Abstract
Physiological occlusal force constitutively exists in the oral environment and is important for periodontal homeostasis and remodeling. Cyclic tensile stress (CTS) triggers the biological response of periodontal ligament (PDL). However, a few reports have studied the correlation between CTS during physiological occlusal force and PDL cell activities such as osteogenic differentiation. In the present study, human PDL cells (hPDLCs) were subjected to 10% elongation CTS loading at 0.5 Hz for 24 h, which represents the physiological conditions of occlusal force. Gene expression microarray was used to investigate the mechano-induced differential gene profile and pathway analysis in vitro. The osteogenic relative factors, that is, SPP1, RUNX2, and SP7, were assessed by real-time PCR and Western blot. The involvement of mitogen-activated protein kinase (MAPK) signaling pathways was investigated by Western blot with a specific inhibitor. The expressions of SPP1, RUNX2, SP7, p-ERK1/2, and p-Elk1 were up-regulated after 10% CTS exposure. However, these up-regulated expressions were prevented by ERK1/2 inhibitor U0126 in the physiological occlusal force-applied hPDLCs. These results showed that 10% CTS could enhance osteogenic differentiation of hPDLCs via ERK1/2-Elk1 MAPK pathway, indicating that CTS during physiological occlusal force is a potent agent for PDL remodeling.
Cyclic tensile stress enhanced osteogenic differentiation of human periodontal ligaments, indicating that this may be a potent agent for tissue remodeling.
Introduction
Mechanical stress of physiological magnitude regulates cellular processes that are critical for normal tissue and organ functions, such as differentiation, proliferation, and migration (Fujihara et al., 2010). In an oral environment, periodontal ligament (PDL) sensitively mediates the transmission of mechanical stress stimuli to the alveolar bone for periodontal tissue remolding. The PDL cells (PDLCs), the most abundant cells in PDL, are capable of regenerating the PDL, which is required for maintaining the PDL integrity (Kasamatsu et al., 2005). This potential of PDLCs is believed to be associated with their property to respond sensitively to physiological and mechanical stress (Yamashiro et al., 2007; Saminathan et al., 2012). In response to such stress, PDLCs perceive mechanical signals and differentiate into connective tissue-forming cells such as cementoblasts and osteoblasts for maintaining periodontal homeostasis and alveolar bone remodeling (Long et al., 2002; Cho et al., 2010; Lin et al., 2010).
To clarify the effect of mechanical stress on PDLCs, many studies have used different systems to mimic the stress in an oral environment to observe the different molecular expression. However, the expression patterns of these molecules in response differed according to the modes of the mechanical stress, such as shear, compression, and tension as well as the magnitudes of the stress (Redlich et al., 2004; Ozaki et al., 2005; Garlet et al., 2007; Kook et al., 2009). Previous evidence has shown that cyclic tensile stress (CTS) during orthodontic tooth movement (OTM) increases the expression of osteogenic genes and proteins such as alkaline phosphatase (ALP), secreted phosphoprotein 1 (SPP1; also known as bone sialoprotein I/osteopontin), transcription factor Sp7 (SP7; also known as osterix), and runt-related transcription factor 2 (RUNX2) of hPDLCs (Wescott et al., 2007; Cho et al., 2010; Tang et al., 2012). However, ignoring physiological occlusal force constitutively exists in the oral environment and is important for periodontal homeostasis and remodeling. So, studying the effect of CTS during physiological occlusal force on the osteogenic differentiation of human PDLCs (hPDLCs) could extend our understanding of the effect of mechanical stress on PDL remodeling.
Previous findings suggested that the mitogen-activated protein kinases (MAPKs) controlled a range of cellular activities, such as embryogenesis, differentiation, proliferation, and death (Zhang and Liu, 2002; Chambard et al., 2007; Kook et al., 2011a). The direct involvement of MAPKs on mechano transduction has been largely suggested in muscle cells (Li et al., 2000; Zampetaki et al., 2005) and in bone cells (Franceschi and Xiao, 2003; Liedert et al., 2006). In periodontium, centrifugal force during OTM up-regulated COL I expression in human periodontal fibroblasts through the activation of MAPK-AP-1signaling (Kook et al., 2009; Hong et al., 2010). However, little knowledge is available about the mechanism of physiological occlusal force on hPDLCs.
In this study, we examined the effect of 10% CTS, which represents physiological occlusal force (Matsuda et al., 1998a; Fujihara et al., 2010) on the osteogenic differentiation of hPDLCs using gene expression microarray. In addition, the molecular mechanism of 10% CTS enhancing the osteogenic differentiation of hPDLCs was also investigated.
Materials and Methods
Cell culture
PDL tissues were gently scraped from the middle third of periodontally healthy, noncarious premolar tooth roots, extracted from donors aged between 12 and 20 years of age for orthodontic reasons with informed consent. Teeth were washed, and fragments of the PDL attached to the middle third of the root were removed with a scalpel. Tissue explants were plated onto 25 cm2 cell culture flasks (Corning) in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal calf serum (Gibco) and antibiotic reagent (10,000 units penicillin,10,000 mg streptomycin; Gibco). In the third passage, cells were stained with vimentin and cytokeratin antibodies for characterization. Cells at passages 3 to 6 were used for mechanical loading. Collection and culture of hPDLCs was approved by the Ethical Committee of Nanjing Medical University. The different batches of cells were applied in the same assays to avoid the effect of the donor on the outcome.
Application of CTS on hPDLCs
hPDLCs suspension (1.5×105 cells/well) were spread into six-well, 35 mm flexible-bottomed BioFlex® Culture Plates coated with type I collagen. When the cells reached 80% confluence, the plates were subjected to an in-plane cyclic deformation of 10% elongation with 0.5 Hz (30 cycles/min) for 12, 24, and 48 h in a standard Bioflex baseplate linked to a Flexcell® FX-5000™ Tension Unit (Flexcell Corp.). This force represented the physiological conditions of occlusal force (Matsuda et al., 1998a; Fujihara et al., 2010). For the control experiments, hPDLCs were cultured in the same plates without any mechanical stress.
