Abstract
We have analyzed the developmental molecular programs of the mouse hippocampus, a cortical structure critical for learning and memory, by means of large-scale DNA microarray techniques. Of 11,000 genes and expressed sequence tags examined, 1,926 showed dynamic changes during hippocampal development from embryonic day 16 to postnatal day 30. Gene-cluster analysis was used to group these genes into 16 distinct clusters with striking patterns that appear to correlate with major developmental hallmarks and cellular events. These include genes involved in neuronal proliferation, differentiation, and synapse formation. A complete list of the transcriptional changes has been compiled into a comprehensive gene profile database (http://BrainGenomics.Princeton.edu), which should prove valuable in advancing our understanding of the molecular and genetic programs underlying both the development and the functions of the mammalian brain.
The hippocampus is a brain structure that plays a critical role in learning and memory in humans and animals (1–3). Extensive electrophysiological studies have demonstrated the existence of activity-dependent synaptic plasticity such as long-term potentiation/long-term depression (LTP/LTD) in hippocampal pathways (4–6). Recent genetic analyses have clearly demonstrated the crucial role of the hippocampal synaptic plasticity in memory formation (7–10). Despite the significance of the hippocampus in learning and memory, our understanding of the genetic programs underlying the developing hippocampus is quite limited. During development, the hippocampus undergoes typical stages involving proliferation, differentiation, synapse formation, and the maturation of synaptic function. Research into the molecular mechanisms that control hippocampal development should enhance our understanding of the hippocampus and its role in various physiological and pathological conditions. To begin to analyze the genome-wide molecular events that occur in the developing hippocampus, we have used Affymetrix oligonucleotide microarrays that contain probes for 11,000 known genes and expressed sequence tags (ESTs) (11–13) to profile comprehensive molecular and genetic programs underlying the mammalian brain development.
Materials and Methods
Matings and RNA Preparation.
Timed matings were set up between adult wild-type mice from strain C57BL6/CBAF1. Females were inspected for plugs on the following day to ensure successful mating and the date of conception was noted so that pups could be collected at the appropriate time. Hippocampi were dissected from pups at the age of embryonic day 16 (E16), postnatal day 1 (P1), P7, P16, and P30. The tissues were immediately frozen in liquid nitrogen, and then stored at −80°C. Total RNA was isolated, using the RNA Extraction Kit (Amersham Pharmacia), from tissue by ultracentrifugation in a cesium trifluoroacetate gradient. RNA concentration was determined spectrophotometrically by taking the optical density at 260 nm before poly(A) RNA isolation (Amersham Pharmacia). Samples were stored at −80°C.
Target Preparation.
Double-stranded DNA was synthesized for each target, with the GIBCO/BRL Superscript Choice System (GIBCO/BRL, 18090-019). Typically, 1 μg of polyA RNA was used in a reverse transcription reaction to synthesize (−) strand cDNA with a primer containing poly T and T7 RNA polymerase promoter sequences. The double-stranded cDNA was isolated by phenol-chloroform extraction, followed by ethanol-precipitation with glycogen as a carrier. The cDNA was resuspended in 3 μl of RNase-free water, and 1 μl of the double-stranded cDNA was used as a template for in vitro transcription in the presence of biotinylated UTP and CTP to generate labeled antisense RNA. The in vitro transcription reaction was performed by using the Enzo BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics, 900182). Purification of the labeled RNA was carried out with the Qiagen RNeasy Mini Kit spin columns (74104 from Qiagen, Chatsworth, CA).
Array Hybridization and Scanning.
The labeled cRNA was fragmented in fragmentation buffer (5× buffer: 200 mM Tris-acetate (pH 8.1)/50 mM KOAc/150 mM MgOAc) and hybridized to the microarrays in 200 μl of hybridization solution containing 10 μg labeled target in 1× Mes buffer [0.1 M Mes/1.0 M NaCl/0.01% Triton X-100 (pH 6.7)] and 0.1 mg/ml herring sperm DNA. The arrays used in this study are the 11K-A (11,000 Affymetrix) mouse expression arrays. Arrays were placed on a rotisserie and rotated at 60 rpm for 16 h at 45°C. Following hybridization, the arrays were washed with 6× SSPE-T [0.9 M NaCl/60 mM NaH2PO4/6 mM EDTA/0.005% Triton X-100 (pH 7.6)] at 22°C on a fluidics station (Affymetrix) for 10 × 2 cycles, and subsequently with 0.1 Mes at 45°C for 30 min. The arrays were then stained with a streptavidin-phycoerythrin conjugate (Molecular Probes), followed by 10 × 2 wash cycles. To enhance the signals, the arrays were further stained with Anti-streptavidin antibody for 30 min followed by a 15 min staining with a streptavidin-phycoerythrin conjugate. After 10 × 2 additional wash cycles, the arrays were scanned at a resolution of 3 μm, using a specifically designed confocal scanner (Affymetrix). To further confirm the reliability of the array data, 16 genes were randomly picked from the list and reverse Northern blot analysis was performed. The dynamic changes in expression are consistent between the two different methods (data not shown).
