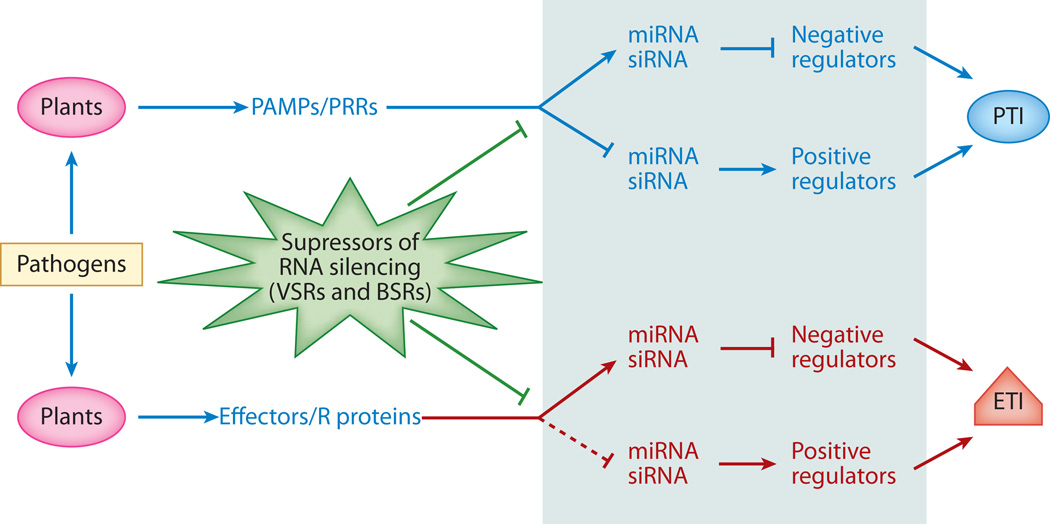
More and more studies have shown that many host miRNAs and siRNAs are induced or suppressed by various pathogen challenges and that modulation of miRNA and siRNA levels plays an important role in gene expression reprogramming and fine-tuning plant responses against a wide range of pathogens (Figure 4). These pathogen-responsive small RNAs induce posttranscriptional gene silencing by guiding mRNA cleavage/degradation or translational repression, or may guide transcriptional gene silencing by direct DNA methylation or chromatin modification. This idea is supported by observations that many components in the small RNA pathways are required for plant defense responses and immunity. As a countermeasure, viruses and bacteria have developed VSRs and BSRs to suppress host RNAi machinery and compromise disease resistance in plants. To combat continuously evolving pathogens, plants have also evolved components, such as R proteins, that can recognize VSRs and BSRs and trigger robust and rapid resistant responses, which are referred to as ETI.
Figure 4.
Immunity against pathogens is regulated by small RNAs in plants. In PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI) regulated by small RNAs, RNA silencing suppressors (VSRs and BSRs) repress the small RNA silencing pathway. PAMPs, pathogen-associated molecular patterns; PRRs, pattern recognition receptors; VSRs, viral suppressors of RNA silencing; BSRs, bacteria-encoded suppressors of RNA silencing.
The study of small RNA–mediated regulatory mechanisms in plant immunity is an emerging field, and we expect that many more pathogen-responsive small RNAs will be identified using new technologies, such as high-throughput deep sequencing. Characterization of these small RNAs and their target genes will reveal new components in plant resistance signaling pathways and help us understand the molecular mechanisms of plant immunity. We also expect that more silencing suppressors will be identified from viruses, bacteria, fungi, and oomycetes, and that such identification will increase our understanding of the interaction and coevolution between pathogens and plant hosts. These studies will elucidate the molecular mechanisms of plant defense responses and will ultimately lead to the development of effective tools for controlling disease in the field.
SUMMARY POINTS.
Endogenous small RNAs play pivotal roles in reprogramming of host gene expression in response to infection by a wide range of microbes, including viruses, bacteria, and fungi.
Small RNAs contribute to PTI or basal defense, as well as ETI or race-specific resistance.
Small RNAs are generated from pathogen-derived nucleic acids as seen in viruses and Agrobacterium. These pathogen-derived small RNAs may regulate the interaction of pathogens with plants.
Endogenous small RNAs are induced or suppressed during legume-rhizobia symbioses. These small RNAs are thought to facilitate the plant-bacterium interaction by regulating different steps in the development of nitrogen-fixing nodules.
Different components of small RNA pathways directly play important roles in mediating host immune responses against pathogens.
RNA silencing is activated in plants to counteract pathogen infection. To counter-counteract effects of silencing and to establish infection, rapidly evolving pathogens secrete suppressors of RNA silencing (VSRs and BSRs) that inhibit different steps of small RNA pathways.