Gene expression microarray analysis
Total RNA in each group was isolated using TRIzol reagent (Invitrogen). The concentration and purity of the RNA samples were determined by the absorbance of RNA at 260 and 280 nm, respectively. O.D. A260/A280 ratios between 1.8 and 2.1 are acceptable. For microarray analysis, cells were subjected to 10% elongation CTS loading at 0.5 Hz for 24 h. The microarray analysis was carried out in accordance with the NimbleGen Gene Expression Analysis protocol (Roche NimbleGen, Inc.). All gene level files were imported into Agilent GeneSpring GX software (version 11.5.1) for further analysis. In this study, a 1.5-fold change was used as the cut-off value, and those genes showing changes that were greater than 1.5 fold were identified as differentially expressed. Then, gene ontology (GO) analysis and pathway analysis based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database were used for further investigation on the differential genes of interest. The microarrays were carried out in triplicate.
Real-time PCR analysis
Real-time PCR was performed using FastStart Universal SYBR Green Master (Roche) in a quantitative PCR System (ABI 7300). Gene expressions were normalized to the housekeeping gene Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was used as an internal control. The primer sequences for SPP1, RUNX2, SP7, and GAPDH were presented in Table 1. The cycling conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 58°C for 1 min. Gene expression levels were calculated using the 2−ΔΔCt method. Data were expressed as mean±standard deviation of three independent experiments.
Table 1.
Primer Sequences of SPP1, RUNX2, SP7, and GAPDH
| Gene name | Primers | Sequences (5′–3′) |
|---|---|---|
| SPP1 | Forward | CCAAGTAAGTCCAACGAAAG |
| Reverse | GGTGATGTCCTCGTCTGTA | |
| RUNX2 | Forward | TCTTAGAACAAATTCTGCCCTTT |
| Reverse | TGCTTTGGTCTTGAAATCACA | |
| SP7 | Forward | CCTCCTCAGCTCACCTTCTC |
| Reverse | GTTGGGAGCCCAAATAGAAA | |
| GAPDH | Forward | GAAGGTGAAGGTCGGAGTC |
| Reverse | GAGATGGTGATGGGATTTC |
Western blot analysis
Cells were collected, washed, and lysed in RIPA lysis buffer (Beyotime) containing 1 mM phenylmethylsulfonyl fluoride (PMSF; Beyotime). After centrifugation, the supernatant was collected and assayed quantitatively with a BCA protein assay kit (Beyotime). Equal amounts of protein extracts (30 μg/sample) were separated by 12% SDS-PAGE and blotted onto polyvinyl difluoride membranes (PVDF; Millipore) at 300 mA for 1 h in a blotting apparatus (Bio-Rad). The blots were incubated, respectively, with primary antibodies (SPP1, 1:800, Abcam; RUNX2, 1:800, Abcam; SP7, 1:800, Abcam; GAPDH, 1:800, Bioworld; ERK1/2, 1:1000, Cell signaling technology; p-ERK1/2, 1:1000, Cell signaling technology; ERK5, 1:1000, Cell signaling technology; p-ERK5, 1:1000, Cell signaling technology; JNK, 1:1000, Cell signaling technology; p-JNK, 1:1000, Cell signaling technology; P38, 1:1000, Cell signaling technology; p-P38, 1:1000, Cell signaling technology; p-Elk, 1:800, Santa Cruz; and GAPDH, 1:800, Bioworld) overnight at 4°C. Finally, the membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (1:10,000; Boster Biotech. Co. Ltd.) at room temperature for an additional 1 h. The membranes were developed with Immobilon Western Chemiluminescent HRP Substrate (Millipore) and were exposed to X-ray film (Eastman-Kodak). Band intensity was calculated using Gel-Pro Analyzer 4. ERK1/2 MAPK inhibitor, and U0126 was purchased from Beyotime. GAPDH serves as the internal control in these experiments. This experiment was repeated in triplicate.
Statistical analysis
Unless specified otherwise, all data are expressed as the mean±standard deviation (SD). A one-way analysis of variance (ANOVA) (SPSS version 17.0 software) was used for multiple comparisons. A value of p<0.05 was considered statistically significant.
Results
Effect of 10% CTS on hPDLCs
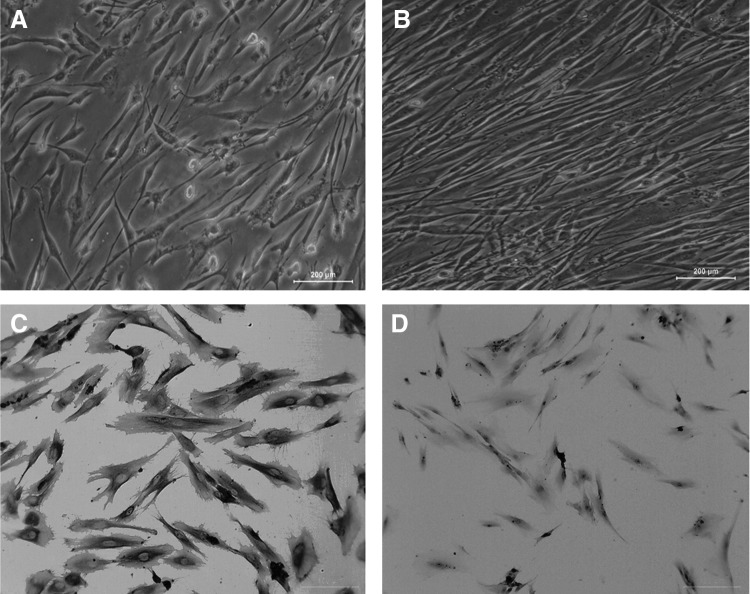
Difference in cellular orientation was observed as early as 12 h after the application of CTS. As shown in Figure 1, control cells remained randomly orientated (Fig. 1A). However, mechanically stretched cells re-orientated along with the direction of the applied tensile force (Fig. 1B). There was no significant difference among cell morphology of 12, 24, and 48 h stress loading (data not shown). After the third passage, hPDLCs stained positively for vimentin, but negatively for cytokeratin, confirming their mesodermal origin (Fig. 1C, D).
FIG. 1.
Effect of cyclic tensile stress on hPDLCs. (A) Control cells, 100×. (B) Cells were exposed to 10% CTS, 100×. (C) Cells were stained with vimentin antibody for characterization, 200×. (D) Cells were stained with cytokeratin antibody for characterization, 200×. hPDLC, human periodontal ligament cells; CTS, cyclic tensile stress.
