Abstract
Our understanding of the bile acid metabolism is limited by the fact that previous analyses have primarily focused on a selected few circulating bile acids; the bile acid profiles of the liver and gastrointestinal tract pools are rarely investigated. Here, we determined how chronic ethanol consumption altered the bile acids in multiple body compartments (liver, gastrointestinal tract, and serum) of rats. Rats were fed a modified Lieber-DeCarli liquid diet with 38% of calories as ethanol (the amount equivalent of 4–5 drinks in humans). While conjugated bile acids predominated in the liver (98.3%), duodenum (97.8%), and ileum (89.7%), unconjugated bile acids comprised the largest proportion of measured bile acids in serum (81.2%), the cecum (97.7%), and the rectum (97.5%). In particular, taurine-conjugated bile acids were significantly decreased in the liver and gastrointestinal tract of ethanol-treated rats, while unconjugated and glycine-conjugated species increased. Ethanol consumption caused increased expression of genes involved in bile acid biosynthesis, efflux transport, and reduced expression of genes regulating bile acid influx transport in the liver. These results provide an improved understanding of the systemic modulations of bile acid metabolism in mammals through the gut-liver axis.—Xie, G., Zhong, W., Li, H., Li, Q., Qiu, Y., Zheng, X., Chen, H., Zhao, X., Zhang, S., Zhou, Z., Zeisel, S. H., Jia, W. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption.
Keywords: gastrointestinal tract, liver, blood, ultraperformance liquid chromatography mass spectrometry, metabolomics
Bile acids are gaining increasing recognition as important metabolic signaling molecules that modulate lipid, glucose, and energy metabolism (1). Also, they regulate their own homeostasis through binding with the nuclear receptor farnesoid X receptor (FXR), the plasma membrane-bound bile acid receptor TGR5/M-BAR (2). Activation of intestinal FXR by bile acids leads to up-regulation of fibroblast growth factor 15 (FGF15) (3, 4), and secreted FGF15 suppresses liver transcription of cholesterol 7α-hydroxylase 1 (CYP7A1), the rate-limiting enzyme for liver bile acid biosynthesis (5). Errors in bile acid metabolism are implicated in several human diseases, including both alcoholic (6) and nonalcoholic fatty liver diseases (7) and colon cancer (8).
The gut microbiome may play a role in the development of a variety of diseases such as obesity, diabetes mellitus, fatty liver disease, and colon cancer (9). Primarily harbored in the large intestine, the gut microbiota not only contribute to the salvage of bile acids that escape reabsorption by active transport in distal ileum but also modify the chemical structure of absorbed bile acids through processes including deconjugation, dehydrogenation, dehydroxylation, and desulfation (10–12). Perturbing the gut microbiome can result in a disturbance of bile acid metabolism and reabsorption, leading to altered bile acid profiles in the blood, liver, kidneys, and heart (2). Moreover, inhibiting intestinal microbiota with ampicillin increases mRNA and protein expression of the apical sodium-dependent bile acid transporter (ASBT/Slc10a2) in the brush-border membrane of the ileum, which in turn increases bile acid transport into portal blood (13).
Chronic ethanol consumption is associated with alcoholic liver diseases and has adverse effects on lipid metabolism, both in hepatic and extrahepatic tissues. This leads to the development of fatty liver, hepatitis, and cirrhosis (14, 15), and ethanol-induced fatty liver is accompanied by alterations in the bile acid profile and in the composition of the gut microbiome (15–17). These changes in the enterohepatic circulation of bile acids are important not only for feedback inhibition of bile acid synthesis but also for whole-body lipid homeostasis. However, there are very few reports that systemically delineate the composition of bile acid profiles in the whole gastrointestinal (GI) tract, liver, and blood pool, nor is there any information available on the dynamic changes of these bile acid profiles during chronic ethanol consumption.
The goal of this study was to use a targeted metabolomics approach to characterize the bile acid profiles of the serum, GI tract (duodenum, jejunum, ileum, cecum, colon, and rectum) and liver in male Sprague Dawley rats consuming ethanol chronically for 8 wk. We quantified a panel of 20–30 bile acids using ultraperformance liquid chromatography–triple-quadrupole mass spectrometry (UPLC-TQMS) and found that ethanol consumption substantially affected the bile acid profiles of different tissues. In addition, we measured the expression of different genes regulating bile acid metabolism in the liver and ileum.
MATERIALS AND METHODS
Chemicals and reagents
Lithocholate (LCA), nordeoxycholate (NDCA), murideoxycholate (MDCA), hyodeoxycholate (HDCA), chenodeoxycholate (CDCA), deoxycholate (DCA), dehydrocholate (DHCA), glycocholate (GCA), taurolithocholate (TLCA), glycolithocholate (GLCA), α-muricholate (α-MCA), β-muricholate (β-MCA), ω-muricholate (ω-MCA), λ-muricholate (λ-MCA), cholate (CA), 7-dehydrocholate (7-DCA), methyl deoxycholate (methyl DCA), 6,7-diketodeoxycholate (6,7-DKDCA), tauro α-muricholate (T α-MCA), tauro β-muricholate (T β-MCA), taurocholate (TCA), tauroursodeoxycholate (TUDCA), taurohyodeoxycholate (THDCA), taurochenodeoxycholate (TCDCA), taurodeoxycholate (TDCA), glycoursodeoxycholate (GUDCA), glycohyodeoxycholate (GHDCA), glycochenodeoxycholate (GCDCA), glycodeoxycholate (GDCA), cholate-d4 (CA-d4; 5β-cholanic acid-3α,7α,12α-triol-2,2,4,4-d4), glycocholate-d4 (GCA-d4; 5β-cholanic acid-3α,7α,12α-triol N-(carboxymethyl)-amide-2,2,4,4-d4), lithocholate-d4 (LCA-d4; 5β-cholanic acid-3α-ol-2,2,4,4-d4), and deoxycholate-d4 (DCA-d4; 5β-cholanic acid-3α, 12α-diol-2,2,4,4-d4) were purchased from Steraloids, Inc. (Newport, RI, USA). HPLC-grade methanol, acetonitrile, water, ammonium acetate, and acetic acid were obtained from Sigma-Aldrich (St. Louis, MO, USA).
Animals and ethanol feeding experiments
Male Sprague-Dawley rats were obtained from Charles River (Wilmington, MA, USA). All rats were treated according to experimental procedures approved by the Institutional Animal Care and Use Committee. Three-month-old rats were pair-fed a modified Lieber-DeCarli liquid diet containing either ethanol or isocaloric maltose dextrin for 8 wk. The calories of the ethanol liquid diet were derived 38% from ethanol, 34% from fat, 16% from protein, and 12% from carbohydrate. The ethanol calories were replaced by maltose dextran in the control liquid diet. All ingredients for the liquid diets were obtained from Dyets (Bethlehem, PA, USA) with the exception of ethanol, which was purchased from Sigma-Aldrich. The ethanol content (%, w/v) was initially 5% for the first 2 wk and was increased by 0.2% every 2 wk up to a concentration of 5.6% during the final 2 wk. The control group was pair-fed the amount that ethanol-fed rats had in the previous day. The rats were denied access to food for 4 h before blood was drawn. Serum samples were collected at wk 2, 4, and 6 as well as at the end of the experiment. Rats (n=5–9/group) were anesthetized with isofluorane and serum, and liver and intestinal contents were harvested for analysis.