Data Analysis.
The image data were analyzed by GENECHIP ANALYSIS SUITE (Affymetrix). Gene clustering analysis was performed by using GENECLUSTER 1.0 (MIT, Cambridge, MA).
Results
Large-Scale Transcriptional Analysis of the Developing Hippocampus.
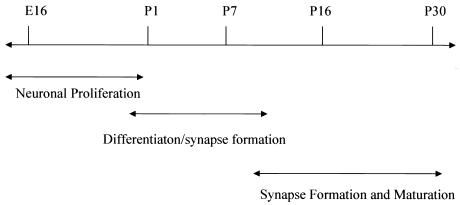
To establish comprehensive gene expression profiles during the development of the mouse hippocampus, five time points were chosen between E16 and P30. The time points, E16, P1, P7, P16, and P30, correspond to the peak periods for which major cellular and physiological events occur during mouse hippocampal development (Fig. 1). It is known that the most active proliferation of neurons occurs during the prenatal period, followed by dynamic outgrowth and differentiation in the neonatal period, and synaptogenesis in the first postnatal week. By the second postnatal week, synaptic connections are established and synaptic activity becomes increasingly active; by the third postnatal week, the fully connected hippocampal circuits exhibit great plasticity including robust long-term potentiation. By the end of the first month, the hippocampus is entering a more mature state (14–17).
Figure 1.
Dissection time line of the developing mouse hippocampus. The arrows illustrate one example of the peak windows for the major neuronal events occurring during the hippocampal development.
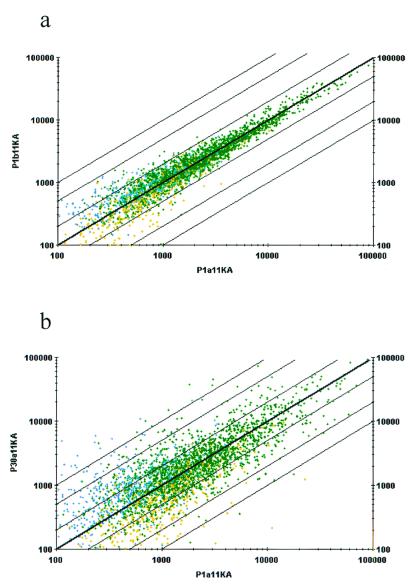
The hippocampi from several dozens of developing mouse brains were dissected at these five time points (E16, P1, P7, P16, and P30) and separated into two independent tissue pools for the extraction of poly(A) mRNA. These duplicate mRNAs were then used for generation of the fluorescently labeled targets for the hybridization to microarrays containing over 11,000 mouse genes and ESTs. To ensure the reliability of the data, all of the hybridization experiments were carried out in duplicate (two independent mRNAs and two sets of duplicate microarrays were used). The hybridization signals in the duplicate experiments were highly consistent and reproducible based on examination of the correlation between the duplicate hybridization signals. For example, from two independent hybridizations onto 11K-A (11,000 Affymetrix) arrays, more than 2,400 genes on this 11K-A array (containing ≈6,500 genes and ESTs) were detectable in the duplicate samples prepared independently from P1 hippocampal mRNA, and showed similar levels of signals in both P1a and P1b samples (Fig. 2a). Only 19 genes whose hybridization signals showed more than 3-fold variation (less than 0.3% of the total genes on the array) differed in the two experiments.
Figure 2.