Identification of genes in response to 10% CTS
The microarray data showed that 453 out of 45,033 genes were differentially expressed. Among the 453 genes, 263 were up-regulated (Fold Change >1.5 and p<0.05), whereas 190 were down-regulated (p<0.05). The differentially expressed genes were partially summarized and listed in Table 2. GO analysis demonstrated that these differentially expressed genes participated in a variety of biological processes and molecular functions, such as regulation of cellular process, response to stimulus, cell proliferation, signal transduction, receptor binding, and multicellular organismal process. SPP1 in the up-regulated genes group with higher expression are involved in most of the biological processes, including response to stimulus, multicellular organismal process, negative regulation of cell proliferation, axon and neuron projection regeneration, regulation of cell growth, cell size, cell adhesion, and anatomical structure morphogenesis. For this reason coupled with SPP1 being one of the main bone-related components, the gene was selected as a gene of interest for further analysis in our study.
Table 2.
Differentially Expressed Genes of hPDLCs Under Cyclic Tensile Stress (Partial Microarray Data)
| Name | Description | p-Value | Fold change | Gene ID |
|---|---|---|---|---|
| Up-regulated genes | ||||
| Response to stimulus | ||||
| APOE | apolipoprotein E | 0.017483 | 3.102327 | 348 |
| SPP1 | secreted phosphoprotein 1 | 0.003096 | 9.22774 | 6696 |
| TAC1 | tachykinin, precursor 1 | 0.023223 | 1.854219 | 6863 |
| THBD | thrombomodulin | 0.017664 | 2.075896 | 7056 |
| CMA1 | chymase 1, mast cell | 0.021977 | 1.60568 | 1215 |
| SFRP2 | secreted frizzled-related protein 2 | 0.022874 | 1.508726 | 6423 |
| LAT2 | linker for activation of T cells family | 0.026814 | 1.767791 | 7462 |
| TPD52L1 | tumor protein D52-like 1 | 0.03251 | 2.121754 | 7164 |
| ASCL2 | achaete-scute complex-like 2 | 0.022832 | 1.926807 | 430 |
| ADRA2A | adrenergic, alpha-2A-, receptor | 0.009503 | 1.896106 | 150 |
| Neg. regulation of cell proliferation | ||||
| DPT | dermatopontin | 0.036332 | 1.579927 | 1805 |
| ASCL2 | achaete-scute complex-like 2 | 0.022832 | 1.926807 | 430 |
| APOE | apolipoprotein E | 0.017483 | 3.102327 | 348 |
| BTG1 | B-cell translocation gene 1 | 0.031139 | 1.523646 | 694 |
| NPPA | natriuretic peptide precursor A | 0.041759 | 2.024056 | 4878 |
| SPP1 | secreted phosphoprotein 1 | 0.003096 | 9.22774 | 6696 |
| Cell adhesion | ||||
| BCL2L11 | BCL2-like 11 | 0.027295 | 1.926735 | 10018 |
| DLL1 | delta-like 1 | 0.022419 | 1.541046 | 28514 |
| SFRP2 | secreted frizzled-related protein 2 | 0.022874 | 1.508726 | 6423 |
| PRPH | Peripherin | 0.047233 | 1.590701 | 5630 |
| SPP1 | secreted phosphoprotein 1 | 0.003096 | 9.22774 | 6696 |
| Cell growth | ||||
| CISH | cytokine-inducible SH2-containing protein | 0.0326 | 1.614761 | 1154 |
| SFRP2 | secreted frizzled-related protein 2 | 0.022874 | 1.508726 | 6423 |
| NPPA | natriuretic peptide precursor A | 0.041759 | 2.024056 | 4878 |
| BTG1 | B-cell translocation gene 1 | 0.031139 | 1.523646 | 694 |
| APOE | apolipoprotein E | 0.017483 | 3.102327 | 348 |
| SPP1 | secreted phosphoprotein 1 | 0.003096 | 9.22774 | 6696 |
| Down-regulated genes | ||||
| SEMA3C | sema domain, immunoglobulin domain (Ig) | 3.03E-04 | 2.899486 | 10512 |
| SYNJ1 | synaptojanin 1 | 0.008957 | 2.348813 | 8867 |
| SLC8A1 | solute carrier family 8, member 1 | 0.019753 | 2.31181 | 6564 |
| GLOXD1 | glyoxalase domain containing 1 | 0.017959 | 2.308117 | 84842 |
| CENTB2 | centaurin, beta 2 | 0.028033 | 2.293343 | 23527 |
| SAC | testicular soluble adenylyl cyclase | 0.008068 | 2.258588 | 55811 |
| WDR51B | WD repeat domain 51B | 0.0117322 | 2.127292 | 282809 |
hPDLC, human periodontal ligament cells; Neg., negative.
Signaling pathway analysis of genes in response to 10% CTS
Based on the KEGG database, 43 signaling pathways associated with genes differentially expressed in response to CTS (p<0.05) were identified. The differentially expressed pathways were partially summarized and listed in Table 3.
Table 3.
Signaling Pathways Involved and Related Differentially Expressed Genes Under Cyclic Tensile Stress (Partial Microarray Data)
| Pathway name | Gene | p-Value |
|---|---|---|
| GnRH signaling pathway | ADCY7 | |
| CAMK2D | ||
| KRAS | ||
| PLCB4 | 0.0004387201 | |
| PRKACB | ||
| SOS2 | ||
| Calcium signaling pathway | ADCY7 | |
| CAMK2D | ||
| PLCB4 | ||
| PPP3CB | 0.001652617 | |
| PRKACB | ||
| SLC8A1 | ||
| SLC8A3 | ||
| MAPK signaling pathway | ATF2 | |
| KRAS | ||
| NF1 | ||
| PPP3CB | 0.004477719 | |
| PRKACB | ||
| RASA2 | ||
| RASGRF2 | ||
| SOS2 | ||
| B-cell receptor signaling pathway | KRAS | |
| PIK3R1 | ||
| PPP3CB | 0.006182614 | |
| SOS2 | ||
| Chemokine signaling pathway | ADCY7 | |
| KRAS | ||
| PIK3R1 | 0.01035684 | |
| PLCB4 | ||
| PRKACB | ||
| SOS2 | ||
| Wnt signaling pathway | CAMK2D | |
| DKK1 | ||
| PLCB4 | 0.01612985 | |
| PPP3CB | ||
| PRKACB |
10% CTS enhanced the osteogenic differentiation of hPDLCs
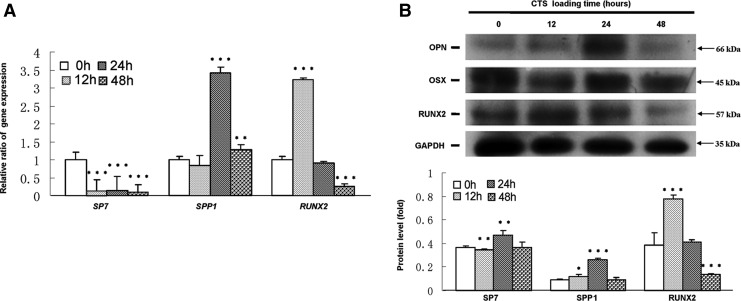
As shown in Figure 2A, RUNX2 mRNA was significantly up-regulated for 12 h CTS exposure, while the increase was significant decreased for 48 h loading as compared with CTS-untreated groups. SPP1 mRNA was significantly increased for 24 and 48 h CTS exposure. However, SP7 mRNA was down-regulated after CTS loading. Western blot results demonstrated that the protein levels of RUNX2 were significantly increased for 12 h and then decreased for 24 and 48 h CTS exposure as compared with CTS-untreated groups. SPP1 levels were elevated for 12 h CTS exposure with a peak increase for 24 h and then declined. Inconsistent with mRNA levels, SP7 level was up-regulated for 24 h CTS loading (Fig. 2B).