To combat continuously evolving pathogens, plants have evolved components such as R proteins that can recognize pathogen-derived suppressors to mount robust and rapid resistance responses.
FUTURE ISSUES.
Deep sequencing technologies will allow small RNA profiling of plants infected with various pathogens and identification of small RNAs involved in plant-microbe interactions.
It is a challenge to identify and characterize the target genes of newly discovered small RNAs that exhibit alteration in their expression level upon pathogen infection. Overcoming this challenge will allow rapid deciphering of new components in plant-pathogen interactions and lead to elucidation of the complex regulatory network of host immune systems.
Identification of pathogen-derived small RNAs and their potential host targets will help us understand how pathogens cause disease. Whether there is any functional interaction between pathogen-derived small RNAs and host mRNAs is an interesting area of research.
Identification of different components of small RNA machinery during disease susceptibility and resistance responses remains an active area of research. It is still not known whether components of the disease resistance pathway also regulate generation of small RNAs.
Studying the changes in the DNA methylation profiles of host plants after infection with various pathogens may help us understand the evolution of new components in plant defense for combating rapidly evolving pathogens.
Microbes such as viruses and bacteria encode suppressors that counteract the plant defense system. The identification of additional silencing suppressors from different pathogens and the elucidation of their functions and how they act as suppressors will shed light on the generality of suppression of disease resistance. It is interesting and useful to identify the targets of these suppressors and see whether these suppressors affect other pathways in plants besides small RNA pathways. Another tempting projection is that the pathogen-encoded suppressors could affect transposon activation and affect plant gene expression. Understanding the action of these suppressors will unravel the diversity and evolution of pathogens as well as RNA silencing pathways.
Information generated from characterization of new small RNAs and the regulatory network of host immune systems needs to be scrutinized for development of tools to enhance plant resistance against pathogens.
ACKNOWLEDGMENTS
We are grateful to Manu Agarwal, Thomas Eulgem, Isgouhi Kaloshian, Shou-Wei Ding, and Padmanabhan Chellappan for critical reading and invaluable comments and to Somya Sinha for helping with the preparation of the manuscript. The Katiyar-Agarwal laboratory receives funding from Department of Biotechnology, Ministry of Science and Technology, India and University Grants Commission, India. Research in the Jin laboratory is supported by National Science Foundation Career Award MCB-0642843, University of California Discovery Grant Bio06-10566, and California Citrus Board grants 5210-131 and 5210-132. We apologize for not citing some publications owing to space limitations.
Glossary
- RISC
RNA-induced silencing complex
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- ETI
effector-triggered immunity
- TTSS
type III secretion system
- R proteins
resistance proteins
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Surekha Katiyar-Agarwal, Email: katiyars@south.du.ac.in.
Hailing Jin, Email: hailingj@ucr.edu.
LITERATURE CITED
- 1.Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, et al. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 2006;16(9):927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 2.Agorio A, Vera P. ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis . Plant Cell. 2007;19:3778–3790. doi: 10.1105/tpc.107.054494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anandalakshmi R, Marathe R, Ge X, Herr JM, Jr, Mau C, et al. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science. 2000;290:142–144. doi: 10.1126/science.290.5489.142. [DOI] [PubMed] [Google Scholar]
- 4.Baulcombe D. Viruses and gene silencing in plants. Arch. Virol. Suppl. 1999;15:189–201. doi: 10.1007/978-3-7091-6425-9_14. [DOI] [PubMed] [Google Scholar]
- 5.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharjee S, Zamora A, Azhar MT, Sacco MA, Lambert LH, Moffett P. Virus resistance induced by NB-LRR proteins involves Argonaute4-dependent translational control. Plant J. 2009;58(6):940–951. doi: 10.1111/j.1365-313X.2009.03832.x. [DOI] [PubMed] [Google Scholar]
- 7.Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis . Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boutet S, Vazquez F, Liu J, Beclin C, Fagard M, et al. Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 2003;13:843–848. doi: 10.1016/s0960-9822(03)00293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 12.Chellappan P, Vanitharani R, Fauquet CM. MicroRNA-binding viral protein interferes with Arabidopsis development. PNAS. 2005;102:10381–10386. doi: 10.1073/pnas.0504439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chellappan P, Zhang X, Jin H. Host small RNAs are big contributors to plant innate immunity. Curr. Opin. Plant Biol. 2009;12:465–472. doi: 10.1016/j.pbi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Li WX, Xie D, Peng JR, Ding SW. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell. 2004;16:1302–1313. doi: 10.1105/tpc.018986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124(4):803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 16.Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, et al. MtHAP2–1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula . Genes Dev. 2006;20:3084–3088. doi: 10.1101/gad.402806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper JE. Early interactions between legumes and rhizobia: disclosing complexity in a molecular dialogue. J. Appl. Microbiol. 2007;103:1355–1365. doi: 10.1111/j.1365-2672.2007.03366.x. [DOI] [PubMed] [Google Scholar]
