Abstract
Hyperexpression of the Saccharomyces cerevisiae multidrug ATP-binding cassette (ABC) transporter Pdr5p was driven by the pdr1-3 mutation in the Pdr1p transcriptional regulator in a strain (AD/PDR5+) with deletions of five other ABC-type multidrug efflux pumps. The strain had high-level fluconazole (FLC) resistance (MIC, 600 μg ml−1), and plasma membrane fractions showed oligomycin-sensitive ATPase activity up to fivefold higher than that shown by fractions from an isogenic PDR5-null mutant (FLC MIC, 0.94 μg ml−1). In vitro inhibition of the Pdr5p ATPase activity and chemosensitization of cells to FLC allowed the systematic screening of a 1.8-million-member designer d-octapeptide combinatorial library for surface-active Pdr5p antagonists with modest toxicity against yeast cells. Library deconvolution identified the 4-methoxy-2,3,6-trimethylbenzensulfonyl-substituted d-octapeptide KN20 as a potent Pdr5p ATPase inhibitor (concentration of drug causing 50% inhibition of enzyme activity [IC50], 4 μM) which chemosensitized AD/PDR5+ to FLC, itraconazole, and ketoconazole. It also inhibited the ATPase activity of other ABC transporters, such as Candida albicans Cdr1p (IC50, 30 μM) and Cdr2p (IC50, 2 μM), and chemosensitized clinical isolates of pathogenic Candida species and S. cerevisiae strains that heterologously hyperexpressed either ABC-type multidrug efflux pumps, the C. albicans major facilitator superfamily-type drug transporter BenRp, or the FLC drug target lanosterol 14α-demethylase (Erg11p). Although KN20 also inhibited the S. cerevisiae plasma membrane proton pump Pma1p (IC50, 1 μM), the peptide concentrations required for chemosensitization made yeast cells permeable to rhodamine 6G. KN20 therefore appears to indirectly chemosensitize cells to FLC by a nonlethal permeabilization of the fungal plasma membrane.
The increased prevalence of superficial and systemic fungal infections over the last two decades has been caused by factors that include the extensive use of broad-spectrum antibiotics, the widespread application of invasive medical procedures, such as cardiovascular surgery, and the increased number of immunocompromised individuals, such as AIDS patients and recipients of organ transplants or cancer chemotherapy. The majority of life-threatening fungal infections are caused by Candida, Aspergillus, and Cryptococcus species, with Candida albicans and non-C. albicans Candida species being prominent in clinics. The emergence of azole resistance in fungi that cause systemic infections has become a serious clinical problem that can limit treatment options (62); for an alternative view on the clinical importance of this development, see Sanglard and Odds (54). Pathogenic fungi can acquire multidrug resistance (MDR; resistance to chemically unrelated compounds), usually in response to long-term antifungal prophylaxis with the azole drugs (62). Some fungal species, e.g., Candida glabrata, Candida krusei, and Aspergillus fumigatus, can show innate resistance to these drugs. Although MDR can occur through changed membrane permeability or because of overproduction of or mutation in the azole drug target lanosterol 14α-demethylase (encoded by ERG11), often the overexpression of multidrug efflux pumps is responsible for the most clinically important forms of resistance (49). Fungal drug efflux is mediated primarily by two groups of membrane-bound transport proteins: the ATP-binding cassette (ABC) transporters and the major facilitator superfamily (MFS) pumps.
Gene families encoding multidrug transporters have been identified for several fungal pathogens. Those most extensively characterized include C. albicans CDR1 (47), CDR2 (52), and BENR (6, 24, 53); C. glabrata CgCDR1 (51) and PDH1 or CgCDR2 (38, 50, 60); C. krusei ABC1 and ABC2 (28); and Aspergillus AfuMDR1, AfuMDR2, AflMDR1, AtrA, and AtrB (12, 58). The ABC transporters use a combination of pump overexpression and the energy of ATP hydrolysis to confer high levels of MDR in the dominant fungal pathogens C. albicans and C. glabrata, while the overexpressed MFS transporters, such as BenRp, use electrochemical energy to obtain intermediate levels of resistance to a more limited range of drugs. A recent survey of over 6,000 fungal clinical isolates suggested that although the new azole drugs voriconazole and ravuconazole appear to be more effective against systemic infections than earlier antifungal drugs, worrisome levels of cross-resistance to these drugs in C. albicans and C. glabrata strains that are capable of expressing MDR were observed (45).
Only a few classes of antifungal agents are used in the treatment of patients with systemic fungal infections. The most prominent of these drugs are the polyene antibiotic amphotericin B and the triazoles, such as fluconazole (FLC) and itraconazole (ITC) (30). Although amphotericin B is a broad-spectrum fungicide, important side effects, including infusion-related problems and severe renal toxicity, have been ameliorated only partially by the use of lipidic formulations (15). The fungistatic triazoles, such as FLC, are often preferred because of their more modest side effects, but they can interact with other drugs (48, 59). New classes of antifungal agents, such as the echinocandins, which inhibit the biosynthesis of fungal cell wall components (3), the aurobasidins, which inhibit sphingolipid biosynthesis (22, 31), compounds that inhibit N-myristoyltransferase (21), and the sordarins, which inhibit fungal protein synthesis (14), are in development. However, their intracellular targets make these drugs susceptible to detoxification or efflux mechanisms that could lead to drug resistance. Similarly, the recently developed triazole derivatives, such as voriconazole, are likely to be affected by multidrug efflux systems (45).
One way to overcome drug resistance is to obtain antifungal chemosensitizers, compounds that potentiate the efficacy of existing azoles, such as FLC. Various milbemycins, for example, chemosensitize Candida species to FLC (32). While FLC-resistant C. albicans clinical isolates and Saccharomyces cerevisiae strains overexpressing C. albicans ABC or MFS transporters were chemosensitized by the immunosuppressive agents FK506 (33) and cyclosporine (34), Del Poeta et al. (11) also showed that the combination of FLC and FK506 was synergistic and made the triazole drug fungicidal against C. albicans and Cryptococcus neoformans. Disruption of either the calcineurin or the FKBP12 gene did not affect chemosensitization in yeast cells, suggesting that FK506 directly affects an FLC efflux pump. Other studies, however, implicated calcineurin in FK506 chemosensitization when calcium was included in the medium (16). Although particular residues in the S. cerevisiae Pdr5p multidrug efflux pump have been implicated in its inhibition by FK506 (17, 18), the mechanism of action of the drug is incompletely resolved and its use in immunocompromised patients is probably inadvisable. Because FLC is a well-tolerated drug with predictable pharmacokinetic properties, it has been a mainstay in the treatment of immunocompromized patients who require long-term drug administration (36). The discovery and development of novel FLC chemosensitizers that increase its potency against both sensitive and resistant fungi may offer alternative ways to combat fungal infections and to extend the commercial life of the drug.
The evolutionarily ancient cationic antimicrobial peptides (CAPs) provide primary antimicrobial defense mechanisms that have been adopted by multicellular organisms (8, 13, 19, 57). Most CAPs present asymmetric patches of positive charge that preferentially interact with negatively charged microbial surfaces rather than the surfaces of higher eukaryotes. With this specificity in mind, researchers have modified, truncated, and produced d-peptide versions of CAPs in efforts to obtain more potent and stable antimicrobial agents. Since their development in the 1990s, peptide combinatorial libraries (44) have been used to screen for antibacterial agents, antifungal agents, and enzyme inhibitors. For example, a cationic decapeptide targeting the plasma membrane had activity against bacteria and C. albicans (27). We have constructed and screened a 1.8-million-member surface-active d-octapeptide combinatorial library for antagonists of fungal ABC transporters. Each member of the d-octapeptide combinatorial library contains a C-terminal amidated triarginine motif designed to mimic the patches of membrane-targeting positive charge that characterize the CAPs. Our unpublished studies show that the motif concentrates the peptide at the fungal surface and stops the peptide from crossing the plasma membrane.
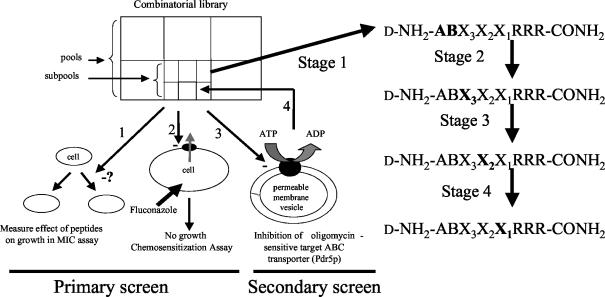
The d-octapeptide combinatorial library was screened for ABC transporter antagonists by using S. cerevisiae strain AD/PDR5+. This strain functionally hyperexpresses (hyperexpression is overexpression at a very high level) the Pdr5p ABC transporter in a background in which five other ABC transporters and the Pdr3p transcriptional regulator are deleted. The hyperexpression of PDR5, driven by the pdr1-3 mutation in the Pdr1p transcriptional regulator (2), confers resistance to structurally and functionally unrelated xenobiotics (29). As previously described (10), AD/PDR5+ had a high level of resistance to FLC (MIC, 600 μg ml−1), while the isogenic null mutant AD/PDR5− was hypersensitive to FLC (MIC, 0.94 μg ml−1). Differential Pdr5p overexpression in this low-noise background allowed both cell-based and in vitro assays of Pdr5p function (Fig. 1). By screening the combinatorial library for chemosensitization of strain AD/PDR5+ to FLC and the inhibition of Pdr5p ATPase activity in vitro, we identified KN20, a potent noncompetitive Pdr5p inhibitor (concentration of drug causing 50% inhibition of enzyme activity [IC50], 4 μM) that requires a substituent attached to the d-peptide for activity. KN20 enhanced the efficacy of agents against model and pathogenic drug-resistant fungi, but chemosensitization appears to be indirect and seems to involve nonlethal cell permeabilization.
FIG. 1.
Screening system used to identify FLC chemosensitizers. Strain AD/PDR5+ was used both for the cell-based primary screen and to provide the plasma membranes used to assay Pdr5p ATPase activity in the in vitro secondary screen. Strain AD/PDR5− was used to provide control plasma membranes for the in vitro assays. Screening was carried out in the following steps. (1) Determine the MIC of each combinatorial library peptide pool for AD/PDR5+ and thus the highest concentration of peptide in each pool that does not affect growth. (2) Determine the degree to which peptide pools chemosensitize AD/PDR5+ to FLC. (3) Measure the concentration-dependent inhibition of Pdr5p ATPase activity by the pools that show the greatest degree of chemosensitization. (4) Select the best candidate pool (stage 1) and repeat the cycle with the resynthesized subpools (stages 2 to 4). Check purified peptides to ensure that a suitable lead has been identified. The letters in bold indicate the amino acid position(s) determined at each stage of library deconvolution.
MATERIALS AND METHODS
Strains and culture conditions.
The S. cerevisiae and Candida species used in this study are listed in Table 1. S. cerevisiae strains AD1002 and AD/CaCDR2 were prepared as described by Nakamura et al. (42); ADCgCDR1-1B and ADCgCDR2-4 were prepared as described by Wada et al. (60); and AD/CaCDR1, AD/BENR, and AD/ERG11 were constructed by using a modification of the method described by Nakamura et al. (42), in which vector pABC3 was used as described by Monk et al. (40). S. cerevisiae strains were routinely maintained on complete synthetic medium without uracil (CSM- ura), which contained 0.67% (wt/vol) yeast nitrogen base (Difco, Becton Dickinson, Sparks, Md.), 0.077% (wt/vol) CSM- ura (Bio 101, Vista, Calif.), 2% (wt/vol) glucose, and 2% (wt/vol) agar (pH 7.0). Candida species were maintained on YPD agar, which contained 1% (wt/vol) yeast extract (Difco), 2% (wt/vol) Bacto Peptone (Difco), 2% (wt/vol) d-glucose, and 2% (wt/vol) agar (pH 5.5). For growth inhibition and chemosensitization assays, liquid CSM- ura was used. This medium contained 0.67% (wt/vol) yeast nitrogen base (Difco), 0.077% (wt/vol) CSM- ura, and 2% (wt/vol) glucose and was buffered to pH 7.0 with 10 mM morpholinoethanesulfonic acid-20 mM HEPES. Cells were grown at 30°C with shaking (150 rpm).