Reproducibility of the genes identified by the microarray technology. (a) Reproducibility of the data obtained from the GeneChip array study. The green dots represent the genes detected in both P1 samples, an estimated 34% of the genes on a single chip. The yellow or blue dots represent the genes detected only in one of the samples, and thus these signals only constitute about 0.25–0.3% of the genes on the chip. (b) Differential gene expression between P1 and P30. An intensity of 400 to 500 corresponds to approximately one copy per cell. The axes represent gene expression intensity. The green dots represent the genes detected in both P1 and P30. The yellow dots represent the genes detected in P1, but not P30. Similarly, the blue dots represent the genes detected in the P30 sample, but not in P1. Five percent of probe sets showed more than 3-fold changes.
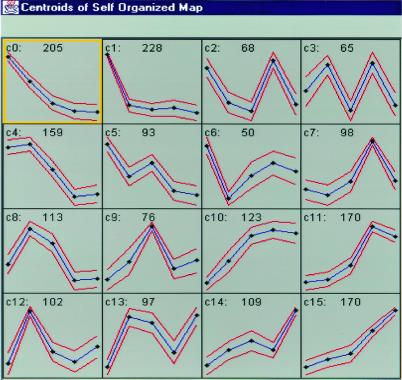
When the expression levels of the same set of these genes were compared between two developmental time points (e.g., P1 vs. P30) we found that 325 genes on the chip were differentially expressed with more than 3-fold changes (Fig. 2b). In total, over 1,926 genes showed dynamic changes in expression level during the hippocampal development period from E16 to P30. For a detailed description of these genes and ESTs, we have created the gene expression profile database that can be accessed via our web (http://BrainGenomics.Princeton.edu). To explore the patterns and possible genetic principles underlying developmentally regulated gene expression, we used the cluster analysis tool SOM (self-organizing map analysis; ref. 18). The 1,926 genes from the 11,000 arrays that showed significant expression changes during development grouped into 16 distinct gene clusters (Fig. 3a). Generally, these 16 clusters can be further classified into four major types. Type I clusters include cluster 0 (c0), c1, and c5, and exhibit overall age-dependent down-regulation, whereas Type II clusters (c10, c11, c14, and c15) show general age-dependent increased expression, reaching peak levels either at P16 or P30. Type III clusters show peak expression at either P1 or P7 (c4, c8, c9, c12, and c13), whereas the Type IV (c2, c3, and c6) clusters exhibit down-regulation at these time points.
Figure 3.
Cluster analysis of probes sets on mouse 11,000 arrays. Self-organizing maps (SOM) were used to group the 4,390 identified genes and ESTs into clusters based on similar expression dynamics over the five time point. Analysis algorithms were used to convert raw data into expression data for these genes before applying SOM analysis. The label at the upper-left corn of each inset represents cluster number (from c0–c15). The number in the top center of each inset represents the number of genes in that particular cluster. The five dots in each inset represent the five developmental time points (E16, P1, P7, P16, and P30).
It appears that these clusters correlate with major cellular changes during hippocampal development. For example, in c1 (Type I) a total of 228 genes exhibited high expression at E16, but were essentially switched off after birth. This genetic switch is correlated with the well known transition of an overwhelming number of neurons from a proliferating to postmitotic state. As expected, most of these 228 genes are known to be involved in control of the cell cycle, histone regulation, and DNA replication. On the other hand, in c12 (Type III) these genes show dramatic up-regulation of expression at P1 with low expression at other time points. This cluster contains genes involved in cell-type development (such as Nkx, homeobox transcription factors; ref. 19), morphogenic events (such as Wnt-3; Wnt-10a), and postmitotic regulation of neuronal differentiation such as Postmitotic Neural Gene-1 (a zinc finger family gene; ref. 20). Therefore, the cluster method appears to reflect the potential underlying molecular and genetic programs during hippocampal development.
Genetic Programs During the Prenatal Period of the Developing Hippocampus.
From the gene cluster analysis of these 1,926 genes and ESTs, overall molecular patterns appear at a genome-wide scale in the prenatal period of the developing hippocampus. To present this comprehensive picture in a concise and coherent manner, we have selected a small list of representative genes (see Tables 1–3, which are published as supplemental data on the PNAS web site, www.pnas.org, and on our web site, http://BrainGenomics.princeton.edu) for the following discussion. For example, from E16 to P1 almost all of the hippocampal neurons switch from a highly active proliferation state to a postmitotic state. As such, expression of proliferative genes involved in cell cycle progression were highly expressed at E16, then subsequently became silent or reduced significantly after birth. These genes were prominently grouped in c0 and c1 (a total of 432 genes; see Table 1 for the representative genes) and appear to correspond to general cellular function, including proliferation, DNA and RNA synthesis, and transcriptional and translational regulation.
