Abstract
Following penetrating injury of the skin, a highly orchestrated and overlapping sequence of events helps to facilitate wound resolution. Inflammation is a hallmark that is initiated early, but the reciprocal relationship between cells and matrix molecules that triggers and maintains inflammation is poorly appreciated. Elastin is enriched in the deep dermis of skin. We propose that deep tissue injury encompasses elastin damage, yielding solubilized elastin that triggers inflammation. As dermal fibroblasts dominate the deep dermis, this means that a direct interaction between elastin sequences and fibroblasts would reveal a proinflammatory signature. Tropoelastin was used as a surrogate for elastin sequences. Tropoelastin triggered fibroblast expression of the metalloelastase MMP-12, which is normally expressed by macrophages. MMP-12 expression increased 1056 ± 286-fold by 6 h and persisted for 24 h. Chemokine expression was more transient, as chemokine C-X-C motif ligand 8 (CXCL8), CXCL1, and CXCL5 transcripts increased 11.8 ± 2.6-, 10.2 ± 0.4-, and 8593 ± 996-fold, respectively, by 6–12 h and then decreased. Through the use of specific inhibitors and protein truncation, we found that transduction of the tropoelastin signal was mediated by the fibroblast elastin binding protein (EBP). In silico modeling using a predictive computational fibroblast model confirmed the up-regulation, and simulations revealed PKA as a key part of the signaling circuit. We tested this prediction with 1 μM PKA inhibitor H-89 and found that 2 h of exposure correspondingly reduced expression of MMP-12 (63.9±12.3%) and all chemokine markers, consistent with the levels seen with EBP inhibition, and validated PKA as a novel node and druggable target to ameliorate the proinflammatory state. A separate trigger that utilized C-terminal RKRK of tropoelastin reduced marker expression to 65.0–76.5% and suggests the parallel involvement of integrin αVβ3. We propose that the solubilization of elastin as a result of dermal damage leads to rapid chemokine up-regulation by fibroblasts that is quenched when exposed elastin is removed by MMP-12.—Almine, J. F., Wise, S. G., Hiob, M., Kumar Singh, N. K., Tiwari, K. K., Vali, S., Abbasi, T., and Weiss, A. S. Elastin sequences trigger transient proinflammatory responses by human dermal fibroblasts.
Keywords: signaling, wound healing
Cutaneous injuries result in the damage and loss of extracellular matrix (ECM) proteins and skin cells, which are fundamental components to the structure of skin. After injury, the wound-healing process is initiated (1). A blood clot is initially formed to stem blood loss, while inflammatory cells infiltrate the wound site to remove foreign cells and materials. The inflammatory stage of wound healing contains an abundance of matrix fragments generated by proteolysis of damaged ECM. These matrix fragments have cell-signaling potential and induce proinflammatory-related cellular responses (2). Elastin fragments are prominent because they are derived from elastic fibers, which are widely distributed throughout the dermis, particularly in the deep layers (3). An intact elastic fiber network is one of the most important elastic facets of skin that is lost due to injury (4). The abundance of solubilized elastin during the early phase of skin damage suggests that they may also be involved in signaling events with human dermal fibroblasts (HDFs), the dominant cell type in the dermis. We studied the interaction of HDFs with exposed elastin sequences with the aim of exploring up-regulated transcripts and candidate cell-signaling pathways induced by solubilized elastin.
Elastin sequences possess potent cell-signaling properties and induce various biological responses in cells, including migration and proliferation (5–9). Because of its mechanical and signaling properties, elastin serves a multifunctional role in wound healing. Tropoelastin is the soluble precursor of elastin, expressed and secreted by elastogenic cells, such as HDFs (10). Mature elastic fibers have little turnover, and tropoelastin production is absent in maturity but is induced by injury. Furthermore, a range of exogenous factors can influence tropoelastin production (11). Despite an occasional short increase in tropoelastin synthesis at the onset of injury, deposition of elastin in the dermis is fleeting, aberrant, and only detectable well after the initial wound-healing stages (12, 13).
The loss of tissue integrity is a direct and immediate result of tissue damage. Re-establishing structural and mechanical integrity is essential to recovery and is aided by inflammation, which is a recognized feature of early wound responses. Inflammation is pivotal to local stabilization, but if left unregulated can chronically delay tissue recovery due to persistent residence of inflammatory cells in the wound site and overproduction of proteases (14).
Only the elastokine signaling capacity of some biologically active small fragments of elastin has been cataloged (15). The most common permutation of elastokines conforms to a GXXPG motif, such as VGVAPG, GVAPGV, and PGAIPG. VGVAPG is the best characterized elastokine and stimulates various biological responses in a range of cells, including monocyte chemotaxis (16); HDF chemotaxis (5), proliferation (17) and protease production (18); keratinocyte migration and differentiation (7); and promotion of an angiogenic phenotype in endothelial cells (19). EBP binds VGVAPG with the highest affinity but can bind other elastin sequences, such as PGIVPG, conforming to the XGXXPG motif (20).
As tropoelastin is the precursor of elastin, tropoelastin encompasses a representative range of the primary sequences of elastin and provides a clean opportunity to systematically examine the full signaling potential of elastin using a precise sequence. Investigating the interaction and cellular response of HDF to tropoelastin contributes to the expanding knowledge base of the dynamic reciprocity paradigm where the bidirectional interaction between cells and ECM is essential to homeostasis and developmental, reparative and regenerative processes (21).
In our cellular response model system, soluble tropoelastin monomers were used for coating substrata to prevent aggregation (22) and exposed to HDFs. From a full microarray study, we found that the only substantially up-regulated transcripts were matrix metalloproteinase 12 (MMP-12) and a chemokine cluster of CXC chemokine ligand 8 (CXCL8), CXCL5, and CXCL1. MMP-12 up-regulation was sustained over 24 h. The chemokine cluster peaked at 6–12 h before declining. To understand the signaling underlying the up-regulation of MMP-12 and the chemokines, we used a predictive computational simulation model of the HDFs. The simulation analysis provided novel insight that tropoelastin-stimulated expression of MMP-12, CXCL8, CXCL5, and CXCL1 was mediated by elastin binding protein (EBP) that included a protein kinase A (PKA) circuit. We further simulated the effect of PKA inhibition in reducing the elastin-mediated inflammation, and this prediction was blindly validated experimentally. A reduced response to tropoelastin lacking the C-terminal RKRK further implicates integrin αVβ3. Through an integration of experimental and predictive approaches, our study reveals atypical up-regulation by MMP-12 and clustered chemokine expression by HDFs, and presents a proof of concept for PKA as a druggable site to regulate the inflammatory trigger.
