Abstract
We analyzed 19 clinical isolates of the family Enterobacteriaceae (16 Escherichia coli isolates and 3 Klebsiella pneumoniae isolates) collected from four different hospitals in Paris, France, from 2000 to 2002. These strains had a particular extended-spectrum cephalosporin resistance profile characterized by a higher level of resistance to cefotaxime and aztreonam than to ceftazidime. The blaCTX-M genes encoding these β-lactamases were involved in this resistance, with a predominance of blaCTX-M-15. Ten of the 19 isolates produced both TEM-1- and CTX-M-type enzymes. One strain (E. coli TN13) expressed CMY-2, TEM-1, and CTX-M-14. blaCTX-M genes were found on large plasmids. In 15 cases the same insertion sequence, ISEcp1, was located upstream of the 5′ end of the blaCTX-M gene. In one case we identified an insertion sequence designated IS26. Examination of the other three blaCTX-M genes by cloning, sequencing, and PCR analysis revealed the presence of a complex sul1-type integron that includes open reading frame ORF513, which carries the bla gene and the surrounding DNA. Five isolates had the same plasmid DNA fingerprint, suggesting clonal dissemination of CTX-M-15-producing strains in the Paris area.
Plasmid-mediated extended-spectrum β-lactamases (ESBLs) are becoming increasingly frequent among clinical isolates of the family Enterobacteriaceae throughout the world. The emergence of new enzyme groups that have a typical ESBL resistance phenotype but that are non-TEM and non-SHV derivatives have recently been reported (11). CTX-M-type β-lactamases constitute a novel group of enzymes encoded by transferable plasmids. This novel family of plasmid-mediated ESBLs has been classified in Ambler class A (2) and in group 2be of the Bush, Jacoby, and Medeiros classification (12). They are capable of hydrolyzing broad-spectrum cephalosporins and are inhibited by clavulanic acid, sulbactam, and tazobactam. They confer a high level of resistance to cefotaxime but have a low level of activity against ceftazidime (37). The first two CTX-M-type enzymes were reported in Europe in 1989 (7, 8). So far, more than 30 CTX-M-type β-lactamases have been identified in various clinical isolates, but mostly in enterobacterial species such as Escherichia coli, Klebsiella pneumoniae, and Salmonella enterica serovar Typhimurium. These enzymes have been classified into four major phylogenetic branches on the basis of their amino acid sequence homologies (10). These clusters are CTX-M-1, CTX-M-2, CTX-M-8, and CTX-M-9. Two of them (the CTX-M-2 and CTX-M-8 types) were recently shown to be similar (95 to 100%) to the chromosomally encoded β-lactamase of Kluyvera ascorbata and Kluyvera georgiana, respectively (19, 29). Although CTX-M-producing strains were initially found in western Europe, they have now been observed over a wide geographic area, including Latin America (9, 10), Asia (14, 20, 23, 24, 27, 40), some parts of eastern Europe (6, 17, 18), and, recently, North America (34). In some countries, CTX-M-type enzymes are the ESBLs most frequently isolated from gram-negative strains (37). They have been involved in several outbreaks (6, 40), although the isolation of CTX-M-producing strains remains sporadic, including in France (16, 30, 32).
bla genes are often associated with transferable plasmids, and some of them are parts of transposons or constitute cassettes in integrons. The association of insertion sequences (e.g., ISEcp1) that mobilize β-lactamase-encoding genes with these β-lactamase genes may be involved in their dissemination and expression (13). Insertion sequences such as IS26 (32) and IS903 (13, 27, 32) have also been associated with blaCTX-M genes. Unusual class 1 integrons, similar to In6 and In7, have been reported to carry antibiotic resistance genes such as blaDHA-1 (39). These sul1-type integrons with similar genetic organizations contain a partial duplication of the 3′ conserved segment (3′-CS) and have an open reading frame (ORF), ORF513, between the two sul1 genes. Part of a novel complex sul1-type integron (In60), including the blaCTX-M-9 gene and its downstream nucleotide sequence, was recently characterized (31). blaCTX-M-2 and the surrounding DNA, which includes ORF513, have been reported in the complex sul1-type integron, designated In35 (3) and InS21 (15). Here we report on the diffusion of various CTX-M-type β-lactamases among clinical isolates of the family Enterobacteriaceae in Paris, France.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The 19 strains of the family Enterobacteriaceae (16 E. coli strains and 3 K. pneumoniae strains) studied here are listed in Table 1; they were isolated in four different hospitals between 2000 and 2002 and were identified with API 20E systems (Bio-Mérieux SA, Marcy l'Etoile, France). E. coli J53-2 (pro met Rifr) and E. coli DH10B (Invitrogen SARL, Cergy-Pontoise, France) were used for resistance transfer assays (conjugation and electroporation).
TABLE 1.
Cefotaxime-resistant clinical strains
| Strain | Date of isolation (mo/day/yr or mo/yr) | Hospital | Specimen | Ward |
|---|---|---|---|---|
| E. coli TN03 | 03/26/2001 | Tenon | Urine | Orthopedic surgery |
| E. coli TN05 | 07/04/2001 | Tenon | Vaginal swab | Obstertics |
| E. coli TN06 | 09/30/2001 | Tenon | Urine | Cardiology |
| E. coli TN07 | 10/03/2001 | Tenon | Urine | Urology |
| E. coli TN08 | 07/31/2001 | Tenon | Urine | Emergency |
| E. coli TN09 | 11/24/2001 | Tenon | Urine | Emergency |
| E. coli TN12 | 12/19/2001 | Tenon | Rectal swab | Nephrology ICUa |
| E. coli TN13 | 02/22/2002 | Tenon | Blood | Digestive surgery |
| E. coli TN14 | 03/12/2002 | Tenon | Urine | Nephrology |
| E. coli TN15 | 04/08/2002 | Tenon | Vaginal swab | Maternity outpatient |
| E. coli TN16 | 05/29/2002 | Tenon | Rectal swab | Nephrology ICU |
| E. coli TN17 | 06/04/2002 | Tenon | Rectal swab | Nephrology ICU |
| E. coli TN18 | 03/05/2002 | Tenon | Urine | Surgical ICU |
| E. coli TN19 | 08/18/2002 | Tenon | Urine | Infectious diseases |
| E. coli RB-01 | 04/2002 | R. Ballanger | Urine | Medical ward |
| E. coli LM-01 | 07/2002 | L. Mourier | Rectal swab | ICU |
| K. pneumoniae KP-LT | 06/2000 | Saint-Michel | Urine | Surgery |
| K. pneumoniae KP-02 | 03/21/2002 | Tenon | Wound | Respiratory ICU |
| K. pneumoniae KP-03 | 06/03/2002 | Tenon | Urine | Nephrology ICU |
ICU, intensive care unit.