The cyclin family of proteins that regulate mitosis were represented in c1 by cyclin-dependent kinase regulatory subunit 2, cyclins B2 and G2, G1/S-specific cyclin D2, and D-type G1 cyclin catalytic subunit, as well as cell division protein kinase 4 (21–23), GADD45 (24), and a growth arrest protein (suggesting a balance of factors that regulate mitosis). Another group of genes whose expression may contribute to the high proliferative activity of embryonic brain encode enzymes essential for DNA and RNA synthesis, such as DNA topoisomerase II (25) and the DEAD family of RNA helicases that regulate ribosome assembly, pre-mRNA splicing, mRNA translation, and RNA (26). Other pre-mRNA splicing factors were also identified, including unwinding protein 1 (27), U2 and U6 snRNPs (28), SRP75 from the SR family, and the myoblast cell surface antigen 24, which has been identified as a pre-mRNA splicing factor (29). In addition, chromosomal proteins H2A.X, H2A.1, and H1 histone subtype H1(0) (30, 31) were also identified in c1.
Genes involved in transcriptional regulation were also identified in c1. These include BTF3, a transcription factor required for transcriptional initiation of RNA polymerase II (32), and NF1-B, which mediates the transcription of several differentiation markers (33). Neurogenin-2, which encodes a neural-specific basic helix–loop–helix protein that is involved in determination of neuronal fate, specification, and differentiation of neuronal cell lineages (34), was also present. In addition to the transcriptional factors, many genes involved in translational regulation were differentially expressed. These genes include the initiation factor 2, elongation factor −1 and −2 (35), and protein translation factor SU1. Many types of ribosomal proteins were also up-regulated at this time (see Table 1). From our profiling analysis, it appears that the high rate of protein synthesis of protein in embryonic hippocampus is also coupled with the up-regulation of genes involved in protein degradation, suggesting that it is a tightly controlled during cell cycle. For example, the expression of ubiquitin-conjugating enzyme E2 (36), which is believed to be involved in ubiquitin-mediated regulation of cyclin regulation at E16, was up-regulated.
Obviously, the above genes represent only a small fraction of the 433 genes in c0 and c1 (see our web site, http://BrainGenomics.Princeton.edu). There are many other genes showing equally interesting features, such as transcriptional coactivator ALY, 11-zinc-finger transcription factor (CTCF), inhibitor of apoptosis protein 1, PEST phosphatase interacting protein, Zinc finger protein 7, zinc finger protein GF-1, and Zinc finger protein 91. These latter genes may be particularly interesting to be examined further for their role in hippocampal development.
Genetic Events During Neonatal and Early Postnatal Development.
The hippocampus undergoes many phenotypic changes after birth. Almost all neurons enter the postmitotic state and show extensive growth and differentiation in the first postnatal week. These cellular changes are marked by rapid cytoskeletal changes, production of cell adhesion molecules, and extracellular matrix formation, as well as expansion of cell membrane. Accordingly, the genes whose activities are highly active during this time are mostly represented by c4 and c8 (see Table 2 for the representative genes). These two clusters have similar, yet distinct profiles, with a peak occurring at P1 that decreased to basal levels by P16.
Among these two clusters of 272 genes, several actin and tubulin isoforms were identified in these clusters, including beta- and gamma-actin, actin-1, and actin-3, as well as several forms of alpha- and beta-tubulin, consistent with the notion that cellular differentiation is associated with the dynamic production of cytoskeletal and structural proteins (37). Interestingly, genes encoding the CCT chaperonin-containing family are prominently expressed in the same cluster (38). These chaperonin proteins are essential for promoting the correct folding of actin and tubulin. We observed that the subunits beta, epsilon, delta, and theta of the CCT chaperonin family are all highly expressed in the neonatal and early postnatal days.
Several cell-adhesion and extracellular-matrix proteins were also up-regulated during this period. These proteins are known to play critical roles during differentiation, pattern formation, and synaptogenesis; they include collagen and fibronectin, L1-like protein (39), a neural cell adhesion molecule, and neural cell adhesion molecule L1 (40), neurophilin, and neural cadherin (41). Morphological differentiation is also accompanied by membrane expansion. Indeed, we found that the expression of several genes involved in fatty acid and membrane synthesis are up-regulated in this period. They include brain fatty acid binding protein (B-FABP), fatty acid binding protein, and fatty acid synthase. These molecules are the building blocks of phospholipids and glycolipids (42), which are important components of the biosynthetic processes occurring at the neonatal time period.