MATERIALS AND METHODS
Tropoelastin and fibroblasts
Recombinant human tropoelastin isoform SHELΔ26A (synthetic human elastin without domain 26A) corresponding to amino acid residues 27–724 of GenBank entry AAC98394 (gi 182020) and ΔRKRK were expressed and purified as described previously (23, 24). ΔRKRK was confirmed by plasmid sequencing and comparative mass spectrometry of the purified protein against wild type (Supplemental Fig. S1). The integrity of all constructs was verified by SDS-PAGE to be full length.
HDFs were obtained from biopsies of donor sites in consenting burn patients in the Burns Unit at Concord Repatriation General Hospital (Concord, New South Wales) in accordance with the approval of the Hospital Research and Ethics Committee.
Cell attachment and spreading
Cell attachment was performed as described previously (25). For cell spreading, triplicate cell culture wells were coated with 30 μg/ml tropoelastin, 2 μg/ml human fibronectin, and 1 mg/ml bovine collagen type I at 4°C overnight and then washed with PBS. Wells were blocked for 1 h with 10 μg/ml denatured BSA in PBS. Trypsinized HDFs were seeded at a density of 5200 cells/cm2. Cells were allowed to attach at 37°C for 1 h. Nonadherent cells were removed with PBS. Cells were fixed with 3.7% formaldehyde. Ten consecutive images of each replicate per treatment were imaged under phase contrast at ×100 view. The total number of attached cells and spreading cells were manually counted and expressed as percentage cell spreading.
Cell proliferation
Triplicate cell culture wells were coated with 30 μg/ml tropoelastin, 2 μg/ml human fibronectin, and 1 mg/ml bovine collagen type I at 4°C overnight, and then washed with PBS. Wells were blocked for 1 h with 10 μg/ml denatured BSA in PBS. Trypsinized HDFs were seeded at a density of 5200 cells/cm2. Cells were allowed to proliferate at 37°C for 1, 3, 5, and 7 d. Nonadherent cells were removed with PBS. Cell viability was assayed using CellTiter 96 Aqueous One Solution (Promega, Madison, WI, USA). CellTiter reagent and DMEM with 10% (v/v) FBS was added to wells in a 5:1 volume ratio. The absorbance at 490 nm of sample wells was measured.
RNA isolation
Triplicate cell culture wells were coated with 30 μg/ml tropoelastin at 4°C overnight and then washed with PBS to remove unbound protein. Wells were blocked for 1 h with 10 μg/ml denatured BSA in PBS. Trypsinized HDFs were seeded at a density of 2.5 × 104 cells/cm2. Cells were allowed to grow for 24 h. Nonadherent cells were removed with PBS, and then total RNA was extracted. RNeasy mini extraction (Qiagen, Valencia, CA, USA) was performed using a modified protocol with the following changes: cells were lysed with TRIzol LS (Life Technologies, Carlsbad, CA, USA). Chloroform was added to the lysate, vortexed for 15 s, and then centrifuged at 12,000 g for 15 min. The upper aqueous layer was extracted and mixed with equal volumes of 70% (v/v) ethanol. RNA concentration and purity were determined by UV absorbance. RNA integrity was assessed by a 1.2% denaturing formaldehyde agarose gel.
Microarray analysis
Duplicate samples of RNA were probed and analyzed by microarray analysis using GeneChip Human Gene 1.0 ST arrays (Affymetrix, Santa Clara, CA, USA). Expression Console 1.0 software (Affymetrix) was used to normalize data using RMA-sketch, which were then annotated using HuGene 1.0 ST v1 library and annotation files. Signal intensities were averaged between duplicates, and sd was determined. Differentially expressed genes were selected on the basis of having a fold change ≥ 2 and a signal intensity above background (i.e., 200) level.
Real-time polymerase chain reaction (PCR)
RNA (200 ng) was reverse transcribed using SuperScript VILO cDNA synthesis kit (Life Technologies). RNA samples without reverse transcriptase (RT) were also included as no-RT controls. Primers were designed by the Primer3Plus online program (http://primer3plus.com/) and are listed in Table 1. Real-time PCR was performed with FAST SYBR Green master mix (Applied Biosystems, Foster City, CA, USA). cDNA samples were analyzed in duplicate at 2 concentrations, 10 and 1.25 ng cDNA/reaction. Negative controls were no-RT and no-template control (NTC). Quantitative comparative (ΔΔCT) real-time PCR was performed using a 7500 fast real-time PCR system (Applied Biosystems) using stage 1, 50°C for 10 min and then 95°C for 20 s, and stage 2, 95°C for 3 s and then 60°C for 30 s, repeated for 40 cycles. A continuous melting curve was performed: 95°C for 15 s, 60°C for 60 s, 95°C for 15 s, and 60°C for 15 s. All data were normalized to 18S rRNA. Fold-change expression relative to the control was calculated using 2−ΔΔCT. Percentage expression was calculated by converting CT values to number of gene copies and comparing expression levels to the control set to 100%.
Table 1.
Real-time PCR primer sequences of target genes
| Target gene and accession number | Primer sequences, 5′ → 3′ | Nucleotide position |
|---|---|---|
| 18S rRNA, NR_003286 | F: CCTGCGGCTTAATTTGACTC | 1230–1347 |
| R: AACTAAGAACGGCCATGCAC | ||
| MMP12, NM_002426 | F: TGCCAAATCCTGACAATTCA | 857–973 |
| R: CCTTCAGCCAGAAGAACCTG | ||
| PAI-2, NM_002575 | F: CAGGCACAAGCTGCAGATAA | 361–480 |
| R: CGCAGACTTCTCACCAAACA | ||
| CXCL8, NM_000584 | F: GTTCCACTGTGCCTTGGTTT | 901–995 |
| R: GCTTCCACATGTCCTCACAA | ||
| CXCL5, NM_002994 | F: GTGTTGAGAGAGCTGCGTTG | 245–332 |
| R: CTATGGCGAACACTTGCAGA | ||
| CXCL1, NM_001511 | F: GAAAGCTTGCCTCAATCCTG | 325–431 |
| R: CACCAGTGAGCTTCCTCCTC | ||
| TFPI2, NM_006528 | F: GCTGTGGAGGGAATGACAAT | 644–760 |
| R: TCCGGATTCTACTGGCAAAG | ||
| SOD2, NM_001024465 | F: GTTGGCCAAGGGAGATGTTA | 370–476 |
| R: TAGGGCTGAGGTTTGTCCAG | ||
| BDKRB1, NM_000710 | F: TGGGTTTCCTCCTACCACTG | 834–924 |
| R: CTTGTCCTGCTGACCTCCTC | ||
| KIF20A, NM_005733 | F: GCAGGAAAACTTCGTCAAGC | 2798–2910 |
| R: TTCCGAAGGTCCAGTTTCAC | ||
| ASPM, NM_018136 | F: AAACGCCATCAGGAGAGAGA | 4116–4226 |
| R: CTGAATGACGAGTGCTGCAT | ||
| HAS2, NM_005328 | F: ACCGGGGTAAAATTTGGAAC | 1734–1820 |
| R: TAAGGCAGCTGGCAAAAGAT |
F, forward; R, reverse.