- 18.Cuellar WJ, Kreuze JF, Rajamäki ML, Cruzado KR, Untiveros M, Valkonen JP. Elimination of antiviral defense by viral RNase III. Proc. Natl. Acad. Sci. USA. 2009;106(25):10354–10358. doi: 10.1073/pnas.0806042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curtin SJ, Watson JM, Smith NA, Eamens AL, Blanchard CL, Waterhouse PM. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett. 2008;582:2753–2760. doi: 10.1016/j.febslet.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 21.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 22.Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 23.Diaz-Pendon JA, Ding SW. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 2008;46:303–326. doi: 10.1146/annurev.phyto.46.081407.104746. [DOI] [PubMed] [Google Scholar]
- 24.Diaz-Pendon JA, Li F, Li WX, Ding SW. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell. 2007;19:2053–2063. doi: 10.1105/tpc.106.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL. A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell. 1997;88(5):685–694. doi: 10.1016/s0092-8674(00)81911-x. [DOI] [PubMed] [Google Scholar]
- 26.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorssers L, Zabel P, Van Der Meer J, van Kammen A. Purification of a host-encoded RNA-dependent RNA polymerase from cowpea mosaic virus–infected cowpea leaves. Virology. 1982;116:236–249. doi: 10.1016/0042-6822(82)90416-0. [DOI] [PubMed] [Google Scholar]
- 28.Dunoyer P, Himber C, Voinnet O. Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat. Genet. 2006;38(2):258–263. doi: 10.1038/ng1722. [DOI] [PubMed] [Google Scholar]
- 29.El Yahyaoui F, Kuster H, Ben Amor B, Hohnjec N, Puhler A, et al. Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 2004;136:3159–3176. doi: 10.1104/pp.104.043612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellendorff U, Fradin EF, de Jonge R, Thomma BP. RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 2009;60:591–602. doi: 10.1093/jxb/ern306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, et al. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis . Curr. Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 33.Feschotte C. Merlin, a new superfamily of DNA transposons identified in diverse animal genomes and related to bacterial IS1016 insertion sequences. Mol. Biol. Evol. 2004;21:1769–1780. doi: 10.1093/molbev/msh188. [DOI] [PubMed] [Google Scholar]
- 34.Finnegan EJ, Genger RK, Peacock WJ, Dennis ES. DNA methylation in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:223–247. doi: 10.1146/annurev.arplant.49.1.223. [DOI] [PubMed] [Google Scholar]
- 35.Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006;7:1168–1175. doi: 10.1038/sj.embor.7400837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 37.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 38.Goto K, Kobori T, Kosaka Y, Natsuaki T, Masuta C. Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities. Plant Cell Physiol. 2007;48(7):1050–1060. doi: 10.1093/pcp/pcm074. [DOI] [PubMed] [Google Scholar]
- 39.Guseinov VA, Kiryanov GI, Vanyushin BF. Intragenome distribution of 5-methylcytosine in DNA of healthy and wilt-infected cotton plants (Gossypium hirsutum L.) Mol. Biol. Rep. 1975;2:59–63. doi: 10.1007/BF00357298. [DOI] [PubMed] [Google Scholar]
- 40.Haas G, Azevedo J, Moissiard G, Geldreich A, Himber C, et al. Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J. 2008;27(15):2102–2112. doi: 10.1038/emboj.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hannon GJ. RNA interference. Nature. 2002;418(6894):244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 42.He X-F, Fang Y-Y, Feng L, Guo H-S. Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica . FEBS Lett. 2008;582:2445–2452. doi: 10.1016/j.febslet.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 43.Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell. 2004;16:241–256. doi: 10.1105/tpc.016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herr AJ. Pathways through the small RNA world of plants. FEBS Lett. 2005;579(26):5879–5888. doi: 10.1016/j.febslet.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 45.Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, et al. Specific interactions between Dicerlike proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana . Plant Mol. Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- 46.Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, et al. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19(3):926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huettel B, Kanno T, Daxinger L, Bucher E, Van Der Winden J, et al. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim. Biophys. Acta. 2007;1769(5–6):358–374. doi: 10.1016/j.bbaexp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Jagadeeswaran G, Saini A, Sunkar R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis . Planta. 2009;229:1009–1014. doi: 10.1007/s00425-009-0889-3. [DOI] [PubMed] [Google Scholar]