Genetic Switches Up-Regulated in the Late Postnatal Hippocampus.
Following the neuronal differentiation and synapse formation in the early postnatal week, the hippocampal synapses and circuits become more active and begin to exhibit increased plasticity. Several clusters show a gradual increase in gene expression and are up-regulated at either P16 or P30. These genes, which are represented in c11 and c15, can be organized into categories of synaptic function, signal transduction, transcriptional and translational control, glucose and oxidative metabolism, and membrane regulation of ionic concentration (see Table 3). These two clusters contain 340 genes in total.
Many genes in c11 and c15 are involved in synaptic function. Some of these are involved in synaptic vesicle trafficking, including clathrin, which is a component of the coat that surrounds vesicles, and synaptogamin, a vesicle-associated protein involved in calcium-mediated release of neurotransmitters. Two synaptic vesicle-associated proteins, VAMP2 and synaptophysin, were also identified (43, 44), as well as UNC-18, which may play some role in vesicle trafficking (45). In addition to the genes involved in vesicular trafficking, several presynaptically released neuromodulators were also identified; these include fractalkine, a type of chemokine (46), cholecystokinin (47), and brain-derived neurotrophic factor (BDNF), a neurotrophin. Furthermore, many neurotransmitter receptors were identified that play postsynaptic functions; these include neurotransmitter receptors for glutamate, including glutamate receptor 1 (GluR1), glutamate receptor 2 (GluR2), and the NMDA receptor, as well as a receptor for acetylcholine. In addition, the neurotensin receptor was identified (48). Finally, several molecules that help to maintain ionic concentrations across membrane were also identified, including potassium channels, vacuolar adenosine triphosphatase (pore-forming subunits B and E), and transporters that catalyze sodium and potassium cotransport.
To ensure correct signal transduction between neurons, proper intracellular signal transduction must be established. Indeed, many signaling molecules, such as ras, ras-related protein RAB-3A, and mitogen-activated protein kinase (erk-1), are differentially expressed. In addition, calcineurin B, a protein phosphatase regulatory subunit, and FK506-binding protein (also known as PKBP-12), which binds to and inhibits calcineurin, are also present in this cluster. These proteins have been shown to be involved in regulating synaptic plasticity (49).
Interestingly, we have also identified a set of transcriptional and translational factors whose expression exhibits a gradual increase during the postnatal weeks, and which are different from the transcriptional and translation factors found in the embryonic hippocampus; these include zif/268, DNA binding protein SMBP2 (50), transcriptional activator FE65 (51), and transcription factor Sox-M. No overlap of factors identified in c1 vs. c11 and c15 was observed, suggesting that these temporally regulated transcription events are distinct and likely control different developmental phenotypes.
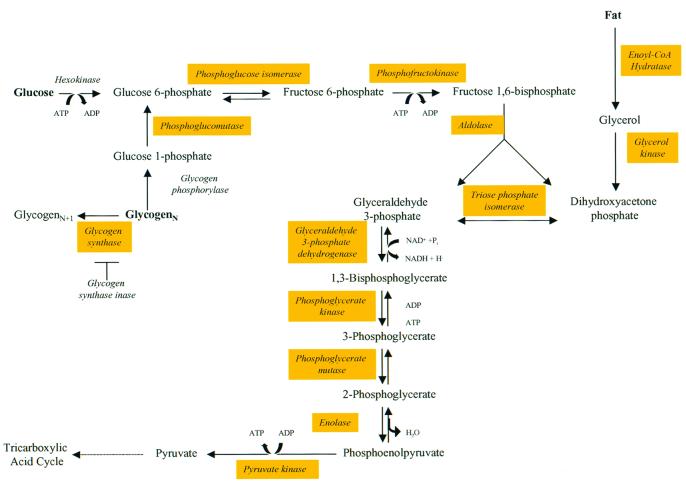
Besides changes in synaptic properties, it is well known that high levels of synaptic activity need to be supported by efficient energy utilization and production. In fact, the brain's energy utilization switches from ketone in neonatal brain to glucose in the adult brain as the major energy source. Interestingly, we have found that many of the enzymes involved in glucose metabolism and oxidative metabolism are gradually up-regulated as a function of postnatal development (see Fig. 4). The identification of all of the key glycolytic enzymes in c11 and c15 supports this transition (Fig. 4). These include two key enzymes that control the pace of glycolysis, phosphofructokinase and pyruvate kinase, as well as glucose-6-phosphate isomerase, fructose bisphosphate aldolase A and C, triose phosphate isomerase, phosphoglycerate kinase, and neural enolase (52). Some of the molecules involved in the mitochondrial respiratory chain that are differentially expressed include NADH-ubiquinone oxidoreductase, cytochrome c oxidase, succinate dehydrogenase, malate dehydrogenase, lactate dehydrogenase, and glycerophosphate dehydrogenase (52).