Predictive experiments on simulation models of human fibroblast cells
Predictive experiments were performed using the human fibroblast cell computational simulation technology (Cellworks Group). This technology has been extensively validated (26, 27).
Simulation technology description
The Cellworks simulation technology platform is implemented using a 3-layered architecture. The top layer is a text user interface/graphic user interface (TUI/GUI)-driven user interface. The middle layer in this context is the human fibroblast cell representation. The bottom layer is the computational back plane, which enables the system to be dynamic and computes all the mathematics in the middle layer.
Among the thousands of markers for skin collagen type I, MMPs and elastin are present in the simulation model. The in silico model encompasses important signaling pathways comprising growth factors like EGF, PDGFA/B, FGF1/2, c-MET, CTGF, and IGF-1; cytokine pathways like IL1A/B, IL4, IL6, IL10, IL15, IL17, IL18, TNF-α/β, IFNA/B/Y, CD40, and TGF-β; GPCR signaling like PGE2, PGI2, EDN1/2/3, S1P, elastin fragments/tropoelastin, and LPA; and cholesterol biosynthesis and oxidative stress pathways. The human fibroblast cell has been modeled with respect to changes in time-dependent fluxes and stimuli, which utilizes modified ordinary differential equations and mass action kinetics. The starting state of the system is based on normal cell physiology. The user can control the transition of the normal system to a triggered state via different triggers. Knockdown or overexpression can be done at the expression or activity levels.
Predictive simulation study experimental protocol
The predictive simulation-based human fibroblast cell system is initialized to normal physiological state, after which the cell is triggered through an increase in transcription of elastin. PKA inhibition was identified and analyzed in this increased transcription of elastin system. Following this, the simulation predictions were validated in the wet laboratory and correlated.
In vitro inhibition studies
Triplicate cell culture wells were coated with 30 μg/ml tropoelastin at 4°C overnight, washed with PBS, and then blocked for 1 h with 10 μg/ml denatured BSA in PBS. Trypsinized HDFs were seeded at a density of 2.5 × 104 cells/cm2 with or without α-lactose (10 mM), β-lactose (10 mM), and glucose (10 mM), and maintained for 3 h. For incubation with or without the pharmacological inhibitor N-[2-((p-bromocinnamyl)amino)ethyl]-5-isoquinolinesulfonamide (H-89; 1 μM), cells were maintained for 2 h. For incubation with the tropoelastin mutant construct ΔRKRK (30 μg/ml), cells were maintained for 3 h.
Statistical analyses
All data in text and figures are expressed as means ± sd. Student's unpaired t test, 1-way ANOVA with Tukey post hoc test or 2-way ANOVA with Bonferroni post hoc test were performed using Prism 5.00 for Windows (GraphPad Software, San Diego, CA, USA). Statistical significance was accepted at values of P < 0.05.
RESULTS AND DISCUSSION
Tropoelastin supports the attachment, spreading, and proliferation of dermal fibroblasts
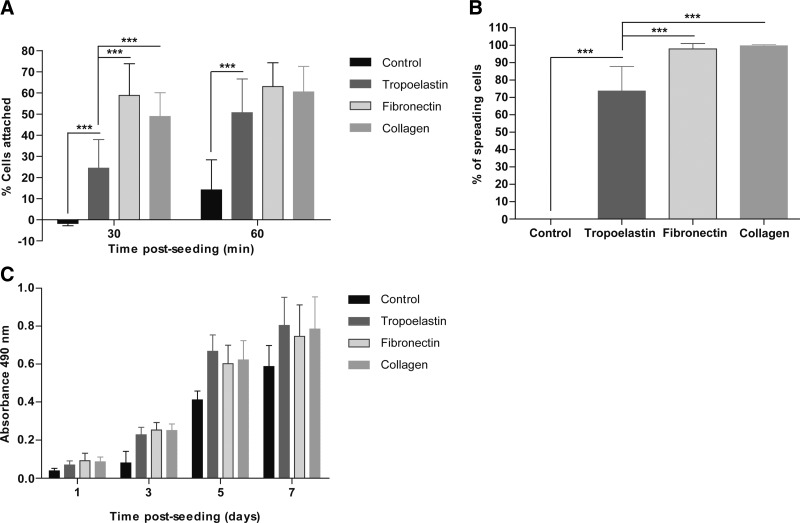
Early attachment of HDF to tropoelastin was less than that of fibronectin and type I collagen, which are known to strongly promote adhesion. After 1 h, this difference was eliminated, and tropoelastin did not significantly differ from either fibronectin or type I collagen (Fig. 1A). Cell attachment on tropoelastin at 1 h (50.6±16.0%) was comparable to fibronectin (63.0±11.3%) and type I collagen (60.5±12.0%). Tropoelastin also promoted HDF spreading, but not to the extent seen for fibronectin and type I collagen. Although these molecules are known to interact with fibroblasts through integrins, tropoelastin lacks the canonical RGD binding sequence and instead uses its C-terminal RKRK to interact with HDF via integrin αVβ3 (Fig. 1B). Differences in binding efficiencies in early attachment and spreading events eventually disappear, leading to proliferation levels of tropoelastin that are not significantly different to fibronectin and type I collagen (Fig. 1C).
Figure 1.
Response of HDFs to tropoelastin-coated surfaces. Tropoelastin promoted the attachment (A), spreading (B), and proliferation (C) of HDFs. Results shown are averaged of 3 independent experiments. ***P < 0.001.
