Abstract
Botulism is a neuroparalytic disease that can occur in all warm-blooded animals, birds, and fishes. The disease in animals is mainly caused by toxins produced by Clostridium botulinum strains belonging to group III, although outbreaks due to toxins produced by group I and II organisms have been recognized. Group III strains are capable of producing botulinum toxins of type C, D, and C/D and D/C mosaic variants. Definitive diagnosis of animal botulism is made by combining clinical findings with laboratory investigations. Detection of toxins in clinical specimens and feed is the gold standard for laboratory diagnosis. Since toxins may be degraded by organisms contained in the gastrointestinal tract or may be present at levels below the detection limit, the recovery of C. botulinum from sick animal specimens is consistent for laboratory confirmation. In this article we report the development and in-house validation of a new multiplex real-time PCR for detecting and typing the neurotoxin genes found in C. botulinum group III organisms. Validation procedures have been carried out according to ISO 16140, using strains and samples recovered from cases of animal botulism in Italy and France.
Botulism is a severe and potentially lethal illness caused by exposure to botulinum neurotoxins (BoNTs) produced by Clostridium botulinum and other BoNT-producing clostridia strains.1,2 All warm-blooded animals (including humans), birds, and some fishes, can be affected by the disease but show different levels of susceptibility to the different types of BoNTs.3 Human botulism is mainly associated with types A, B, E, and F toxins, whereas animal botulism is primarily associated with types C, D, C/D, and D/C toxins, although some outbreaks due to types A, B, and E have also been recognized.4-8 Mammals, such as cattle, horses, and sheep, as well as waterfowl and poultry are the animals most often involved in outbreaks.4,9 Cattle botulism is most frequently caused by types D or D/C toxin, followed by types C, A, B, and C/D toxins.4,8,10 Equine botulism is caused by types B, C, and A toxins.11,12 Fur farm animals, such as foxes and minks, seem to be susceptible to types C and C/D toxin.13,14 Birds are very sensitive to type C and C/D toxins, although type A outbreaks have been recognized among poultry and type E outbreaks among fish-eating waterfowl.15-19 Fishes seem vulnerable to type E toxin.4,20
Animal botulism is suspected to be an emerging disease in Europe, and awareness among veterinarians and all competent authorities has increased in the past decade because of the impact its high mortality rates can have on animal welfare and on the economy of zoo-technical divisions.21 An additional concern may be related to the intentional release of BoNTs and BoNT-producing organisms as biological weapons.22-24
Diagnosis of the disease is based on the observation of clinical signs, although they are often strongly indicative but not specific. Laboratory confirmation of clinical suspicion is required for definitive diagnosis. The criteria for laboratory confirmation have been extensively reported elsewhere.4,9,25,26 Among BoNT-producing clostridia strains, C. botulinum strains that produce types C, D, C/D, and D/C toxins (group III organisms) are the organisms mainly responsible for animal botulism outbreaks.6,7 Rapid detection and typing of these organisms and their toxins are a prerequisite for a prompt diagnosis, for treatment of sick animals, and to prevent or minimize further cases. Moreover, the knowledge of toxin subtype is valuable, not only for epidemiologic reasons but also because the mosaic toxins have shown a higher toxic activity in animals compared to nonmosaic toxins.8
Although several diagnostic methods have been published up to now, only one PCR-based method is capable of discriminating between mosaic and nonmosaic toxins.6 This method is fast, sensitive, and robust but requires special equipment even if it may be optimized and used with open real-time PCR platforms. In this article we report on a new open platform PCR-based method that can detect the neurotoxin genes of group III C. botulinum strains and discriminate between mosaic variants and nonmosaics. These protocols have been developed and validated in-house within the framework of the AniBioThreat research project.