FIG. 2.
CTS up-regulated the expression of RUNX2, SP7, and SPP1 in hPDLCs. (A) Gene expression of RUNX2, SP7, and SPP1 by real-time PCR. (B) Protein expression of RUNX2, SP7, and SPP1 by Western blot. GAPDH was used as an internal control for each group. Protein expression was described as a fold change relative to the control group. Values are the mean±SD, n=3. *p<0.05, **p<0.01, and ***p<0.001 versus the control values.
10% CTS could activate ERK1/2-Elk1 MAPK pathway in hPDLCs
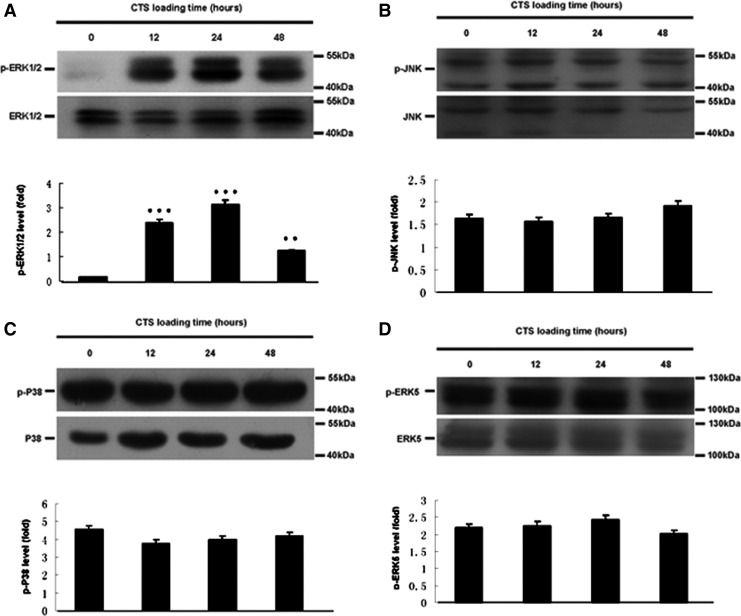
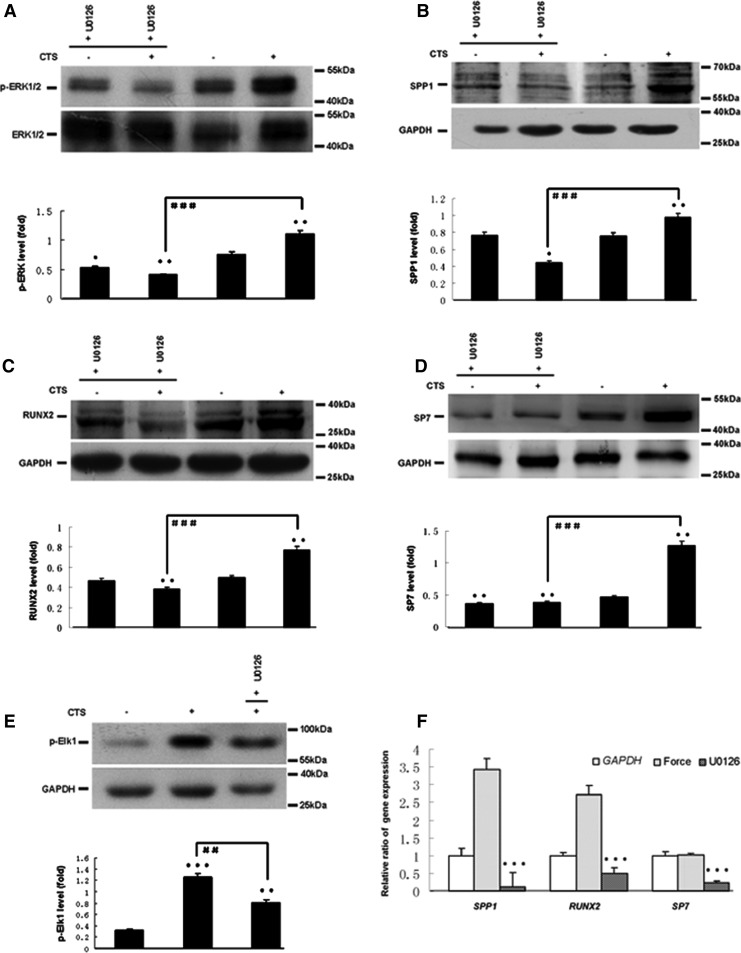
Based on pathway analysis, MAPK pathway was found to be related to osteogenic differentiation of hPDLCs induced by CTS. As shown in Figure 3, the levels of p-ERK1/2 (Fig. 3A) in CTS groups were up-regulated. Quantitatively, the p-ERK1/2 increased approximately 18-fold for 24 h, 14-fold for 12 h, and seven-fold for 48 h in CTS groups as compared with control groups. However, the levels of p-JNK, p-P38, and p-ERK5 were not affected (Fig. 3B–D). 10 μM ERK1/2 MAPK inhibitor U0126 was treated with hPDLCs for 1 h before the cells' exposure to CTS. Pretreatment with U0126 significantly prevented the CTS-induced increases of SPP1, RUNX2, SP7, p-ERK1/2, and p-Elk1 expression (Fig. 4).