TABLE 1.
Yeast strains used in this study
| Species | Strain | Characteristics (genotype) | Reference or sourcea |
|---|---|---|---|
| Saccharomyces cerevisiae | AD/PDR5− | MATα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr5Δ::hisG | 10 (designated strain AD1234567) |
| AD/PDR5+ | MATα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG PDR5 | 10 (designated strain AD124567) | |
| AD1-8u− | MATα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ::hisG pdr5Δ::hisG | 40 | |
| AD1002 | MATα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ::hisG pdr5Δ::CDR1-URA3 | 42 | |
| AD/CaCDR1 | MATα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ::hisG pdr5Δ::CDR1-URA3 | 40 | |
| AD/CaCDR2 | MATα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ::hisG pdr5Δ::CDR2-URA3 | S. Wada, National Institute of Infectious Diseases, Tokyo, Japan | |
| AD/CgCDR1-1B | MATα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ::hisG pdr5Δ::CgCDR1-URA3 | 60 | |
| AD/CgCDR2-4 | MATα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ::hisG pdr5Δ::CgCDR2-URA3 | 60 | |
| AD/BENR | MATα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ::hisG pdr5Δ::BENR-URA3 | 40 | |
| AD/ERG11 | MATα pdr1-3 ura3 his1 yor1Δ::hisG snq2Δ::hisG pdr10Δ::hisG pdr11Δ::hisG ycf1Δ::hisG pdr3Δ::hisG pdr15Δ::hisG pdr5Δ::CaERG11-URA3 | 40 | |
| T48 | HO/HO MATa/MATα ade6-1/ade6-1 trp5-1/trp5-1 leu2-1/leu2-1 lys1-1/lys1-1 ura3-1/ura3-1 PMA1-URA3/PMA1-URA3 | 37 | |
| Candida albicans | ATCC 10261 | American Type Culture Collection, Manassas, Va. | |
| FR2 | 1 (derived from SGY-243) | ||
| KB | Pfizer, Sandwich, United Kingdom | ||
| C. dubliniensis | CD36 | D. C. Coleman, School of Dental Science, University of Dublin, Dublin, Republic of Ireland | |
| CD43 | D. C. Coleman (clinical isolate) | ||
| C. glabrata | CBS138 | Schimmelcultures, Baarn, The Netherlands | |
| 850821 | ESR: Health, NZCDC, Wellington, New Zealand | ||
| 850920 | ESR: Health, NZCDC | ||
| C. krusei | B2399 | ESR: Health, NZCDC | |
| C. parapsilosis | 90.493 | ESR: Health, NZCDC | |
| 425 | School of Dentistry, Kagoshima University, Kagoshima, Japan | ||
| C. tropicalis | IFO0618 | Institute for Fermentation, Osaka, Japan | |
| Cryptococcus neoformans | ATCC 90112 | New Zealand Reference Culture Collection, Institute of Environmental Sciences and Research Limited, Wellington, New Zealand |
ESR, Institute of Environmental Science and Research; NZCDC, New Zealand Centre for Disease Control.
Chemicals and antifungal agents.
The chemicals and antifungal agents used in this study were obtained from the following sources: FLC (Diflucan; Pfizer Laboratories Limited, Auckland, New Zealand); ITC (Janssen Research Foundation, Beerse, Belgium); ketoconazole (KTC) (Janssen Pharmaceutica Limited, Sydney, Australia); oligomycin, aurovertin B, sodium metavanadate, sodium azide, ATP, UTP, and tetramethyl rhodamine isothiocyanate (TRITC) (Sigma, St. Louis, Mo.); phenylmethylsulfonyl fluoride (Roche Diagnostics NZ Limited, Auckland, New Zealand); and l-ascorbic acid, ammonium molybdate, and dimethyl sulfoxide (DMSO) (BDH, Poole, United Kingdom). ITC, KTC, oligomycin, and the peptide combinatorial library pools and subpools were prepared as stock solutions dissolved in DMSO. Agarose, minimum essential medium with Earle's salts, fetal bovine serum, and l-glutamine for tissue cultures were obtained from Gibco (Invitrogen Corporation, Auckland, New Zealand). Calcein AM and ethidium homodimer were from a live/dead viability/cytotoxicity kit (Molecular Probes, Eugene, Oreg.).
Combinatorial library.
The d-octapeptide combinatorial library was synthesized manually by using solid-phase 9-fluorenylmethoxy carbonyl chemistry (44). The combinatorial library is comprised of 324 peptide pools (each theoretically containing 5,832 separate peptides), with all possible peptides being present in approximately equimolar amounts. Each peptide can be represented by the formula d-NH2-ABX3X2X1RRR-CONH2 where, for each pool, A and B are known and X represents any of 18 amino acids (cysteine was excluded because a cysteine-targeting sublibrary was constructed, and glycine was excluded because it lacks a side chain). The most active peptide library pools were identified by bioassays (Fig. 1, stage 1). The most suitable pools were deconvoluted by cycles of resynthesis and bioassays to sequentially determine the optimal amino acids present at positions X3 (stage 2), X2 (stage 3), and X1 (stage 4). Finally, the most potent peptides identified in stage 4 were manually resynthesized and purified by high-pressure liquid chromatography (HPLC) with a Shimadzu LC-6 system equipped with 15-μm, 300-Å Phenomenex guard and separation (250 by 21.20 mm) columns. The HPLC buffers used were as follows: buffer A, 2% acetonitrile and 0.1% trifluoroacetic acid; and buffer B, 90% acetonitrile and 0.1% trifluoroacetic acid. As an example, KN20 was purified as follows. The crude peptide was loaded at 5% acetonitrile, and the column was run at 5 to 15% acetonitrile over 10 min and then at 15 to 45% acetonitrile over 2 h. KN20 eluted at 32% acetonitrile under these conditions. The peptides were analyzed by HPLC with a Waters Alliance HT 2790 system coupled to a Micromass ZMD 4000 electrospray mass spectrometer. The purification yielded either the naked peptide or the monosubstituted 4-methoxy-2,3,6-trimethylbenzensulfonyl (Mtr) derivative.
In vivo assays. (i) Peptide MIC as a measure of toxicity against S. cerevisiae.
Prior to chemosensitization assays, the MIC of each peptide pool, peptide subpool, or peptide for Pdr5p-hyperexpressing strain AD/PDR5+ and isogenic null mutant AD/PDR5− was determined by a microdilution method with 96-well microplates. Cells in CSM-ura (200 μl) were inoculated at 4 × 103 per well and incubated at 30°C for 48 h with shaking (150 rpm) in the presence of a series of twofold dilutions of individual peptide pools or selected peptides. Cell growth was monitored at 590 nm with an EL340 Bio Kinetics reader (BioTek Instruments).
(ii) FLC chemosensitization assay.
Pdr5p-hyperexpressing strain AD/PDR5+ was grown in the presence of the highest concentration (sub-MIC) of each peptide pool that provided less than 10% inhibition of cell growth after 48 h, together with FLC at 10 or 40 μg ml−1 (1/60 or 1/15 the MIC for Pdr5p-hyperexpressing cells in the absence of peptides, respectively) (Fig. 2). This procedure identified peptide pools that sensitized these cells to FLC. Checkerboard drug chemosensitization assays then were performed with selected pools to more fully evaluate the effects on AD/PDR5+ of both the peptide pool and the FLC concentration. In brief, six rows each comprising five samples of twofold dilutions of individual peptide pools were prepared in a microplate to yield final concentrations in the test plate of 200, 100, 50, 25, and 0 μg ml−1. In a separate microplate, five columns each comprising six samples of twofold dilutions of FLC were prepared to yield final concentrations in the test plate of 80, 40, 20, 10, 5, and 0 μg ml−1. The FLC dilutions were added to wells containing the peptide dilutions, and 4 × 103 cells in a final volume of 200 μl were inoculated per well. The microplates were incubated at 30°C with shaking (150 rpm), and cell growth was monitored at 24, 42, and 48 h. After 48 h, 5-μl samples from wells with no visible growth were spotted on YPD agar and incubated at 30°C for 48 h to test for cell viability. The concentrations of peptides to be used in the checkerboard assays were suitably decreased to take into account improved potency as the deconvolution progressed from library pools through subpools to purified peptides.
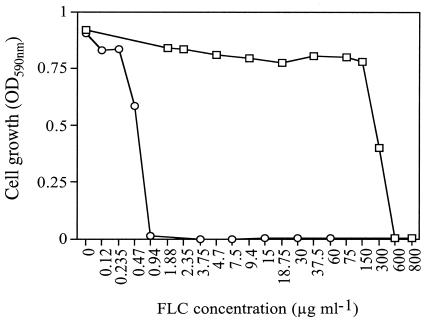
FIG. 2.
MICs of FLC for S. cerevisiae strains. Strains AD/PDR5+ (□) and AD/PDR5− (○) were incubated at 30°C for 48 h in the presence of the indicated concentrations of FLC as described in Materials and Methods.
(iii) Agarose diffusion chemosensitization assay.
CSM-ura (20 ml) containing 120 μg of FLC/ml was solidified with 0.6% agarose in an Omnitray (126 by 86 by 19 mm; Nunc, Roskilde, Denmark). AD/PDR5+ cells (1 × 105 to 2 × 105) in 20 ml of melted top CSM-ura containing 0.4% agarose and 120 μg of FLC/ml were placed over the agarose. An appropriate volume of peptide (<10 μl) was applied to 3MM blotting paper disks (5-mm diameter; Whatman), dried at 37°C for 1 to 2 h, and placed on the solidified medium surface. The tray was incubated at 30°C for 48 h. FLC-free medium was also used in the disk assay to assess the effects of test peptides alone on the growth of strains AD/PDR5+ and AD/PDR5−.
In vitro assays. (i) Isolation of plasma membranes.
Plasma membranes from strains AD/PDR5+ and AD/PDR5− were used for in vitro Pdr5p ATPase assays. S. cerevisiae strain T48 was used as a source of membranes for plasma membrane ATPase (Pma1p) assays. Cells were grown in YPD liquid medium, which contained 1% (wt/vol) yeast extract (Difco), 2% (wt/vol) Bacto Peptone, and 2% (wt/vol) glucose, at 30°C with shaking (200 rpm) and harvested in the diauxic phase of growth (optical density at 600 nm [OD600], ∼7). AD/PDR5+ and AD/PDR5− cells were washed twice and starved for glucose on ice for 30 min in distilled water, while T48 cells were harvested without the washing steps. The glucose starvation step minimized Pma1p activity in the AD/PDR5+ and AD/PDR5− cells, while the T48 cells were harvested to maximize the ATPase activity of Pma1p (37). AD/PDR5+ and AD/PDR5− cells were resuspended in homogenization medium, which contained 50 mM Tris (pH 7.5), 2 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride, and disrupted by using a Braun homogenizer. The homogenization medium for the T48 cells was supplemented with 2% glucose. Cell debris and unbroken cells were removed by centrifugation at 2,000 × g for 10 min. A crude membrane fraction was isolated from the cell-free supernatant by centrifugation at 30,000 × g for 45 min. The plasma membrane fraction was enriched by selective precipitation of mitochondria at pH 5.1 as described by Goffeau and Dufour (23), pelleted by centrifugation at 30,000 × g for 45 min, resuspended in 10 mM Tris (pH 7.0)-0.5 mM EDTA-20% (vol/vol) glycerol, and stored at −80°C. Protein concentrations were determined by a micro-Bradford assay (Bio-Rad Laboratories, Hercules, Calif.) with bovine gamma globulin as the standard. Protein profiles of membrane samples were examined after electrophoresis through sodium dodecyl sulfate (SDS)-8% polyacrylamide gels and staining with Coomassie blue.