Materials and Methods
Bacterial Strains and Culture Conditions
The bacterial strains used in this study were from the Italian National Reference Centre for Botulism (NRCB), from the French Agency for Food, Environment and Occupational Health Safety (ANSES), and from Istituto Zooprofilattico Sperimentale delle Venezie (IZSVe) strains collections (Table 1). Clostridia strains were cultured in trypticase-peptone-glucose-yeast extract (TPGY) broth or fortified cooked meat medium (CMM) and incubated under anaerobic conditions for 24h±2h at 30°C±1°C. Campylobacter jejuni was cultured in Bolton broth (Oxoid, UK) and incubated in microaerophilic condition for 48h±2h at 41.5°C±1°C. All remaining strains were cultured in brain heart infusion (BHI) broth (Oxoid, UK) and incubated under aerobic conditions for 24h±2h at 37°C±1°C. DNA extraction was performed using 1 ml of each cultured strain, as reported elsewhere.26 Positive real-time PCR controls were generated by cloning each PCR product into 4 plasmids. Cloning procedures were carried out by MWG Eurofins (Ebersber, Germany).
Table 1.
Selectivity Study: Strains Extracted DNAs and Results
|
Strain |
No. of Tested Strains |
3-plex Real-Time PCR bont/type |
5-plex Real-Time PCR bont/type |
||||||
|---|---|---|---|---|---|---|---|---|---|
| C | D | CD | DC | C | D | CD | DC | ||
| Clostridium botulinum type C | 9 | 9 | – | – | – | 9 | – | – | – |
| Clostridium botulinum type CD | 24 | 24 | – | 24 | – | 24 | – | 22 | – |
| Clostridium botulinum type D | 5 | – | 5 | 5 | – | – | 5 | 5 | – |
| Clostridium botulinum type DC | 24 | – | 24 | – | 24 | – | 24 | – | 24 |
| Clostridium botulinum type A | 22 | – | – | – | – | – | – | – | – |
| Clostridium botulinum type A(B) | 1 | – | – | – | – | – | – | – | – |
| Clostridium botulinum type Ab | 4 | – | – | – | – | – | – | – | – |
| Clostridium botulinum type B | 21 | – | – | – | – | – | – | – | – |
| Clostridium botulinum type F | 4 | – | – | – | – | – | – | – | – |
| Clostridium butyricum type E | 7 | – | – | – | – | – | – | – | – |
| Clostridium butyricum | 1 | – | – | – | – | – | – | – | – |
| Clostridium carnis | 1 | – | – | – | – | – | – | – | – |
| Clostridium difficile | 1 | – | – | – | – | – | – | – | – |
| Clostridium histolyticum | 1 | – | – | – | – | – | – | – | – |
| Clostridium novyi | 1 | – | – | – | – | – | – | – | – |
| Clostridium paraputrificum | 1 | – | – | – | – | – | – | – | – |
| Clostridium perfringens | 1 | – | – | – | – | – | – | – | – |
| Clostridium sordelli | 1 | – | – | – | – | – | – | – | – |
| Clostridium sporogenes | 1 | – | – | – | – | – | – | – | – |
| Bacillus cereus | 1 | – | – | – | – | – | – | – | – |
| Bacillus coagulans | 1 | – | – | – | – | – | – | – | – |
| Bacillus subtilis | 1 | – | – | – | – | – | – | – | – |
| Campylobacter jejuni | 1 | – | – | – | – | – | – | – | – |
| Citrobacter fruendii | 1 | – | – | – | – | – | – | – | – |
| Cronobacter zakazakii | 1 | – | – | – | – | – | – | – | – |
| Enterocuccos faecalis | 1 | – | – | – | – | – | – | – | – |
| Escherichia coli | 1 | – | – | – | – | – | – | – | – |
| Listeria innocua | 1 | – | – | – | – | – | – | – | – |
| Listeria monocytogenes | 1 | – | – | – | – | – | – | – | – |
| Micrococcus luteus | 1 | – | – | – | – | – | – | – | – |
| Pseudomonas aeruginosa | 1 | – | – | – | – | – | – | – | – |
| Rodococcus equi | 1 | – | – | – | – | – | – | – | – |
| Salmonella Choleraesuis | 1 | – | – | – | – | – | – | – | – |
| Staphylococcus aureus | 1 | – | – | – | – | – | – | – | – |
| Streptococcus thermophylus | 1 | – | – | – | – | – | – | – | – |
Sample Collection, Culture Conditions, and DNA Extraction
Samples collected during animal botulism surveillance programs performed in Italy and in France were tested in this study (Table 2). An amount of 1 to 2 grams of each sample was cultured in prereduced TPGY broth or in fortified CMM and incubated under anaerobic conditions at 30°C±1°C. After 24h±2h of incubation, a 1-ml aliquot of each enrichment broth was subjected to DNA extraction while the remaining broth was incubated again for a total of 96h±2h. At the end of the last incubation period, all enrichment broths were centrifuged at 12,000×g for 20 min at 4°C, and the upper phase was subjected to mouse bioassay for the confirmation of BoNT production.