FIG. 3.
Effect of CTS on the MAPK pathway of hPDLCs. (A) Protein expression of p-ERK 1/2 and ERK 1/2 in different groups by Western blot. (B) Protein expression of p-JNK and JNK in different groups by Western blot. (C) Protein expression of p-P38 and P38 in different groups by Western blot. (D) Protein expression of p-ERK5 and ERK5 in different groups by Western blot. The results were normalizing the bands to their whole protein bands and described as a fold change. Values are the mean±SD, n=3. **p<0.01 and ***p<0.001 versus the control values.
FIG. 4.
Effect of ERK1/2 inhibitor on the phosphorylation of ERK1/2, Elk and the expression of SPP1, RUNX2, and SP7 in CTS-exposed hPDLCs. (A) Protein expression of p-ERK 1/2 by Western blot. (B) Protein expression of p-Elk by Western blot. (C) Protein expression of SPP1 by Western blot. (D) Protein expression of RUNX2 by Western blot. (E) Protein expression of SP7 by Western blot. (F) Gene expression of SPP1, RUNX2, and SP7 by real-time PCR. The results were normalizing to their whole protein bands or GAPDH and described as a fold change. Values are the mean±SD, n=3. *p<0.05, **p<0.01, and ***p<0.001 versus the control values. ##p<0.01 and ###p<0.001 versus the experiments.+U0126 ERK1/2 inhibitor U0126 treated.
Discussion
Cultured hPDLCs have been widely used to study the effects of mechanical stress on the expression of individual genes of interest (Saito et al., 1991; Shimizu et al., 1994; Howard et al., 1998; Chiba and Mitani, 2004; Yang et al., 2006; Nishijima et al., 2006; Wescott et al., 2007; Pavlidis et al., 2009; Tang et al., 2012). The results are equivocal depending on the culture conditions, stress regime, and the design of the mechanical deformation apparatus (Ozaki et al., 2005; Wescott et al., 2007; Saminathan et al., 2012). For example, cyclic tension has been reported to both inhibit (Chiba and Mitani, 2004) and stimulate (Yang et al., 2006) the synthesis of ALP. In the Flexcell system, a silicone membrane is stretched across a loading post by the application of vacuum pressure and depending on the shape of the loading post, either a biaxial or a uniaxial strain (as in the present study) may be applied to the cells. The silicone membrane is coated with type I collagen, which is more suitable for cell adhesion and simulates the in vivo environment. This system can have a better control of waveforms at low and high amplitudes through a computer-regulated bioreactor. Although the Flexcell system does have some limitations (Vande Geest et al., 2004; Wall et al., 2007; Milne et al., 2009; Saminathan et al., 2012), this system still remains the most effective way to screen cells for the expression of mechano-responsive gene in vitro methodology.
Laboratory evidence has already affirmed the significance of tensile strain during OTM with regard to the osteogenic differentiation of PDLCs. 3,000 μstrain cyclic uniaxial tensile stress significantly increased mRNA and protein expressions of STAB2, RUNX2, and OSX using a self-made system (Tang et al., 2012). 12% uniaxial tensile during OTM could express multiple genes involved in osteogenesis in hPDLCs (Wescott et al., 2007). However, a few researches are involved in physiological occlusal force, which constitutively exists in a normal oral environment and is important for PDL homeostasis and remodeling. Physiological occlusal force is an important symbol of the stomatognathic system. The homeostasis of periodontal tissue depends on the stimulation of occlusal force. On the contrary, loss of occlusal function caused atrophic changes in the PDL, such as narrowing of the space, disorientation of collagen fibers, and decreases in proteoglycans (Kaneko et al., 2001). Thus, it is important to clarify the physiological functions of mechanical stress with regard to the PDL, and our study aims at investigating the effect of CTS during physiological occlusal force on the behavior of hPDLCs.
Human gene expression microarray was employed in this study, which enables us to assess the whole genome transcriptome changes even though it would take us more time to clarify the whole content in the results. Previous studies have shown that cyclic tension loaded to hPDLCs would either increase (Yousefian et al., 1995; Matsuda et al., 1998b) or inhibit proliferation (Hao et al., 2009). In our microarray analysis, DPT, ASCL2, APOE, BTG1, NPPA, and SPP1, which were involved in negative regulation of cell proliferation, were up-regulated. As we know, slowing down proliferation will benefit cells to respond to external signals and then to affect differentiation (Lee et al., 2007; Wang et al., 2011). Therefore, from another aspect, the microarray results confirm that cells tend to differentiate rather than proliferate on being exposed to 10% CTS.
In the present microarray, SPP1 in the up-regulated genes group with higher expression was involved in most of the biological processes. SPP1 is an early marker of osteoblast differentiation and plays an important role in bone formation, resorption, and remodeling (McKee et al., 1993; Yang et al., 2010). SPP1 is thought to promote or regulate the adhesion, attachment, and spreading of osteoclasts to the bone surface during bone resorption (Standal et al., 2004). Previous studies have demonstrated that mechanical stress-induced expression of SPP1 is mostly composed of, or is present in, almost all osteocytes and in some osteoblasts and bone-lining cells at the resorption site in the early stage of experimental tooth movement (Terai et al., 1999; Kim et al., 2012). Therefore, we chose SPP1 as the major indicators to observe the osteogenic differentiation of hPDLCs under CTS. We demonstrated that the mRNA and protein expressions of SPP1 were up-regulated under 10% CTS in vitro. So, we hypothesized that SPP1 might act as a trigger for bone and cementum remodeling and is involved in osteogenic differentiation caused by 10% CTS.