(ii) Pdr5p ATPase inhibition assay.
Pdr5p ATPase assays were optimized according to the standard method described previously (9). Pdr5p has nucleoside triphosphatase activity over a broad pH range, with optimal vanadate- and oligomycin-sensitive activities between pHs 7 and 8. Peptides that sensitized strain AD/PDR5+ to FLC were assessed for their inhibition of oligomycin-sensitive ATPase activity at pH 7.5. Pdr5p ATPase activity was measured in the presence or absence of test peptides by incubating plasma membrane fractions (5 μg) at 30°C for 30 min in a final volume of 120 μl containing 6 mM ATP and 7 mM MgSO4 in 59 mM morpholinoethanesulfonic acid-Tris as previously described (42). In ATP protection assays, 6 mM ATP was added 5 min prior to the addition of peptides, and the reaction was initiated by the addition of 7 mM MgSO4 after an additional 5 min of incubation. The ATPase specific activity of hyperexpressed Pdr5p was corrected for the activities of other ATPases by subtraction of the ∼5-fold-lower background oligomycin-sensitive specific activity detected in plasma membranes from strain AD/PDR5−.
(iii) Pma1p inhibition assay.
Plasma membrane fractions were prepared from S. cerevisiae strain T48 by the method described above. ATPase assays were carried out at 30°C and pH 7.0 for 20 min with 2 μg of membrane protein per assay in the presence of oligomycin (10 μM) (37).
(iv) Inhibition of heterologously expressed ABC transporter ATPase activity.
Plasma membrane fractions were isolated as described above from S. cerevisiae strain AD/CaCDR1 or AD/CaCDR2. ATPase assays were carried out at 30°C and pH 7.5 for 30 min with 5 to 10 μg of membrane protein per assay. The oligomycin-sensitive ATPase specific activities of the heterologously hyperexpressed C. albicans ABC transporters were corrected for background activity by subtraction of the oligomycin-sensitive specific activity detected in membranes from isogenic null strain AD1-8u−.
(v) Rhodamine uptake and efflux.
Log-phase (OD600, 1.5) AD/PDR5+ or AD/PDR5− cells were stored overnight on ice. The cells were harvested by centrifugation, washed twice with distilled water, resuspended in HEPES buffer (50 mM HEPES-NaOH [pH 7.0]), and incubated with 5 mM 2-deoxyglucose at 30°C for 30 min to deplete intracellular energy levels and then with 15 μM rhodamine 6G (Rh6G). For uptake experiments, 1-ml samples of cells (OD600, 1.0) were harvested by centrifugation at various time intervals after the addition of Rh6G with or without test peptides and boiled for 10 min with 250 μl of SDS lysis buffer (2% SDS, 2% β-mercaptoethanol, 125 mM Tris-HCl [pH 6.8]) to release cell-associated Rh6G. After centrifugation, the amount of Rh6G in each supernatant was quantitated fluorimetrically by using a standard curve generated from known amounts of Rh6G in SDS lysis buffer. For Rh6G efflux experiments, cells preloaded with 15 μM Rh6G for 30 min were washed twice and resuspended in HEPES buffer at an OD600 of 10. Cell samples (400 μl) were incubated at 30°C for 5 min with or without test peptides and with 40 μl of 2% glucose added to start the reaction. After 8 min, the cells were pelleted by centrifugation, and the amount of Rh6G in the supernatant was quantitated fluorimetrically by using a standard curve for Rh6G in HEPES buffer.
(vi) Preparation of TRITC-labeled KN20 and confocal microscopy of labeled cells.
KN20 (200 nmol) dissolved in 400 μl of NaHCO3 (pH 8.5) was incubated with 100 μl of TRITC (10 mg ml−1 dissolved in dimethylformamide) for 1 h at room temperature. Fifty microliters of 1.5 M NH2OH-HCl adjusted to pH 8.5 with NaOH was added, incubation was continued for 1 h, and then the reaction mixture was freeze-dried. Half the sample was dissolved in 1 ml of 20% acetonitrile containing 0.1% trifluoroacetic acid (TFA) and then loaded onto a Waters Sep-Pak C18 minicolumn that had been prewashed with 3 ml of acetonitrile and 3 ml of 20% acetonitrile in 0.1% TFA. The column was eluted with a step gradient comprised of sequential 15-ml washes containing 20, 25, 30, 35, and 40% acetonitrile in 0.1% TFA. The labeled peptide was eluted with 30% acetonitrile in 0.1% TFA. The TRITC-labeled peptide was distinguished from TRITC by its absorbance peak at 554 nm rather than 550 nm. TRITC-KN20 was freeze-dried and dissolved in DMSO. Yeast cells (OD600, 1.0) were incubated at room temperature for 30 min in 50 μl of CSM-ura (pH 7.0) containing 10 μM TRITC-KN20. Some samples were also harvested by centrifugation (5,600 × g) and washed with 100 μl of CSM-ura. Cells in 10-μl samples were visualized by confocal microscopy with a Zeiss LSM510 confocal laser scanning microscope in the Zeiss Axiovert 200 M inverted microscope configuration (Zeiss, Jena, Germany).
Toxicity for human cells. (i) Hemolysis.
The lytic effects of peptides on human erythrocytes were assessed by the method of Helmerhorst et al. (25). Human erythrocytes were washed three times and resuspended in phosphate-buffered saline (PBS [pH 7.2]) at 1% (vol/vol). Twofold dilutions of peptide in PBS were mixed with the erythrocyte suspension (final density, 0.5%) in a final volume of 200 μl in a 96-well U-bottom microplate and incubated at 37°C for 1 h. The erythrocytes were pelleted by centrifugation at 3,000 × g for 5 min, and 100 μl of the supernatant was transferred to the wells of a flat-bottom microplate. The absorbance of the hemoglobin released from the erythrocytes was measured at 540 nm. Measurements for a nonhemolyzed control and for 100% hemolysis were determined with PBS alone and with PBS containing 1% Triton X-100, respectively. The polyene antifungal agent amphotericin B provided a reference hemolysis value.
(ii) Toxicity of peptides for cultured human epithelial cells.
HEp-2 cells were grown in the presence of 5% CO2 at 37°C in minimum essential medium with Earle's salts, supplemented with 10% fetal bovine serum and 2 mM l-glutamine (tissue culture medium). A cell suspension was prepared by treating the seed culture with trypsin-EDTA in PBS (pH 7.2), and 100-μl portions were transferred to a 96-well microplate. Once the cells had formed a monolayer on the plastic surfaces (after about 24 h of incubation), peptide diluted in tissue culture medium (100 μl) was added to the wells, and incubation was continued for an additional 3 or 24 h. HEp-2 cells were washed three times with PBS and stained with 2 μM calcein AM and 8 μM ethidium homodimer in PBS. Cell viability was assessed by viewing individual microplate wells with a confocal microscope as described above. Color-separated Photoshop images were used for manual counting of ethidium homodimer-stained dead cells and calcein AM-stained live cells in duplicate, and the percentage of dead cells was calculated.
RESULTS
Screening system.
Figure 1 outlines our system for screening a combinatorial peptide library for chemosensitizers of Pdr5p-mediated FLC efflux. It utilizes the Pdr1p transcriptional regulator-mediated hyperexpression in S. cerevisiae of the Pdr5p multidrug efflux pump to confer FLC resistance in a low-noise background achieved by the deletion of five other drug efflux pumps. A 1.8-million-member d-octapeptide combinatorial library was screened by using iterative cycles of a cell-based primary screen (steps 1 and 2) and an enzyme-based secondary screen (step 3) to identify FLC chemosensitizers that were Pdr5p inhibitors. The library members (general formula, d-NH2-ABX3X2X1RRR-CONH2) each contained a C-terminal triarginine motif that was designed to confer membrane impermeability and concentrate the peptide at the fungal cell surface. After identification of the most active pool, defined by the N-terminal sequence AB (stage 1), three cycles of resynthesis and screening of subpools (stages 2 to 4) were used to select the best residues at positions X3, X2, and X1. Lead peptides identified in this way were resynthesized and purified to identify the active moiety.
Cell-based primary screen. (i) MICs of peptides and FLC.
As expected, the FLC MIC for Pdr5p-overexpressing strain AD/PDR5+ was 600 μg ml−1, while that for the isogenic null mutant AD/PDR5−, which was hypersensitive, was 0.94 μg ml−1 (Fig. 2) (10). The MIC of each of the 324 stage 1 peptide pools was determined for strains AD/PDR5+ and AD/PDR5− by using the microplate assay described in Materials and Methods. Peptide pools were diluted with CSM-ura to final concentrations of 200, 150, 100, 50, 25, 12.5, and 0 μg ml−1. Most peptide pools in the library did not inhibit cell growth at 200 μg ml−1, and only 40 peptide pools inhibited growth at less than 150 μg ml−1. Both yeast strains were equally susceptible to each peptide pool tested (data not shown), indicating that the effects on growth were independent of Pdr5p hyperexpression.
(ii) FLC chemosensitization assay.
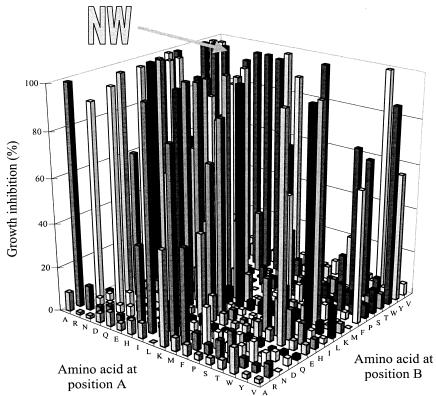
The FLC chemosensitization assay measured the effect of the highest concentration of each stage 1 peptide pool (100, 150, or 200 μg ml−1), which on its own did not significantly inhibit growth after 48 h, on the growth of AD/PDR5+ when used in combination with FLC at 10 or 40 μg ml−1. The Growth of Pdr5p-hyperexpressing cells was inhibited by a limited number of stage 1 peptide pools in the presence of FLC at 40 μg ml−1. Only 23 peptide pools gave greater than 99% growth inhibition (Fig. 3). This response was dose dependent, with far fewer pools showing extensive chemosensitization at an FLC concentration of 10 μg ml−1 than at an FLC concentration of 40 μg ml−1. The sequence of the two N-terminal amino acids in d-NH2-ABX3X2X1RRR-CONH2 was important for chemosensitization. For example, even at 40 μg of FLC ml−1, no members of some peptide pools with an amino acid in common at position A chemosensitized the cells to FLC (e.g., amino acids P, S, and Y), and not all members of peptide pools with other amino acids in common at position A chemosensitized the cells to FLC. Similar observations were made for amino acids at position B. Furthermore, no pools with N at position B gave significant chemosensitization at 40 μg of FLC ml−1, while the peptide pool with NW at positions A and B was a potent chemosensitizer.
FIG. 3.
Chemosensization of strain AD/PDR5+ to FLC (40 μg ml−1) by 324 stage 1 peptide pools. NW represents the d-NH2-NWX3X2X1RRR-CONH2 peptide pool that was chosen for deconvolution. Chemosensitization of cell growth was measured as described in Materials and Methods.
(iii) Checkerboard chemosensitization assay.