Table 2.
List of Naturally Contaminated Samples
| |
Reference Method |
Real-Time PCR Positive |
||
|---|---|---|---|---|
| Samples | No. Tested | No. Positive | 3-plex | 5-plex |
| Intestinal content/feces | 62 | 26 C | 26 C | 26 C |
| 1 D | 1 D | 1 D | ||
| 15 CD | 15 CD | 13 CD | ||
| 13 DC | 13 DC | 13 DC | ||
| Liver/spleen | 14 | 3 C | 3 C | 3 C |
| 1 D | 1 D | 1 D | ||
| 8 CD | 8 CD | 7 CD | ||
| 2 DC | 2 DC | 2 DC | ||
| Feed/water | 6 | 1 CD | 1 CD | 1 CD |
| 1 DC | 1 DC | 1 DC | ||
| Soil/manure/wipe | 12 | 9 CD | 9 CD | 5 CD |
| Maggots | 2 | 2 CD | 2 CD | 1 CD |
Before DNA extraction, which was performed as described elsewhere,26 each aliquot of enrichment broth was supplemented with an amount of approximately 100 cells of process control (PC). PC consisted of a 4-day-old culture of a C. botulinum type B–like strain (a C. botulinum strain that had naturally lost the gene encoding the toxin) or, alternatively, a 4-day-old culture of Clostridium sporogenes strain ATCC 19404. The added amount of PC generated a Cq value in the range of 30 to 33.
Standard Mouse Bioassay
The detection and typing of BoNTs in the supernatants of samples and bacterial cultures were performed by standard mouse bioassay (SMB) as described elsewhere.9 The SMB was performed in accordance with European Directive 86/609/EEC on the protection of animals used for experimental and other scientific purposes, and approved by Istituto Superiore di Sanità, ANSES, and the IZSVe Committee on the use of animals in research and teaching.
Primers and Probes
Primer and probe sets used in this study are listed in Table 3. Alignments of the neurotoxin gene sequences of bont/C, bont/D, bont/CD, and bont/DC, available in GenBank (http://www.ncbi.nih.gov/GenBank), were performed using the multialignment program CLUSTAL W (http://www.align.genome.jp). The sequences used in the alignment studies were the same as those reported elsewhere.26 Primers and probes were designed on consensus sequences generated by alignment studies, using the software Beacon Designer version 7.91 (Premier Biosoft International, USA). PC primers and probes were designed from gyrB of C. botulinum type B strain Okra.
Table 3.