- 49.Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, et al. An active DNA transposon family in rice. Nature. 2003;421:163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- 50.Jiang N, Feschotte C, Zhang X, Wessler SR. Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs) Curr. Opin. Plant Biol. 2004;7:115–119. doi: 10.1016/j.pbi.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 51.Jin H. Endogenous small RNAs and antibacterial immunity in plants. FEBS Lett. 2008;582:2679–2684. doi: 10.1016/j.febslet.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 53.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant. Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 54.Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis . Genes Dev. 2007;21:3123–3134. doi: 10.1101/gad.1595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr, et al. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuang H, Padmanabhan C, Li F, Kamei A, Bhaskar PB, et al. Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: new functional implications for MITEs. Genome Res. 2009;19:42–56. doi: 10.1101/gr.078196.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumakura N, Takeda A, Fujioka Y, Motose H, Takano R, Watanabe Y. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 2009;583:1261–1266. doi: 10.1016/j.febslet.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 58.Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 59.Lelandais-Brière C, Naya L, Sallet E, Calenge F, Frugier F, et al. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell. 2009;21(9):2780–2796. doi: 10.1105/tpc.109.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li CF, Henderson IR, Song L, Fedoroff N, Lagrange T, Jacobsen SE. Dynamic regulation of ARGONAUTE4 within multiple nuclear bodies in Arabidopsis thaliana . PLoS Genet. 2008;4:e27. doi: 10.1371/journal.pgen.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu S, Sun YH, Amerson H, Chiang VL. MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J. 2007;51:1077–1098. doi: 10.1111/j.1365-313X.2007.03208.x. [DOI] [PubMed] [Google Scholar]
- 62.Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006;38(Suppl):S31–S36. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- 64.Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- 65.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6(1):24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 66.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 2009;21(3):367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 67.Moissiard G, Voinnet O. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl. Acad. Sci. USA. 2006;103:19593–19598. doi: 10.1073/pnas.0604627103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 69.Nakazawa Y, Hiraguri A, Moriyama H, Fukuhara T. The dsRNA-binding protein DRB4 interacts with the Dicer-like protein DCL4 in vivo and functions in the trans-acting siRNA pathway. Plant Mol. Biol. 2007;63:777–785. doi: 10.1007/s11103-006-9125-8. [DOI] [PubMed] [Google Scholar]
- 70.Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 71.Navarro L, Jay F, Nomura K, He SY, Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neill SJ, Desikan R, Clarke A, Hurst RD, Hancock JT. Hydrogen peroxide and nitric oxide as signalling molecules in plants. J. Exp. Bot. 2002;53:1237–1247. [PubMed] [Google Scholar]
- 73.Noel L, Moores TL, Van Der Biezen EA, Parniske M, Daniels MJ, et al. Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis . Plant Cell. 1999;11:2099–2112. [PMC free article] [PubMed] [Google Scholar]
- 74.Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, et al. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 75.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana . Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pavet V, Quintero C, Cecchini NM, Rosa AL, Alvarez ME. Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by Pseudomonas syringae . Mol. Plant-Microbe Interact. 2006;19:577–587. doi: 10.1094/MPMI-19-0577. [DOI] [PubMed] [Google Scholar]
- 77.Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, et al. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. USA. 2006;103:1994–1999. doi: 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pikaard CS, Haag JR, Ream T, Wierzbicki AT. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci. 2008;13(7):390–397. doi: 10.1016/j.tplants.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pruss GJ, Nester EW, Vance V. Infiltration with Agrobacterium tumefaciens induces host defense and development-dependent responses in the infiltrated zone. Mol. Plant-Microbe Interact. 2008;21:1528–1538. doi: 10.1094/MPMI-21-12-1528. [DOI] [PubMed] [Google Scholar]
- 80.Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;4437:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 81.Qu F, Ye X, Hou G, Sato S, Clemente TE, Morris TJ. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana . J. Virol. 2005;79:15209–15217. doi: 10.1128/JVI.79.24.15209-15217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qu F, Ye X, Morris TJ. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. USA. 2008;105:14732–14737. doi: 10.1073/pnas.0805760105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raja P, Sanville BC, Buchmann RC, Bisaro DM. Viral genome methylation as an epigenetic defense against geminiviruses. J Virol. 2008;82:8997–9007. doi: 10.1128/JVI.00719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rakhshandehroo F, Takeshita M, Squires J, Palukaitis P. The influence of RNA-dependent RNA polymerase 1 on potato virus Y infection and on other antiviral response genes. Mol. Plant-Microbe Interact. 2009;22:1312–1318. doi: 10.1094/MPMI-22-10-1312. [DOI] [PubMed] [Google Scholar]