Antibiotic susceptibility.
Susceptibilities to antimicrobial agents that are usually active against the Enterobactericaeae were determined by an antibiotic disk (Bio-Rad, Marnes-la-Coquette, France) diffusion method on Mueller-Hinton (MH) agar (Bio-Rad). The MICs of the antibiotics, including penicillins and cephalosporins with and without β-lactamase inhibitors (clavulanic acid at 2 μg/ml or tazobactam at 4 μg/ml), were determined by a dilution method on MH agar. An inoculum of 104 CFU per spot was delivered with a multipoint inoculator. ESBLs were detected by using the standard disk synergy test (21).
β-Lactam resistance transfer assays.
Mating experiments were performed with E. coli J53-2 (met pro Rifr). One-milliliter volumes of cultures of each donor and the rifampin-resistant E. coli recipient strain grown in Trypticase soy broth (Bio-Rad) were mixed and incubated for 18 h at 37°C. Transconjugants were then selected on Drigalski (Bio-Rad) agar plates containing rifampin (250 μg · ml−1) and cefotaxime (2.5 μg · ml−1).
E. coli strains liable to produce colicins were plated on MH agar that had been plated with E. coli J53-2 (32).
Plasmid DNA was isolated by the method of Takahashi and Nagano (35), and 2 μl was transformed into 20 μl of E. coli DH10B cells by electroporation according to the instructions of the manufacturer (Bio-Rad). Transformants were incubated for 1.5 h at 37°C and mated on Drigalski medium supplemented with cefotaxime (2.5 μg · ml−1) or cefoxitin (40 μg · ml−1).
Plasmid DNA analysis.
Plasmid DNA from the K. pneumoniae and E. coli isolates and their corresponding E. coli transconjugants and transformants was obtained as described by Takahashi and Nagano (35) and Kado and Liu (22). Plasmid DNA was detected by electrophoresis in a 0.8% agarose gel. The molecular sizes of plasmid DNAs were estimated by comparison with the following plasmids of known sizes from the Institute Pasteur Collection: pIP112 (100.5 kb), pCFF04 (85 kb), and pIP173 (125.8 kb).
IEF of β-lactamases.
Bacteria growing exponentially at 37°C in 50 ml of Trypticase soy broth were pelleted and sonicated with a Vibracell apparatus (twice for 30 s each time, 60 W). Isoelectric focusing (IEF) was performed on a pH 3.5 to 10 ampholine polyacrylamide gel. β-Lactamase activity was detected with the chromogen nitrocefin (Oxoid, Dardilly, France) (25). The pI values used as standards were those of TEM-1 (pI 5.4), TEM-2 (pI 5.6), SHV-4 (pI 7.8), and CMY-2b (pI 9.3).
Characterization of β-lactamase-encoding (bla) genes.
Detection of gene sequences coding for the TEM, CMY, and CTX-M-type enzymes was performed by PCR with genomic DNA. The oligonucleotide primer sets specific for the β-lactamase genes used in the PCR assays are listed in Table 2. To characterize the 3′ end of blaCTX-M-15, we used primers ORF1 pol M3 and M3 int upp (Table 2); primer ORF1 pol M3 was designed by using the sequence of ORF1 described downstream of the CTX-M-3 gene of Citrobacter freundii (GenBank accession no. AF550415). The blaTEM genes were characterized by PCR-restriction fragment length polymorphism (RFLP) analysis (4).
TABLE 2.