Tropoelastin induces the expression of MMP12 and a cluster of chemokines
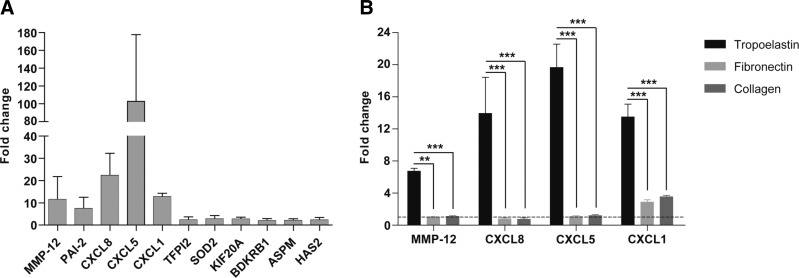
A collection of genes up-regulated by tropoelastin after 24 h of exposure was identified by microarray analysis: MMP-12 had the highest fold change of 9, and a cluster of chemokines, CXCL8, CXCL5, and CXCL1, had fold changes of 5.0, 4.5, and 3.4, respectively (Table 2). Real-time PCR validated the consistent up-regulation across multiple patient cells tested (Fig. 2A) of MMP-12 (6.7±0.4-fold), CXCL8 (13.9±4.5-fold), CXCL5 (19.6±2.9-fold), and CXCL1 (13.5±1.6-fold) (Fig. 2B). In contrast, fibronectin and type I collagen did not elicit these effects. MMP-12 is a matrix metalloelastase and is known to generate elastokines (28), which are broadly released following MMP exposure (8). CXCL8, CXCL5, and CXCL1 are chemokines that participate in proinflammatory cell signaling (29, 30).
Table 2.
Genes up-regulated in HDF grown on tropoelastin-coated surfaces relative to a control surface as identified by microarray analysis
| Accession number | Gene | Gene name | Fold change | Signal intensity on control | Signal intensity on tropoelastin |
|---|---|---|---|---|---|
| NM_002426 | MMP-12 | Matrix metalloproteinase 12 (metalloelastase) | 9.0 | 60.6 ± 9.0 | 545.2 ± 82.0 |
| NM_002575 | PAI-2 | Plasminogen activator inhibitor 2 | 7.5 | 62.4 ± 4.5 | 469.6 ± 80.3 |
| NM_000584 | CXCL8 | Interleukin-8 (IL-8) | 5.0 | 45.6 ± 6.1 | 227.8 ± 4.4 |
| NM_002994 | CXCL5 | CXC chemokine ligand 5 | 4.5 | 107.6 ± 7.9 | 482.2 ± 5.6 |
| NM_001511 | CXCL1 | CXC chemokine ligand 1 | 3.4 | 121.7 ± 23.2 | 415.2 ± 9.3 |
| NM_006528 | TFPI2 | Tissue factor plasminogen inactivator 2 | 3.0 | 110.9 ± 9.8 | 334.2 ± 11.6 |
| NM_001024465 | SOD2 | Superoxide dismutase 2 | 2.7 | 164.8 ± 6.3 | 438.0 ± 6.4 |
| NM_005733 | KIF20A | Kinesin family member 20A | 2.4 | 310.2 ± 14.5 | 734.8 ± 66.1 |
| NM_000710 | BDKRB1 | Bradykinin receptor B1 | 2.1 | 189.6 ± 18.1 | 403.4 ± 42.4 |
| NM_018136 | ASPM | Abnormal spindle homolog | 2.1 | 273.5 ± 26.9 | 581.6 ± 50.0 |
| NM_005328 | HAS2 | Hyaluronan synthase 2 | 2.1 | 980.0 ± 26.5 | 2049.5 ± 157.6 |
Figure 2.
A) MMP-12 and a cluster of chemokines (CXCL8, CXCL5, and CXCL1) exhibited the highest fold changes in real-time PCR validation of microarray analysis. B) MMP-12, CXCL8, CXCL5, and CXCL1 expression was specifically up-regulated by tropoelastin signaling. Dotted line represents a fold change of 1. Results shown are averages of 3 independent experiments. **P < 0.01; ***P < 0.001.
Up-regulated transcripts display temporal expression
Tropoelastin signaling induced distinct temporal expression profiles. In each case, the expression level was compared to the amount at 1 h. MMP-12 expression levels peaked at 6 h (1056±286-fold) and were sustained for at least 24 h (Fig. 3A). In contrast, CXCL8 (11.8±2.6-fold; Fig. 3B) peaked at 6 h, but expression levels were reduced to no significant fold change by 24 h. CXCL5 expression levels peaked later at 12 h (8593±996-fold), with levels significantly declining by 24 h (5534±1262-fold; P<0.01) (Fig. 3C). CXCL1 (10.2±0.4-fold) followed the pattern for CXCL8 (Fig. 3D).
Figure 3.
Temporal gene expression profiles of MMP-12 (A), CXCL8 (B), CXCL5 (C), and CXCL1 (D). Expression level in each case was normalized to the value at 1 h. Dotted line represents a fold change of 1 (i.e., no change in expression). Results shown are representative of 3 independent experiments. ns, not significant. **P < 0.01; ***P < 0.001.
In silico simulation modeling of HDFs accounts for up-regulation and proposes an intracellular circuit
We used a predictive fibroblast cell system (PFCS), which served as a computational simulation model of HDFs for studying exposure to tropoelastin/elastin fragments. This type of in silico approach and technology has been previously extensively validated in the context of cancer and other autoimmune disorder indications (26, 27). The PFCS is a comprehensive representation of signaling and metabolic pathways and also includes tropoelastin/elastin fragment stimulation of multiple signaling cascades starting with the receptor EBP leading to key intracellular mediators PKA, ERK, and Src, which ultimately regulate cAMP responsive element binding protein 1 (CREB1), nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB), and activator protein 1 (AP-1) (Fig. 4A). On this basis, EBP is the main cell receptor for tropoelastin/elastin fragment signal transduction, with PKA identified as an intracellular mediator for several pathways. This EBP-PKA pathway in elastin signal transduction is demonstrated in vitro by elastin peptide stimulation of HDFs (31). Stimulation of EBP in the in silico PFCS up-regulated MMP-12 (70%), CXCL8 (60%), CXCL5 (200%), and CXCL1 (130%) expression compared to unstimulated cells (Fig. 4B). The involvement of PKA in tropoelastin signal transduction and its potential as a druggable target was predicted by the PFCS, where expression levels of MMP-12 (30%), CXCL8 (40%), CXCL5 (120%), and CXCL1 (80%) were reduced in the presence of a PKA-specific inhibitor compared to untreated cells (Fig. 4C).