The 23 most potent chemosensitizing stage 1 peptide pools were evaluated in detail for their synergistic inhibition of cell growth in the presence of FLC. A checkerboard chemosensitization assay, which also gave data on the time course of growth, helped us to select peptide pools suitable for further study. Each peptide pool tested, either alone or in combination with FLC, caused a significant delay in cell growth during the first 24 h of incubation (Fig. 4A). After the lag in the absence of FLC, the apparent growth rates and the final growth yield were the same for all peptides. In most instances, cells treated with a low FLC concentration showed growth yields comparable to those of control cells after 42 to 48 h. At higher FLC concentrations (>40 μg/ml), the highest non-growth-inhibiting concentrations of the peptide pools totally blocked cell growth. Fungicidal effects were seen in several instances. The five most potent chemosensitizing d-octapeptide pools (N-terminal sequences ND, NW, EW, TW, and WE) were selected for secondary screening.
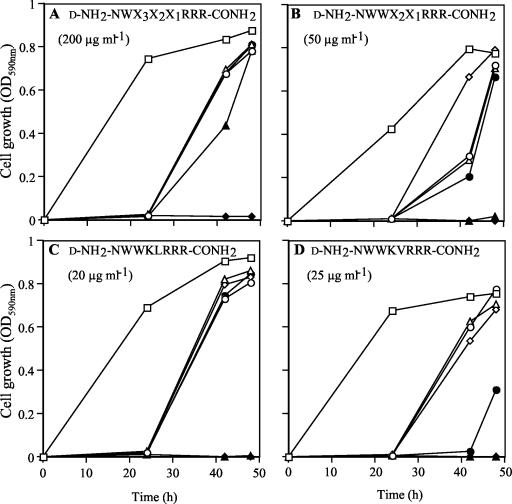
FIG. 4.
Chemosensitization of strain AD/PDR5+ to FLC measured at stages of library deconvolution. Stage 1 peptide pool d-NH2-NWX3X2X1RRR-CONH2 (A), stage 2 peptide subpool d-NH2-NWWX2X1RRR-CONH2 (B), stage 4 subpool d-NH2-NWWKLRRR-CONH2 (C), and stage 4 subpool d-NH2-NWWKVRRR-CONH2 (D) were tested with FLC at 0 (○), 5 (▵), 10 (⋄), 20 (•), 40 (▴), and 80 (♦) μg/ml. Growth was measured in the presence of peptide (concentration given in parentheses) and compared to that in control incubations in the absence of both peptide and FLC (□).
Enzyme-based secondary screen for inhibition of Pdr5p ATPase activity.
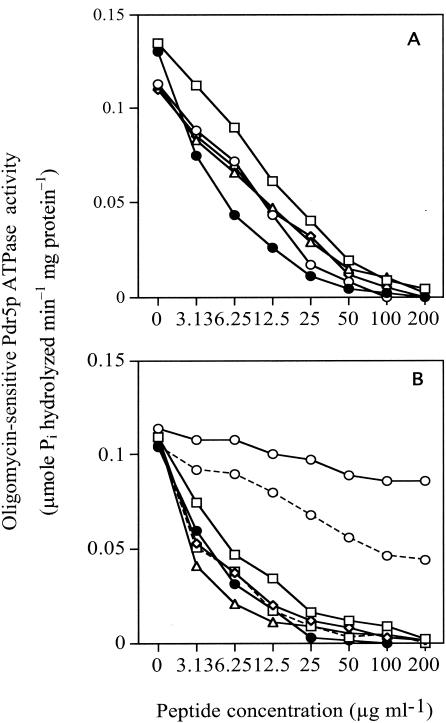
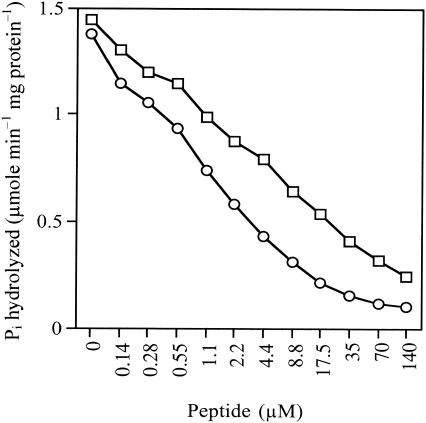
An assay of Pdr5p nucleoside triphosphatase activity in purified plasma membranes was used as the in vitro secondary screen for identifying candidate inhibitors that directly affected the intended molecular target of the whole-cell screen. Nucleoside triphosphatase activity, in particular, ATPase activity, is required for the pumping activity of the fungal ABC proteins (29). In addition, yeast plasma membranes form vesicles that are randomly oriented and permeable to ATP (41). The Pdr5p ATPase assay with membranes from Pdr5p-hyperexpressing strain AD/PDR5+ (with membranes from strain AD/PDR5− serving as a negative control) therefore is suited to the discovery of inhibitors acting via the Pdr5p ectodomain. The assay was conducted at pH 7.5, the optimum pH for Pdr5p ATPase but not for Pma1p (9). Membranes from strain AD/PDR5+ contained oligomycin-sensitive ATPase specific activity that was up to fivefold higher than the residual oligomycin-sensitive activity of plasma membranes from strain AD/PDR5−. Aurovertin B (20 μM) did not inhibit the oligomycin-sensitive Pdr5p ATPase, suggesting negligible mitochondrial contamination of the membrane preparations. Of the five peptide pools selected for whole-cell chemosensitization, the NW pool showed the strongest Pdr5p ATPase inhibition (IC50, 6.25 μg ml−1) (Fig. 5A).
FIG. 5.
Inhibition of oligomycin-sensitive plasma membrane Pdr5p ATPase activity by selected peptide pools. ATPase assays were carried out at 30°C for 30 min without ATP protection and in the presence of the indicated concentrations of peptide pools as described in Materials and Methods. Values for each data point are the means of two separate determinations that varied by less than 10%. (A) Inhibition by peptide library pools. The first two amino acids of the d-NH2-ABX3X2X1RRR-CONH2 pools were ND (□), NW (•), EW (○), TW (▵), and WE (⋄). (B) Inhibition by the deconvoluted combinatorial library pool d-NH2-NWX3X2X1RRR-CONH2 (□—□), subpools d-NH2-NWWX2X1RRR-CONH2 (⋄), d-NH2-NWWKX1RRR-CONH2 (□---□), and d-NH2-NWWKVRRR-CONH2 (▵), and purified peptides KN0 (○—○), KN1 (○---○), and KN20 (•).
Deconvolution of peptide pools and subpools.
The peptide pool d-NH2-NWX3X2X1RRR-CONH2 was selected for deconvolution because it strongly inhibited the molecular target Pdr5p and was a potent chemosensitizer of AD/PDR5+ cells to FLC. Its intrinsic toxicity for fungal cells appeared to be lower than that of the other peptide pools (data not shown). This property was considered important for specific chemosensitization of the nonessential Pdr5p transporter; i.e., when Pdr5p is inhibited, cell growth should not be affected, except in the presence of toxic Pdr5p substrates, such as FLC. In stage 2 of library deconvolution, 18 subpools containing alanine (A) through valine (V) at amino acid position X3 of d-NH2-NWX3X2X1RRR-CONH2 were assessed by using the cell-based and in vitro screens. d-NH2-NWWX2X1RRR-CONH2 (Fig. 4B) was selected for stage 3 deconvolution, and d-NH2-NWWKX1RRR-CONH2 was selected for stage 4 deconvolution. At this stage, peptide subpools containing peptides with the sequences d-NH2-NWWKLRRR-CONH2 (Fig. 4C) and d-NH2-NWWKVRRR-CONH2 (Fig. 4D) were selected. The peptides were active at concentrations 8- to 10-fold lower than that of the stage 1 pool d-NH2-NWX3X2X1RRR-CONH2 over the 48-h time course of growth (compare Fig. 4A with Fig. 4C and D).
These peptide sequences then were synthesized by using solid-phase 9-fluorenylmethoxy carbonyl chemistry and were purified by HPLC. However, both pure d-NH2-NWWKVRRR-CONH2 (KN0) and pure d-NH2-NWWKLRRRCONH2 (KN1) were poor FLC chemosensitizers (Table 2) and weak inhibitors of Pdr5p ATPase activity at concentrations at which the equivalent stage 4 subpools were strongly inhibitory (Fig. 5B). KN0 and KN1 chemosensitized AD/PDR5+ cells to FLC, but only at extremely high peptide concentrations (80 to 160 μM). Mass spectrometry of the HPLC-separated stage 4 subpools showed that these preparations were highly sensitive to the method of peptide cleavage from the solid matrix. About two-thirds of the product in the d-NH2-NWWKVRRR-CONH2 subpool, obtained under standard cleavage conditions with TFA, was comprised of a peptide containing a single Mtr group substitution. The Mtr group is used as a side-chain-blocking agent for arginine. The conditions for acid-catalyzed peptide release from the solid phase allowed the blocking agent to migrate and form a substituent linked to either one of the tryptophan side chains, possibly via the nitrogen or the adjacent carbon (C-2) of the indole ring (56), although other modes of attachment to the peptide are possible. This analysis indicated that either a modified version of the peptide or some other contaminant(s) in the peptide pools was responsible for their inhibitory and chemosensitizing activities.
TABLE 2.
Properties of HPLC-purified peptides
| Peptide | MIC (μM) | Chemosensitization of cells to FLCa | IC50 (μM)
|
|
|---|---|---|---|---|
| With ATP protection | Without ATP protection | |||
| KN0 | 160 | + | NTb | >140 |
| KN20 | 40 | ++ | 4.0 | 4.4 |
| KN20p | 10 | − | 8.8 | 7.4 |
| KN1 | 80 | + | NT | >140 |
| KN21 | 20 | ++ | 3.0 | 2.0 |
| KN21p | 10 | − | 7.4 | 7.6 |
−, none; +, moderate; ++, strong.
NT, not tested.
We therefore resynthesized KN0 with an Mtr group (KN20) and tested its activity in comparison with those of the deconvoluted combinatorial library pool and subpools (Fig. 5B). The results demonstrated that the Mtr group in KN20 is required for the inhibition of Pdr5p ATPase activity. A comparison of the parent peptides with the HPLC-purified derivatives containing single Mtr substitutions (KN20 and KN21 [d-NH2-NWWKLRRR-CONH2 + Mtr]) showed that the Mtr derivatives delayed cell growth in the absence of FLC, that 20 to 30 μM KN20 and 10 to 20 μM KN21 caused FLC sensitization, and that both derivatives inhibited Pdr5p ATPase activity at 2 to 4 μM (Table 2). The in vitro inhibition of Pdr5p ATPase activity by KN20 and KN21 was not ATP protected (Table 2). This result indicated that the antagonism was not affected by (i.e., did not compete with) ATP and therefore did not involve the cytoplasmic active site of Pdr5p. ATP protection has been used with the MRP6 ABC transporter expressed in yeast cells to demonstrate the specificity of photolabeling with [8-azido-α-32P]ATP (7).
To further examine the specificity of the side-chain substitutions, monosubstituted 2,2,5,7,8-pentamethylchroman-6-sulfonyl (Pmc) derivatives (KN20p [NH2-NWWKVRRR-CONH2 + Pmc] and KN21p [NH2-NWWKLRRR-CONH2 + Pmc]) of KN0 and KN1 were prepared, HPLC purified, and verified by mass spectroscopy. The Pmc group is used as an arginine side-chain blocker in t-butyloxy-carbonyl solid-phase peptide synthesis and can migrate like the Mtr group during acid cleavage. KN20p and KN21p were more toxic to yeast cells than were KN20 and KN21, but at sub-MICs, they showed no FLC chemosensitization (Table 2). These data indicate that the more compact Mtr group is more effective in conferring chemosensitization.