Figure 4.
Biochemical pathway of glycolysis. In Type II clusters (primarily in the c15 of SOM), all of the major enzymes involved in glycolysis were identified by gene-cluster analysis. This observation not only validates the approach, but also fits nicely with the well known fact that the energy utilization of the mammalian brain switches from ketone in neonatal stages to glucose at more mature stages. The genes identified from gene-cluster analysis are shown as marked in yellow.
Discussion
In an effort to dissect the molecular and genetic basis underlying the development of the hippocampus, we have used DNA microarray technology. This technology offers the unique advantage of being able to assess gene expression on a genome-wide scale. It has allowed us to establish the first genetic atlas of the developing mouse hippocampus, a region crucial for learning and memory.
Using Affymetrix GeneChip arrays, we have screened over 11,000 mouse genes and ESTs, and identified 1,926 genes that exhibited dynamic changes during the developmental period of the mouse hippocampus. We have further grouped these genes into 16 different categories by using SOM, a gene cluster program. This cluster analysis allows us to identify genes according to similar expression dynamics. The coordinated expression of genes in each cluster suggests that their function may be similar or may underlie a specific biological process. In fact, there are many clusters whose expression dynamics can be easily associated with known cellular phenotypes.
To further study the relationship between genes or gene clusters and their cellular function, a covariance analysis was used that is based on the principle that expression of genes that participate in a common function are coordinately regulated within a defined time window, and can be collectively changed upon genetic or environmental manipulation. Our analysis of the known genes in these clusters has clearly suggested that such a covariance strategy of linking a gene cluster to functional phenotypes based on their common time windows is reasonable and valid. For example, genes in c1, which are elevated at the embryonic brain but switched off after birth, may contribute to the particular phenotypes during the period, such as active neuronal proliferation. A large number of known genes in this cluster are involved in cell cycle and DNA replication.
The transcriptional analysis described here of the developing hippocampus may contribute to the understanding of the genetic program underlying the development of the mouse hippocampus. It is expected that a systematic and detailed cluster analysis of these genetic expression profiles will be valuable and should provide molecular insights into cellular and physiological hallmarks of hippocampal development, such as neuronal proliferation, differentiation, and synaptic formation and maturation. In fact, as a part of our comprehensive effort to understand the molecular mechanisms underlying mammalian brain development, function, and dysfunction, we have already conducted a series of gene profiling studies under various behavioral conditions, including exposure to enriched environments and aging (12, 53), which are well known to influence brain cognition. It is conceivable that the establishment of a variety of neurogenomic databases will provide an unprecedented opportunity for discovering key brain molecules, thereby potentially leading to new therapeutics for the treatment of both developmental and adult central nervous system disorders.
Supplementary Material
Acknowledgments
We thank Jaime Cochran for secretarial assistance and Shuqing Zhang for valuable technical assistance. This research was supported by funding from the W. M. Keck Foundation, the Burroughs Welcome Fund, and the National Institutes of Health (to J.Z.T.). J.Z.T. is a Beckman, Burroughs Welcome, and Keck Scholar.