Preparation of samples and standards
Serum sample preparation
An aliquot of 100 μl of serum was mixed with 400 μl of a mixture of methanol and acetonitrile [5:3, contains 5 μg/ml of aqueous 4-chlorophenylalanine, used as the internal standard (IS)]. The mixture was then vortexed for 2 min, allowed to stand for 10 min, and centrifuged at 13,000 rpm for 20 min. The supernatant was used for UPLC-MS/MS analysis.
Liver sample preparation
Liver tissue samples (100 mg) were homogenized on ice in 500 μl of a mixture of chloroform, methanol and water (1:2.5:1, v/v/v). The samples were then centrifuged at 13,000 rpm for 10 min at 4°C, and a 150-μl aliquot of the supernatant was transferred to an LC sampling vial containing an IS (10 μl L-4-chloro-phenylalanine in water, 5 μg/ml). The deposit was rehomogenized with 500 μl of methanol, and a 150-μl aliquot of supernatant was added to the same vial for drying prior to reconstitution with acetonitrile/H2O (6:4, v/v) to a final volume of 500 μl.
Intestinal content sample preparation
Intestinal contents (100 mg) were mixed with 500 μl of ice-cold water. The mixture was vortexed for 4 min and then centrifuged at 13,200 rpm for 10 min at 4°C. A 300-μl aliquot of supernatant was transferred to a 2-ml tube, and the pellets were further extracted with ice-cold methanol using the same protocol. Another 300-μl aliquot of supernatant was added to the same tube as the initial aliquot, and 10 μl of IS (p-chlorophenylalanine in water, 5 μg/ml) was added. The extraction was vortexed for 30 s and centrifuged at 13,000 rpm for 20 min. The resulting supernatant was used for UPLC-MS analysis.
Standard solution
Each of the 32 standards was individually dissolved in methanol or water and prepared as a stock solution at a concentration of 5 mg/ml.
Method validation
Each aliquot of standard stock solution was mixed to obtain a mixed stock solution. The resulting mixed solution was diluted to generate a series of concentrations of 0.01, 0.1, 0.5, 1, 5, 10, 50, 100, 500, 1000, and 10,000 ng/ml. LCA-d4 was used as an IS, and the calibration curve and the corresponding regression coefficients were obtained by IS adjustment (Supplemental Table S1). All bile acids were found to be linear over the measured range.
Instrumentation
A Waters Acquity UPLC system equipped with a binary solvent delivery manager and a sample manager (Waters, Milford, MA, USA) was used throughout the study. The mass spectrometer was a Waters TQ instrument with an electrospray ionization (ESI) source (Waters). The entire LC-MS system was controlled by MassLynx 4.1 software (Waters). All chromatographic separations were performed with an Acquity UPLC C18 column (1.7 μm, 50 mm×2.1 mm internal dimensions; Waters).
LC and MS conditions
The mobile phase consisted of 10 mM ammonium acetate adjusted to pH 4 using acetic acid (mobile phase A) and methanol (MeOH; mobile phase B) run at a flow rate of 0.3 ml/min. The LC elution conditions were optimized as follows: isocratic at 40% B (0–0.5 min), linear gradient from 40 to 80% B (0.5–9.0 min), 80 to 100% B (9.0–12.0 min), isocratic at 100% B (12.0–12.5 min); and isocratic at 40% B (12.5–15.0 min). The column was maintained at 40°C, and the injection volume of all samples was 10 μl.
The mass spectrometer was operated with source and desolvation temperatures set at 120 and 350°C, respectively. Bile acids were detected in the negative mode. The capillary, extractor, and RF voltages were 3000, 4, and 0 V, respectively. The desolvation gas (nitrogen) was set at a flow rate of 650 L/h. The cone voltages, collision energies, and multiple reaction monitoring (MRM) transitions are listed in Supplemental Table S2.
qRT-PCR analysis
Total RNA was isolated from liver or ileum mucosa using TRIzol reagent (Invitrogen, Life Technologies, Grand Island, NY, USA) and reverse transcribed with TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, CA, USA). Expression of target mRNA was measured in triplicate by the comparative cycle threshold method on the Applied Biosystems 7500 Real Time PCR System. The forward and reverse primers were purchased from Integrated DNA Technologies (Coralville, IA, USA), and sequences are shown in Table 1. Target gene expression was normalized to 18 s rRNA levels and presented as fold changes relative to control values (which were set at 1).
Table 1.
Primer sets for quantitative RT-PCR analysis
| Gene | Full name | GeneBank ID | Sequences, forward/reverse 5′–3′ | Amplicon size (bp) |
|---|---|---|---|---|
| CYP7A1 | Cytochrome P450 7A1 | NM_012942 | TGAAAGCGGGAAAGCAAAGACCAC/TCTTGGACGGCAAAGAGTCTTCCA | 117 |
| CYP8B1 | Cytochrome P450 8B1 | NM_031241 | TCCATATGTCCCGGCAGGTTCTTT/TGTCAGGGTCCACCAGTTCAAAGT | 97 |
| CYP27A1 | Cytochrome P450 27A1 | NM_178847 | ACCTTTCCTGAGCTGATCTTGGCT/GCATGTGGGCAAAGTCCTTGTTCT | 169 |
| StAR | Steroidogenic acute regulatory protein | NM_031558 | AAGCCAGCAGGAGAATGGAGATGA/TGACATTTGGGTTCCACTCTCCCA | 158 |
| Fatp2 | Fatty acid transport protein 2 | NM_031736 | AGTACATCGGTGAACTGCTTCGGT/CAATGTTGCCTTCAGTGGAAGCGT | 180 |
| BACS/Fatp5 | Bile acid CoA synthase | NM_024143 | TTCAGGGACCACTGGACTTCCAAA/ACCACATCATCAGCTGTTCTCCCA | 105 |
| BAAT | Bile acid CoA: amino acid N-acyltransferase | NM_017300 | GGTTGGCATCCTTTCTGTGTGCAT/ATTCTTCACTGCAGGGTGTAGGCT | 167 |
| CYP3A11 | Cytochrome P450 3A11 | NM_153312 | TCCAGCTGATGCTGAACGCTCATA/AGGGTGAGTGGCCAGGAAATACAA | 164 |
| SULT2A1 | Sulfotransferase family 2A1 | NM_131903 | TCAGTTCCAAGGCCAAGGTGATCT/AGTTCCCAGTGAGTCTGGCTTCTT | 116 |
| NTCP/Slc10a1 | Na+-taurocholate cotransporting polypeptide | NM_017047 | AGGATGGAGGTGCACAACGTATCA/AGCCCAGTGAGAGCATGATAAGCA | 133 |
| BSEP/Abcb11 | Bile salt export pump | NM_031760 | ATCTGTTAATCCTGGGCAGACGCT/TGGGAGACAATCCCGATGTTGGAA | 180 |
| MRP2/Abcc2 | Multidrug resistance-associated protein 2 | NM_012833 | AACCGGGAAGGTCAAGTTCTCCAT/TTGTCAGAGTCACTGGTCCAAGCA | 153 |
| MRP4/Abcc4 | Multidrug resistance-associated protein 4 | NM_133411 | ATTGTGGGAAGAACTGGAGCTGGA/TGGTTCCAGTGAACAGGACAGGTT | 175 |
| OSTα | Organic solute transporter alpha | NM_001107087 | AGCAATTTCCTTGCTGTGTCCACC/AGGATGACAAGCACCTGGAACAGA | 131 |
| OSTβ | Organic solute transporter beta | XM_238546 | TCCGTTCAGAGGATGCAACTCCTT/CATTCCGTTGTCTTGTGGCTGCTT | 140 |
| ASBT/Slc10a2 | Apical sodium dependent bile acid transporter | NM_017222 | TGGGCTTCCTCTGTCAGTTTGGAA/AGTGTGGAGCAAGTGGTCATGCTA | 196 |
| TGR5/Gpbar1 | G protein-coupled bile acid receptor 1 | NM_177936 | TTGCTCCTGTCAGTCTTGGCCTAT/TTGGGTCTTCCTCGAAGCACTTGT | 191 |
| FGF15 | Fibroblast growth factor 15 | NM_130753 | ACAATTGCCATCAAGGACGTCAGC/TGAAGATGATGTGGAGGTGGTGCT | 172 |
| FGFR4 | Fibroblast growth factor receptor 4 | NM_001109904 | TCACAGTTGAGGCCTTCTGTTCCA/TGCTTCGCTCCTTTGAGGATGAGT | 108 |
| 18s rRNA | 18s ribosomal RNA | NR_046237 | ACGGACCAGAGCGAAAGCAT/TGTCAATCCTGTCCGTGTCC | 120 |
Data analysis
UPLC-MS raw data obtained with negative mode were analyzed using QuanLynx 4.1 applications manager (Waters). A Student's t test was used to investigate differences between the groups in bile acids measurements. Principal component analysis (PCA) was performed using SIMCA-P software (Umetrics, San Jose, CA, USA).