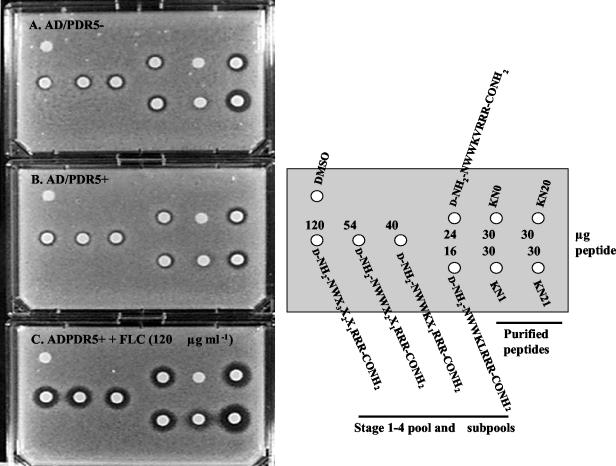
Chemosensitization was also demonstrated by agarose diffusion in the presence or absence of FLC (Fig. 6). As the deconvolution progressed, smaller amounts of deconvoluted peptides had to be applied to disks to obtain similar levels of chemosensitization, indicating purification of the active peptide. In the absence of FLC, however, the toxicity of the subpools for the yeast cells also increased during deconvolution. This toxicity was independent of Pdr5p expression because strains AD/PDR5+ (Fig. 6B) and AD/PDR5− (Fig. 6A) were equally sensitive to the peptides during library deconvolution. This assay also confirmed that chemosensitization required the Mtr substituent in both KN20 and KN21 (Fig. 6B and C) and that KN21 was more toxic to yeast cells than was KN20 (Fig. 6A and B).
FIG. 6.
Agar diffusion chemosensitization of S. cerevisiae strains AD/PDR5− and AD/PDR5+ to FLC by the deconvoluted combinatorial library pool, subpools, and purified peptides. Strain AD/PDR5− was grown on CSM-ura with agarose in the absence of FLC (A), and strain AD/PDR5+ was grown in the absence of FLC (B) or in the presence of FLC at 120 μg ml−1 (C) with DMSO (5 μl; control), the deconvoluted peptide pool, subpools, and HPLC-purified peptides. The agar diffusion chemosensitization assay was conducted as described in Materials and Methods.
KN20 toxicity for human cells. (i) Hemolytic effect of peptides on human erythrocytes.
Purified peptides KN0, KN1, KN20, and KN21 were tested for their hemolytic activity on human erythrocytes. At the highest peptide concentration used in the assay (40 μM), the parent peptides KN0 and KN1 released less than 2% of erythrocyte hemoglobin. The monosubstituted peptides KN20 and KN21 (also 40 μM) were only slightly more hemolytic (3% and 5 to 6% hemolysis, respectively) than were their parent peptides. All peptides were much less hemolytic than the polyene antifungal agent amphotericin B, which at 1.5 μM gave >50% hemolysis.
(ii) Peptide cytotoxicity for cultured HEp-2 cells.
At the highest concentration of KN20 used to chemosensitize a range of fungi (80 μM), there was no effect on HEp-2 cell viability. After 24 h of incubation in the presence and in the absence of the peptide, 3.1 and 3.7% of the populations were dead, respectively. These data showed that, under the tissue culture conditions used for this study, no cytoxicity was attributable to the peptide. The solvent DMSO (up to 4%) did not affect cell viability.
Specificity of the Pdr5p inhibitor KN20.
Individual drug efflux pumps from C. albicans (CaCdr1p, CaCdr2p, and BenRp) and from C. glabrata (CgCdr1p and CgCdr2p) and the azole drug target from C. albicans (Erg11p) were functionally hyperexpressed in AD1-8u−. The hyperexpressed proteins were detected in plasma membrane preparations as intense Coomassie blue-stained bands and were verified by mass spectroscopy of tryptic fingerprints or with specific antibodies. Each recombinant also gave the expected patterns of resistance and sensitivity to azoles and other antifungal agents.
(i) Broad-spectrum activity of KN20.
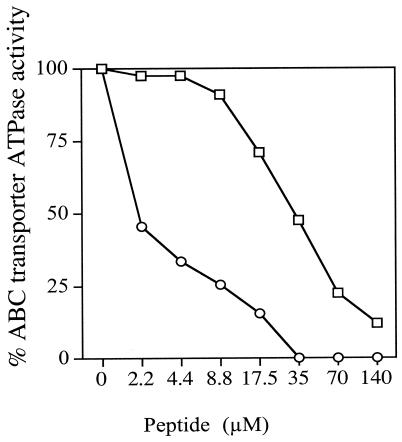
The nucleoside triphosphatase activities of plasma membrane fractions isolated from S. cerevisiae strains hyperexpressing C. albicans Cdr1p and Cdr2p allowed the in vitro inhibition of these enzymes by KN20 to be measured. The peptide inhibited the ATPase activities of CaCdr1p and CaCdr2p at IC50s of 30 and 2 μM, respectively (Fig. 7). These measurements were both within 1 order of magnitude of the IC50 for Pdr5p ATPase inhibition and indicated some broad-spectrum activity.
FIG. 7.
Inhibition of heterologously expressed oligomycin-sensitive C. albicans Cdr1p and Cdr2p ATPases by KN20. Membrane fractions were prepared from S. cerevisiae strains hyperexpressing Cdr1p (□) and Cdr2p (○). Background ATPase activities in membrane fractions from the null parental strain AD1-8u− have been subtracted. ATPase inhibition assays were carried out with 5 μg (Cdr1p) and 10 μg (Cdr2p) of membrane proteins without ATP protection as described in Materials and Methods. The results are the means of two separate determinations in which the values that contributed to individual data points varied by less than 10%.
(ii) Chemosensitization of AD/PDR5+ to other azoles.
The overexpression of ABC transporters confers cross-resistance to several azole antifungal drugs in C. albicans (1) and S. cerevisiae (42). KN20 chemosensitized AD/PDR5+ cells to both the triazole ITC and the azole KTC (Table 3).
TABLE 3.
Chemosensitization of S. cerevisiae AD/PDR5+ to other azole antifungal agents by KN20
| Azole | Concn used for chemosensitization
|
MIC
|
Fold chemosensitization by KN20 (for azole MIC) | ||
|---|---|---|---|---|---|
| KN20 (μM) | Azole (μg ml−1) | KN20 (μM) | Azole (μg ml−1) | ||
| ITC | 20 | 10 | 30 | >64 | >6.4 |
| KTC | 20 | 2.5 | 30 | 40 | 16 |
(iii) Chemosensitization of a range of yeast species to FLC.
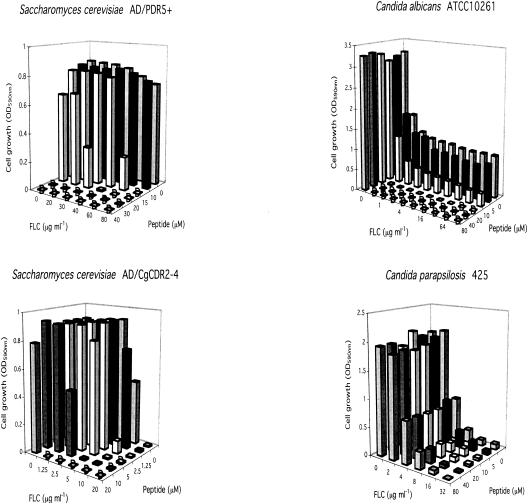
Checkerboard assays (Fig. 8 and Table 4) showed that KN20 chemosensitized yeast cells hyperexpressing the ABC transporters (MICs of FLC, 40 to 400 μg/ml), the MFS transporter BenRp (MICs of FLC, 60 to 80 μg/ml), and lanosterol 14α-demethylase Erg11p (MICs of FLC, 2 to 4 μg/ml) to FLC. KN20 was also a broad-spectrum chemosensitizer of most of the resistant fungal species tested, including the azole-resistant C. albicans strains FR2 and KB (Table 4). The exceptions were C. glabrata strain 850821, Candida parapsilosis strain 425, and C. neoformans ATCC 90112. Athough FLC was not fungicidal and the extent of chemosensitization varied between species and strains, the combination of KN20 and FLC was often fungicidal. In addition, KN20 eliminated the trailing tail of resistance seen in C. albicans ATCC 10261 (Fig. 8), Candida tropicalis IFO 0618, and Candida dubliniensis CD36 (Table 4). The diverse effects of KN20 suggested that instead of being specific for Pdr5p, it might act via Pma1p, a surface-exposed controller of fungal metabolism, or more generally affect membrane function.
FIG. 8.
Chemosensitization of Candida sp. and S. cerevisiae strains hyperexpressing ABC multidrug transporters to FLC by KN20. The checkerboard chemosensitization assays were carried out as described in Materials and Methods. D, cells were dead (fungicidal activity).
TABLE 4.
Chemosensitization of yeast clinical isolates and S. cerevisae strains hyper expressing individual transporters or Erg11p to FLC by KN20
| Species | Strain (hyperexpressed transporter) | Concn used for chemosensitization
|
MIC
|
Fold chemosensitization by KN20 (for FLC MIC) | ||
|---|---|---|---|---|---|---|
| KN20 (μM) | FLC (μg ml−1) | KN20 (μM) | FLC (μg ml−1) | |||
| S. cerevisiae | AD/PDR5− | 20 | 0.25 | 30 | 1 | 4 |
| AD/PDR5+ (Pdr5p) | 20 | 60 | 30 | 600 | 10 | |
| AD1-8u− | 7.5 | 0.125 | 30 | 0.5 | 4 | |
| AD1002 (CaCdr1p) | 10 | 10 | 30 | 40 | 4 | |
| AD/CaCDR1 (CaCdr1p) | 20 | 20 | 30 | 400 | 20 | |
| AD/CaCDR2 (CaCdr2p) | 20 | 20 | >20 | 128 | 6.2 | |
| AD/CgCDR1-1B (CgCdr1p) | 15 | 40 | 20 | 320 | 8 | |
| AD/CgCDR2-4 (CgCdr2p) | 20 | 1.25 | 30 | 20 | 16 | |
| AD/BENR (BenRp) | 20 | 3.75 | 40 | 60 | 16 | |
| AD/ERG11 (Erg11p) | 20 | 0.25 | 40 | 2 | 8 | |
| C. albicans | ATCC 10261 | 20 | 0.5 | >80 | >128 | >256a |
| FR2 | 20 | 8 | 80 | 32 | 4 | |
| KB | 80 | 20 | >80 | >160 | >8a | |
| C. dubliniensis | CD36 | 40 | 0.31 | >80 | >20 | >64.5a |
| CD43 | 20 | 1.25 | >80 | >20 | >16a | |
| C. glabrata | CBS138 | 20 | 20 | 80 | 80 | 4 |
| 850821 | 0-80 | 0-160 | 80 | 160 | 1b | |
| 850920 | 40 | 2.5 | >80 | 40 | 16a | |
| C. krusei | B2399 | 80 | 80 | 160 | 200 | 2.5 |
| C. parapsilosis | 90.403 | 80 | 0.5 | >80 | 4 | 8a |
| 425 | 0-80 | 0-32 | 80 | 32 | 1b | |
| C. tropicalis | IFO0618 | 10 | 10 | 80 | >160 | >16a |
| C. neoformans | ATCC 90112 | 10 | 8 | 15 | 16 | 2 |
The trailing tail of resistance was eliminated.
No chemosensitization.
(iv) Inhibition of the yeast plasma membrane proton pump ATPase.