Sequences of the primers used to detect bla genes and their promoter regions
| PCR target | Primer name | Primer sequencea |
|---|---|---|
| blaCTX-M (CTX-M-1 group) | M13 upper | 5′-GGT TAA AAA ATC ACT GCG TC-3′ |
| M13 lower | 5′-TTG GTG ACG ATT TTA GCC GC-3′ | |
| blaCTX-M (CTX-M-2 group) | M25 upper | 5′-ATG ATG ACT CAG AGC ATT CG-3′ |
| M25 lower | 5′-TGG GTT ACG ATT TTC GCC GC-3′ | |
| blaCTX-M (CTX-M-9 group) | M9 upper | 5′-ATG GTG ACA AAG AGA GTG CA-3′ |
| M9 lower | 5′-CCC TTC GGC GAT GAT TCT C-3′ | |
| blaTEM | OT3 | 5′-ATG AGT ATT CAA CAT TTC CG-3′ |
| OT4 | 5′-CCA ATG CTT AAT CAG TGA GG-3′ | |
| blaCMY | CF1 | 5′-ATG ATG AAA AAA TCG TTA TGC-3′ |
| CF2 | 5′-TTG TAG CTT TTC AAG AAT GCG C-3′ | |
| tnpISEcp1b | ISEcp1 | 5′-AAA AAT GAT TGA AAG GTG GT-3′ |
| MA1 reverse | 5′-ACY TTA CTG GTR CTG CAC AT-3′ | |
| tnpIS26 | IS26 | 5′-AGC GGT AAA TCG TGG AGT GA-3′ |
| MA1 reverse | 5′-ACY TTA CTG GTR CTG CAC AT-3′ | |
| IS903 | IS903 reverse | 5′-CGG TTG TAA TCT GTT GTC CA-3′ |
| M1 IS903 | 5′-CGT CGG CGG CTA AAA TCG TC-3′ | |
| M9 IS903 | 5′-CTA CGG CAC CAC CAA TGA TA-3′ | |
| ORF513 | ORF513 | 5′-TGG AAG AGG GCG AAG ACG AT-3′ |
| M2 reverse upper | 5′-CGA ATG CTC TGA GTC ATC AT-3′ | |
| M9 reverse upper | 5′-TGC ACT CTC TTT GTC ACC AT-3′ | |
| ORF1005 | ORF1005 reverse | 5′-ATC CAT AAT AGC ATC CAT CAT-3′ |
| M9 reverse lower | 5′-GAG AAT CAT CGC CGA AGG G-3′ | |
| sul1 | sul1 reverse | 5′-GCT CAA GAA AAA TCC CAT CCC C-3′ |
| M2 reverse lower | 5′-GCG GCG AAA ATC GTA ACC CA-3′ | |
| M9 reverse lower | 5′-GAG AAT CAT CGC CGA AGG G-3′ | |
| ORF1 | M3 int upp | 5′-TCA CCC AGC CTC AAC CTA AG-3′ |
| ORF1 pol M3 | 5′-GCA CCG ACA CCC TCA CAC CT-3′ |
Y = C or T; R = A or G.
tnp, transposase.
The PCR products of blaCMY and blaCTX-M were subjected to direct sequencing by the dideoxy chain termination method of Sanger et al. (33). Both strands of the PCR products were sequenced twice with an Applied Biosystems sequencer (model ABI 377).
The nucleotide sequences and deduced protein sequences were analyzed with the BLAST and Clustal W programs (for preparation of multiple-sequence alignments, pairwise comparisons of sequences, and preparation of dendrograms) (1, 36).
Genetic environment of blaCTX-M genes.
The genetic organization of the blaCTX-M genes was investigated by PCR, cloning, and sequencing of the regions surrounding these genes. The internal IS26 and ISEcp1 forward primers and the CTX-M reverse consensus primer (MA1 reverse) were used to investigate the promoter regions of the blaCTX-M genes. PCR primers corresponding to sequences upstream of the blaCTX-M genes (ORF513) and downstream of the blaCTX-M genes (IS903, ORF1005, ORF1, and sul1) were also used.
β-Lactamase gene cloning was performed with plasmid DNA digested and ligated in the EcoRI or HindIII (Ozyme; New England Biolabs Inc., Saint Quentin en Yvelines, France) site of phagemid pBK-CMV (Stratagene, La Jolla, Calif.). E. coli DH10B was transformed by electroporation. The transformants harboring the recombinant CTX-M-encoding plasmids were selected on MH agar supplemented with 2.5 μg of cefotaxime per ml and 25 μg of kanamycin per ml. The molecular sizes of the inserts were estimated from the results of restriction enzyme digestion and electrophoresis in 1% agarose gels. Finally, inserts were investigated by sequencing the ends and then by PCR.
Rep-PCR and ERIC-PCR.
DNA was extracted by using the Qiagen Mini kit (Qiagen, Courtaboeuf, France). Repetitive extragenic palindromic sequence PCR (Rep-PCR) was performed with primers rep-1R and rep-2T; enterobacterial repetitive intergenic repetitive consensus sequence PCR (ERIC-PCR) was performed with primer ERIC-2, as described previously (26). The resulting products were run in 1.5% agarose gels.
Plasmid DNA fingerprinting.
Plasmid DNA was purified from transformant cells with the Qiagen Plasmid Midi kit (Qiagen), according to the recommendations of the manufacturer. For fingerprinting analysis, plasmid DNA was digested with the BamHI restriction enzyme (New England Biolabs Inc.) and subjected to electrophoresis in a 1% agarose gel at 80 V for 4 h.
RESULTS
Description of clinical isolates.
Sixteen E. coli strains and 3 K. pneumoniae strains were recovered between June 2000 and August 2002. Most isolates were isolated at Tenon Hospital. The strains were associated with urinary tract infections (11 strains), blood infection (1 strain), a wound infection (1 strain), and vaginal or gastrointestinal colonization (6 strains) (Table 1).
β-Lactam susceptibility profile and associated resistance (Table 3).
TABLE 3.