RESULTS
General information about the animal experiment
A number of serum parameters, including serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphate (ALP), albumin, ammonia, and γ-glutamyl transpeptidase (GGT), were measured, and the values were listed in Table 2. ALT activity was markedly increased due to ethanol consumption (P<0.05; Table 2).
Table 2.
Serum values of alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphate (ALP), albumin, ammonia, and γ-glutamyl transpeptidase (GGT) in control rats and rats under ethanol consumption for 8 wk
| Group | ALT (U/L) | AST (U/L) | ALP (U/ml) | Albumin (mg/ml) | Ammonia (μM) | GGT (mU/ml) |
|---|---|---|---|---|---|---|
| Control | 25.02 ± 3.41 | 38.12 ± 6.38 | 39.98 ± 5.96 | 30.00 ± 0.94 | 815.73 ± 60.78 | 43.87 ± 6.48 |
| Ethanol | 41.90 ± 7.03* | 44.83 ± 7.07 | 39.21 ± 4.11 | 29.90 ± 0.97 | 825.29 ± 41.80 | 39.49 ± 6.96 |
P < 0.05 vs. control group.
GI tract bile acid profiles of control rats
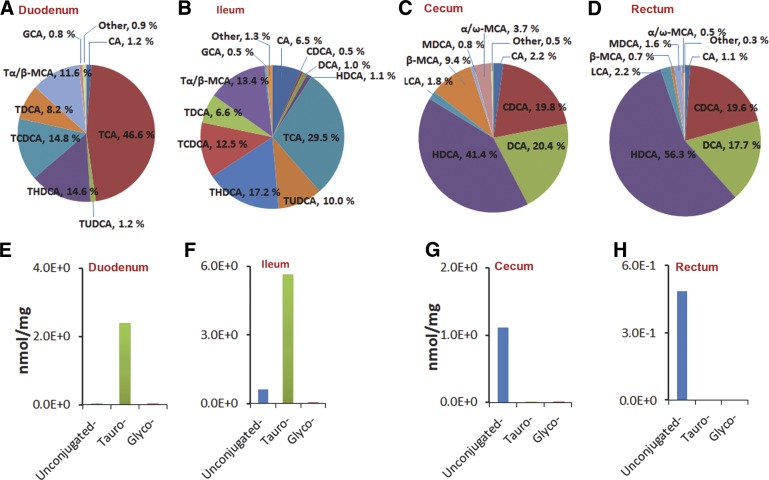
To systemically characterize the bile acid profile in the GI tract of control rats, we quantitatively measured the abundance of bile acids in intestinal contents from the duodenum, ileum, cecum, and rectum (Table 3). The representative UPLC-MS/MS chromatograms of the authentic standards of bile acids under the final chromatography and detection conditions were shown in Fig. 1. The duodenum is the first site where bile acids are secreted into the GI tract, and most of the bile acids will be reabsorbed through enterohepatic circulation before reaching the cecum. The concentrations of unconjugated bile acids, such as CA, CDCA, DCA, and HDCA, changed significantly from the duodenum to the ileum, the cecum, and the rectum. Taurine-conjugated bile acids accounted for the greatest portion of all measured bile acids from the duodenum and ileum, whereas their levels decreased dramatically in the cecum and rectum. For example, the concentration of TCA was the highest in the duodenum (1.15 nmol/mg) and ileum (1.87 nmol/mg), but only a very low level of TCA was detected in the cecum (0.0007 nmol/mg) and rectum (0.000004 nmol/mg). In general, the abundance of glycine-conjugated bile acids was very low. GCA was the most abundant glycine-conjugated bile acid in the duodenum (0.02 nmol/mg) and ileum (0.03 nmol/mg), and its levels decreased to 0.00005 and 0.000009 nmol/mg in the cecum and rectum, respectively.
Table 3.