Primer and Probe Sets
| Target | Primer/Probe | Sequences (5′-3′)a | Position | GeneBank Acc. No. |
|---|---|---|---|---|
| bont/C | Forward | cta cgt tta aat taa cta ac | 76176–76195 | AP008983 |
| Reverse | ggt gat att gaa att att g | 76233–76251 | ||
| Probe | cacCaaAtcCttCttgtg | 76209–76226 | ||
| bont/D | Forward | cag gaa att ttg ttg taa | 8745–8762 | AB745669 |
| Reverse | cga aga ata aac aac ttc | 8813–8830 | ||
| Probe | acaTaaCatTagTcaagtct | 8791–8810 | ||
| bont/CD | Forward | aca gga tat aca aat aaa tg | 2998–3017 | FN436022 |
| Reverse | cct cat cta aat ctt caa | 3098–3115 | ||
| Probe | ttcActCtgCttTaattct | 3075–3093 | ||
| bont/DC | Forward | gtt cgt tta taa tac aac c | 3620–3638 | EF378947 |
| Reverse | cca agt ttg aaa cta taa c | 3725–3743 | ||
| Probe | tgaTagTatTccAagccta | 3699–3717 | ||
| PC | Forward | agc agt tat ttc agt aaa | 5396–5413 | CP000939 |
| Reverse | cac cat ttc cta att ttg | 5449–5466 | ||
| Probe | tgtCtgTccTtcAaattg | 5427–5444 |
Bases written in capital letters in probe sequences represent LNA modifications.
The specificity of primers and probes was evaluated using Basic Local Alignment Search Tool, BLAST (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Primers were purchased from MWG Eurofins (Ebersber, Germany) and probes from Eurogentec (Ougrèe, Belgium).
Multiplex Real-Time PCR
Multiplex real-time PCR was optimized for Stratagene Mx3005P spectro-fluorometric thermal cycler (Agilent, USA) and Bio-Rad CFX96 thermal cycler (Bio-Rad Laboratories, USA) using 2 3-plex (bont/C+bont/D+PC; bont/CD+bont/DC+CP) and 5-plex (bont/C+bont/D+bont/CD+bont/DC+CP) schemes. Each assay was performed in a total volume of 25 μL containing 3 μL of DNA template, 12.5 μL of 2×QuantiTect Multiplex PCR no ROX kit (Qiagen, Germany), and a final concentration of 9.5 mM MgCl2. Primer and probe concentrations as well as the reporters used in the hydrolysis probes are summarized in Table 4. Thermal profile was the same for the 2 instruments and for both schemes. It consisted of 15 min at 95°C, followed by 40 cycles of denaturing at 94°C for 30 s and annealing extension at 56°C for 90 s.
Table 4.
Primer and Probe Concentration and Reporter Labeling
| |
|
Mx3005P |
CFX96 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| |
|
3-plex |
5-plex |
3-plex |
5-plex |
||||
| Target | Primer/Probe | Conc. (nM) | Rep. | Conc. (nM) | Rep. | Conc. (nM) | Rep. | Conc. (nM) | Rep. |
| bont/C | Forward | 600 | – | 200 | – | 300 | – | 200 | – |
| Reverse | 600 | – | 200 | – | 300 | – | 200 | – | |
| Probe | 100 | HEX | 100 | HEX | 100 | HEX | 100 | HEX | |
| bont/D | Forward | 600 | – | 300 | – | 300 | – | 300 | – |
| Reverse | 600 | – | 300 | – | 300 | – | 300 | – | |
| Probe | 100 | FAM | 100 | FAM | 100 | FAM | 100 | FAM | |
| bont/CD | Forward | 600 | – | 600 | – | 300 | – | 600 | – |
| Reverse | 600 | – | 600 | – | 300 | – | 600 | – | |
| Probe | 100 | HEX | 100 | Cy5 | 100 | HEX | 100 | Cy5 | |
| bont/DC | Forward | 600 | – | 600 | – | 300 | – | 300 | – |
| Reverse | 600 | – | 600 | – | 300 | – | 300 | – | |
| Probe | 100 | FAM | 300 | Alexa350 | 100 | FAM | 600 | Cy5.5 | |
| PC | Forward | 600 | – | 600 | – | 200 | – | 600 | – |
| Reverse | 600 | – | 600 | – | 200 | – | 200 | – | |
| Probe | 50 | ROX | 50 | ROX | 50 | ROX | 50 | ROX | |
Each PCR run was carried out at least twice and included both positive controls for all targets and a no-template control (NTC). NTC was used as negative control to evaluate for no reagent contamination, and PC was used to evaluate the Cq value without inhibitors.