Abbreviations
- EST
expressed sequence tag
- Pn
postnatal day n
- En
embryonic day n
- cn
cluster n
References
- 1.Squire L R. Memory and Brain. New York: Oxford Univ. Press; 1997. [Google Scholar]
- 2.O'keefe J, Nadel L. The Hippocampus as a Cognitive Map. Oxford: Clarendon; 1978. [Google Scholar]
- 3.Cohen N J, Eichenbaum H. Memory, Amnesia, and Hippocampal System. Cambridge, MA: MIT Press; 1993. [Google Scholar]
- 4.Bear M F, Malenka R L. Curr Opin Neurobiol. 1994;4:389–399. doi: 10.1016/0959-4388(94)90101-5. [DOI] [PubMed] [Google Scholar]
- 5.Bliss T V P, Collingridge G L. Nature (London) 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 6.Malenka R C, Nicoll R A. Science. 1999;285:1870–1874. doi: 10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- 7.Tsien J Z. Sci Am. 2000;282:62–68. doi: 10.1038/scientificamerican0400-62. [DOI] [PubMed] [Google Scholar]
- 8.Tsien J Z, Huerta P T, Tonegawa S. Cell. 1996;87:1327–1338. doi: 10.1016/s0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- 9.Rampon C, Tang Y P, Goodhouse J, Shimizu E, Kyin M, Tsien J Z. Nat Neurosci. 2000;3:238–244. doi: 10.1038/72945. [DOI] [PubMed] [Google Scholar]
- 10.Tsien J Z. Curr Opin Neurobiol. 2000;10:266–273. doi: 10.1016/s0959-4388(00)00070-2. [DOI] [PubMed] [Google Scholar]
- 11.Duggan D J, Bittner M, Chen Y, Meltzer P, Trent J M. Nat Genet. 1999;21, Suppl. 1:10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- 12.Rampon C, Jiang C H, Dong H, Tang Y P, Lockhart D, Schultz P G, Tsien J Z, Hu Y. Proc Natl Acad Sci USA. 2000;97:12880–12884. doi: 10.1073/pnas.97.23.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandberg R, Yasuda R, Pankratz D G, Carter T A, Del Rio J A, Wodicka L, Mayford M, Lockhart D J, Barlow C. Proc Natl Acad Sci USA. 2000;97:11038–11043. doi: 10.1073/pnas.97.20.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson M. Developmental Neurobiology. 3rd Ed. New York: Plenum; 1991. [Google Scholar]
- 15.Isaacson R L, Pribram K H. The Hippocampus. New York: Plenum; 1975. [Google Scholar]
- 16.Pokorny J, Yamamoto T. Brain Res Bull. 1981;7:113–120. doi: 10.1016/0361-9230(81)90075-7. [DOI] [PubMed] [Google Scholar]
- 17.Pokorny J, Yamamoto T. Brain Res Bull. 1981;7:121–130. doi: 10.1016/0361-9230(81)90076-9. [DOI] [PubMed] [Google Scholar]
- 18.Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander E S, Golub T R. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pabst O, Herbrand H, Takuma N, Arnold H H. Dev Genes Evol. 2000;210:47–50. doi: 10.1007/pl00008188. [DOI] [PubMed] [Google Scholar]
- 20.Weiner J A, Chun J. J Comp Neurol. 1997;381:130–142. doi: 10.1002/(sici)1096-9861(19970505)381:2<130::aid-cne2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Endicott J A, Noble M E, Tucker J A. Curr Opin Struct Biol. 1999;9:738–744. doi: 10.1016/s0959-440x(99)00038-x. [DOI] [PubMed] [Google Scholar]
- 22.Horne M C, Goolsby G L, Donaldson K L, Tran D, Neubauer M, Wahl A F. J Biol Chem. 1996;271:6050–6061. doi: 10.1074/jbc.271.11.6050. [DOI] [PubMed] [Google Scholar]
- 23.Sherr C J. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 24.Sheikh M S, Hollander M C, Fornance A J., Jr Biochem Pharmacol. 2000;59:43–45. doi: 10.1016/s0006-2952(99)00291-9. [DOI] [PubMed] [Google Scholar]
- 25.Berger J M. Curr Opin Struct Biol. 1998;8:26–32. doi: 10.1016/s0959-440x(98)80006-7. [DOI] [PubMed] [Google Scholar]
- 26.Luking A, Stahl U, Schmidt U. Crit Rev Biochem Mol Biol. 1998;33:59–96. doi: 10.1080/10409239891204233. [DOI] [PubMed] [Google Scholar]