In the current study, 10% CTS can exert some influence on the expression of osteoblast markers (RUNX2 and SP7) in hPDLCs, indicating that 10% CTS could enhance the osteogenic differentiation of these cells. RUNX2 and SP7 are necessary for the osteogenic differentiation (Komori, 2006; Baek et al., 2009). Genetic and molecular studies have shown that RUNX2 plays a key role in the osteogenesis and serves as an early transcriptional regulator of osteogenic differentiation (Takeda et al., 2001; Karsenty and Wagner, 2002). 10% CTS-treated hPDLCs exhibited higher gene and protein expression levels of RUNX2 for 12 h as well as RUNX2-positive cells were detected in the PDL during the first 3 days, suggesting that CTS can trigger the early-stage osteogenic differentiation of hPDLCs in vitro and in vivo. SP7 is vital in osteogenic differentiation as a result of its ability to regulate the expression of a number of important osteoblast marker genes such as SPP1, osteonectin, osteocalcin (OCN), bone sialoprotein (BSP), and collagen type I (COL I) (Nakashima et al., 2002). We detected that 10% CTS-treated hPDLCs exhibited higher protein expression levels of SP7 for 12, 24, and 48 h along with a large number of SP7-positive cells compared with RUNX2 and KI67 from day 3 to day 10, suggesting that CTS plays an important role not only in the early stage of osteogenesis but also in the later stage of bone formation. Interestingly, we observed that SP7 mRNA was decreased after 10% CTS loading from 12 to 48 h, which was inconsistent with other studies (Zhao et al., 2008; Tang et al., 2012). We hypothesized that 10% CTS might regulate SP7 mRNA, thus affecting its stability and leading to inconsistent transcript and protein levels. The relationship between mRNA and protein is not strictly linear, but has a more intrinsic and complex dependence, deviating from the classical view referred to as the molecular dogma. Different regulation mechanisms such as post-transcriptional processing, the degradation of the transcripts, translation, post-translational processing and modification, acting on both the synthesized mRNA and the synthesized protein, affect the amount of the two molecules differentially. So, it is not exactly the same as the transcription and translation levels. In addition, we noted that the significant expressions of SPP1, RUNX2, and SP7 were in a different CTS loading time in vitro and in vivo. It is speculated that these three factors were affected by CTS in different ways, and the specific mechanisms involved need further investigations.
The most important kinases involved in mechano transduction are the MAPKs, because most mechanical signals are transmitted to the nucleus through the activation of these kinases (Whitmarsh and Davis, 1996; Liedert et al., 2006). In our microarray analysis, an MAPK pathway was identified. Kook and Lee (2012) demonstrated that 1.5% elongation tension-stimulated expression of COL I and MMP-1 was controlled by the ERK/JNK-AP-1 and ERK-NF-κB signaling pathways and inhibited the proliferation of human PDL fibroblasts through Ras-p38 MAPK up-regulation. Human PDL fibroblasts stimulate osteoclastogenesis in response to compression force through TNF-alpha-mediated activation of CD4+ T cells (Kook et al., 2011b). Osteopontin expression in mechanical force-subjected PDL fibroblasts was activated by the ERK pathway (Hong et al., 2010). The inconsistent findings show that the mechanisms involved in force-applied hPDLCs also depend on the culture conditions and stress regime. Unlike the previous studies, our current findings showed that ERK1/2 not the JNK, P38, and ERK5 was a central mediator of mechanical signals in hPDLCs under 10% CTS during physiological occlusal force. Elk1, one of the Ets family of transcription factors, was a DNA-binding protein that activates transcription through its phosphorylation (Cruzalegui et al., 1999). ERK inhibitor blocked Elk1 phosphorylation as well as SPP1 gene expression stimulated by oscillatory fluid flow in human osteoblast-like cells (Wu et al., 2006). Therefore, we supposed whether the downstream mechano sensor of ERK1/2 was Elk1 or not. Our present results revealed that when CTS was applied to hPDLCs, the phosphorylation of Elk1 was increased, and this increase was significantly prevented by ERK1/2 inhibitor, suggesting that the cascade phosphorylation of ERK1/2 and Elk1 appeared to be, at least in part, associated with SPP1 expression in 10% CTS-exposed hPDLCs. Further studies are needed to clarify the relationship between ERK1/2/Elk1 and their target transcription factors in CTS-induced up-regulation of SPP1. In general, the microarray data is complex, and will take more time to be clarified. Next, more extensive studies are required to investigate whether other pathway mechanisms that are associated with physiological occlusal force enhance osteogenic differentiation of hPDLCs.
In conclusion, we may first demonstrate that physiological occlusal force induces ERK1/2-Elk1 MAPK signaling in the PDL, resulting in enhancement of the osteogenic differentiation of hPDLCs. These findings suggest that physiological occlusal force-induced ERK1/2-Elk1 MAPK signaling is involved in the homeostasis, remodeling, and regeneration of periodontal tissue.
Acknowledgments
This study was supported in part by grants from the Graduate Student Innovation Project of Jiangsu province (Grant No. CXZZ11_0732), National Natural Science Foundations of China (Grant No. 81170962, 81170981, and 81230022), Project of Science and Technology Department of Jiangsu province (Grant No. BK2011763), Ph.D. Programs Foundation of Ministry of Education of China (20113234110003), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Disclosure Statement
No competing financial interests exist.