Figure 4.
In silico signaling pathways induced in simulation model of human fibroblast cells by tropoelastin and elastin fragments. A) Intracellular pathways induced by elastin-based signaling. B) Elastin fragments induce the expression of CXCL1, CXCL5, CXCL8/IL8, and MMP12 compared to unstimulated cells. C) PKA inhibition reduces the expression of CXCL1, CXCL5, CXCL8/IL8, and MMP12 induced by elastin fragments.
Inhibition of EBP and PKA reduces tropoelastin-induced expression levels
The in silico prediction of EBP and PKA in tropoelastin signaling was validated by in vitro inhibition studies. EBP is inhibited by β-lactose, while α-lactose can partially inhibit due to its partial conversion to β-lactose by anomerization (32). The importance of the EBP to tropoelastin signaling was demonstrated by the lactose-mediated reduction of the expression levels of MMP-12 (α-lactose 65.5±7.6%; β-lactose 56.9±3.0%; Fig. 5A), CXCL8 (α-lactose 87.7±1.9%; β-lactose 79.9±2.4%; Fig. 5B), CXCL5 (α-lactose 70.8±10.8%; β-lactose 61.7±2.3%; Fig. 5C), and CXCL1 (α-lactose 83.7±2.9%; β-lactose 78.1±6.2%; Fig. 5D).
Figure 5.
Tropoelastin-induced signaling pathways involve EBP and PKA. Tropoelastin signaling is mediated through the EBP (A–E) and involves the intracellular mediator PKA (F–J). MMP-12 (A, F), CXCL8 (B, G), CXCL5 (C, H), and CXCL1 (D, I) expression was significantly reduced in the presence of lactose, a competitive binding ligand for EBP and H-89, a specific inhibitor of PKA. GAPDH (E, J) was used as a control gene for both EBP and PKA inhibition studies. Results shown are representative of 2 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served to control the effect of inhibition on signaling receptors and intracellular mediators not stimulated by tropoelastin signaling. Expression levels of GAPDH in the presence of lactose (α-lactose 87.9±2.4%; β-lactose 83.3±10.0%) were not significantly different from those of the control (Fig. 5E). On this basis, lactose inhibition confirmed the participation of EBP.
Simulation modeling pointed to PKA as a mediator of elastin-induced intracellular signaling. H-89 inhibition of PKA was used to assess the putative role of PKA as a key intracellular signaling mediator downstream of EBP. The inhibited expression by H-89 for MMP-12 (63.9±12.3%; Fig. 5F), CXCL8 (73.3±9.7%; Fig. 5G), CXCL5 (58.2±31.2%; Fig. 5H), and CXCL1 (81.5±2.3%; Fig. 5I) was consistent with lower levels seen following lactose inhibition (Fig. 5A–D) in contrast to the GAPDH control (Fig. 5J). cAMP-dependent PKA is an integral intracellular mediator implicated in multiple signaling pathways and cellular processes (33), so it may be worth exploring additional control sites proposed by the PFCS model. Nevertheless, the reduction of MMP-12, CXCL8, CXCL5, and CXCL1 expression by lactose and H-89 correlates with a paradigm of EBP and PKA involvement in signaling cascades induced by tropoelastin/elastin fragments, and accounts for the identified transcriptional responses.
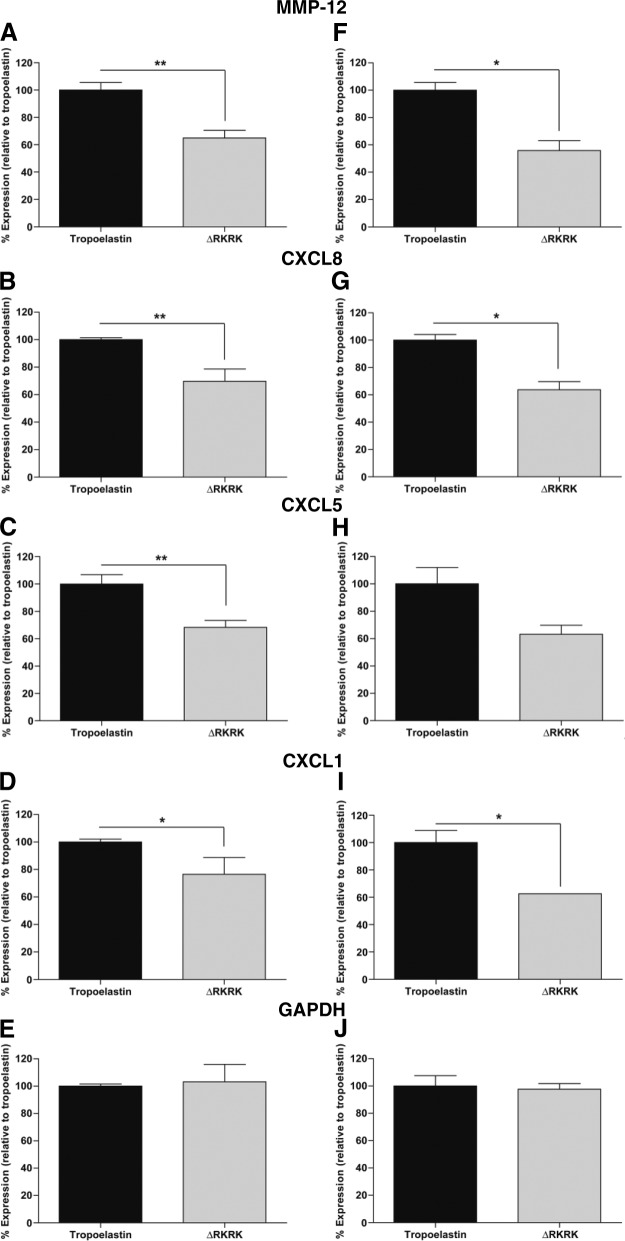
Truncated tropoelastin implicates integrin αVβ3
Integrins serve dual roles as cell adhesion receptors that anchor cells to matrix molecules and function as signaling mediators. The involvement of integrin αVβ3 in tropoelastin signaling transduction was assessed through the use of a tropoelastin mutant construct ΔRKRK, which lacks the integrin αVβ3 binding RKRK sequence. The expression levels for MMP-12 (65.0±5.6%; Fig. 6A), CXCL8 (69.8±8.9%; Fig. 6B), CXCL5 (68.3±5.2%; Fig. 6C), and CXCL1 (76.5±12.2%; Fig. 6D) on ΔRKRK were significantly lower than on tropoelastin in patient HDFs. These expression levels were reproduced with a second set of patient HDFs for MMP-12 (55.8±7.3%; Fig. 6F), CXCL8 (63.7±5.9%; Fig. 6G), CXCL5 (63.3±6.6%; Fig. 6H), and CXCL1 (62.5±0.02%; Fig. 6I). The expression level of control GAPDH in the presence of ΔRKRK did not differ from tropoelastin in either the first (103.1±12.9%; Fig. 6E) or second study (97.66±4.1%; Fig. 6J) cells.