Characteristics of ESBL-producing clinical isolates and their transformants, including β-lactam MICs for the clinical strains, their E. coli J53-2 transconjugants, and E. coli DH10B transformants and non β-lactam antibiotic resistance markers
| Straina | pI by IEF | Result by:
|
CTX-M identified by sequencing | MIC (μg/ml)b
|
Associated antibiotic resistance markersc | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR for blaCTX-M | PCR for blaCMY | PCR for blaTEM | PCR-RFLP analysis | CTX | CTX-CA | CAZ | CAZ-CA | FEP | FEP-CA | ATM | ATM-CA | IPM | ||||
| TN03 | 5.4, 7.6 | M-1/M-3 | + | TEM-1 | CTX-M-15 | 128 | 0.06 | 64 | 0.5 | 128 | 0.12 | 128 | 0.06 | 0.12 | G, T, Nt, Te | |
| Ep TN03 | 5.4, 7.6 | + | 64 | 32 | 32 | 64 | G, T, Nt, Te | |||||||||
| TN05 | 8.4 | M-9 | − | CTX-M-9 | 32 | 0.12 | 0.5 | 0.03 | 32 | 0.12 | 4 | 0.06 | 0.12 | G, T, Nt, Te, Tp, Su | ||
| Ep TN05 | 8.4 | 32 | 0.5 | 4 | 4 | G, T, Nt, Tp, Su | ||||||||||
| TN06 | 5.4, 8.2 | M-2/M-5 | − | + | TEM-1 | CTX-M-2 | 128 | 0.5 | 64 | 0.5 | 64 | 1 | 128 | 1 | 0.12 | G, T, Nt, Te, C, Tp, Su |
| Ep TN06 | 5.4, 8.2 | + | 64 | 0.5 | 32 | 64 | G, T, Nt, Su | |||||||||
| TN07 | 8.4 | M-9 | − | CTX-M-14 | 32 | 0.12 | 0.5 | 0.12 | 4 | 0.12 | 8 | 0.06 | 0.12 | G, T, Te, C, Tp, Su | ||
| Ep TN07 | 8.4 | 4 | 0.5 | 4 | 4 | None | ||||||||||
| TN08 | 5.4, 7.6 | M-1/M-3 | + | TEM-1 | CTX-M-15 | 128 | 0.25 | 64 | 2 | 128 | 0.12 | 64 | 0.25 | 0.12 | T, Nt, A, Te, Tp, Su | |
| Tc TN08 | 5.4, 7.6 | + | 32 | 32 | 16 | 16 | T, Nt, A, Te, Tp, Su | |||||||||
| TN09 | 5.4, 7.6 | M-1/M-3 | + | TEM-1 | CTX-M-15 | 128 | 0.25 | 64 | 2 | 64 | 0.12 | 128 | 0.25 | 0.12 | G, T, Nt, Te | |
| Ep TN09 | 5.4, 7.6 | + | 128 | 64 | 64 | 128 | G, T, Nt, Te | |||||||||
| TN12 | 5.4, 7.6 | M-1/M-3 | + | TEM-1 | CTX-M-15 | 128 | 0.25 | 64 | 1 | 32 | 0.5 | 64 | 0.5 | 0.12 | G, T, Nt, Te, C, Tp, Su | |
| Tc TN12 | 5.4, 7.6 | + | 64 | 32 | 32 | 64 | C | |||||||||
| TN13 | 5.4, 8.4, >9 | M-9 | + | + | TEM-1 | CTX-M-14 | 128 | 32 | 128 | 64 | 16 | 1 | 32 | 32 | 0.25 | Te, C |
| Tc TN13 | 8.4 | − | − | 4 | 0.12 | 0.5 | 0.5 | C | ||||||||
| TN14 | 5.4, 7.6 | M-1/M-3 | + | TEM-1 | CTX-M-15 | 128 | 0.5 | 64 | 2 | 128 | 0.25 | 128 | 0.5 | 0.12 | G, T, Nt, Te | |
| Ep TN14 | 5.4, 7.6 | + | 128 | 64 | 64 | 128 | G, T, Nt, Te | |||||||||
| TN15 | 5.4, 7.6 | M-1/M-3 | + | TEM-1 | CTX-M-15 | 128 | 0.25 | 64 | 1 | 32 | 0.5 | 64 | 0.5 | 0.12 | G, T, Nt, Te, C, Tp, Su | |
| Tc TN15 | 5.4, 7.6 | + | 32 | 32 | 16 | 16 | G, T, Nt, Te, Tp, Su | |||||||||
| TN16 | 7.8 | M-1/M-3 | − | CTX-M-1 | 32 | 0.25 | 2 | 0.25 | 8 | 0.12 | 16 | 0.12 | 0.12 | Tp, Su | ||
| Ep TN16 | 7.8 | 32 | 2 | 8 | 16 | Tp, Su | ||||||||||
| TN17 | 5.4, 7.6 | M-1/M-3 | + | TEM-1 | CTX-M-15 | 64 | 16 | 32 | 8 | 32 | 16 | 64 | 32 | 0.12 | G, T, Nt, Te, C, Tp, Su | |
| Tc TN17 | 7.6 | − | 8 | 2 | 4 | 8 | G, T, Nt, Te | |||||||||
| TN18 | 5.4, 7.6 | M-1/M-3 | + | TEM-1 | CTX-M-15 | 128 | 32 | 64 | 32 | 64 | 16 | 128 | 32 | 0.12 | G, T, Nt, Te | |
| Ep TN18 | 5.4, 7.6 | + | 64 | 32 | 64 | 64 | G, T, Nt, Te | |||||||||
| TN19 | 8.2 | M-2/M-5 | − | CTX-M-2 | 128 | 4 | 16 | 2 | 64 | 4 | 128 | 4 | 0.25 | Te, Tp, Su | ||
| Ep TN19 | 8.2 | 64 | 16 | 64 | 64 | Te, Tp, Su | ||||||||||
| RB-01 | 7.8 | M-1/M-3 | − | CTX-M-1 | 64 | 0.06 | 4 | 0.12 | 64 | 0.12 | 32 | 0.06 | 0.25 | Te, C, Tp, Su | ||
| Tc RB-01 | 7.8 | 4 | 0.12 | 4 | 2 | Tp, Su | ||||||||||
| LM-01 | 7.6 | M-1/M-3 | − | CTX-M-15 | >128 | 4 | 64 | 2 | 64 | 2 | 128 | 2 | 0.12 | T, Nt, A, Te | ||
| Tc LM-01 | 7.6 | 64 | 16 | 32 | 32 | T, Nt, A, Te | ||||||||||
| KP-LT | 8 | M-1/M-3 | − | CTX-M-3 | 32 | 2 | 1 | 4 | 16 | 0.25 | 32 | 0.5 | 0.12 | G, T, Nt, Te, C, Tp, Su | ||
| Tc KP-LT | 8 | 4 | 1 | 1 | 1 | G, T, Nt, Tp | ||||||||||
| KP-02 | 7.8 | M-1/M-3 | − | CTX-M-1 | 4 | 0.06 | 0.25 | 0.25 | 2 | 0.12 | 1 | 0.03 | 0.12 | Tp, Su | ||
| Tc KP-02 | 7.8 | 4 | 1 | 1 | 1 | Tp | ||||||||||
| KP-03 | 5.4, 7.6 | M-1/M-3 | + | TEM-1 | CTX-M-15 | >128 | 4 | >128 | 32 | 128 | 4 | >128 | 8 | 0.25 | G, T, Nt, A, C, Tp, Su | |
| Ep KP-03 | 5.4, 7.6 | + | 128 | 64 | 16 | 128 | T, Nt, A, Tp, Su | |||||||||
Ep, electroporant; Tc, transconjugant.