Bile acid profiles in the GI tract (duodenum, ileum, cecum, and rectum) of control rats
| Bile acid | Duodenum | Ileum | Cecum | Rectum |
|---|---|---|---|---|
| CA | 3.03E-02 ± 1.97E-02 | 4.15E-01 ± 2.42E-01* | 2.42E-02 ± 7.08E-03 | 5.41E-03 ± 2.41E-03* |
| CDCA | 1.14E-03 ± 3.83E-04 | 3.14E-02 ± 1.59E-02 | 2.20E-01 ± 9.03E-02 | 9.53E-02 ± 2.04E-02* |
| DCA | 1.17E-03 ± 4.00E-04 | 6.08E-02 ± 3.13E-02 | 2.28E-01 ± 8.97E-02 | 8.61E-02 ± 1.98E-02* |
| HDCA | 2.75E-03 ± 9.37E-04 | 7.00E-02 ± 3.63E-02 | 4.61E-01 ± 1.15E-01* | 2.74E-01 ± 5.85E-02* |
| LCA | 0.00E+00 ± 0.00E+00 | 1.27E-03 ± 4.71E-04* | 2.04E-02 ± 4.12E-03* | 1.06E-02 ± 2.10E-03* |
| β-MCA | 2.29E-03 ± 1.05E-03 | 2.25E-02 ± 8.74E-03 | 1.05E-01 ± 1.85E-02* | 3.34E-03 ± 1.34E-03 |
| MDCA | 1.05E-04 ± 4.19E-05 | 2.03E-03 ± 9.07E-04 | 8.48E-03 ± 1.95E-03* | 7.74E-03 ± 2.66E-03 |
| α/ω-MCA | 1.77E-03 ± 7.37E-04 | 2.39E-02 ± 1.15E-02 | 4.15E-02 ± 1.25E-02 | 2.59E-03 ± 6.72E-04 |
| TCA | 1.15E+00 ± 3.37E-01 | 1.87E+00 ± 6.11E-01 | 6.62E-04 ± 5.90E-04* | 3.66E-06 ± 2.25E-06* |
| TLCA | 1.22E-03 ± 3.58E-04 | 5.35E-03 ± 6.15E-04* | 5.52E-05 ± 3.52E-05 | 9.59E-06 ± 2.25E-06* |
| TUDCA | 3.06E-02 ± 2.72E-03 | 6.33E-01 ± 2.75E-01 | 3.33E-05 ± 1.49E-05* | 2.53E-06 ± 1.76E-06* |
| THDCA | 3.61E-01 ± 1.13E-01 | 1.09E+00 ± 1.95E-01* | 2.45E-04 ± 5.11E-05* | 1.13E-04 ± 3.21E-05* |
| TCDCA | 3.65E-01 ± 1.47E-01 | 7.93E-01 ± 2.14E-01 | 2.71E-04 ± 7.71E-05 | 9.14E-05 ± 1.85E-05 |
| TDCA | 2.03E-01 ± 8.48E-02 | 4.16E-01 ± 1.30E-01 | 1.68E-03 ± 1.37E-03 | 5.87E-05 ± 9.76E-06 |
| T α/β-MCA | 2.86E-01 ± 5.88E-02 | 8.51E-01 ± 2.66E-01 | 5.44E-05 ± 1.44E-05* | 6.74E-06 ± 1.60E-06* |
| GCA | 2.03E-02 ± 5.09E-03 | 3.00E-02 ± 3.96E-03 | 4.89E-05 ± 1.02E-05* | 9.06E-06 ± 1.11E-06* |
| GLCA | 0.00E+00 ± 0.00E+00 | 4.35E-05 ± 1.29E-05* | 3.52E-05 ± 2.16E-05 | 2.19E-05 ± 1.00E-05 |
| GUDCA | 2.76E-03 ± 1.53E-03 | 7.08E-03 ± 2.69E-03 | 5.13E-04 ± 1.61E-04 | 2.23E-04 ± 8.44E-05 |
| GHDCA | 3.90E-03 ± 2.01E-03 | 6.87E-03 ± 3.82E-03 | 7.53E-04 ± 3.23E-04 | 2.29E-04 ± 1.15E-04 |
| GCDCA | 2.03E-03 ± 4.69E-04 | 4.96E-03 ± 9.12E-04* | 8.51E-04 ± 4.41E-04 | 3.91E-04 ± 1.64E-04* |
| GDCA | 3.02E-03 ± 5.38E-04 | 5.86E-03 ± 8.58E-04* | 8.03E-04 ± 5.19E-04* | 4.08E-04 ± 1.70E-04* |
Values are mean ± se concentrations (nmol/mg intestinal contents) measured using UPLC-MS/MS.
P < 0.05 vs. duodenum concentration of same bile acid.
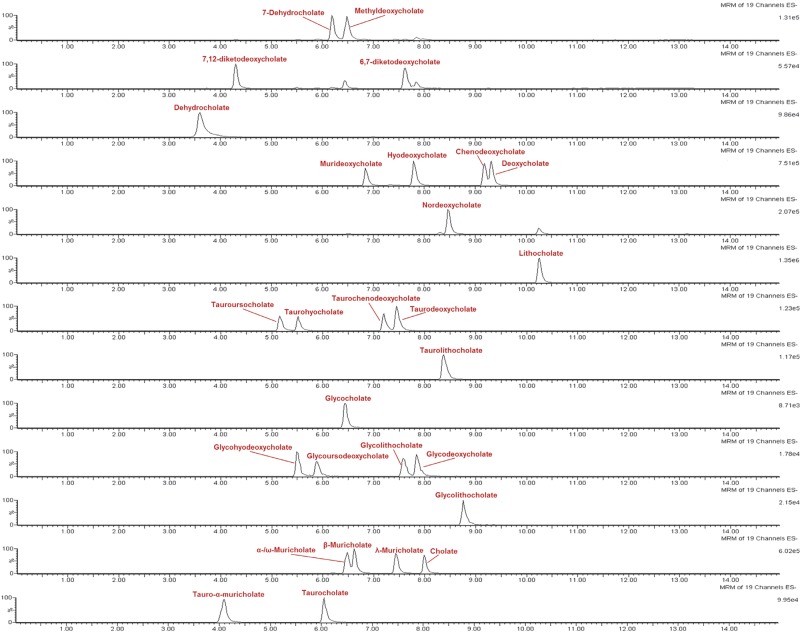
Figure 1.
Representative LC-MS/MS chromatogram of the authentic standards of bile acids under the final chromatography and detection conditions.
The relative composition of bile acids differed by region of the GI tract (Fig. 2). Taurine-conjugated bile acids accounted for 97.1 and 89.1% of all detected bile acids in the duodenum and ileum, but were much less prevalent in the cecum and rectum (<0.5% in each location). In contrast, unconjugated bile acids constituted the largest portion of all measured bile acids in the cecum and rectum. TCA levels decreased (46.6 to 29.5%), whereas TUDCA (1.2 to 10.0%), THDCA (14.6 to 17.2%), and CA (1.2 to 6.5%) levels all increased between the duodenum and ileum. The predominant unconjugated bile acids in the cecum and rectum included CDCA, DCA, and HDCA; these all had comparable abundance between the cecum and rectum. However, the proportion of β-MCA and α/ω-MCA decreased significantly from the cecum to the rectum (β-MCA: 9.4 to 0.7%, P < 0.05; α/ω-MCA: 3.7 to 0.5%, P < 0.05). The levels of these bile acids in the jejunum and colon (Supplemental Fig. S1) were also measured.
Figure 2.
Composition of bile acids in gastrointestinal tract of control rats. Proportions of individual bile acids in GI tract (A–D) and concentrations of unconjugated, taurine-conjugated, and glycine-conjugated bile acids (E–H) in GI tract in rats. A, E) duodenum; B, F) ileum; C, G) cecum; D, H) rectum.
Chronic ethanol consumption alters the bile acid metabolome of liver, gastrointestinal tract, and serum
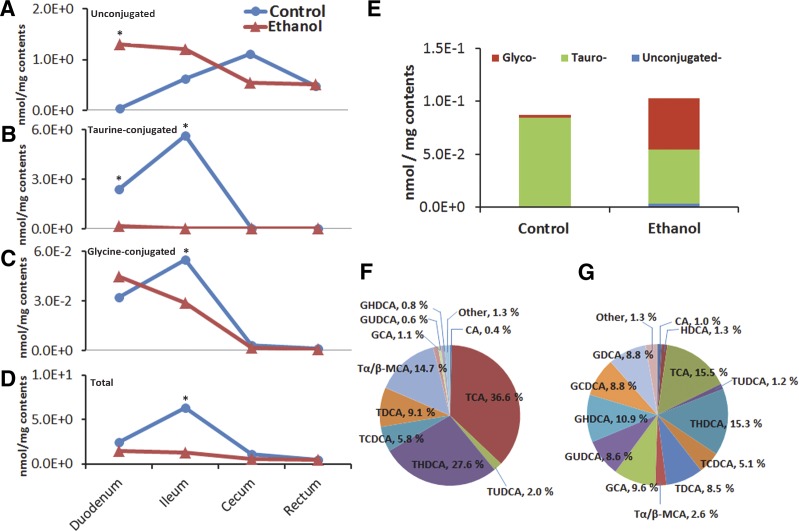
We compared the bile acid profile of the contents of GI tract, liver, and serum from control and ethanol-treated rats. As described above, the concentration of unconjugated bile acids in control rats increased steadily from the duodenum to cecum (0.04 to 1.11 nmol/mg), whereas a decreasing trend was observed in the ethanol group (1.30 to 0.54 nmol/mg) (Fig. 3). However, the concentration of unconjugated bile acids in the rectum was comparable between the control and ethanol groups. Ethanol consumption led to lower levels of taurine-conjugated bile acids in the duodenum and ileum (0.15 and 0.02 nmol/mg, respectively) relative to control rats (2.39 and 5.66 nmol/mg). Finally, a very low level of taurine-conjugated bile acids was detected in the cecums and rectums of both control and ethanol-treated rats.