Figure 6.
Tropoelastin-induced signaling pathways implicate integrin αVβ3. MMP-12 (A, F), CXCL8 (B, G), CXCL5 (C, H), and CXCL1 (D, I) expression was significantly reduced when HDFs were grown on the tropoelastin mutant construct ΔRKRK, which lacks the binding site for integrin αVβ3. There was strong consistency in reduced expression levels in HDF from patient 1 (A–E) and patient 2 (F–J). GAPDH (E, J) served as an unaffected control. *P < 0.05; **P < 0.01.
Examples of known signaling functions by integrin αVβ3 include MMP-2 expression induction by periostin in human periodontal ligament cells through ERK (34), increased Bcl-2 expression and cell survival in fibroblasts attached to vitronectin (35), and increased tenasin-C expression in smooth muscle cells binding to denatured collagen type I (36). Reduced MMP-12, CXCL8, CXCL5, and CXCL1 expression by integrin αVβ3 interference is the first demonstration of this function as a candidate signaling receptor in tropoelastin/elastin fragment signal transduction. These results are also the first evidence of an alternative signaling pathway for tropoelastin/elastin fragment signal transduction that functions in parallel with the EBP-PKA pathway.
A model for tropoelastin/elastin fragment signaling by HDFs
Two mechanisms for early stage signaling are proposed (Fig. 7). Cell surface EBP occupancy by tropoelastin/elastin fragment is the initial stimulus that promotes cell proliferation and the up-regulation of MMP-12, CXCL8, CXCL5, and CXCL1.
Figure 7.

Model for elastin sequence signaling. HDF interactions with elastin sequences are mediated by the EBP and integrin αVβ3, which facilitates cell adhesion. PKA is a main intracellular mediator of elastin sequence signaling, as it directly regulates CREB1 and is an upstream kinase common to the NF-κB and AP1 pathways. Elastin sequence signaling induces 1) MMP-12 expression, which is postulated to generate elastokines for as long as an elastin source is available and thus propagate signaling; 2) expression of a chemokine cluster of CXCL8, CXCL5, and CXCL1; and 3) cell spreading and proliferation.
The temporal profile of the chemokine cluster showed an early (6–12 h) peak in expression before declining. This reflects the natural progression of an inflammatory stage in wound healing, which typically lasts for 2 d in normal healing wounds (1). CXCL8, CXCL5, and CXCL1 are temporally associated with neutrophil migration into the wound site, which peaks in the first day of inflammation (37, 38). CXCL8, CXCL1, and, to a lesser extent, CXCL5 recruit neutrophils, and the vital role of these chemokines in wound healing is demonstrated in CXC receptor 2 (CXCR2)-deficient mice. CXCR2 is the main receptor for CXCL8, CXCL5, and CXCL1 (39), and antagonizing this interaction impairs neutrophil migration, among other wound-healing abnormalities (40). These results demonstrate that elastin sequences, represented by tropoelastin in this model, have the capacity to contribute to temporal regulation of this early inflammatory stage.
In contrast, the temporal prolife of MMP-12 exhibited a peak in expression by 6 h that was sustained for 24 h. This temporal pattern difference aligns with the differing role of MMP-12 in wound healing compared to the chemokine cluster. MMP-12 is typically expressed by macrophages and functions in the inflammatory and repair stages of wound healing (41). It is remarkable that MMP-12 is so effectively switched on in HDF on exposure to tropoelastin/elastin fragments. It is possible that the sustained expression of MMP-12 may be due to a positive feedback loop generated through elastokine formation by MMP-12, as surface-bound tropoelastin is slowly susceptible to proteolysis (42) and signal modulation by chemokines (30, 37). These results suggest a novel function for MMP-12 following elastin exposure and fragmentation in the early stages of wound healing. As MMP-12 is a potent elastase that can generate bioactive elastokines (43), it may be that the transcriptional response is propagated by elastokines that are generated by MMP-12. This model allows for the attractive possibility that MMP-12 may temporally regulate chemokine expression by altering the elastin signaling environment, which is important to the overall regulation of the proinflammatory state. Signaling by elastin sequences has in vivo relevance as elastin fragmentation occurs on significant injury to the skin and during the inflammatory stage of wound healing.
Central to the tropoelastin-stimulated expression of MMP-12 and the chemokines is PKA, which acts a key junction between the elastin signaling receptors and transcription factors. The partial inhibition of PKA by H-89 provides a proof of concept that druggable targets can be used to modulate an early stage proinflammatory response by HDFs.
CONCLUSIONS
Tropoelastin is a cell-interactive ECM protein that supports HDF attachment, spreading, and proliferation and induces transcriptional responses. This elastin signaling involves EBP through PKA, where this was identified in silico and prospectively validated in vitro. This study also provides the first in vitro indication of integrin αVβ3's parallel role as a signaling receptor in tropoelastin signal transduction. These results reveal that elastin sequences contribute inherent cell signaling properties pertinent to inflammatory regulation that complement their unique elastic function. The elastin-integrated model described here points to a reciprocal relationship between elastin sequences and HDFs and points to a model of solubilized elastin sequences in early stage inflammatory responses by HDFs.
Supplementary Material
Acknowledgments
A.S.W. acknowledges support from the Australian Research Council, Australian Defense Health Foundation, Australian National Health and Medical Research Council, and the U.S. National Institutes of Health (EB014283). A.S.W. is the scientific founder of Elastagen Pty. Ltd. J.F.A received an Australian Postgraduate Award. The authors thank their colleagues at Concord Hospital for the generous donation of human dermal fibroblasts; A/Prof. Gareth Denyer, Dr. Dale Hancock, Mrs. Sarah Martinez, Ms. Yannie Poon, and Ms. Kate Nieuwendyk for their technical assistance; and Dr. Suzanne Mithieux and Dr. Daniel Bax for advice on the text of this article.