Method Performance
The following performance parameters have been evaluated as requested by ISO 16140:2003: selectivity, linearity, limit of detection, relative accuracy, relative sensitivity, relative specificity, and repeatability. Definitions adopted in the present study were those reported in ISO 16140:2003.27
The study of selectivity (inclusivity and exclusivity) was performed twice with DNA extracted from pure cultures of bacterial strains. Inclusivity was calculated as the ratio between positive results by multiplex real-time PCR and positive results obtained by the reference method. Exclusivity was calculated as the ratio between negative results obtained by multiplex real-time PCR and by the reference method. Linearity and limit of detection (LOD) studies were performed testing 20 replicates of each serial dilution of DNA extracted from positive controls. Amplification efficiency for the different targets was calculated according to the following equation:
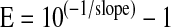 |
Relative accuracy (AC), relative sensitivity (SE), and relative specificity (SP) were determined by comparing the results obtained by the analysis of the samples with the reference method and multiplex real-time PCR.
AC was calculated for positive and negative samples by the following equation: AC=[(PA+NA)/N]×100%, where PA represents the positive results and NA represents the negative results obtained by both reference method and multiplex real-time PCR methods, and N represents the total number of tested samples.
SE was calculated for negative samples with the following equation: SE=(NA/N−)×100%, where NA represent the negative results obtained by both reference method and multiplex real-time PCR methods, and N− represents negative results obtained by the reference method.
SP was calculated for positive samples with the equation: SP=(PA/N+)×100%, where PA represents the positive results obtained by both the reference method and multiplex real-time PCR methods, and N+ represents positive results obtained by reference method.
Repeatability of the multiplex real-time PCR protocols was evaluated by determination of intra-assay variation of Cq values obtained testing the samples reported in Table 2. The coefficient of variance (CV=SD/arithmetic mean of Cq) was calculated for results of 2 replicated in the same run.
In this study the protocol published by Woudstra and colleagues was used as reference method.26 The capability to correctly detect 1 target when the others are present in the same well at a higher level (disproportion among the targets) was evaluated using plasmid positive controls as template DNA.
Results
Primer and probe sets for detecting bont/C and bont/D were designed, respectively, on the light chain of type C and D neurotoxin genes. Primer and probe set for detecting bont/CD was designed in the heavy chain of the type D neurotoxin gene. Primer and probe set for detecting bont/DC was designed in the heavy chain of the type C neurotoxin gene, in the region specific of DC mosaic toxin.26 C. botulinum strains harboring type C toxin gene produce positive results for bont/C. C. botulinum strains harboring type D toxin gene produce positive results for bont/D and for bont/CD. C. botulinum strains harboring type CD mosaic gene produce positive results for bont/C and bont/CD. C. botulinum strains harboring type DC mosaic gene produce positive results for bont/D and bont/DC.
All bacterial strains and cultured samples tested by means of SMB showed the expected result and matched perfectly with the data from the reference method (data not shown). A selectivity study conducted on 62 target strains and on 84 nontarget strains showed 100% inclusivity and 100% exclusivity for both 3-plex schemes and 96.8% inclusivity and 100% exclusivity for the 5-plex scheme (Table 1). Although discrimination of 2 type CD mosaics failed, these strains were correctly detected for bont/C target.
Serial dilutions of positive real-time PCR controls were used to test the response linearity and LOD of the 2 3-plex schemes. A standard curve for each target was plotted, and the linearity of the response was demonstrated in a range of 8 orders of magnitude for all targets (data not shown). Limit of detection, correlation coefficients (R2), slopes of the curves, and PCR efficiency for all targets are reported in Table 5.
Table 5.