Figure 3.
Alteration of bile acid profiles due to ethanol consumption. Ethanol consumption resulted in significant alteration in bile acid profiles. Unconjugated bile acids (A); taurine-conjugated bile acids (B); glycine-conjugated bile acids (C); and total bile acids in GI tract (D). Liver concentrations of unconjugated, taurine-conjugated, and glycine-conjugated bile acids in rats (E) and proportions of bile acids in liver of normal rats (F) and ethanol-treated rats (G). *P < 0.05 vs. control group.
The liver is the critical site where bile acids are synthesized and secreted. In the livers of control rats, taurine-conjugated bile acids predominated; however, ethanol consumption resulted in a significant increase in both glycine-conjugated as well as unconjugated bile acids (Fig. 3). In addition, the levels of several bile acids were different between the livers of control and ethanol-treated rats, including TCA (36.6 vs. 15.5% in control and ethanol-treated, respectively), THDCA (27.6 vs. 15.3%), T α/β-MCA (14.7 vs. 2.6%), GCA (1.1 vs. 9.6%), GUDCA (0.6 vs. 8.6%), and GHDCA (0.8 vs. 10.9%).
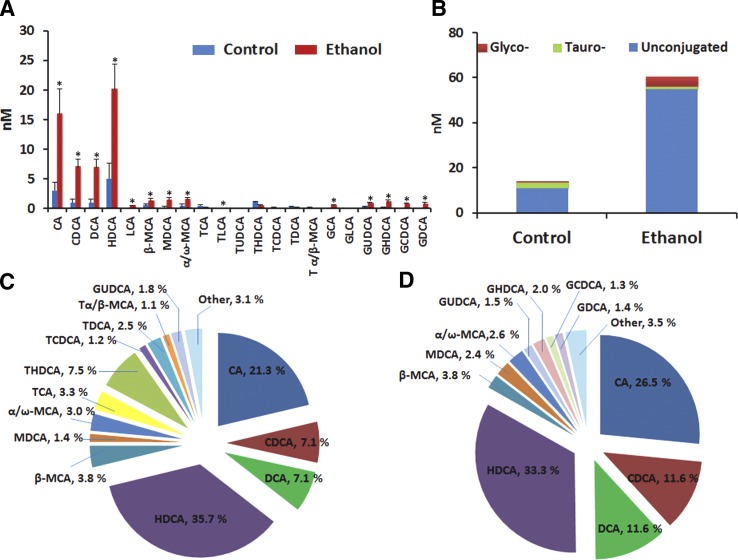
In serum from control rats, unconjugated bile acids were most abundant, particularly CA and HDCA. In contrast with the liver and intestinal contents, the serum levels of glycine-conjugated bile acids were greater than those of taurine-conjugated bile acids (Fig. 4). The serum bile acid profile was also different from that of the liver or GI tract; this is potentially because the bile acid concentrations measured in tissues predominantly derive from tissue bile acids and not from residual blood contained within the tissue (2). Unconjugated and glycine-conjugated bile acids were significantly elevated in the serum following chronic ethanol consumption, similar to what was observed in tissue samples (Fig. 3E). Furthermore, significant (P<0.05) alterations in taurine-conjugated bile acids and DCA were observed between serum from control and ethanol-treated rats (from 15.6% to 0 and from 7.1 to 11.6%, respectively, Fig. 4).
Figure 4.
Ethanol consumption induced significant alteration of bile acids profile in serum. Concentrations of individual bile acids in serum (A); concentrations of unconjugated, taurine-conjugated and glycine-conjugated bile acids in serum in rats (B); and proportions of them in serum of control rats (C) and ethanol treated rats (D). Data are presented as means ± se of 5–9 rats. *P < 0.05 vs. control group.
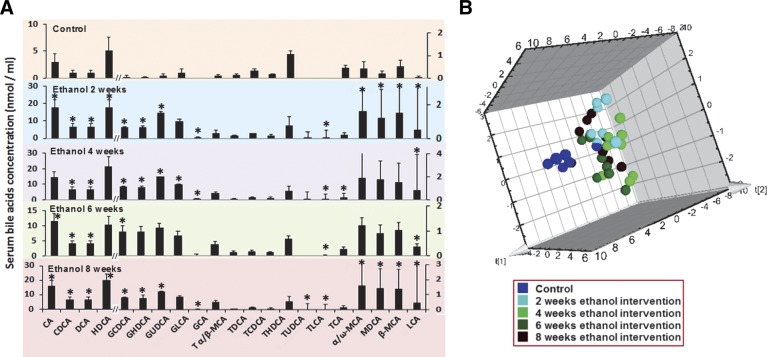
Ethanol-induced changes in serum bile acids over time
The time course of changes in serum bile acids before, during, and after 8 wk of ethanol consumption are presented in Fig. 5A. Unconjugated bile acids are most prevalent in control rats, with conjugated bile acids present in negligible amounts. We found that chronic ethanol consumption caused a marked perturbation of the bile acid profiles. PCA scores plot are shown in Fig. 5B. There were significant (P<0.01) increases in unconjugated bile acids and glycine-conjugated bile acids, whereas taurine-conjugated bile acids were markedly decreased after 2 wk and each point afterward. It is worth noting that the bile acid levels were almost constant after short-term (2 wk) ethanol consumption.
Figure 5.
Time course of the altered serum bile acid profile in response to ethanol consumption. Concentrations of individual bile acids in serum from rats in the control group and the ethanol intervention group at 2, 4, 6, and 8 wk postdose (A) and 3D principal component analysis (PCA) scores plot of serum bile acid profiles at predose and at 2, 4, 6, and 8 wk postdose of the ethanol intervention (B). *P < 0.05 vs. control group.