Pma1p, which comprises 10 to 20% of the plasma membrane protein in S. cerevisiae, dominates the fungal plasma membrane. It is an essential primary pump that generates the plasma membrane electrochemical gradient required for nutrient uptake and metabolic flux in yeast cells (46). Extensive inhibition of Pma1p is lethal, while a significant reduction in its activity should limit metabolic flux, restrict the availability of intracellular ATP, and indirectly modify the activity of the Pdr5p multidrug efflux pump. KN20 was, in fact, a stronger in vitro inhibitor of the Pma1p ATPase activity of S. cerevisiae T48 cells (IC50, 1 μM) than of the Pdr5p ATPase activity of AD/PDR5+ cells (IC50, ∼3 μM) (compare Fig. 9 with Fig. 5B). Interestingly, the inhibition of Pma1p ATPase activity also required the Mtr group in KN20. The IC50 of KN0 was about sevenfold higher than that of KN20, and KN0 incompletely inhibited Pma1p-ATPase activity even at the highest concentration (140 μM) tested. Furthermore, the IC50 of KN20 for Pma1p ATPase was unaltered when its cytoplasmic active site was protected by ATP. These results suggested that KN20 might inhibit both Pdr5p and Pma1p in S. cerevisiae cells, either by direct effects on both enzymes or through an indirect mechanism solely involving an interaction with Pma1p. KN20, however, showed poor inhibition of C. albicans Pma1p (IC50, >100 μM).
FIG. 9.
Inhibition of plasma membrane H+-ATPase activity by KN0 and KN20. Membrane fractions were prepared from S. cerevisiae T48, and ATPase assays were conducted without ATP protection as described in Materials and Methods. Oligomycin (10 μM) was included in the assays to eliminate non-Pma1p ATPase activity. The results are the means of two separate determinations in which the values that contributed to individual data points varied by less than 10%. Symbols: □, KN0; ○, KN20.
(v) Effects on Rh6G efflux and uptake.
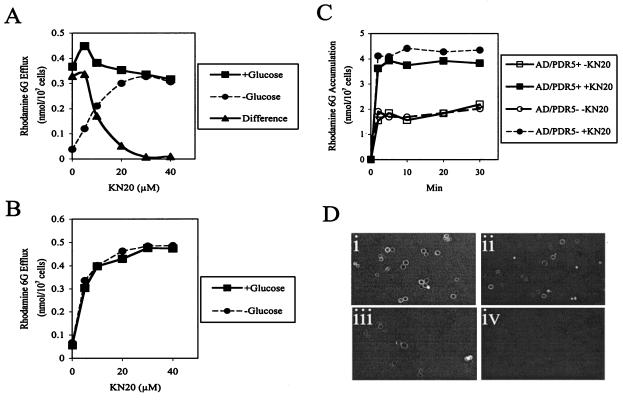
Rh6G is a fluorescent Pdr5p substrate, and in the absence of KN20, Rh6G efflux from preloaded AD/PDR5+ cells was glucose dependent, while AD/PDR5− cells showed no efflux. KN20 had an apparent IC50 of about 10 μM for glucose-dependent Rh6G efflux from AD/PDR5+ cells (difference between the cells in the presence and in the absence of glucose) (Fig. 10A). The inhibitor had little effect on dye efflux from Rh6G-preloaded AD/PDR5+ cells in the presence of glucose and instead caused concentration-dependent efflux in the absence of glucose. KN20 also caused glucose-independent efflux of Rh6G from preloaded AD/PDR5− cells (Fig. 10B). Rh6G efflux therefore was independent of both energy and hyperexpressed Pdr5p, suggesting that KN20 permeabilized the yeast cells. This suggestion was confirmed by showing that KN20 (20 μM) increased Rh6G accumulation by starved AD/PDR5+ or AD/PDR5− cells by at least twofold (Fig. 10C).
FIG. 10.
Effects ofKN20 on Rh6G efflux and uptake and TRITC-KN20 binding to cell surfaces. (A) The efflux of Rh6G from preloaded AD/PDR5+cells in the presence or absence of 0.2% glucose was measured as described in Materials and Methods. (B) The efflux of Rh6G from preloaded AD/PDR5−cells in the presence or absence of 0.2% glucose was measured as described in Materials and Methods. (C) The accumulation of 15 μM Rh6G in 2-deoxyglucose-treated AD/PDR5+or AD/PDR5−cells was measured in the presence or absence of KN20 (20 μM) as described in Materials and Methods. (D) Log-phase cells were incubated in CSM-ura(pH 7.0) containing 10 μM TRITC-KN20 for 30 min and viewed by confocal microscopy as described in Materials and Methods immediately or after being washed in 100 μl of CSM-ura. (i) AD/PDR5+. (ii) Washed AD/PDR5+. (iii) AD/PDR5−. (iv) Washed AD/PDR5−.
(vi) Binding of TRITC-labeled KN20 to yeast cells.
Confocal microscopy showed that TRITC-KN20 (10 μM) specifically associated with the surface of AD/PDR5+ cells (Fig. 10D, panel i) and that only a portion of the bound material was eluted by washing (Fig. 10D, panel ii). AD/CaCDR1 cells gave identical results. TRITC-KN20 also associated with the surface of AD/PDR5− cells (Fig. 10D, panel iii), albeit more weakly than with that of AD/PDR5+ cells, and was completely removed by washing (Fig. 10D, panel iv). S. cerevisiae T48 and C. albicans ATCC 10261 cells gave the same results as AD/PDR5− cells. The fluorescent peptide penetrated and heavily labeled only a few cells, which were probably not intact. TRITC-KN20 therefore does not permeate membranes. It strongly interacts with surface components, the expression of which coincides with the hyperexpression of Pdr5p or Cdr1p, but TRITC-KN20 also has a lower affinity for other cell surface components.
Optimization of KN20 by amino acid substitution and side-chain modification.
A total of 29 derivatives of KN20 were synthesized in order to obtain a peptide with improved potency. The amino acids at positions A, B, X3, and X2 in the KN20 sequence were substituted with ones that appeared to have potencies similar to those of the amino acids selected during the deconvolution. Other changes introduced into the KN20 sequence included the addition of up to two further Mtr substituents, the insertion of up to three extra d-arginines at the peptide N terminus, and carboxymethylation of the N terminus. Some of the modified peptides (KN5 [d-NH2-RNWWKVRRR-CONH2], KN25 [KN5 + Mtr], d-NH2-RRNWWKVRRR-CONH2, and d-NH2-IWWKVRRR-CONH2) chemosensitized AD/PDR5+ cells and strongly but incompletely inhibited Pdr5p ATPase activity (IC50s, 2.2 to 17.5 μM), yet none was as potent as KN20 (data not shown). All of these peptides extensively inhibited Pma1p ATPase activity at IC50s in the range of 2 to 4 μM. The other peptides either failed to chemosensitize cells or affected yeast cell viability so strongly that chemosensitization could not be assessed. These data emphasize the robustness of the deconvolution process used to identify KN20.
DISCUSSION
Combination therapy with different classes of antifungal agents has long been proposed by clinicians to enhance efficacy and to reduce toxic effects in the treatment of refractory fungal infections (20), but the suggestion that inhibitors of fungal drug efflux pathways could serve as antifungal chemosensitizers is more recent (32, 39, 55). We have obtained from a d-octapeptide combinatorial library a chemosensitizer that accumulates at the fungal cell surface, probably via its C-terminal amidated triarginine motif. The accumulation of library peptides as a reservoir of non-membrane-permeating reagents was designed to focus their activity on cell surface targets, avoid cellular detoxification processes, and minimize host toxicity. The 1.8-million-member combinatorial library was screened for surface-active Pdr5p inhibitors by using Pdr5p-hyperexpressing S. cerevisiae strain AD/PDR5+. This strain, which required the Pdr5p ABC transporter for growth in the presence of FLC but not in its absence, was used in a cell-based FLC chemosensitization assay that identified candidate pump inhibitors with the expected modest toxicity for yeast cells, as determined by their effect on yeast growth in the absence of FLC. Although the FLC chemosensitization assay identified active peptide pools that diminished FLC resistance, the cell-based assay did not discriminate between direct inhibition of Pdr5p and indirect inhibition via other cellular properties that affect pump function. An in vitro secondary screen therefore was used to identify inhibitory pools, subpools, and peptides that most strongly and directly inhibited the target enzyme. However, because purified plasma membrane fractions were used, the assay failed to exclude inhibitors that also indirectly affected whole cells, e.g., through Pma1p or limited permeabilization.
Assays with purified plasma membrane preparations from physiologically conditioned AD/PDR5+ and AD/PDR5− cells were used to discriminate the ATPase activity of overexpressed Pdr5p from that of plasma membrane H+-ATPase (Pma1p), normally the dominant plasma membrane protein. The ATPase activity of Pma1p is tightly regulated by phosphorylation mechanisms that are sensitive to glucose availability and cold temperatures (37). The activity of this enzyme is reduced at least 10-fold in cold-treated glucose-starved cells. The residual activity of Pma1p under these conditions is optimal below pH 6 and rapidly decreases with increasing pH. At pH 7.0, only about 10% of this vanadate-sensitive and oligomycin-insensitive activity remains (9). The optimum pHs for the ATPase activity of membranes from strain AD/PDR5+ were 7.0 to 8.0, and most of this activity was sensitive to both oligomycin and vanadate in a system where azide plus aurovertin, nitrate, and molybdate were used to minimize contributions from mitochondrial, vacuolar, and nonspecific phosphatases, respectively. By growing cells hyperexpressing Pdr5p to late log phase, deactivating the 1-order-of-magnitude-higher activity of Pma1p (by washing to remove the remaining glucose and incubating the cells on ice), and conducting assays at the optimum pH for Pdr5p ATPase (pH 7.5), the expected oligomycin- and vanadate-sensitive Pdr5p ATPase activity of purified plasma membranes was found to be up to fivefold higher than the background oligomycin-sensitive ATPase activity detected in plasma membranes from the null mutant AD/PDR5− (data not shown). Although the Pdr5p efflux pump requires ATP as a substrate, the enzyme showed a broad nucleoside triphosphatase specificity, with UTP, GTP, and CTP also serving as substrates in vitro. In contrast, yeast Pma1p is highly specific for ATP (9). This approach allowed specific quantitative measurements of the inhibitory effects of candidate library pools, subpools, and peptides on Pdr5p ATPase activity.
A combination of the cell-based primary screen and the in vitro secondary screen was used for deconvolution of the combinatorial library and for determination of the primary sequences of two potent peptides (KN20 and KN21). Both peptides were found to be derivatized by a single Mtr group during acid-catalyzed release from the solid-phase matrix. Either of the two tryptophans in KN20 and KN21 is a possible candidate for derivatization by Mtr, so there may be two separate forms of each inhibitor. KN20 and KN21 were noncompetitive Pdr5p ATPase inhibitors that affected the activity of the enzyme at a site other than the ATP-binding site. These in vitro observations are consistent with inhibition via surface-exposed components of the pump but do not exclude inhibition via a cytoplasmic site on the enzyme outside the ATP-binding site. KN20 was chosen for further study because it appeared to be less toxic for S. cerevisiae cells than KN21 and therefore more likely to target the nonessential Pdr5p pump.