The authors declare no conflicts of interest.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- CREB1
- cAMP responsive element binding protein 1
- CXCL
- CXC chemokine ligand
- CXCR2
- CXC receptor 2
- EBP
- elastin binding protein
- ECM
- extracellular matrix
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- H-89
- N-[2-((p-bromocinnamyl)amino)ethyl]-5-isoquinolinesulfonamide
- HDF
- human dermal fibroblast
- MMP-12
- matrix metalloproteinase 12
- NF-κB
- nuclear factor κ-light-chain-enhancer of activated B cells
- PCR
- polymerase chain reaction
- PFCS
- predictive fibroblast cell system
- PKA
- protein kinase A
- RT
- reverse transcriptase
REFERENCES
- 1. Gurtner G. C., Werner S., Barrandon Y., Longaker M. T. (2008) Wound repair and regeneration. Nature 453, 314–321 [DOI] [PubMed] [Google Scholar]
- 2. Adair-Kirk T. L., Senior R. M. (2008) Fragments of extracellular matrix as mediators of inflammation. Int. J. Biochem. Cell Biol. 40, 1101–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roten S. V., Bhat S., Bhawan J. (1996) Elastic fibers in scar tissue. J. Cutan. Pathol. 23, 37–42 [DOI] [PubMed] [Google Scholar]
- 4. Kielty C. M., Sherratt M. J., Shuttleworth C. A. (2002) Elastic fibres. J. Cell Sci. 115, 2817–2828 [DOI] [PubMed] [Google Scholar]
- 5. Senior R. M., Griffin G. L., Mecham R. P. (1982) Chemotactic responses of fibroblasts to tropoelastin and elastin-derived peptides. J. Clin. Invest. 70, 614–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamoun A., Landeau J. M., Godeau G., Wallach J., Duchesnay A., Pellat B., Hornebeck W. (1995) Growth stimulation of human skin fibroblasts by elastin-derived peptides. Cell Adhes. Commun. 3, 273–281 [DOI] [PubMed] [Google Scholar]
- 7. Fujimoto N., Tajima S., Ishibashi A. (2000) Elastin peptides induce migration and terminal differentiation of cultured keratinocytes via 67 kDa elastin receptor in vitro: 67 kDa elastin receptor is expressed in the keratinocytes eliminating elastic materials in elastosis perforans serpiginosa. J. Invest. Dermatol. 115, 633–639 [DOI] [PubMed] [Google Scholar]
- 8. Duca L., Debelle L., Debret R., Antonicelli F., Hornebeck W., Haye B. (2002) The elastin peptides-mediated induction of pro-collagenase-1 production by human fibroblasts involves activation of MEK/ERK pathway via PKA- and PI(3)K-dependent signaling. FEBS Lett. 524, 193–198 [DOI] [PubMed] [Google Scholar]
- 9. Cantarelli B., Duca L., Blanchevoye C., Poitevin S., Martiny L., Debelle L. (2009) Elastin peptides antagonize ceramide-induced apoptosis. FEBS Lett. 583, 2385–2391 [DOI] [PubMed] [Google Scholar]
- 10. Wise S. G., Weiss A. S. (2009) Tropoelastin. Int. J. Biochem. Cell Biol. 41, 494–497 [DOI] [PubMed] [Google Scholar]
- 11. Pierce R. A., Moore C. H., Arikan M. C. (2006) Positive transcriptional regulatory element located within exon 1 of elastin gene. Am. J. Physiol. Lung Cell. Mol. Physiol. 291, L391–L399 [DOI] [PubMed] [Google Scholar]
- 12. Amadeu T. P., Braune A. S., Porto L. C., Desmouliere A., Costa A. M. (2004) Fibrillin-1 and elastin are differentially expressed in hypertrophic scars and keloids. Wound Repair Regen. 12, 169–174 [DOI] [PubMed] [Google Scholar]
- 13. Raghunath M., Bächi T., Meuli M., Altermatt S., Gobet R, Bruckner-Tuderman L, Steinmann B. (1996) Fibrillin and elastin expression in skin regenerating from cultured keratinocyte autografts: morphogenesis of microfibrils begins at the dermo-epidermal junction and precedes elastic fiber formation. J. Invest. Dermatol. 106, 1090–1095 [DOI] [PubMed] [Google Scholar]
- 14. Diegelmann R. F., Evans M. C. (2004) Wound healing: an overview of acute, fibrotic and delayed healing. Front. Biosci. 9, 283–289 [DOI] [PubMed] [Google Scholar]
- 15. Duca L., Floquet N., Alix A. J., Haye B., Debelle L. (2004) Elastin as a matrikine. Crit. Rev. Oncol. Hematol. 49, 235–244 [DOI] [PubMed] [Google Scholar]
- 16. Senior R. M., Bächi T., Meuli M., Altermatt S., Gobet R, Bruckner-Tuderman L, Steinmann B. (1984) Val-Gly-Val-Ala-Pro-Gly, a repeating peptide in elastin, is chemotactic for fibroblasts and monocytes. J. Cell Biol. 99, 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tajima S., Wachi H., Uemura Y., Okamoto K. (1997) Modulation by elastin peptide VGVAPG of cell proliferation and elastin expression in human skin fibroblasts. Arch. Dermatol. Res. 289, 489–492 [DOI] [PubMed] [Google Scholar]
- 18. Brassart B., Fuchs P., Huet E., Alix A. J., Wallach J., Tamburro A. M., Delacoux F., Haye B., Emonard H., Hornebeck W., Debelle L. (2001) Conformational dependence of collagenase (matrix metalloproteinase-1) up-regulation by elastin peptides in cultured fibroblasts. J. Biol. Chem. 276, 5222–5227 [DOI] [PubMed] [Google Scholar]
- 19. Robinet A., Fahem A., Cauchard J. H., Huet E., Vincent L., Lorimier S., Antonicelli F., Soria C., Crepin M., Hornebeck W., Bellon G. (2005) Elastin-derived peptides enhance angiogenesis by promoting endothelial cell migration and tubulogenesis through upregulation of MT1-MMP. J. Cell Sci. 118, 343–356 [DOI] [PubMed] [Google Scholar]
- 20. Antonicelli F., Bellon G., Debelle L., Hornebeck W. (2007) Elastin-elastases and inflamm-aging. Curr. Top. Dev. Biol. 79, 99–155 [DOI] [PubMed] [Google Scholar]