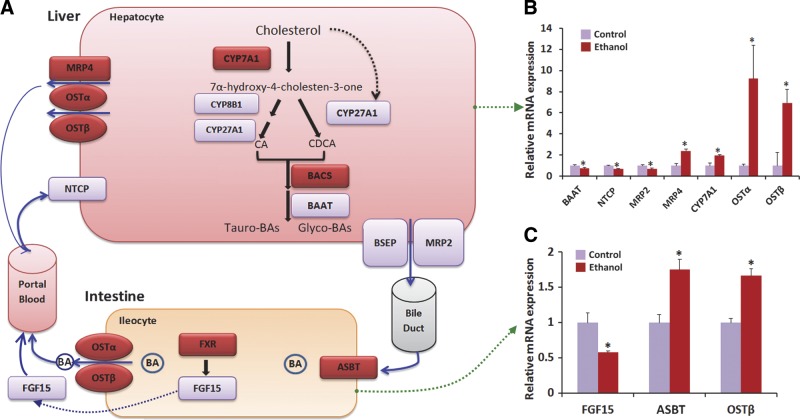
Ethanol consumption altered expression of genes related to bile acid metabolism and bile acid transport in liver and ileum
Given the significant effect of ethanol consumption on bile acid metabolism, we hypothesized that ethanol consumption modulated transcription of genes involved in bile acid metabolism and transport in the liver and ileum. We used qRT-PCR to analyze the expression of 20 genes involved in the regulation of bile acid synthesis, secretion, metabolism, and transport in the liver and ileum (Fig. 6 and Supplemental Fig. S2). Expression of CYP7A1, the rate-limiting enzyme for the “neutral” pathway of bile acid synthesis in the liver, was significantly up-regulated in ethanol-treated rats. This is consistent with the elevated levels of the primary bile acids CA and CDCA in ethanol-treated rats, as well as with the suppression of hepatic fibroblast growth factor receptor 4 (FGFR4), a transcriptional inhibitor of CYP7A1. However, sterol 27-hydroxylase (CYP27A1) mRNA was down-regulated by ethanol treatment, suggesting that the increase of primary bile acids in liver of ethanol-treated rats was not associated with the “acidic” pathway of bile acid synthesis. In addition, the mRNA levels of bile acid CoA: amino acid N-acyltransferase (BAAT), the gene encoding the enzyme that modulates bile acid conjugation, was significantly down-regulated by ethanol consumption, while expression of bile acid CoA synthetase (BACS) was slightly increased. The transport of bile acids between hepatocytes and blood is critical for maintaining bile acid homeostasis in liver and blood. The mRNA levels of genes encoding bile acid efflux transporters, the multidrug resistance protein 4 (MRP4) and the organic solute transporter α/β (OSTα/β), were significantly increased in ethanol-treated rats. In contrast, expression of the uptake transporter for bile acids, the sodium-taurocholate cotransporting polypeptide (NTCP), was suppressed by ethanol consumption. The ileum is the primary site for bile acid reabsorption in the GI tract. Ethanol consumption significantly up-regulated the expression of bile acid transporters in the ileum, including OSTβ and the apical sodium dependent bile acid transporter (ASBT) and also down-regulated expression of FGF15. Overall, our analysis of gene expression indicates that ethanol consumption caused an up-regulation of genes involved in bile acid synthesis and efflux transport as well as a down-regulation of genes regulating influx transport in the liver. In addition, expression of genes involved in reabsorption of bile acids in the ileum was also stimulated by ethanol treatment.
Figure 6.
Ethanol-induced altered expression of genes involved in bile acid metabolism and transport. Pathways of bile acid synthesis and metabolism in rats (A); and the mRNA expression of genes in the control group and the chronic ethanol intervention group with qRT-PCR analysis in liver (B) and ileum (C). Target gene expression was normalized to 18 s rRNA levels (which were set at 1) and presented as fold changes relative to control values. Each measure was performed with 3 duplicates; values are expressed as means ± se. The neutral and alternative pathways of bile acid biosynthesis are initiated by cholesterol 7α-hydroxylase (CYP7A1) and mitochondrial sterol 27-hydroxylase (CYP27A1), respectively. Sterol 12α-hydroxylase (CYP8B1) determines the ratio of cholate to chenodeoxycholate synthesized. Bile acids are secreted into the gallbladder via bile salt export pump (BSEP). At the terminal ileum, most of the bile acids are reabsorbed by ASBT into the enterocytes, and secreted into the portal circulation via basolateral bile acid transporters Ostα/Ostβ. At the basolateral membrane of the hepatocytes, bile acids are taken up by the NTCP transporter for resecretion into the gallbladder. ASBT, apical sodium dependent bile acid transporter; BACS, bile acid CoA synthetase; BAAT, bile acid CoA: amino acid N-acyltransferase; FGF15, fibroblast growth factor 15; FXR, farnesoid X receptor; MRP2, multidrug resistance protein 2; MRP4, multidrug resistance protein 4; NTCP, sodium-taurocholate cotransporting polypeptide; OSTα/β, organic solute transporter α/β. *P < 0.05 vs. control group.
Analysis of bile acid contents in experimental diets
To control for unintentional introduction of bile acids through the diet, we analyzed the two types of liquid diets used in this study and detected no bile acids in the dietary samples.
DISCUSSION
In this study, we examined the bile acid profiles of the GI tract, liver, and serum in normal rats and determined how those profiles were impacted by ethanol consumption. Serum ALT, one of the widely used markers in evaluating the degree of liver injury (18), was markedly increased due to ethanol consumption. Chronic ethanol consumption resulted in a global alteration of bile acid profiles in the liver, blood, and GI tract. The bile acid profiles for all tissues in the ethanol-treated rats were dominated by a small subset of unconjugated and glycine-conjugated bile acids, with taurine-conjugated bile acids present in negligible amounts. In contrast; in control rats, taurine-conjugated bile acids were predominant.
In control rats, taurine-conjugated bile acids were most abundant in the contents of the duodenum, jejunum, and ileum, as well as in the liver. Nearly 95% of bile acids in rodents are reabsorbed into bloodstream in the ileum. In both control and ethanol-treated rats, we observed a dramatic decrease in the concentrations of taurine- and glycine-conjugated bile acids in the cecum, colon, and rectum, while concentrations of unconjugated bile acids increased. Since the large intestine (the cecum and colon) has the highest abundance of microbes, the increase in unconjugated bile acids from the ileum to the cecum in control rats is primarily due to the deconjugation of taurine- and glycine-conjugated bile acids by the gut microbiota (19, 20). However, since the concentrations of taurine-conjugated bile acids were greatly reduced by ethanol consumption, unconjugated bile acids accounted for a large proportion of the total bile acids in the entire gastrointestinal tract in ethanol-treated rats.
Generally, ethanol consumption led to a substantial decrease in taurine-conjugated bile acids in all tissues examined. The concentration of unconjugated and glycine-conjugated bile acids in the liver was increased in ethanol-treated rats, even during short-term consumption of ethanol (2 wk). In contrast, taurine-conjugated bile acids in the serum were reduced under the same conditions, and the alterations in the bile acid profile in the blood were maintained during the entire experiment. Gene expression analysis indicated that ethanol consumption led to up-regulation of genes involved in bile acid synthesis and efflux transport, whereas the expression of genes regulating bile acid uptake by the liver were down-regulated. The hepatic extraction of free bile acids is less efficient than the extraction of conjugated forms (21), thus potentially explaining the greater proportion of unconjugated bile acids in the systemic circulation.
Taurine-conjugated bile acids are more hydrophilic and less toxic than either unconjugated or glycine-conjugated bile acids. Liver and intestinal bile acids in control rats were predominantly taurine conjugated, consistent with the findings of Dupont et al. (22), who saw that one-half of the liver bile acids in miniature swine were conjugated with taurine. In our study, the ethanol-induced decrease in taurine-conjugated bile acids in the contents of the duodenum, jejunum, and ileum may impair lipid emulsification, and subsequently may promote the development of ethanol-induced fatty liver. Since ethanol consumption can have a profound effect on the gut microbiota (23, 24), it is possible that the reduced abundance of taurine-conjugated bile acids in ethanol-treated rats is at least partially due to an ethanol-induced disturbance of the gut microbiota.