- 85.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 86.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shikanai T. RNA editing in plant organelles: machinery, physiological function and evolution. Cell Mol. Life Sci. 2006;63:698–708. doi: 10.1007/s00018-005-5449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Steward N, Ito M, Yamaguchi Y, Koizumi N, Sano H. Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J. Biol. Chem. 2002;277:37741–37746. doi: 10.1074/jbc.M204050200. [DOI] [PubMed] [Google Scholar]
- 89.Stokes TL, Richards EJ. Induced instability of two Arabidopsis constitutive pathogen-response alleles. Proc. Natl. Acad. Sci. USA. 2002;99:7792–7796. doi: 10.1073/pnas.112040999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O. Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics. 2008;9:160. doi: 10.1186/1471-2164-9-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sunkar R, Zhu JK. MicroRNAs and short-interfering RNAs in plants. J. Integr. Plant Biol. 2007;49:817–826. [Google Scholar]
- 93.Szittya G, Silhavy D, Molnar A, Havelda Z, Lovas A, et al. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 2003;22:633–640. doi: 10.1093/emboj/cdg74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Torres MA, Jones JD, Dangl JL. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006;141(2):373–378. doi: 10.1104/pp.106.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Trinks D, Rajeswaran R, Shivaprasad PV, Akbergenov R, Oakeley EJ, et al. Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 2005;79:2517–2527. doi: 10.1128/JVI.79.4.2517-2527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vaistij FE, Jones L, Baulcombe DC. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Van Der Biezen EA, Freddie CT, Kahn K, Parker JE, Jones JD. Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 2002;29:439–451. doi: 10.1046/j.0960-7412.2001.01229.x. [DOI] [PubMed] [Google Scholar]
- 98.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 99.Vazquez F. Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci. 2006;11(9):460–468. doi: 10.1016/j.tplants.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 100.Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 2005;79:7410–7418. doi: 10.1128/JVI.79.12.7410-7418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Y, Li P, Cao X, Wang X, Zhang A, Li X. Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochem. Biophys. Res. Commun. 2009;378:799–803. doi: 10.1016/j.bbrc.2008.11.140. [DOI] [PubMed] [Google Scholar]
- 102.Wang X-B, Wu Q, Ito T, Cillo F, Li W-X, et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA. 2010;107:484–489. doi: 10.1073/pnas.0904086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Z, Zou Y, Li X, Zhang Q, Chen L, et al. Cytoplasmic male sterility of rice with Boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell. 2006;18:676–687. doi: 10.1105/tpc.105.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wendehenne D, Durner J, Klessig DF. Nitric oxide: a new player in plant signalling and defence responses. Curr. Opin. Plant Biol. 2004;7:449–455. doi: 10.1016/j.pbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 105.Wu HW, Lin SS, Chen KC, Yeh SD, Chua NH. Discriminating mutations of HC-pro of zucchini yellow mosaic virus with differential effects on small RNA pathways involved in viral pathogenicity and symptom development. Mol. Plant-Microbe Interact. 2010;23(1):17–28. doi: 10.1094/MPMI-23-1-0017. [DOI] [PubMed] [Google Scholar]
- 106.Xie Z, Fan B, Chen C, Chen Z. An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. USA. 2001;98:6516–6521. doi: 10.1073/pnas.111440998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie Z, Qi X. Diverse small RNA-directed silencing pathways in plants. Biochim. Biophys. Acta. 2008;1779(11):720–724. doi: 10.1016/j.bbagrm.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 108.Yang S, Hua J. A haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis . Plant Cell. 2004;16:1060–1071. doi: 10.1105/tpc.020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yi H, Richards EJ. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell. 2007;19:2929–2939. doi: 10.1105/tpc.107.051821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yu B, Bi L, Zheng B, Ji L, Chevalier D, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. USA. 2008;105:10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu B, Chapman EJ, Yang Z, Carrington JC, Chen X. Transgenically expressed viral RNA silencing suppressors interfere with microRNA methylation in Arabidopsis . FEBS Lett. 2006;580:3117–3120. doi: 10.1016/j.febslet.2006.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu D, Fan B, MacFarlane SA, Chen Z. Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol. Plant-Microbe Interact. 2003;16:206–216. doi: 10.1094/MPMI.2003.16.3.206. [DOI] [PubMed] [Google Scholar]
- 113.Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang Y, Goritschnig S, Dong X, Li X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1–1, constitutive 1 . Plant Cell. 2003;15:2636–2646. doi: 10.1105/tpc.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]