- 27.Jiang J, Horowitz D S, Xu R M. Proc Natl Acad Sci USA. 2000;97:3022–3027. doi: 10.1073/pnas.97.7.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ro-Choi T S. Crit Rev Eukaryotic Gene Expression. 1999;9:107–158. doi: 10.1615/critreveukargeneexpr.v9.i2.20. [DOI] [PubMed] [Google Scholar]
- 29.Gower H J, Moore S E, Dickson G, Elsom V L, Nayak R, Walsh F S. Development (Cambridge, UK) 1989;105:723–731. doi: 10.1242/dev.105.4.723. [DOI] [PubMed] [Google Scholar]
- 30.Doenecke D, Albig W, Bode C, Drabent B, Franke K, Gavenis K, Witt O. Histochem Cell Biol. 1997;107:1–10. doi: 10.1007/s004180050083. [DOI] [PubMed] [Google Scholar]
- 31.Doenecke D, Albig W, Bouterfa H, Drabent B. J Cell Biochem. 1994;54:423–431. doi: 10.1002/jcb.240540409. [DOI] [PubMed] [Google Scholar]
- 32.Grein S, Pyerin W. Mol Cell Biochem. 1999;191:121–128. [PubMed] [Google Scholar]
- 33.Osada S, Matsubara T, Daimon S, Terazu Y, Xu M, Nishihara T, Imagawa M. Biochem J. 1999;342:189–198. [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Q, Fode C, Guillemot F, Anderson D J. Genes Dev. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark B F, Thirup S, Kjeldgaard M, Nyborg J. FEBS Lett. 1999;452:41–46. doi: 10.1016/s0014-5793(99)00562-1. [DOI] [PubMed] [Google Scholar]
- 36.Yamao F. J Biochem (Tokyo) 1999;25:223–229. doi: 10.1093/oxfordjournals.jbchem.a022277. [DOI] [PubMed] [Google Scholar]
- 37.Bradke F, Dotti C G. Microsc Res Tech. 2000;48:3–11. doi: 10.1002/(SICI)1097-0029(20000101)48:1<3::AID-JEMT2>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 38.Lund P A. Essays Biochem. 1995;29:113–123. [PubMed] [Google Scholar]
- 39.Hillenbrand R, Molthagen M, Montag D, Schachner M. Eur J Neurosci. 1999;11:813–826. doi: 10.1046/j.1460-9568.1999.00496.x. [DOI] [PubMed] [Google Scholar]
- 40.Baldwin T J, Fazeli M S, Doherty P, Walsh F S. J Cell Biochem. 1996;61:502–513. doi: 10.1002/(sici)1097-4644(19960616)61:4<502::aid-jcb3>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 41.Jessell T M, Hynes M A, Dodd J. Annu Rev Neurosci. 1990;13:227–255. doi: 10.1146/annurev.ne.13.030190.001303. [DOI] [PubMed] [Google Scholar]
- 42.Glatz J F, van der Vusse G J. Prog Lipid Res. 1996;35:243–282. doi: 10.1016/s0163-7827(96)00006-9. [DOI] [PubMed] [Google Scholar]
- 43.Bennett M K, Calakos N, Kreiner T, Scheller R H. J Cell Biol. 1992;116:761–775. doi: 10.1083/jcb.116.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calakos N, Scheller R H. Physiol Rev. 1996;76:1–29. doi: 10.1152/physrev.1996.76.1.1. [DOI] [PubMed] [Google Scholar]
- 45.Hata Y, Slaughter C A, Sudhof T C. Nature (London) 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- 46.Meucci O, Fatatis A, Simen A A, Bushell T J, Gray P W, Miller R J. Proc Natl Acad Sci USA. 1998;95:14500–14505. doi: 10.1073/pnas.95.24.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly J S, Dodd J. Adv Biochem Psychopharmacol. 1981;28:133–144. [PubMed] [Google Scholar]
- 48.Vincent J P, Mazella J, Kitabgi P. Trends Pharmacol Sci. 1999;20:302–309. doi: 10.1016/s0165-6147(99)01357-7. [DOI] [PubMed] [Google Scholar]
- 49.Mulkey R M, Endo S, Shenolikar S, Malenka R C. Nature (London) 1994;369:486–488. doi: 10.1038/369486a0. [DOI] [PubMed] [Google Scholar]
- 50.Cox G A, Mahaffey C L, Frankel W N. Neuron. 1998;21:1327–1337. doi: 10.1016/s0896-6273(00)80652-2. [DOI] [PubMed] [Google Scholar]
- 51.Faraonio R, Minopoli G, Porcellini A, Costanzo F, Cimino F, Russo T. Nucleic Acids Res. 1994;22:4876–4883. doi: 10.1093/nar/22.23.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stryer L. Biochemistry. New York: Freeman; 1995. [Google Scholar]
- 53.Jiang C H, Tsien J Z, Schultz P G, Hu Y. Proc Natl Acad Sci USA. 2001;98:1930–1934. doi: 10.1073/pnas.98.4.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.