Bile acid metabolism is codependent on the biological activities of the gut microflora, and both bacterial and hepatic enzymes further modify bile acids during enterohepatic circulation (19, 25). Interestingly, germ-free mice and rats have a higher proportion of taurine-conjugated bile acids in their livers and intestines (25, 26), demonstrating the close association between gut microbiota and bile acid composition. The primary bile acids CA and CDCA are converted to the secondary bile acids DCA and LCA, respectively, by microbial biotransformation in the large intestine, cecum and colon. DCA, and, to a lesser extent, LCA, accumulate in the bile acid pool through the combination of passive absorption through the colonic mucosa and the inability of the human liver to hydroxylate DCA and LCA back to their respective primary bile acids (20). LCA is sulfated in the human liver at the 3-hydroxy position, conjugated at C-24, and excreted back into bile (27). In the intestinal lumen, conjugated bile acids are deconjugated by gut microbiota (19), and ethanol consumption promotes the overgrowth of gram-negative bacteria in the small intestine (16). An overgrowth of gut microbiota in ethanol-treated rats could accelerate microbial degradation of taurine, therefore decreasing taurine bioavailability and thus providing a potential explanation for the ethanol-induced decrease in taurine-conjugated bile acids in our study. Moderate alcohol consumption has also been seen to correlate with small intestinal bacterial overgrowth (28), thus the increased DCA, LCA, UDCA, and HDCA levels in all the compartments may be due to increased intestinal bacterial activity.
It has also been reported that the ratio of glycine-conjugated to taurine-conjugated bile acids is dependent on the hepatic taurine concentration (29). In our study, we found that the hepatic bile salt taurine to glycine ratio was 30:1 in control rats but equivalent in ethanol-treated rats. The majority of taurine is usually acquired through the diet, biosynthesized in hepatocytes, and degraded by the gut microbiota to inorganic sulfate (30). For this reason, an overgrowth of gut microbiota would be expected to cause a reduction in taurine-conjugated bile acids, similar to what we observed in ethanol-treated rats in our study. In addition, a recent investigation suggested that the reduction of taurine in the liver in ethanol-treated mice is due to the formation of N-acetyltaurine, a novel metabolite synthesized from taurine and acetate and excreted in urine (31). This metabolite may contribute to the lower concentration of taurine-conjugated bile acids we observed in the liver.
In hepatocytes, conjugation of bile acids with glycine or taurine is catalyzed by two sequential enzymatic reactions involving BAAT and BACS (32). BAAT expression in the liver was reduced in ethanol-treated mice. Since BAAT has a much higher affinity for taurine than glycine in rats (33), reduced BAAT expression could contribute to the lower concentration of taurine-conjugated bile acids in the livers of ethanol-treated rats. Conjugation of bile acids with either taurine or glycine has minimal effect on FXR activation; strong activation of FXR was observed despite a significant decrease in taurine- and glycine- conjugated bile acids in the ileum of normal rats (34). Instead, unconjugated bile acids induced a stronger activation of FXR than did conjugated bile acids. Although the exact mechanisms underlying the reduction of taurine-conjugated bile acids in ethanol-treated rats are unclear, we propose that ethanol-induced alterations in the gut microbiota may play a critical role in this process.
In summary, in order to characterize the effect of ethanol consumption on bile acid profiles in the serum, GI tract, and liver, we profiled a panel of bile acids in each of those tissues and found unique bile acid profiles characteristic for each tissue. Conjugated bile acids predominated in the liver, duodenum, and ileum, while unconjugated bile acids comprised the largest proportion of the bile acid profile in the serum, cecum, and rectum. Ethanol consumption had a profound effect on the bile acid profiles in the liver, GI tract, and serum. The ethanol-modulated bile acid profiles were characterized by a dramatic decrease in taurine-conjugated bile acids and a marked increase in unconjugated and glycine-conjugated bile acids. In support of these alterations, gene expression analysis revealed increased expression of CYP7A1, MRP4, OSTα, and OSTβ and decreased CYP27A1 and BAAT expression. This altered bile acid profile may be due to ethanol-induced changes in the gut microbiota and may indicate another mechanism by which the gut microbiota influence host metabolism.
Supplementary Material
Acknowledgments
This work was financially supported by U.S. National Institutes of Health/National Institute on Alcohol Abuse and Alcoholism grant R01 AA020212.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- 6,7-DKDCA
- 6,7-diketodeoxycholate
- 7-DCA
- 7-dehydrocholate
- α-MCA
- α-muricholate
- β-MCA
- β-muricholate
- λ-MCA
- λ-muricholate
- ω-MCA
- ω-muricholate
- ALT
- alanine aminotransferase
- ASBT
- apical sodium dependent bile acid transporter
- BAAT
- bile acid CoA: amino acid N-acyltransferase
- BACS
- bile acid CoA synthetase
- BSEP
- bile salt export pump
- CA
- cholate
- CDCA
- chenodeoxycholate
- CYP7A1
- cholesterol 7α-hydroxylase 1
- CYP8B1
- sterol 12α-hydroxylase
- CYP27A1
- sterol 27-hydroxylase
- DCA
- deoxycholate
- DHCA
- dehydrocholate
- FGF15
- fibroblast growth factor 15
- FGFR4
- fibroblast growth factor receptor 4
- FXR
- farnesoid X receptor
- GCA
- glycocholate
- GCDCA
- glycochenodeoxycholate
- GDCA
- glycodeoxycholate
- GHDCA
- glycohyodeoxycholate
- GI
- gastrointestinal
- GLCA
- glycolithocholate
- GUDCA
- glycoursodeoxycholate
- HDCA
- hyodeoxycholate
- IS
- internal standard
- LCA
- lithocholate
- MDCA
- murideoxycholate
- methyl DCA
- methyl deoxycholate
- MRP2
- multidrug resistance-associated protein 2
- MRP4
- multidrug resistance-associated protein 4
- MS
- mass sprectrometry
- NDCA
- nordeoxycholate
- NTCP
- sodium-taurocholate cotransporting polypeptide
- OSTα/β
- organic solute transporter α/β
- PCA
- principal component analysis
- T α-MCA
- tauro α-muricholate
- T β-MCA
- tauro β-muricholate
- TCA
- taurocholate
- TCDCA
- taurochenodeoxycholate
- TDCA
- taurodeoxycholate
- THDCA
- taurohyodeoxycholate
- TLCA
- taurolithocholate
- TUDCA
- tauroursodeoxycholate
- UPLC
- ultraperformance liquid chromatography
REFERENCES
- 1. Watanabe M., Houten S. M., Mataki C., Christoffolete M. A., Kim B. W., Sato H., Messaddeq N., Harney J. W., Ezaki O., Kodama T., Schoonjans K., Bianco A. C., Auwerx J. (2006) Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439, 484–489 [DOI] [PubMed] [Google Scholar]
- 2. Swann J. R., Want E. J., Geier F. M., Spagou K., Wilson I. D., Sidaway J. E., Nicholson J. K., Holmes E. (2011) Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl. 1), 4523–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim I., Ahn S. H., Inagaki T., Choi M., Ito S., Guo G. L., Kliewer S. A., Gonzalez F. J. (2007) Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 48, 2664–2672 [DOI] [PubMed] [Google Scholar]