References
- Baek W.Y. Lee M.A., et al. Positive regulation of adult bone formation by osteoblast-specific transcription factor osterix. J Bone Miner Res. 2009;24:1055–1065. doi: 10.1359/jbmr.081248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambard J.C. Lefloch R., et al. ERK implication in cell cycle regulation. Biochim Biophys Acta. 2007;1773:1299–1310. doi: 10.1016/j.bbamcr.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Chiba M. Mitani H. Cytoskeletal changes and the system of regulation of alkaline phosphatase activity in human periodontal ligament cells induced by mechanical stress. Cell Biochem Funct. 2004;22:249–256. doi: 10.1002/cbf.1097. [DOI] [PubMed] [Google Scholar]
- Cho J.H. Lee S.K., et al. The role of heme oxygenase-1 in mechanical stress- and lipopolysaccharide-induced osteogenic differentiation in human periodontal ligament cells. Angle Orthod. 2010;80:552–559. doi: 10.2319/091509-520.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruzalegui F.H. Cano E., et al. ERK activation induces phosphorylation of Elk-1 at multiple S/T-P motifs to high stoichiometry. Oncogene. 1999;18:7948–7957. doi: 10.1038/sj.onc.1203362. [DOI] [PubMed] [Google Scholar]
- Franceschi R.T. Xiao G. Regulation of the osteoblast-specific transcription factor, Runx2: responsiveness to multiple signal transduction pathways. J Cell Biochem. 2003;88:446–454. doi: 10.1002/jcb.10369. [DOI] [PubMed] [Google Scholar]
- Fujihara C. Yamada S., et al. Role of mechanical stress-induced glutamate signaling-associated molecules in cytodifferentiation of periodontal ligament cells. J Biol Chem. 2010;285:28286–28297. doi: 10.1074/jbc.M109.097303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlet T.P. Coelho U., et al. Cytokine expression pattern in compression and tension sides of the periodontal ligament during orthodontic tooth movement in humans. Eur J Oral Sci. 2007;115:355–362. doi: 10.1111/j.1600-0722.2007.00469.x. [DOI] [PubMed] [Google Scholar]
- Hao Y. Xu C., et al. Cyclic stretching force induces apoptosis in human periodontal ligament cells via caspase-9. Arch Oral Biol. 2009;54:864–870. doi: 10.1016/j.archoralbio.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Hong S.Y. Jeon Y.M., et al. Activation of RhoA and FAK induces ERK-mediated osteopontin expression in mechanical force-subjected periodontal ligament fibroblasts. Mol Cell Biochem. 2010;335:263–272. doi: 10.1007/s11010-009-0276-1. [DOI] [PubMed] [Google Scholar]
- Howard P.S. Kucich U., et al. Mechanical forces alter extracellular matrix synthesis by human periodontal ligament fibroblasts. J Periodontal Res. 1998;33:500–508. doi: 10.1111/j.1600-0765.1998.tb02350.x. [DOI] [PubMed] [Google Scholar]
- Kaneko S. Ohashi K. Soma K. Yanagishita M. Occlusal hypofunction causes changes of proteoglycan content in the rat periodontal ligament. J Periodontal Res. 2001;36:9–17. doi: 10.1034/j.1600-0765.2001.00607.x. [DOI] [PubMed] [Google Scholar]
- Karsenty G. Wagner E.F. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- Kasamatsu A. Uzawa K., et al. Elevation of galectin-9 as an inflammatory response in the periodontal ligament cells exposed to Porphylomonas gingivalis lipopolysaccharide in vitro and in vivo. Int J Biochem Cell Biol. 2005;37:397–408. doi: 10.1016/j.biocel.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Kim J.Y. Kim B.I., et al. Localization of osteopontin and osterix in periodontal tissue during orthodontic tooth movement in rats. Angle Orthod. 2012;82:107–114. doi: 10.2319/030911-173.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T. Regulation of osteoblast differentiation by transcription factors. J Cell Biochem. 2006;99:1233–1239. doi: 10.1002/jcb.20958. [DOI] [PubMed] [Google Scholar]
- Kook S.H. Hwang J.M., et al. Mechanical force induces type I collagen expression in human periodontal ligament fibroblasts through activation of ERK/JNK and AP-1. J Cell Biochem. 2009;106:1060–1067. doi: 10.1002/jcb.22085. [DOI] [PubMed] [Google Scholar]
- Kook S.H. Lee J.C. Tensile force inhibits the proliferation of human periodontal ligament fibroblasts through Ras-p38 MAPK up-regulation. J Cell Physiol. 2012;227:1098–1106. doi: 10.1002/jcp.22829. [DOI] [PubMed] [Google Scholar]
- Kook S.H. Jang Y.S., et al. Involvement of JNK-AP-1 and ERK-NF-kappaB signaling in tension-stimulated expression of type I collagen and MMP-1 in human periodontal ligament fibroblasts. J Appl Physiol. 2011a;111:1575–1583. doi: 10.1152/japplphysiol.00348.2011. [DOI] [PubMed] [Google Scholar]
- Kook S.H. Jang Y.S., et al. Human periodontal ligament fibroblasts stimulate osteoclastogenesis in response to compression force through TNF-alpha-mediated activation of CD4+ T cells. J Cell Biochem. 2011b;112:2891–2901. doi: 10.1002/jcb.23205. [DOI] [PubMed] [Google Scholar]
- Lee Y.H. Nahm D.S., et al. Differential gene expression of periodontal ligament cells after loading of static compressive force. J Periodontol. 2007;78:446–452. doi: 10.1902/jop.2007.060240. [DOI] [PubMed] [Google Scholar]
- Li C. Hu Y., et al. Ras/Rac-Dependent activation of p38 mitogen-activated protein kinases in smooth muscle cells stimulated by cyclic strain stress. Arterioscler Thromb Vasc Biol. 2000;20:E1–E9. doi: 10.1161/01.atv.20.3.e1. [DOI] [PubMed] [Google Scholar]
- Liedert A. Kaspar D., et al. Signal transduction pathways involved in mechanotransduction in bone cells. Biochem Biophys Res Commun. 2006;349:1–5. doi: 10.1016/j.bbrc.2006.07.214. [DOI] [PubMed] [Google Scholar]
- Lin Z. Navarro V.P., et al. LMP1 regulates periodontal ligament progenitor cell proliferation and differentiation. Bone. 2010;47:55–64. doi: 10.1016/j.bone.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long P. Liu F., et al. Signaling by mechanical strain involves transcriptional regulation of proinflammatory genes in human periodontal ligament cells in vitro. Bone. 2002;30:547–552. doi: 10.1016/s8756-3282(02)00673-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N. Yokoyama K., et al. Role of epidermal growth factor, its receptor in mechanical stress-induced differentiation of human periodontal ligament cells in vitro. Arch Oral Biol. 1998a;43:987–997. doi: 10.1016/s0003-9969(98)00079-x. [DOI] [PubMed] [Google Scholar]