The FLC chemosensitizer KN20 fulfilled the expectations of noncompetitive inhibition of Pdr5p and of the related molecules CaCdr1p and CaCdr2p, it preferentially bound (as TRITC-KN20) to the surface of cells hyperexpressing Pdr5p and CaCdr1p, and it chemosensitized cells to the Pdr5p triazole substrates FLC and ITC and the azole substrate KTC. TRITC-KN20 also bound more weakly to an additional component of the surface of AD/PDR5+ and AD/PDR5− cells, while KN20 caused glucose- and Pdr5p-independent diffusion of Rh6G across the yeast cell membrane. At 20 μM, KN20 increased Rh6G accumulation by cells and caused efflux from Rh6G-preloaded cells, without requiring energy or ABC pump hyperexpression. The concentrations of KN20 that caused Rh6G diffusion correlated well with the sub-MIC concentrations of the compound required to chemosensitize pathogenic Candida species and a range of S. cerevisiae strains hyperexpressing ABC transporters, the MFS transporter BenRp, and Erg11p to FLC. This correlation suggests that KN20-mediated Rh6G permeabilization and FLC chemosensitization are related. Experiments which measured the lysis of human erythrocytes or the killing of cultured human epithelial cell line HEp-2 showed that KN20 is not cytotoxic for mammalian cells at concentrations that chemosensitize or stop the growth of model yeast strains and pathogenic Candida species. After a lag of up to 24 h, yeast cells treated with the sub-MIC of KN20 grew normally and gave normal growth yields, unless FLC was present. Thus, drug-resistant yeast cells were unable to overcome the combined stress of KN20 and FLC exposure. Finally, the toxicity of KN20 to yeast cells was independent of the overexpression of drug resistance determinants, because the KN20 MICs for parental null mutant AD/PDR5− and the Pdr5p-overexpressing strain AD/PDR5+, as well as strain AD1-8u− and its derivative membrane protein-overexpressing strains, were essentially identical (∼30 μM). A simple explanation of these properties is that low-affinity binding of KN20 to the cell surface causes concentration-dependent cell permeabilization that is not lethal to yeast cells at chemosensitizing concentrations. The pluronic block copolymer P85, a nonionic detergent drug formulation component, may act in a similar but not identical fashion. P85 is thought to modulate the activity of P-glycoprotein through direct inhibition, diminution of intracellular ATP levels, and modification of membrane fluidity in human cells that preferentially overexpress P-glycoprotein (4, 5).
KN20 chemosensitized a range of pathogenic Candida species, including several FLC-resistant clinical isolates and some wild-type strains of C. albicans, C. tropicalis, and C. dubliniensis which had a trailing tail of resistance to FLC. Although not all strains showing FLC resistance were chemosensitized, the spectrum of strains affected indicates useful broad-spectrum activity. Strains chemosensitized included C. glabrata strains that have been characterized by mass spectrometry as overexpressing CgCdr1p or lanosterol 14α-demethylase in their plasma membranes (43). The trailing tail of resistance, thought to consist of a cell population with low-level resistance to FLC, is commonly seen in C. albicans clinical isolates (35) and may be important for the subsequent evolution of clinical resistance in patients subjected to long-term treatment with FLC. Elimination of the trailing tail of FLC-resistant organisms in such populations could reduce the potential for treatment failure in infected patients. In several instances, KN20 converted FLC from a fungistatic to a fungicidal drug, further minimizing opportunity for the evolution of drug resistance. Since yeast cells stop growing but do not die when sterol metabolism is limiting, chemosensitization to FLC probably involves both cell permeabilization and membrane modification.
Does Pma1p of S. cerevisiae also mediate indirect chemosensitization? KN20 completely inhibited the in vitro activity of S. cerevisiae Pma1p (IC50, 1 μM). Like Pdr5p inhibition, Pma1p inhibition required monosubstitution of the peptide with the Mtr group. Furthermore, KN20 chemosensitized FLC-resistant yeast cells hyperexpressing either the MFS BenRp drug efflux pump, which requires the plasma membrane electrochemical gradient for function, or lanosterol 14α-demethylase, which requires electrons from reduced nicotine adenine nucleotides for function. Pma1p is required to maintain the electrochemical gradient of the plasma membrane and powers the uptake of nutrients required for energy metabolism, including the production of NADPH, NADH, and ATP. Partial inhibition of Pma1p by 10 to 20 μM KN20 therefore might limit cellular energy supply and indirectly chemosensitize multidrug efflux. The inhibition of Pma1p might also explain why KN20 blocks growth and is fungicidal for yeast cells at higher concentrations. However, C. albicans Pma1p was essentially insensitive to KN20 (IC50, >100 μM), suggesting that chemosensitization of the trailing tail of resistance in the otherwise sensitive C. albicans strain ATCC 10261 at 20 μM KN20 is independent of Pma1p inhibition. Although the CaCdr1p (IC50, 30 μM) and CaCdr2p (IC50, 2 μM) ATPases were fully inhibited by KN20, the differential in vitro sensitivities of the two enzymes suggest significant structural differences in their binding sites for KN20. These differences were not reflected in cellular chemosensitization by KN20, because similar concentrations of KN20 (10 to 20 μM) chemosensitized S. cerevisiae strains hyperexpressing the ABC transporters Pdr5p, CaCdr1p, CaCdr2p, CgCdr1p, and CgCdr2p. It is therefore reasonable to conclude that their chemosensitization was indirect. While we cannot exclude indirect effects on drug efflux pumps and Erg11p due to Pma1p inhibition, it is more likely that the sub-MIC of KN20 partially permeabilizes the plasma membrane, depleting ions and metabolites to levels that cannot be tolerated in the presence of elevated FLC concentrations. Thus, while the KN20 inhibitory sites of Pdr5p and Pma1p are accessible in membrane preparations, their accessibility may be limited in intact cells or their affinity for KN20 may be insufficient to compete with its adsorption to other cell surface sites.
In hindsight, we suggest that counterscreens using chemosensitization of BenRp-mediated FLC resistance, in vitro assays of Pma1p inhibition, assays of glucose-dependent and glucose-independent Rh6G efflux, and scrupulous avoidance of yeast cell lethality could eliminate chemosensitizers that operate via either Pma1p or membrane permeabilization. On the other hand, the essential ATPase Pma1p may be a more robust target in fungi than the ABC transporter family, because Pma1p inhibitors should be fungicidal and, at sub-MICs, should indirectly block the antifungal resistance mechanisms mediated by Erg11p, BenRp, and the ABC transporters. This prediction is supported by the properties of a more potent Pma1p inhibitor that was recently discovered (B. C. Monk, unpublished data).
The pairing of whole-cell and in vitro screens with functionally hyperexpressed ABC transporters, together with the accessory counterscreens that we have noted, should identify drug efflux pump antagonists in resources such as combinatorial libraries, natural or synthetic compound libraries, culture supernatants, or traditional medicines. The discovery of fungal multidrug transporter inhibitors is gaining importance (26, 61), and Essential Therapeutics has been developing the milbemycins as potential antifungal chemosensitizers (32). With the functional hyperexpression in S. cerevisiae of ABC pumps from other medically important pathogens, such as C. albicans and C. glabrata, having been achieved (42, 60), together with the potential to clone and express similar pumps from less tractable organisms, such as C. neoformans and A. fumigatus, and from humans (e.g., P-glycoprotein) in a hypersensitive yeast background (40), it should be possible to develop screens that select for broad-spectrum antifungal chemosensitizers with minimal yeast and human toxicity. We predict that many such compounds will be discovered in existing compound libraries, because most previous screens have sought compounds that are toxic for yeast cells. Futhermore, the hyperexpressing constructs could also provide counterscreens for the selection of antifungal agents that are not susceptible to multidrug efflux.
Although the screening of 29 KN20 derivatives failed to identify more potent or specific chemosensitizers with lower levels of yeast toxicity, KN20 will still be of value in structural studies of the Pdr5p ABC transporter. The in vitro affinity of KN20 for Pdr5p appears to be sufficiently high for it to be used to stabilize the structure of this multidomain polytopic integral membrane protein and to provide information about an inhibited state for future structure-directed drug design efforts.
Acknowledgments
We acknowledge the financial support of the Health Research Council of New Zealand, The New Zealand Lottery Grants Board, the University of Otago, Université Catholique de Louvain (Interuniversity Poles of Attraction Program from the Belgian State Scientific, Technical and Cultural Services), and the Japan Health Science Foundation.
We are grateful for the assistance of J. Hislop in the synthesis and purification of KN20 and analogues in the later phases of this study and to E. Lamping, A. R. Holmes (University of Otago), and S. Wada (National Institute of Infectious Diseases) for providing S. cerevisiae constructs.
REFERENCES
- 1.Albertson, G. D., M. Niimi, R. D. Cannon, and H. F. Jenkinson. 1996. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob. Agents Chemother. 40:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balzi, E., M. Wang, S. Leterme, L. Van Dyck, and A. Goffeau. 1994. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcriptional regulator PDR1. J. Biol. Chem. 269:2206-2214. [PubMed] [Google Scholar]
- 3.Bartizal, K., C. J. Gill, G. K. Abruzzo, A. M. Flattery, L. Kong, P. M. Scott, J. G. Smith, C. E. Leighton, A. Bouffard, J. F. Dropinski, and J. Balkovec. 1997. In vitro preclinical evaluation studies with the echinocandin antifungal MK-0991 (L-743,872). Antimicrob. Agents Chemother. 41:2326-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batrakova, E. V., S. Li, W. F. Elmquist, D. W. Miller, V. Y. Alakhov, and A. V. Kabanov. 2001. Mechanism of sensitization of MDR cancer cells by Pluronic block copolymers: selective energy depletion. Br. J. Cancer 85:1987-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batrakova, E. V., S. Li, S. V. Vinogradov, V. Y. Alakhov, D. W. Miller, and A. V. Kabanov. 2001. Mechanism of pluronic effect on P-glycoprotein efflux system in blood-brain barrier: contributions of energy depletion and membrane fluidization. J. Pharmacol. Exp. Ther. 299:483-493. [PubMed] [Google Scholar]
- 6.Ben-Yaacov, R., S. Knoller, G. A. Caldwell, J. M. Becker, and Y. Koltin. 1994. Candida albicans gene encoding resistance to benomyl and methotrexate is a multidrug resistance gene. Antimicrob. Agents Chemother. 38:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, J., R. Daoud, O. Alqawi, E. Georges, J. Pelletier, and P. Gros. 2002. Nucleotide binding and nucleotide hydrolysis properties of the ABC transporter MRP6 (ABCC6). Biochemistry 41:8058-8067. [DOI] [PubMed] [Google Scholar]
- 8.Cole, A. M., R. O. Darouiche, D. Legarda, N. Connell, and G. Diamond. 2000. Characterization of a fish antimicrobial peptide: gene expression, subcellular localization, and spectrum of activity. Antimicrob. Agents Chemother. 44:2039-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decottignies, A., M. Kolaczkowski, E. Balzi, and A. Goffeau. 1994. Solubilization and characterization of the overexpressed multidrug resistance nucleotide triphosphatase of yeast. J. Biol. Chem. 269:12797-12803. [PubMed] [Google Scholar]
- 10.Decottignies, A., B. Rogers, M. Kowlaczkowski, E. Carvajal, E. Balzi, G. Conseil, K. Niimi, A. Di Pietro, B. C. Monk, and A. Goffeau. 2002. The pleiotropic drug ABC transporters from Saccharomyces cerevisiae, p. 157-176. In I. T. Paulsen and K. Lewis (ed.), Microbial multidrug efflux. Horizon Scientific Press, Wymondham, United Kingdom.