CTX, cefotaxime; CA, clavulanic acid (2 μg/ml); CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; IPM, imipenem.
G, gentamicin; T, tobramycin; Nt, netilmicin,; A, amikacin; Te, tetracycline; C, chloramphenicol; Tp, trimethoprim; Su, sulfamides.
All the strains were resistant to penicillins at high concentrations (MICs, ≥128 μg/ml), but clavulanic acid and tazobactam partially restored the activities of amoxicillin (MICs, 4 to 16 μg/ml) and piperacillin (MICs, 2 to 8 μg/ml) against all except three strains (strains TN13, TN17, and TN18). One of these strains (strain TN13) was also resistant to cefoxitin (MIC, 128 g/ml), but its transconjugant was not. All but one of the strains (strain TN13) had a higher level of resistance to cefotaxime, cefepime, and aztreonam than to ceftazidime. Similar results were observed with the transconjugants or electroporants. The disk diffusion method showed synergy between ceftazidime, cefotaxime, aztreonam, cefepime, and clavulanic acid against all the strains and their transconjugants or electroporants: these results agreed with the MICs of the extended-spectrum cephalosporins and aztreonam combined with clavulanic acid. The non-β-lactam antibiotic resistance markers are also listed in Table 3. Fourteen isolates were resistant to aminoglycosides, and this resistance was transferred for 12 of them.
Transfer of resistance.
Cefotaxime resistance transfer by conjugation was obtained for only seven E. coli isolates and two K. pneumoniae isolates. The probable production of colicins by three E. coli isolates (isolates TN07, TN16, and TN19), suggested by observation of a growth inhibition zone, possibly explained why the mating-out assays failed to yield transconjugants of these three strains. Electroporation of plasmid DNA from the other 10 strains into E. coli DH10B successfully transferred cefotaxime resistance. Cefoxitin resistance was not cotransferred with the ESBL in E. coli TN13 but was independently transferred by electroporation by selection on cefoxitin.
Plasmids encoding ESBLs.
Plasmid DNA was isolated from all the strains and their transconjugants or electroporants. All 19 wild-type isolates had one or more plasmids. Analysis of E. coli transconjugants or electroporants expressing ESBLs revealed the presence of large plasmids, with estimated molecular sizes of 80 to >130 kb (data not shown).
IEF.
All clinical strains and their transconjugants or electroporants exhibited a band of β-lactamase activity with an alkaline pI (7.6 to 8.4). In addition to enzymes with these pIs, 11 isolates had another band of β-lactamase activity with a pI of 5.4. One strain (strain TN13) also had an additional band of β-lactamase activity with a pI of >9. IEF of sonic extracts of E. coli transconjugants or electroporants revealed that all but two strains (strains TN13 and TN17) transferred the β-lactamase activity with a pI of 5.4 (Table 3).
Characterization of β-lactamase-encoding (bla) genes.
The results of PCR and sequence analysis are summarized in Table 3. PCR experiments were positive for blaCTX-M for all isolates and their transconjugants or electroporants. These results corresponded to pI values of 7.6 to 8.4 for CTX-M-type enzymes.
Sequence analysis of the deduced amino acid sequences showed the presence of various CTX-M-type enzymes: CTX-M-1, -2, -3, -9, and -14 and particularly CTX-M-15 (with a glycine at position 240) (23), produced by TN03, TN08, TN09, TN12, TN14, TN15, TN17, TN18, LM-01, and KP-03.
The 11 isolates harboring the enzyme with a pI of 5.4 were found to carry TEM-1 by PCR-RFLP analysis. Amplification was obtained with the CMY-specific primers for E. coli TN13 producing a β-lactamase of pI >9, and sequencing showed that this β-lactamase was CMY-2.
Exploration of the regions surrounding blaCTX-M genes.
PCR identified the insertion sequence ISEcp1 upstream of the blaCTX-M gene in 15 strains (Table 4). The sizes of the PCR products were about 0.8 kb in all except one strain (strain TN13). For this strain, the PCR fragment was about 2.4 kb, suggesting the insertion of an additional DNA fragment. The upstream region of blaCTX-M-15 in E. coli strain TN17, analyzed by PCR, contained the transposase gene of the insertion sequence IS26. PCR with the IS903-specific primer produced no amplicons.
TABLE 4.