LOD, Coefficient of Correlation (R2), Slope of Linearity Curve, PCR Efficiency for Each Target
| PCR Scheme | Parameter | bont/C | bont/D | bont/CD | bont/DC |
|---|---|---|---|---|---|
| 3-plex | LOD (genomes equivalent) | 58 | 43 | 58 | 221 |
| Coefficient of correlation (R2) | 0.999 | 0.998 | 0.995 | 0.995 | |
| Slope of linearity curve | −3.24 | −3.26 | −3.41 | −3.33 | |
| PCR efficiency | 103.4% | 102.4% | 96.45% | 99.66% | |
| 5-plex | LOD (genomes equivalent) | 58 | 43 | 58 | 221 |
| Coefficient of correlation (R2) | 0.996 | 0.999 | 0.999 | 0.988 | |
| Slope of linearity curve | −3.32 | −3.29 | −3.41 | −3.39 | |
| PCR efficiency | 100.1% | 101.0% | 96.61% | 96.88% |
Ninety-six samples were tested to evaluate AC, SE, and SP. The results showed 100% AC, 100% SE, and 100% SP with both of the 3-plex schemes and 91.6% AC, 100% SE, and 90.2% SP with the 5-plex scheme. No inhibition was observed testing the samples.
Repeatability was evaluated testing all samples at least twice, reporting a coefficient of variation (CV) of Cq values ranging from 0.00% to 3.71% with a midpoint of 0.61% (data not shown). Results of disproportion among targets study are reported in Table 6.
Table 6.
Results of Disproportion Among Target Study
|
Target at Low Concentration |
Target at High Concentration (105×LOD) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Target | Concentration | C | D | CD | DC | C+D | C+CD | C+DC | D+CD | D+DC | CD+DC | C+D+CD | C+D+DC | C+CD+DC | D+CD+DC |
| C | LOD | + | + | + | + | + | + | + | |||||||
| 10×LOD | + | + | + | + | + | + | + | ||||||||
| D | LOD | + | + | + | + | − | − | − | |||||||
| 10×LOD | + | + | + | + | + | + | + | ||||||||
| CD | LOD | − | + | + | − | − | − | − | |||||||
| 10×LOD | + | + | + | + | + | + | + | ||||||||
| DC | LOD | − | − | + | + | + | − | − | – | ||||||
| 10×LOD | + | + | + | + | + | + | + | + | |||||||
+=target at low concentration correctly detected; −=target at low concentration not detected.
Discussion
Laboratory confirmation of animal botulism outbreaks diagnosed on the basis of clinical signs represents a crucial step in definitive diagnosis. The lack of laboratory diagnosis may represent a significant drawback for the management of animal botulism outbreaks and may hamper our ability to minimize further cases, especially in terms of detecting the source of toxins and/or BoNT-producing clostridia. Given the toxico-infection forms of botulism, the detection of the source of BoNT-producing clostridia becomes essential.3 Although SMB represents the gold standard method for laboratory diagnosis, several alternative and rapid methods such as PCR-based protocols have been reported in the literature.9,26 Among them, only a few methods are capable of simultaneously detecting the different BoNT-producing clostridia strains mainly involved in animal botulism outbreaks, and only 1 method is capable of discriminating mosaic from nonmosaic variants.26 This latter method is rapid, robust, and may be easily used in a field investigation because it does not require specialized personnel. On the other hand, it does require special equipment, although it may be easily optimized using open real-time PCR platforms.
Since BoNT-producing clostridia genomes are low in GC content, we have chosen to use locked nucleic acid (LNA) hydrolysis probes. In fact, previous researchers suggested that probes containing LNA bases have strong affinities for their DNA complementary targets. In addition, DNA-LNA interaction compared with DNA-DNA interaction shows higher thermal stability and melting temperature, allowing the use of short probes that simplify the design of multiplex real-time PCR assay.28 A multiplex real-time PCR approach was chosen to save consumable material, reagents, work load, and time, considering that during animal botulism outbreaks it may be necessary to test a large number of samples, especially for identifying the source of contamination.
Environmental and food samples may harbor several PCR inhibitors,29 and to avoid false-negative results, we have used a PC included in each sample before the DNA extraction. The method reported in this study represents a new diagnostic tool that can improve and facilitate the laboratory confirmation procedures in cases of animal botulism outbreaks.