- 11.Del Poeta, M., M. C. Cruz, M. E. Cardenas, J. R. Perfect, and J. Heitman. 2000. Synergistic antifungal activities of bafilomycin A(1), fluconazole, and the pneumocandin MK-0991/caspofungin acetate (L-743,873) with calcineurin inhibitors FK506 and L-685,818 against Cryptococcus neoformans. Antimicrob. Agents Chemother. 44:739-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Sorbo, G., A. C. Andrade, J. G. Van Nistelrooy, J. A. Van Kan, E. Balzi, and M. A. De Waard. 1997. Multidrug resistance in Aspergillus nidulans involves novel ATP-binding cassette transporters. Mol. Gen. Genet. 254:417-426. [DOI] [PubMed] [Google Scholar]
- 13.De Lucca, A. J., and T. J. Walsh. 1999. Antifungal peptides: novel therapeutic compounds against emerging pathogens. Antimicrob. Agents Chemother. 43:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominguez, J. M., and J. J. Martin. 1998. Identification of elongation factor 2 as the essential protein targeted by sordarins in Candida albicans. Antimicrob. Agents Chemother. 42:2279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupont, B. 2002. Overview of the lipid formulations of amphotericin B. J. Antimicrob. Chemother. 49(Suppl. 1):31-36. [DOI] [PubMed] [Google Scholar]
- 16.Edlind, T., L. Smith, K. Henry, S. Katiyar, and J. Nickels. 2002. Antifungal activity in Saccharomyces cerevisiae is modulated by calcium signalling. Mol. Microbiol. 46:257-268. [DOI] [PubMed] [Google Scholar]
- 17.Egner, R., B. E. Bauer, and K. Kuchler. 2000. The transmembrane domain 10 of the yeast Pdr5p ABC antifungal efflux pump determines both substrate specificity and inhibitor susceptibility. Mol. Microbiol. 35:1255-1263. [DOI] [PubMed] [Google Scholar]
- 18.Egner, R., F. E. Rosenthal, A. Kralli, D. Sanglard, and K. Kuchler. 1998. Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Mol. Biol. Cell 9:523-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fehlbaum, P., P. Bulet, L. Michaut, M. Lagueux, W. F. Broekaert, C. Hetru, and J. A. Hoffmann. 1994. Insect immunity. Septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J. Biol. Chem. 269:33159-33163. [PubMed] [Google Scholar]
- 20.Francis, P., and T. J. Walsh. 1992. Evolving role of flucytosine in immunocompromised patients: new insights into safety, pharmacokinetics, and antifungal therapy. Clin. Infect. Dis. 15:1003-1018. [DOI] [PubMed] [Google Scholar]
- 21.Georgopapadakou, N. H. 2002. Antifungals targeted to protein modification: focus on protein N-myristoyltransferase. Exp. Opin. Investig. Drugs 11:1117-1125. [DOI] [PubMed] [Google Scholar]
- 22.Georgopapadakou, N. H. 2000. Antifungals targeted to sphingolipid synthesis: focus on inositol phosphorylceramide synthase. Exp. Opin. Investig. Drugs 9:1787-1796. [DOI] [PubMed] [Google Scholar]
- 23.Goffeau, A., and J. P. Dufour. 1988. Plasma membrane ATPase from the yeast Saccharomyces cerevisiae. Methods Enzymol. 157:528-533. [DOI] [PubMed] [Google Scholar]
- 24.Goldway, M., D. Teff, R. Schmidt, A. B. Oppenheim, and Y. Koltin. 1995. Multidrug resistance in Candida albicans: disruption of the BENr gene. Antimicrob. Agents Chemother. 39:422-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helmerhorst, E. J., W. Van't Hof, E. C. Veerman, I. Simoons-Smit, and A. V. Nieuw Amerongen. 1997. Synthetic histatin analogues with broad-spectrum antimicrobial activity. Biochem. J. 326:39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiraga, K., A. Wanigasekera, H. Sugi, N. Hamanaka, and K. Oda. 2001. A novel screening for inhibitors of a pleiotropic drug resistant pump, Pdr5, in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 65:1589-1595. [DOI] [PubMed] [Google Scholar]
- 27.Hong, S. Y., J. E. Oh, M. Y. Kwon, M. J. Choi, J. H. Lee, B. L. Lee, H. O. Moon, and K. H. Lee. 1998. Identification and characterization of novel antimicrobial decapeptides generated by combinatorial chemistry. Antimicrob. Agents Chemother. 42:2534-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katiyar, S. K., and T. D. Edlind. 2001. Identification and expression of multidrug resistance-related ABC transporter genes in Candida krusei. Med. Mycol. 39:109-116. [DOI] [PubMed] [Google Scholar]
- 29.Kolaczkowski, M., M. van der Rest, A. Cybularz-Kolaczkowski, J.-P. Soumillion, W. N. Koning, and A. Goffeau. 1996. Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. J. Biol. Chem. 271:31543-31548. [DOI] [PubMed] [Google Scholar]
- 30.Kontoyiannis, D. P., and R. E. Lewis. 2002. Antifungal drug resistance of pathogenic fungi. Lancet 359:1135-1144. [DOI] [PubMed] [Google Scholar]
- 31.Kuroda, M., T. Hashida-Okado, R. Yasumoto, K. Gomi, I. Kato, and K. Takesako. 1999. An aurobasidin A resistance gene isolated from Aspergillus is a homolog of yeast AUR1, a gene responsible for inositol phosphoceramide (IPC) synthase activity. Mol. Gen. Genet. 261:290-296. [DOI] [PubMed] [Google Scholar]
- 32.Lee, M. D., J. L. Galazzo, A. L. Staley, J. C. Lee, M. S. Warren, H. Fuernkranz, S. Chamberland, O. Lomovskaya, and G. H. Miller. 2001. Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. Farmaco 56:81-85. [DOI] [PubMed] [Google Scholar]
- 33.Maesaki, S., P. Marichal, M. A. Hossain, D. Sanglard, H. Vanden Bossche, and S. Kohno. 1998. Synergic effects of tactolimus and azole antifungal agents against azole-resistant Candida albicans strains. J. Antimicrob. Chemother. 42:747-753. [DOI] [PubMed] [Google Scholar]
- 34.Marchetti, O., J. M. Entenza, D. Sanglard, J. Bille, M. P. Glauser, and P. Moreillon. 2000. Fluconazole plus cyclosporine: a fungicidal combination effective against experimental endocarditis due to Candida albicans. Antimicrob. Agents Chemother. 44:2932-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchetti, O., P. Moreillon, M. P. Glauser, J. Bille, and D. Sanglard. 2000. Potent synergism of the combination of fluconazole and cyclosporine in Candida albicans. Antimicrob. Agents Chemother. 44:2373-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin, M. V. 1999. The use of fluconazole and itraconazole in the treatment of Candida albicans infections: a review. J. Antimicrob. Chemother. 44:429-437. [DOI] [PubMed] [Google Scholar]
- 37.Mason, A. B., T. B. Kardos, and B. C. Monk. 1998. Regulation and pH-dependent expression of a bilaterally truncated yeast plasma membrane H+-ATPase. Biochim. Biophys. Acta 1372:261-271. [DOI] [PubMed] [Google Scholar]
- 38.Miyazaki, H., Y. Miyazaki, A. Geber, T. Parkinson, C. Hitchcock, D. J. Falconer, D. J. Ward, K. Marsden, and J. E. Bennett. 1998. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob. Agents Chemother. 42:1695-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monk, B. C., and R. D. Cannon. 2002. Genomic pathways to antifungal discovery. Curr. Drug Targets Infect. Disord. 2:309-329. [DOI] [PubMed] [Google Scholar]
- 40.Monk, B. C., R. D. Cannon, K. Nakamura, M. Niimi, K. Niimi, D. R. K. Harding, A. R. Holmes, E. Lamping, A. Goffeau, and A. Decottignies. June 2003. Membrane protein expression system and its application in drug screening. International patent PCT/NZ02/00163.
- 41.Monk, B. C., C. Montesinos, K. Leonard, and R. Serrano. 1989. Sidedness of yeast plasma membrane vesicles and mechanisms of activation of the ATPase by detergents. Biochim. Biophys. Acta 981:226-234. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura, K., M. Niimi, K. Niimi, A. R. Holmes, J. E. Yates, A. Decottignies, B. C. Monk, A. Goffeau, and R. D. Cannon. 2001. Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrob. Agents Chemother. 45:3366-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niimi, M., Y. Nagai, K. Niimi, S. Wada, R. D. Cannon, Y. Uehara, and B. C. Monk. 2002. Identification of two proteins induced by exposure of the pathogenic fungus Candida glabrata to fluconazole. J. Chromatogr. B 782:245-252. [DOI] [PubMed] [Google Scholar]
- 44.Ostresh, J. M., S. E. Blondelle, B. Dorner, and R. A. Houghten. 1996. Generation and use of nonsupport-bound peptide and peptidomimetic combinatorial libraries. Methods Enzymol. 267:220-234. [DOI] [PubMed] [Google Scholar]
- 45.Pfaller, M. A., S. A. Messer, R. J. Hollis, R. N. Jones, and D. J. Diekema. 2002. In vitro activities of ravuconazole and voriconazole compared with those of four approved systemic antifungal agents against 6,970 clinical isolates of Candida spp. Antimicrob. Agents Chemother. 46:1723-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Portillo, F., and R. Serrano. 1989. Growth control strength and active site of yeast plasma membrane ATPase studied by site-directed mutagenesis. Eur. J. Biochem. 186:501-507. [DOI] [PubMed] [Google Scholar]
- 47.Prasad, R., P. De Wergifosse, A. Goffeau, and E. Balzi. 1995. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27:320-329. [DOI] [PubMed] [Google Scholar]
- 48.Romero, A. J., P. L. Pogamp, L. G. Nilsson, and N. Wood. 2002. Effect of voriconazole on the pharmacokinetics of cyclosporine in renal transplant patients. Clin. Pharmacol. Ther. 71:226-234. [DOI] [PubMed] [Google Scholar]
- 49.Sanglard, D., and J. Bille. 2002. Current understanding of the modes of action and resistance mechanisms to conventional and emerging antifungal agents for treatment of Candida infection, p. 349-383. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 50.Sanglard, D., F. Ischer, and J. Bille. 2001. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45:1174-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanglard, D., F. Ischer, D. Calabrese, P. A. Majcherczyk, and J. Bille. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43:2753-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 53.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1996. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 40:2300-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 55.Schuetzer-Muehlbauer, M., B. Willinger, R. Egner, G. Ecker, and K. Kuchler. 2003. Reversal of antifungal resistance mediated by ABC efflux pumps from Candida albicans functionally expressed in yeast. Int. J. Antimicrob. Agents 22:291-300. [DOI] [PubMed] [Google Scholar]
- 56.Sieber, P. 1987. Modification of tryptophan residues during acidolysis of 4-methoxy-2,3,6-trimethylbenzenesulfonyl groups. Effects of scavengers. Tetrahedron Lett. 28:1637-1640. [Google Scholar]
- 57.Tang, Y. Q., J. Yuan, G. Osapay, K. Osapay, D. Tran, C. J. Miller, A. J. Ouellette, and M. E. Selsted. 1999. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science 286:498-502. [DOI] [PubMed] [Google Scholar]
- 58.Tobin, M. B., R. B. Peery, and P. L. Skatrud. 1997. Genes encoding multiple drug resistance-like proteins in Aspergillus fumigatus and Aspergillus flavus. Gene 20:11-23. [DOI] [PubMed] [Google Scholar]
- 59.Venkataramanan, R., S. Zang, T. Gayowski, and N. Singh. 2002. Voriconazole inhibition of the metabolism of tacrolimus in a liver transplant recipient and in human liver microsomes. Antimicrob. Agents Chemother. 46:3091-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wada, S., M. Niimi, K. Niimi, A. R. Holmes, B. C. Monk, R. D. Cannon, and Y. Uehara. 2002. Candida glabrata ABC transporters Cdr1p and Pdh1p expressed in a Saccharomyces cerevisiae strain deficient in membrane transporters show phosphorylation-dependent pumping properties. J. Biol. Chem. 277:46809-46821. [DOI] [PubMed] [Google Scholar]
- 61.Wanigasekera, A., K. Hiraga, N. Hamanaka, and K. Oda. 2001. Purification and some properties of an inhibitor for a yeast pleiotropic drug resistant pump from Kitasatospora sp. E-420. Biosci. Biotechnol. Biochem. 65:2353-2357. [DOI] [PubMed] [Google Scholar]
- 62.White, T., K. Marr, and R. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]