Genotypic characterization of blaCTX-M surrounding DNA
| Strain | Distance (kb) by PCR with the indicated primer:
|
||||||
|---|---|---|---|---|---|---|---|
| Upstream
|
Downstream
|
||||||
| ISEcpl | IS26 | ORF513 | IS903 | ORF1 | sul1 reverse | ORF1005 | |
| E. coli TN03 | +0.8 | +0.45 | |||||
| E. coli TN05 | +1.1 | +1.6 | |||||
| E. coli TN06 | +1.3 | +2 | |||||
| E. coli TN07 | +0.8 | ||||||
| E. coli TN08 | +0.8 | +0.45 | |||||
| E. coli TN09 | +0.8 | +0.45 | |||||
| E. coli TN12 | +0.8 | +0.45 | |||||
| E. coli TN13 | +2.4 | ||||||
| E. coli TN14 | +0.8 | +0.45 | |||||
| E. coli TN15 | +0.8 | +0.45 | |||||
| E. coli TN16 | +0.8 | ||||||
| E. coli TN17 | +0.6 kb | +0.45 | |||||
| E. coli TN18 | +0.8 kb | +0.45 | |||||
| E. coli TN19 | +2 | ||||||
| E. coli RB-01 | +0.8 kb | +0.45 | |||||
| E. coli LM-01 | +0.8 kb | +0.45 | |||||
| K. pneumoniae KP-LT | +0.8 kb | +0.45 | |||||
| K. pneumoniae KP-02 | +0.8 kb | ||||||
| K. pneumoniae KP-03 | +0.8 kb | +0.45 | |||||
The genetic organization of the blaCTX-M genes of E. coli TN05, TN06, and TN19 was investigated by cloning, partial sequencing of the regions surrounding these genes, and PCR analysis. Upstream of the blaCTX-M gene, these strains harbored a region common to the complex sul1-type integron (38, 39). This region includes ORF513, which is present in In6 and In7. Analysis of the nucleotide sequence of the region downstream of the blaCTX-M-9 gene from E. coli TN05 revealed the presence of ORF1005. In strains TN06 and TN19, we found part of the 3′-CS complex sul1-type integron downstream of the blaCTX-M-2 stop codon.
Epidemiological analysis.
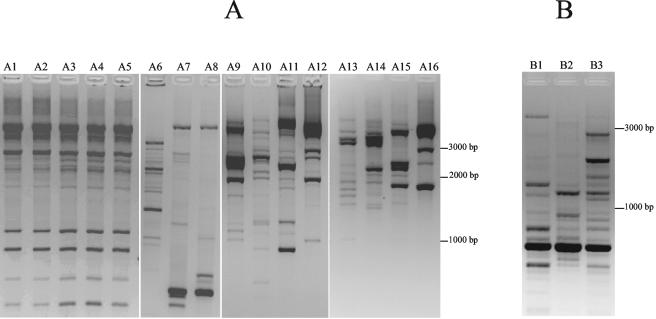
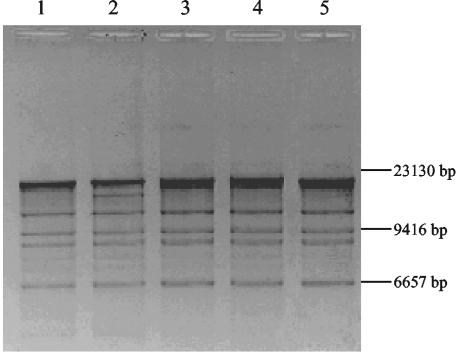
ERIC-PCR and Rep-PCR analyses were used to analyze the molecular epidemiology of the 19 clinical isolates. Rep-PCR showed that five E. coli clinical isolates were clonally related (Fig. 1A). Plasmids isolated from transconjugants or electroporants of these five E. coli strains yielded similar restriction patterns after digestion with BamHI (Fig. 2) These strains were isolated from five different patients with no apparent relationships in time or space. ERIC-PCR of the three K. pneumoniae strains gave different restriction patterns (Fig. 1B).
FIG. 1.
Fingerprinting analysis of ESBL-producing strains. (A) Rep-PCR of E. coli clinical isolates carrying blaCTX-M genes. Lane A1, E. coli TN18; lane A2, E. coli TN03; lane A3, E. coli TN08; lane A4, E. coli TN09; lane A5, E. coli TN14; lane A6, E. coli TN15; lane A7, E. coli RB-01; lane A8, E. coli TN12; lane A9, E. coli TN13; lane A10, E. coli TN05; lane A11, E. coli TN06; lane A12, E. coli TN07; lane A13, E. coli TN16; lane A14, E. coli TN17; lane A15, E. coli LM-01; lane A16, E. coli TN19. (B) ERIC-PCR of K. pneumoniae isolates. Lane B1, K. pneumoniae KP-LT; lane B2, K. pneumoniae KP-02; lane B3, K. pneumoniae KP-03.
FIG. 2.
BamHI-digested plasmid profiles of transconjugants or electroporants producing CTX-M-type β-lactamases. Lane 1, E. coli TN03; lane 2, E. coli TN08; lane 3, E. coli TN09; lane 4, E. coli TN14; lane 5, E. coli TN18.
DISCUSSION
We studied 19 enterobacterial strains collected in four Paris hospitals between June 2000 and August 2002. The antimicrobial susceptibility patterns showed that the strains harbored ESBLs responsible for resistance to penicillin, broad-spectrum cephalosporins, and aztreonam. The ESBLs were inhibited by clavulanate and tazobactam. Importantly, the isolates were more resistant to cefotaxime and aztreonam than to ceftazidime, suggesting that they were CTX-M producers. Some CTX-M ESBLs confer high-level resistance to ceftazidime, and this was the case for 10 of our strains. Cefotaxime resistance could be transferred by conjugation for 9 of the 19 isolates and by electroporation for 10 isolates. The transconjugants or electroporants showed similar resistance profiles.
We used primers specific to each cluster of CTX-M-type enzymes. We obtained amplification of all the strains, confirming the presence of such enzymes. Analysis of the deduced amino acid sequences showed the diversity of the CTX-M-type enzymes. Indeed, we found six different enzymes, including CTX-M-15 (23), produced by 10 isolates. This member of the CTX-M family showed increased activity against ceftazidime. Sequence analysis revealed an Asp-240 Gly substitution. This substitution has already been reported in CTX-M-16 and is known to confer high-level resistance to ceftazidime (9, 23). Another substitution known to increase the hydrolyzing activity of ceftazidime is due to the Pro-167 Ser substitution in CTX-M-19 (30).