Acknowledgments
This research was supported by the framework of the EU project AniBioThreat (Grant Agreement: Home/2009/ISEC/AG/191) with the financial support from the Prevention of and Fight against Crime Programme of the European Union, European Commission—Directorate General Home Affairs. This publication reflects the views only of the authors, and the European Commission cannot be held responsible for any use that may be made of the information contained therein.
References
- 1.Hatheway CL. Clostridium botulinum and other clostridia that produce botulinum neurotoxins. In: Hauschild AHW, editor; Dodds KL, editor. Clostridium botulinum: Ecology and Control in Food. New York: Marcel Dekker Inc.; 1992. pp. 3–20. [Google Scholar]
- 2.Peck MW. Biology and genetic analysis of Clostridium botulinum. Adv Microb Physiol. 2009;75:183–265. doi: 10.1016/S0065-2911(09)05503-9. 320. [DOI] [PubMed] [Google Scholar]
- 3.Critchley EM. A comparison of human and animal botulism: a review. J R Soc Med. 1991;84:295–298. doi: 10.1177/014107689108400516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deprez PR. Tetanus and botulism in animals. In: Mainil J, editor. Clostridia in Medical, Veterinary and Food Microbiology—Diagnosis and Typing. Luxembourg: European Commission; 2006. pp. 27–36. [Google Scholar]
- 5.Sobel J. Botulism. Clin Infect Dis. 2005;41:1167–1173. doi: 10.1086/444507. [DOI] [PubMed] [Google Scholar]
- 6.Takeda M. Tsukamoto K. Kohda T. Matsui M. Mukamoto M. Kozaki S. Characterization of the neurotoxin produced by isolates associated with avian botulism. Avian Dis. 2005;49:376–381. doi: 10.1637/7347-022305R1.1. [DOI] [PubMed] [Google Scholar]
- 7.Takeda M. Kasai H. Torii Y, et al. Protective effect of botulinum C/D mosaic toxoid against avian botulism. J Vet Med Sci. 2006;68:325–330. doi: 10.1292/jvms.68.325. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura K. Kohda T. Umeda K. Yamamoto H. Mukamoto M. Kozaki S. Characterization of the D/C mosaic neurotoxin produced by Clostridium botulinum associated with bovine botulism in Japan. Vet Microbiol. 2010;140:147–154. doi: 10.1016/j.vetmic.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 9.Anniballi F. Auricchio B. Delibato E. Antonacci M. De Medici D. Fenicia L. Multiplex real-time PCR SYBR Green for detection and typing of group III Clostridium botulinum. Vet Microbiol. 2012;154:332–338. doi: 10.1016/j.vetmic.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 10.Lindström M. Myllykoski J. Sivelä S. Korkeala H. Clostridium botulinum in cattle and dairy products. Crit Rev Food Sci Nutr. 2010;50:281–304. doi: 10.1080/10408390802544405. [DOI] [PubMed] [Google Scholar]
- 11.Johnson AL. McAdams SC. Whitlock RH. Type A botulism in horses in the United States: a review of the past ten years (1998-2008) J Vet Diagn Invest. 2010;22:165–173. doi: 10.1177/104063871002200201. [DOI] [PubMed] [Google Scholar]
- 12.Johnson AL. Sweeney RW. McAdams SC. Whitlock RH. Quantitative real-time PCR for detection of neurotoxin gene of Clostridium botulinum type B in equine and bovine samples. Vet J. 2012;194:118–120. doi: 10.1016/j.tvjl.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Lindström M. Nevas M. Kurki J, et al. Type C botulism due to toxic feed affecting 52,000 farmed foxes and minks in Finland. J Clin Microbiol. 2004;42:4718–4725. doi: 10.1128/JCM.42.10.4718-4725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myllykoski J. Lindström M. Bekema E. Pölöne I. Korkeala H. Fur animal botulism hazard due to feed. Res Vet Sci. 2011;90:412–418. doi: 10.1016/j.rvsc.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 15.Rocke TE. The global importance of avian botulism. In: Boere GC, editor; Galbraith CA, editor; Stroud DA, editor. Waterbirds Around the World. Edinburgh: Stationery Office; 2006. pp. 422–426. [Google Scholar]