- 4. Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., Gerard R. D., Repa J. J., Mangelsdorf D. J., Kliewer S. A. (2005) Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2, 217–225 [DOI] [PubMed] [Google Scholar]
- 5. Chiang J. Y. (1998) Regulation of bile acid synthesis. Front. Biosci. 3, d176–193 [DOI] [PubMed] [Google Scholar]
- 6. Oliva L., Beauge F., Choquart D., Montet A. M., Guitaoui M., Montet J. C. (1998) Ursodeoxycholate alleviates alcoholic fatty liver damage in rats. Alcohol. Clin. Exp. Res. 22, 1538–1543 [PubMed] [Google Scholar]
- 7. Wei J., Qiu de K., Ma X. (2009) Bile acids and insulin resistance: implications for treating nonalcoholic fatty liver disease. J. Dig. Dis. 10, 85–90 [DOI] [PubMed] [Google Scholar]
- 8. Reddy B. S. (1981) Diet and excretion of bile acids. Cancer Res. 41, 3766–3768 [PubMed] [Google Scholar]
- 9. Chu F. F., Esworthy R. S., Chu P. G., Longmate J. A., Huycke M. M., Wilczynski S., Doroshow J. H. (2004) Bacteria-induced intestinal cancer in mice with disrupted Gpx1 and Gpx2 genes. Cancer Res. 64, 962–968 [DOI] [PubMed] [Google Scholar]
- 10. Hylemon P. B., Zhou H., Pandak W. M., Ren S., Gil G., Dent P. (2009) Bile acids as regulatory molecules. J. Lipid Res. 50, 1509–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Midtvedt T. (1974) Microbial bile acid transformation. Am. J. Clin. Nutr. 27, 1341–1347 [DOI] [PubMed] [Google Scholar]
- 12. Robben J., Eldere J., Eyssen H. (1987) Bile acid desulfation by the gut microflora. In Frontiers in Microbiology (Clercq E., ed) Vol. 13, pp. 241–243, Springer, The Netherlands [Google Scholar]
- 13. Miyata M., Yamakawa H., Hamatsu M., Kuribayashi H., Takamatsu Y., Yamazoe Y. (2011) Enterobacteria modulate intestinal bile acid transport and homeostasis through apical sodium-dependent bile acid transporter (SLC10A2) expression. J. Pharmacol. Exp. Ther. 336, 188–196 [DOI] [PubMed] [Google Scholar]
- 14. Zhong Wei W., Zhao Y., Tang Y., Wei X., Shi X. (2012) Chronic alcohol exposure stimulates adipose tissue lipolysis in mice: role of reverse triglyceride transport in the pathogenesis of alcoholic steatosis. Am. J. Pathol. 180, 998–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jesus Delgado-Villa M., Luisa Ojeda M., Maria Rubio J., Luisa Murillo M., Carreras Sanchez O. (2009) Beneficial role of dietary folic acid on cholesterol and bile acid metabolism in ethanol-fed rats. J. Studies Alcohol Drugs 70, 615–622 [DOI] [PubMed] [Google Scholar]
- 16. Purohit V., Bode J. C., Bode C., Brenner D. A., Choudhry M. A., Hamilton F., Kang Y. J., Keshavarzian A., Rao R., Sartor R. B., Swanson C., Turner J. R. (2008) Alcohol, intestinal bacterial growth, intestinal permeability to endotoxin, and medical consequences: summary of a symposium. Alcohol 42, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dumas M. E., Barton R. H., Toye A., Cloarec O., Blancher C., Rothwell A., Fearnside J., Tatoud R., Blanc V., Lindon J. C., Mitchell S. C., Holmes E., McCarthy M. I., Scott J., Gauguier D., Nicholson J. K. (2006) Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. U. S. A. 103, 12511–12516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Niemela O. (2007) Biomarkers in alcoholism. Clin. Chim. Acta 377, 39–49 [DOI] [PubMed] [Google Scholar]
- 19. Martin F. P. J., Dumas M. E., Wang Y. L., Legido-Quigley C., Yap I. K. S., Tang H. R., Zirah S., Murphy G. M., Cloarec O., Lindon J. C., Sprenger N., Fay L. B., Kochhar S., van Bladeren P., Holmes E., Nicholson J. K. (2007) A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol. Syst. Biol. 3, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ridlon J. M., Kang D. J., Hylemon P. B. (2006) Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–259 [DOI] [PubMed] [Google Scholar]
- 21. Iga T., Klaassen C. D. (1982) Hepatic extraction of bile acids in rats. Biochem. Pharmacol. 31, 205–209 [PubMed] [Google Scholar]
- 22. Dupont J., Oh S. Y., O'Deen L. A., Geller S. (1974) Cholanoic (bile) acids in hepatic and nonhepatic tissues of miniature swine. Lipids 9, 294–297 [DOI] [PubMed] [Google Scholar]
- 23. Isabel Queipo-Ortuno M., Boto-Ordonez M., Murri M., Miguel Gomez-Zumaquero J., Clemente-Postigo M., Estruch R., Cardona Diaz F., Andres-Lacueva C., Tinahones F. J. (2012) Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am. J. Clin. Nutr. 95, 1323–1334 [DOI] [PubMed] [Google Scholar]
- 24. Mutlu E. A., Gillevet P. M., Rangwala H., Sikaroodi M., Naqvi A., Engen P. A., Kwasny M., Lau C. K., Keshavarzian A. (2012) Colonic microbiome is altered in alcoholism. Am. J. Physiol. Gastrointest. Liver Physiol. 302, G966–G978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Claus S. P., Tsang T. M., Wang Y., Cloarec O., Skordi E., Martin F. P., Rezzi S., Ross A., Kochhar S., Holmes E., Nicholson J. K. (2008) Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol. Syst. Biol. 4, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wostmann B. S. (1973) Intestinal bile acids and cholesterol absorption in the germfree rat. J. Nutr. 103, 982–990 [DOI] [PubMed] [Google Scholar]
- 27. Vlahcevic Z. R., Heuman D. M., Hylemon P. B. (1996) Physiology and pathophysiology of enterohepatic circulation of bile acids. In Hepatology: A Textbook of Liver Disease (Zakim D., Boyer T., eds) Vol. 1, pp. 376–417, Saunders, Philadelphia [Google Scholar]
- 28. Corrao G., Ferrari P., Zambon A., Torchio P. (1997) Are the recent trends in liver cirrhosis mortality affected by the changes in alcohol consumption? Analysis of latency period in European countries. J. Stud. Alcohol. 58, 486–494 [DOI] [PubMed] [Google Scholar]
- 29. Hardison W. G., Proffitt J. H. (1977) Influence of hepatic taurine concentration on bile acid conjugation with taurine. Am. J. Physiol. 232, E75–79 [DOI] [PubMed] [Google Scholar]
- 30. Hepner G. W., Sturman J. A., Hofmann A. F., Thomas P. J. (1973) Metabolism of steroid and amino acid moieties of conjugated bile acids in man. 3. Cholyltaurine (taurocholic acid). J. Clin. Invest. 52, 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shi X., Yao D., Chen C. (2012) Identification of N-acetyltaurine as a novel metabolite of ethanol through metabolomics-guided biochemical analysis. J. Biol. Chem. 287, 6336–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Falany C. N., Johnson M. R., Barnes S., Diasio R. B. (1994) Glycine and taurine conjugation of bile acids by a single enzyme. Molecular cloning and expression of human liver bile acid CoA: amino acid N-acyltransferase. J. Biol. Chem. 269, 19375–19379 [PubMed] [Google Scholar]
- 33. Killenberg P. G., Jordan J. T. (1978) Purification and characterization of bile acid-CoA: amino acid N-acyltransferase from rat liver. J. Biol. Chem. 253, 1005–1010 [PubMed] [Google Scholar]
- 34. Reschly E. J., Ai N., Ekins S., Welsh W. J., Hagey L. R., Hofmann A. F., Krasowski M. D. (2008) Evolution of the bile salt nuclear receptor FXR in vertebrates. J. Lipid Res. 49, 1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.