Of the 19 strains producing cefotaximases and screened for the presence of TEM β-lactamases genes, 11 were found to express TEM-1. This association is frequent and has already been described in the literature (9, 30, 32). Lastly, E. coli strain TN13, which had a high level of resistance to cefoxitin (MIC, 128 mg · liter−1), harbored both CTX-M-14 and CMY-2; to our knowledge this is only the third report of such a combination (5, 40). All the strains yielded an enzyme with an alkaline pI, as observed for the CTX-M-type enzymes. Some had an additional band of β-lactamase activity with a pI of 5.4 (11 strains) and, in one case (strain TN13), a third band of pI >9; these bands correspond to TEM-1 and CMY-2, respectively.
Previously reported analyses of the surrounding regions have shown the frequent association of β-lactamase genes with the insertion sequence ISEcp1. This element was first described upstream of blaCMY-4 (P. D. Stapleton, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1457, 1999), and then ISEcp1 was detected upstream of several blaCTX-M genes (13, 14, 16, 23, 32). Of the 19 clinical isolates studied here, 15 had this same insertion sequence upstream of the blaCTX-M gene. Another insertion sequence, IS26, was described by Saladin et al. (32) to be upstream of a blaCTX-M-1 gene. This structure was found upstream of the blaCTX-M gene in one of our strains (strain TN17). It is worth mentioning that none of the 19 strains harbored the IS903 insertion sequence. Interestingly, PCR amplification with primers specific for the tnpA genes of ISEcp1 or IS26 was negative for three strains. Analysis of the surrounding regions after cloning and end sequencing allowed us to draw some primers. PCR analysis showed the presence of a structure-type integron. The sizes of the fragments were compatible with the genetic organization described in the literature for two blaCTX-M-2 sequences and one blaCTX-M-9 sequence inserted in novel complex sul1-type integrons In35, InS21, and In60, respectively (3, 15, 31). Our three blaCTX-M genes (two blaCTX-M-2 genes and one blaCTX-M-9 gene) were located in unusual class 1 integrons. ORF1005 was found in the 3′ end of blaCTX-M-9 (strain TN05) when PCR amplification with the sul1-specific primer was positive with strains TN06 and TN19. ORF513 was found at the 5′ ends of these bla genes in these three strains and has already been reported; ORF513 may be a transposase (38). This is the first report of a blaCTX-M gene in a complex sul1-type integron in France. These findings confirm the diversity of transposable elements and integrons associated with blaCTX-M genes.
Ten of the 19 strains harbored a CTX-M-15 enzyme, enabling us to determine whether these clinical isolates were genetically related. The results of Rep-PCR strongly suggested that five E. coli isolates with no epidemiological relationship were identical. In addition, plasmids extracted from transconjugants or electroporants yielded similar restriction patterns, supporting evidence of the dissemination of these enterobacterial strains. The corresponding patients were from three long-term-care facilities near Tenon Hospital, suggesting the presence of this enzyme in these institutions (28).
CTX-M β-lactamases constitute a novel and rapidly growing family of plasmid-mediated ESBLs. Outbreaks have been described in several countries (6, 40). These enzymes have been reported at a much lower frequency in France (16, 30, 32). However, CTX-M β-lactamase producers represented 50% of the ESBL-harboring E. coli clinical isolates in Tenon Hospital in 2001 and 2002 (unpublished data). A previous study described 9 CTX-M-producing strains collected over 11 years (32), while we found 19 CTX-M-producing isolates of the Enterobacteriaceae during a 2-year survey of four Paris hospitals. This work confirms the emergence of CTX-M-type enzymes and their spread in the Paris area.
Acknowledgments
This work was financed by grants from Ministère de la Recherche (réseau bêta-lactamase) and Faculté de Médecine Saint-Antoine, Université Paris VI.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. GappedBLAST and PSI-LAST: a new generation of protein data-base search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambler, R. P. 1980. The structure of β-lactamase. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 289:321-331. [DOI] [PubMed] [Google Scholar]
- 3.Arduino, S. M., P. H. Roy, G. A. Jacoby, B. E. Orman, S. A. Pineiro, and D. Centron. 2002. blaCTX-M is located in an unusual class 1 integron (In35) which includes Orf513. Antimicrob. Agents Chemother. 46:2303-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arlet, G., G. Brami, D. Decrè, A. Flippo, O. Gaillot, P. H. Lagrange, and A. Philippon. 1995. Molecular characterization by PCR-restriction fragment length polymorphism of TEM β-lactamases. FEMS Microbiol. Lett. 134:203-208. [DOI] [PubMed] [Google Scholar]
- 5.Armand-Lefèvre, L., V. Leflon-Guibout, J. Bredin, F. Barguellil, A. Amor, J. M. Pagès, and M. H. Nicolas-Chanoine. 2003. Imipenem resistance in Salmonella enterica serovar Wien related to porin loss and CMY-4 β-lactamase production. Antimicrob. Agents Chemother. 47:1165-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baraniak, A., J. Fiett, A. Sulikowska, W. Hryniewicz, and M. Gniadkowski. 2002. Countrywide spread of CTX-M-3 extended-spectrum β-lactamase-producing microorganisms of the family Enterobacteriaceae in Poland. Antimicrob. Agents Chemother. 46:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barthélémy, M., J. Péduzzi, H. Bernard, C. Tancrède, and R. Labia. 1992. Close amino-acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim. Biophys. Acta 1122:15-22. [DOI] [PubMed] [Google Scholar]
- 8.Bauerfeind, A., H. Grimm, and S. Schweighart. 1990. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18:294-298. [DOI] [PubMed] [Google Scholar]