- 21. Schultz G. S., Davidson J. M., Kirsner R. S., Bornstein P., Herman I. M. (2011) Dynamic reciprocity in the wound microenvironment. Wound Repair Regen. 19, 134–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yeo G. C., Keeley F. W., Weiss A. S. (2011) Coacervation of tropoelastin. Adv. Colloid Interface Sci. 167, 94–103 [DOI] [PubMed] [Google Scholar]
- 23. Wu W. J., Weiss A. S. (1999) Deficient coacervation of two forms of human tropoelastin associated with supravalvular aortic stenosis. Eur. J. Biochem. 266, 308–314 [DOI] [PubMed] [Google Scholar]
- 24. Martin S. L., Vrhovski B., Weiss A. S. (1995) Total synthesis and expression in Escherichia coli of a gene-encoding human tropoelastin. Gene 154, 159–166 [DOI] [PubMed] [Google Scholar]
- 25. Humphries M. J. (2001) Cell-substrate adhesion assays. Curr. Protoc. Cell Biol. Chap. 9, Unit 9.1 [DOI] [PubMed] [Google Scholar]
- 26. Harvey L. E., Kohlgraf K. G., Mehalick L. A., Raina M., Recker E. N., Radhakrishnan S., Prasad S. A., Vidva R., Progulske-Fox A., Cavanaugh J. E., Vali S., Brogden K. A. (2013) Defensin DEFB103 bidirectionally regulates chemokine and cytokine responses to a pro-inflammatory stimulus. Sci. Rep. 3, 1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rajendran P., Li F., Shanmugam M. K., Vali S., Abbasi T., Kapoor S., Ahn K. S., Kumar A. P., Sethi G. (2012) Honokiol inhibits signal transducer and activator of transcription-3 signaling, proliferation, and survival of hepatocellular carcinoma cells via the protein tyrosine phosphatase SHP-1. J. Cell. Physiol. 227, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 28. Taddese S., Weiss A. S., Jahreis G., Neubert R. H., Schmelzer C. E. (2009) In vitro degradation of human tropoelastin by MMP-12 and the generation of matrikines from domain 24. Matrix Biol. 28, 84–91 [DOI] [PubMed] [Google Scholar]
- 29. Flavell S. J., Hou T. Z., Lax S., Filer A. D., Salmon M., Buckley C. D. (2008) Fibroblasts as novel therapeutic targets in chronic inflammation. Br. J. Pharmacol. 153(Suppl. 1), S241–S246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith R. S., Smith T. J., Blieden T. M., Phipps R. P. (1997) Fibroblasts as sentinel cells. Synthesis of chemokines and regulation of inflammation. Am. J. Pathol. 151, 317–322 [PMC free article] [PubMed] [Google Scholar]
- 31. Duca L., Lambert E., Debret R., Rothhut B., Blanchevoye C., Delacoux F., Hornebeck W., Martiny L., Debelle L. (2005) Elastin peptides activate extracellular signal-regulated kinase 1/2 via a Ras-independent mechanism requiring both p110γ/Raf-1 and protein kinase A/B-Raf signaling in human skin fibroblasts. Mol. Pharmacol. 67, 1315–1324 [DOI] [PubMed] [Google Scholar]
- 32. Jawad R., Elleman C., Vermeer L., Drake A. F., Woodhead B., Martin G. P., Royall P. G. (2012) The measurement of the β/α anomer composition within amorphous lactose prepared by spray and freeze drying using a simple (1)H-NMR method. Pharm. Res. 29, 511–524 [DOI] [PubMed] [Google Scholar]
- 33. Tasken K., Aandahl E. M. (2004) Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol. Rev. 84, 137–167 [DOI] [PubMed] [Google Scholar]
- 34. Watanabe T., Yasue A., Fujihara S., Tanaka E. (2012) PERIOSTIN regulates MMP-2 expression via the alphavbeta3 integrin/ERK pathway in human periodontal ligament cells. Arch. Oral Biol. 57, 52–59 [DOI] [PubMed] [Google Scholar]
- 35. Matter M. L., Ruoslahti E. (2001) A signaling pathway from the α5β1 and α(v)β3 integrins that elevates bcl-2 transcription. J. Biol. Chem. 276, 27757–27763 [DOI] [PubMed] [Google Scholar]
- 36. Jones P. L., Jones F. S., Zhou B., Rabinovitch M. (1999) Induction of vascular smooth muscle cell tenascin-C gene expression by denatured type I collagen is dependent upon a β3 integrin-mediated mitogen-activated protein kinase pathway and a 122-base pair promoter element. J. Cell Sci. 112, 435–445 [DOI] [PubMed] [Google Scholar]
- 37. Gillitzer R., Goebeler M. (2001) Chemokines in cutaneous wound healing. J. Leukoc. Biol. 69, 513–521 [PubMed] [Google Scholar]
- 38. Engelhardt E., Toksoy A, Goebeler M, Debus S, Bröcker EB, Gillitzer R. (1998) Chemokines IL-8, GROalpha, MCP-1, IP-10, and Mig are sequentially and differentially expressed during phase-specific infiltration of leukocyte subsets in human wound healing. Am. J. Pathol. 153, 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murdoch C., Finn A. (2000) Chemokine receptors and their role in inflammation and infectious diseases. Blood 95, 3032–3043 [PubMed] [Google Scholar]
- 40. Devalaraja R. M., Nanney L B., Du J., Qian Q., Yu Y., Devalaraja M. N., Richmond A. (2000) Delayed wound healing in CXCR2 knockout mice. J. Invest. Dermatol. 115, 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nenan S., Boichot E., Lagente V., Bertrand C. P. (2005) Macrophage elastase (MMP-12): a pro-inflammatory mediator? Mem. Inst. Oswaldo Cruz 100(Suppl. 1), 167–172 [DOI] [PubMed] [Google Scholar]
- 42. Waterhouse A., Bax D. V., Wise S. G., Yin Y., Dunn L. L., Yeo G. C., Ng M. K. C., Bilek M. M. M., Weiss A. S. (2011) Stability of a therapeutic layer of immobilized recombinant human tropoelastin on a plasma-activated coated surface. Pharm. Res. 28, 1415–1421 [DOI] [PubMed] [Google Scholar]
- 43. Heinz A., Jung M. C., Duca L., Sippl W., Taddese S., Ihling C., Rusciani A., Jahreis G., Weiss A. S., Neubert R. H., Schmelzer C. E. (2010) Degradation of tropoelastin by matrix metalloproteinases–cleavage site specificities and release of matrikines. FEBS J. 277, 1939–1956 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.