- 16.Roberts TA. Collings DF. An outbreak of type C botulism in broiler chickens. Avian Dis. 1973;17:650–658. [PubMed] [Google Scholar]
- 17.Neimans A. Gavier-Widén D. Leighton F. Bollinger T. Rocke T. Mörner T. An outbreak of type C botulism in herring gulls (Larus argentatus) in southern Sweden. J Wild Dis. 2007;43:327–336. doi: 10.7589/0090-3558-43.3.327. [DOI] [PubMed] [Google Scholar]
- 18.Dohms JE. Allen PH. Rosenberger JK. Case of type C botulism in broiler chickens. Avian Dis. 1981;26:204–210. [PubMed] [Google Scholar]
- 19.Sharpe AE. Sharpe EJ. Ryan ED. Clarke HJ. McGettrick SA. Outbreak of type C botulism in laying hens. Vet Rec. 2011;168:669a. doi: 10.1136/vr.d1090. [DOI] [PubMed] [Google Scholar]
- 20.Khoo LH. Goodwin AE. Wise GJ, et al. The pathology associated with visceral toxicosis of catfish. J Vet Diagn Invest. 2011;6:1217–1221. doi: 10.1177/1040638711425577. [DOI] [PubMed] [Google Scholar]
- 21.AniBioThreat. Abstracts of reports of botulism in animals over the last decade written by participants of the Animal Botulism Workshop. Animal Botulism in Europe: Current Status of an Emerging Disease. 2012 Report No. 17. Uppsala; [Google Scholar]
- 22.Arnon SS. Schechter R. Inglesby TV, et al. Botulinum toxin as a biological weapon. JAMA. 2001;285:1059–1070. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- 23.Dembek ZF. Smith LA. Rusnak JM. Botulism: cause, effects, diagnosis, clinical and laboratory identification, and treatment modalities. Disaster Med Public Health Prep. 2007;1:122–134. doi: 10.1097/DMP.0b013e318158c5fd. [DOI] [PubMed] [Google Scholar]
- 24.Yeh JY. Seo HJ. Cho YS, et al. Livestock agroterrorism: the deliberate introduction of a highly infectious animal pathogen. Foodborne Pathog Dis. 2012;9:869–877. doi: 10.1089/fpd.2012.1146. [DOI] [PubMed] [Google Scholar]
- 25.Lindberg A. Skarin H. Knutsson R. Blomqvist G. Båverud V. Real-time PCR for Clostridium botulinum type C neurotoxin (BoNTC) gene, also covering a chimeric C/D sequence: application on outbreak of botulism in poultry. Vet Microbiol. 2010;146:118–123. doi: 10.1016/j.vetmic.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 26.Woudstra C. Skarin H. Anniballi F, et al. Neurotoxin gene profiling of Clostridium botulinum types C and D native to different countries within Europe. Appl Environ Microbiol. 2012;78:3120–3127. doi: 10.1128/AEM.07568-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.ISO 16140;2003. Microbiology of food and animal feeding stuffs – Protocol for the validation of alternative methods. ISO website. 2003. http://www.iso.org/iso/iso_catalogue/catalogue_tc/catalogue_detail.htm?csnumber=30158. [Jun 19;2013 ]. http://www.iso.org/iso/iso_catalogue/catalogue_tc/catalogue_detail.htm?csnumber=30158
- 28.Sun Z. Zhou L. Zeng H. Chen Z. Zhu H. Multiplex locked nucleic acid probes for analysis of hepatitis B virus mutants using real-time PCR. Genomics. 2007;89:151–159. doi: 10.1016/j.ygeno.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 29.Braconnier A. Broussolle V. Perelle S. Fach P. Nguyen-The C. Carlin F. Screening for Clostridium botulinum type A, B, and E in cooked chilled foods containing vegetable and raw material using polymerase chain reaction and molecular probes. J Food Prot. 2001;64:201–207. doi: 10.4315/0362-028x-64.2.201. [DOI] [PubMed] [Google Scholar]