- 9.Bonnet, R., C. Dutour, J. L. M. Sampaio, C. Chanal, D. Sirot, R. Labia, C. De Champs, and J. Sirot. 2001. Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240 Gly. Antimicrob. Agents Chemother. 45:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnet, R., J. L. M. Sampaio, R. Labia, C. De Champs, D. Sirot, C. Chanal, and J. Sirot. 2000. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao, V., T. Lambert, and P. Courvalin. 2002. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum β-lactamase CTX-M-17. Antimicrob. Agents Chemother. 46:1212-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chanawong, A., F. Hannachi M'Zali, J. Heritage, J. H. Xiong, and P. M. Hawkey. 2002. Three cefotaximases, CTX-M-9, CTX-M-13, and CTX-M-14, among Enterobacteriaceae in the People's Republic of China. Antimicrob. Agents Chemother. 46:630-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Conza, J., J. A. Ayala, P. Power, M. Mollerach, and G. Gutkind. 2002. Novel class 1 integron (InS21) carrying blaCTX-M-2 in Salmonella enterica serovar Infantis. Antimicrob. Agents Chemother. 46:2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutour, C., R. Bonnet, H. Marchandin, M. Boyer, C. Chanal, D. Sirot, and J. Sirot. 2002. CTX-M-1, CTX-M-3, and CTX-M-14 β-lactamases from Enterobacteriaceae isolated in France. Antimicrob. Agents Chemother. 46:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gazouli, M., S. V. Sidorenko, E. Tzelepi, N. S. Kozlova, D. P. Gladin, and L. S. Tzouvelekis. 1998. A plasmid-mediated β-lactamase conferring resistance to cefotaxime in a Samonella typhimurium clone found in St Petersbourg, Russia. J. Antimicrob. Chemother. 41:119-121. [DOI] [PubMed] [Google Scholar]
- 18.Gniadkowski, M., I. Schneider, A. Palucha, R. Jungwirth, B. Mikiewicz, and A. Bauernfeind. 1998. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishii, Y., A. Ohno, H. Taguchi, S. Imajo, M. Ishiguro, and H. Matsuzawa. 1995. Cloning and sequence of the gene encoding a cefotaximase-hydrolyzing class A β-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 39:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended-broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 22.Kado, C. I., and S. T. Liu. 1981. A rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 24.Ma, L., Y. Ishii, M. Hishiguro, H. Matsuzawa, and K. Yamaguchi. 1998. Cloning and sequencing of the gene encoding Toho-2, a class A β-lactamase preferentially inhibited by tazobactam. Antimicrob. Agents Chemother. 42:1181-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthew, M., and R. W. Hedges. 1976. Analytical isoelectric focusing of R factor-determined β-lactamases: correlation with plasmid compatibility. J. Bacteriol. 125:713-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pal, H., E. H. Choi, H. J. Lee, J. Y. Hong, and G. A. Jacoby. 2001. Identification of CTX-M-14 extended-spectrum β-lactamase in clinical isolates of Shigella sonnei, Escherichia coli, and Klebsiella pneumoniae in Korea. J. Clin. Microbiol. 39:3747-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Philippon, A., G. Arlet, and P. H. Lagrange. 1994. Origin and impact of plasmid-mediated extended-spectrum β-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 13(Suppl. 1):17-29. [DOI] [PubMed] [Google Scholar]
- 29.Poirel, L., P. Kampfer, and P. Nordmann. 2002. Chromosome-encoded Ambler class A β-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum β-lactamase. Antimicrob. Agents Chemother. 46:4038-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poirel, L., T. Naas, I. Le Thomas, A. Karim, E. Bingen, and P. Nordmann. 2001. CTX-M-type extended-spectrum β-lactamase that hydrolyses ceftazidime through a single amino acid substitution in the omega loop. Antimicrob. Agents Chemother. 45:3355-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabate, M., F. Navarro, E. Miro, S. Campoy, B. Mirelis, J. Barbé, and G. Prats. 2002. Novel complex sul1-type integron in Escherichia coli carrying blaCTX-M-9. Antimicrob. Agents Chemother. 46:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saladin, M., V. T. B. Cao, T. Lambert, J.-L. Donay, J.-L. Herrmann, Z. Ould-Hocine, C. Verdet, F. Delisle, A. Philippon, and G. Arlet. 2002. Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161-168. [DOI] [PubMed] [Google Scholar]
- 33.Sanger, T., S. Nicklen, and A. R. Coulsen. 1977. DNA sequencing with chain-terrminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith Moland, E., J. A. Black, A. Hossain, N. D. Hanson, K. S. Thomson, and S. Pottumarthy. 2003. Discovery of CTX-M-like extended-spectrum β-lactamases in Escherichia coli isolates from five U.S. states. Antimicrob. Agents Chemother. 47:2382-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi, S., and Y. Nagano. 1984. Rapid procedure for isolation of plasmid DNA and application to epidemiological analysis. J. Clin. Microbiol. 20:608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W; improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzouvelekis, L. S., E. Tzelepi, P. T. Tassios, and N. J. Legakis. 2000. CTX-M-type β-lactamases: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 14:137-142. [DOI] [PubMed] [Google Scholar]
- 38.Valentine, C. R., M. J. Heinrichs, S. L. Chissoe, and A. Roe. 1994. DNA sequence of direct repeats of the sul1 gene of plasmid pSa. Plasmid 32:222-227. [DOI] [PubMed] [Google Scholar]
- 39.Verdet, C., G. Arlet, G. Barnaud, P. H. Lagrange, and A. Philippon. 2000. A novel integron in Salmonella enterica serovar Enteritidis, carrying the blaDHA-1 gene and its regulator gene ampR, originated from Morganella morganii. Antimicrob. Agents Chemother. 44:222-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan, J. J., W. C. Ko, S. H. Tsal, H. M. Wu, Y. T. Jin, and J. J. Wu. 2000. Dissemination of CTX-M-3 and CMY-2 β-lactamases among clinical isolates of Escherichia coli in southern Taiwan. J. Clin. Microbiol. 38:4320-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]