- Matsuda N. Morita N., et al. Proliferation and differentiation of human osteoblastic cells associated with differential activation of MAP kinases in response to epidermal growth factor, hypoxia, and mechanical stress in vitro. Biochem Biophys Res Commun. 1998b;249:350–354. doi: 10.1006/bbrc.1998.9151. [DOI] [PubMed] [Google Scholar]
- McKee M.D. Farach-Carson M.C., et al. Ultrastructural immunolocalization of noncollagenous (osteopontin and osteocalcin) and plasma (albumin and alpha 2HS-glycoprotein) proteins in rat bone. J Bone Miner Res. 1993;8:485–496. doi: 10.1002/jbmr.5650080413. [DOI] [PubMed] [Google Scholar]
- Milne T.J. Ichim I., et al. Induction of osteopenia during experimental tooth movement in the rat: alveolar bone remodelling and the mechanostat theory. Eur J Orthod. 2009;31:221–231. doi: 10.1093/ejo/cjp032. [DOI] [PubMed] [Google Scholar]
- Nakashima K. Zhou X., et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Nishijima Y. Yamaguchi M., et al. Levels of RANKL and OPG in gingival crevicular fluid during orthodontic tooth movement and effect of compression force on releases from periodontal ligament cells in vitro. Orthod Craniofac Res. 2006;9:63–70. doi: 10.1111/j.1601-6343.2006.00340.x. [DOI] [PubMed] [Google Scholar]
- Ozaki S. Kaneko S., et al. Modulation of extracellular matrix synthesis and alkaline phosphatase activity of periodontal ligament cells by mechanical stress. J Periodontal Res. 2005;40:110–117. doi: 10.1111/j.1600-0765.2004.00782.x. [DOI] [PubMed] [Google Scholar]
- Pavlidis D. Bourauel C., et al. Proliferation and differentiation of periodontal ligament cells following short-term tooth movement in the rat using different regimens of loading. Eur J Orthod. 2009;31:565–571. doi: 10.1093/ejo/cjp053. [DOI] [PubMed] [Google Scholar]
- Redlich M. Roos H., et al. The effect of centrifugal force on mRNA levels of collagenase, collagen type-I, tissue inhibitors of metalloproteinases and beta-actin in cultured human periodontal ligament fibroblasts. J Periodontal Res. 2004;39:27–32. doi: 10.1111/j.1600-0765.2004.00700.x. [DOI] [PubMed] [Google Scholar]
- Saito M. Saito S., et al. Interleukin 1 beta and prostaglandin E are involved in the response of periodontal cells to mechanical stress in vivo and in vitro. Am J Orthod Dentofacial Orthop. 1991;99:226–240. doi: 10.1016/0889-5406(91)70005-H. [DOI] [PubMed] [Google Scholar]
- Saminathan A. Vinoth K.J., et al. The effect of cyclic mechanical strain on the expression of adhesion-related genes by periodontal ligament cells in two-dimensional culture. J Periodontal Res. 2012;47:212–221. doi: 10.1111/j.1600-0765.2011.01423.x. [DOI] [PubMed] [Google Scholar]
- Shimizu N. Yamaguchi M., et al. Cyclic-tension force stimulates interleukin-1 beta production by human periodontal ligament cells. J Periodontal Res. 1994;29:328–333. doi: 10.1111/j.1600-0765.1994.tb01230.x. [DOI] [PubMed] [Google Scholar]
- Standal T. Borset M., et al. Role of osteopontin in adhesion, migration, cell survival and bone remodeling. Exp Oncol. 2004;26:179–184. [PubMed] [Google Scholar]
- Takeda S. Bonnamy J.P., et al. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 2001;15:467–481. doi: 10.1101/gad.845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N. Zhao Z., et al. Up-regulated osteogenic transcription factors during early response of human periodontal ligament stem cells to cyclic tensile strain. 2012. pp. 422–430. [DOI] [PMC free article] [PubMed]
- Terai K. Takano-Yamamoto T., et al. Role of osteopontin in bone remodeling caused by mechanical stress. J Bone Miner Res. 1999;14:839–849. doi: 10.1359/jbmr.1999.14.6.839. [DOI] [PubMed] [Google Scholar]
- Vande Geest J.P. Di Martino E.S., et al. An analysis of the complete strain field within Flexercell membranes. J Biomech. 2004;37:1923–1928. doi: 10.1016/j.jbiomech.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Wall M.E. Weinhold P.S., et al. Comparison of cellular strain with applied substrate strain in vitro. J Biomech. 2007;40:173–181. doi: 10.1016/j.jbiomech.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Wang Y. Li Y., et al. Early proliferation alteration and differential gene expression in human periodontal ligament cells subjected to cyclic tensile stress. Arch Oral Biol. 2011;56:177–186. doi: 10.1016/j.archoralbio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Wescott D.C. Pinkerton M.N., et al. Osteogenic gene expression by human periodontal ligament cells under cyclic tension. J Dent Res. 2007;86:1212–1216. doi: 10.1177/154405910708601214. [DOI] [PubMed] [Google Scholar]
- Whitmarsh A.J. Davis R.J. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J Mol Med (Berl) 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- Wu C.C. Li Y.S., et al. Roles of MAP kinases in the regulation of bone matrix gene expressions in human osteoblasts by oscillatory fluid flow. J Cell Biochem. 2006;98:632–641. doi: 10.1002/jcb.20697. [DOI] [PubMed] [Google Scholar]
- Yamashiro K. Myokai F., et al. Oligonucleotide array analysis of cyclic tension-responsive genes in human periodontal ligament fibroblasts. Int J Biochem Cell Biol. 2007;39:910–921. doi: 10.1016/j.biocel.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Yang Y.Q. Li X.T., et al. Human periodontal ligament cells express osteoblastic phenotypes under intermittent force loading in vitro. Front Biosci. 2006;11:776–781. doi: 10.2741/1835. [DOI] [PubMed] [Google Scholar]
- Yang Y. Li X., et al. Functional analysis of core binding factor a1 and its relationship with related genes expressed by human periodontal ligament cells exposed to mechanical stress. Eur J Orthod. 2010;32:698–705. doi: 10.1093/ejo/cjq010. [DOI] [PubMed] [Google Scholar]
- Yousefian J. Firouzian F., et al. A new experimental model for studying the response of periodontal ligament cells to hydrostatic pressure. Am J Orthod Dentofacial Orthop. 1995;108:402–409. doi: 10.1016/s0889-5406(95)70038-2. [DOI] [PubMed] [Google Scholar]
- Zampetaki A. Zhang Z., et al. Biomechanical stress induces IL-6 expression in smooth muscle cells via Ras/Rac1-p38 MAPK-NF-kappaB signaling pathways. Am J Physiol Heart Circ Physiol. 2005;288:H2946–H2954. doi: 10.1152/ajpheart.00919.2004. [DOI] [PubMed] [Google Scholar]
- Zhang W. Liu H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- Zhao Y. Wang C., et al. Expression of Osterix in mechanical stress-induced osteogenic differentiation of periodontal ligament cells in vitro. Eur J Oral Sci. 2008;116:199–206. doi: 10.1111/j.1600-0722.2008.00533.x. [DOI] [PubMed] [Google Scholar]