Abstract
Kidney is a major target for adverse effects associated with corticosteroids. A microarray dataset was generated to examine changes in gene expression in rat kidney in response to methylprednisolone. Four control and 48 drug-treated animals were killed at 16 times after drug administration. Kidney RNA was used to query 52 individual Affymetrix chips, generating data for 15,967 different probe sets for each chip. Mining techniques applicable to time series data that identify drug-regulated changes in gene expression were applied. Four sequential filters eliminated probe sets that were not expressed in the tissue, not regulated by drug, or did not meet defined quality control standards. These filters eliminated 14,890 probe sets (94%) from further consideration. Application of judiciously chosen filters is an effective tool for data mining of time series datasets. The remaining data can then be further analyzed by clustering and mathematical modeling. Initial analysis of this filtered dataset identified a group of genes whose pattern of regulation was highly correlated with prototype corticosteroid enhanced genes. Twenty genes in this group, as well as selected genes exhibiting either downregulation or no regulation, were analyzed for 5′ GRE half-sites conserved across species. In general, the results support the hypothesis that the existence of conserved DNA binding sites can serve as an important adjunct to purely analytic approaches to clustering genes into groups with common mechanisms of regulation. This dataset, as well as similar datasets on liver and muscle, are available online in a format amenable to further analysis by others.
Keywords: data mining, gene arrays, glucocorticoids, pharmacogenomics, evolutionary conservation
Glucocorticoids are a class of steroid hormones that play a central role in regulating carbohydrate, protein, and lipid metabolism. They also modulate immune function. Most tissues are targets for glucocorticoid action and contribute in some way to their wide-ranging physiological effects. Corticosteroids are synthetic glucocorticoids with potent anti-inflammatory and immunosuppressive effects that define their pharmacological use (6, 50). They are used therapeutically for a variety of conditions, including transplantation (bone marrow, liver, and kidney), asthma, nephrotic syndrome, Crohn’s disease, lupus, multiple sclerosis, dermatomyositis, arthritis, inflammatory bowl disease, leukemia, and non-Hodgkin’s lymphoma. Beneficial effects deriving from inhibition of the immune system are accompanied by adverse effects that include hyperglycemia, dyslipidemia, muscle wasting, hypertension, nephropathy, fatty liver, and an increased risk of arteriosclerosis (8, 18, 32, 44, 50).
The generally accepted mechanism for most glucocorticoid effects involves binding of free steroid to a cytoplasmically localized receptor, translocation of ligand-bound receptor to the nucleus, binding to specific DNA sites (GREs), and modulation of the amounts of selective mRNAs (39, 48). Although some effects on mRNA stability have been noted, a common mechanism involves increasing or decreasing the rate of transcription of particular genes. Modulation of transcriptional rate can entail direct interaction of ligand-bound receptor with GREs in the 5′ regulatory regions of the gene, interaction with other transcriptional enhancers or suppressors, or by regulation of other biosignals that in turn modulate gene expression. Understanding the complex patterns of mRNA changes induced by these steroids in various target tissues, as well as commonalities in their regulation, is important to understanding both the beneficial and adverse effects of corticosteroids.
The kidney has a major impact on blood volume, content, and distribution. Effects of pharmacological doses of corticosteroids on the kidney contribute to several of the adverse systemic effects of corticosteroids such as hyperglycemia and hypertension (28, 49). Corticosteroids have both direct and indirect adverse effects on the kidney. There are direct changes in gene expression in the kidney that promote hypertrophy and fibrosis (13, 21, 25, 60). Indirectly, hyperlipidemia and hyperglycemia caused by corticosteroids contribute to the development of nephropathy. Corticosteroids are capable of binding to both the glucocorticoid and the mineralocorticoid receptors in the kidney (15, 19). Physiologically, the mineralocorticoid receptor is protected from endogenous glucocorticoid binding by the enzyme 11β-hydroxysteroid dehydrogenase type 2 (11β-HSD2). However, in conditions in which corticosteroids are excessive and the capacity of the enzyme is exceeded, there are potent mineralocorticoid effects resulting in sodium retention and increased blood volume (38, 51).
A major difficulty in parsing causal molecular relationships for the complex systemic effects caused by corticosteroids is the diverse origins of the available data. A large proportion of the data is derived from cell culture studies in which corticosteroids are applied for a period of time and changes in the expression of particular genes are noted. Although useful, these studies do not consider the fact that other factors in the environmental milieu, such as other hormones and substrates, often modulate the cellular response to corticosteroids. Even when these factors are added, they are not presented in the temporal sequence extant in vivo. Similarly, the available in vivo data are derived from different species subjected to a wide variety of dosing regimens and are often studied at a single time point.
Our approach to studying multitissue, multigene effects of corticosteroids involves in vivo administration coupled with a rich time series design that allows us to capture the dynamic patterns of molecular changes that occur in response to the drug in a population of animals. For many years we have used this time series design to evaluate cascades of molecular events in time initiated by an acute dose of the corticosteroid methylprednisolone (MPL) (34, 40, 43, 52–54, 58). Complex systemic phenomena such as diabetes, arteriosclerosis, and hypertension do not develop as the result of a single dose of drug. However, each dose of a drug initiates molecular events in time that, if perpetuated by repeated dosing, results in the development of the complex adverse systemic phenomena. Our studies were initiated with adrenalectomized (ADX) rats, where the expression of the corticosteroid-responsive genes have a stable baseline. Therefore, in such a time series, the drug is a stimulus causing divergence from baseline followed by return to the initial value. The results of those studies were used to develop pharmacokinetic/pharmacodynamic (PK/PD) models that describe limited cascades of molecular events initiated by MPL in liver and skeletal muscle. More recently, mRNA prepared from both liver and skeletal muscle from the same population of animals were analyzed using Affymetrix RU_34A rat gene chips that contain 8,799 probe sets. Initial approaches to data mining, clustering, and PK/PD modeling of data from those datasets have been published (1, 2, 4, 26, 27). In the present report, we describe the development and data mining of a parallel dataset developed using the kidneys from these same animals. However, in this case, the samples (52) were applied to the newer Affymetric RAE230A chip, which contains 15,967 probe sets. (All genes represented on the RU_34A chips are also present on the newer chip.) This report describes the 977 probe sets potentially regulated by corticosteroids in kidney (6%), which merit further attention with respect to functional clustering and PK/PD modeling. We also describe an initial test of the hypothesis that evolutionary conservation of specific transcription factor binding sites can provide a biologically relevant approach to clustering genes into groups with a common mechanism of regulation. These results, along with the probe sets identified in liver and skeletal muscle, provide a dynamic multitissue picture of the origins of the adverse systemic effects caused by corticosteroids.
METHODS
Experimental design
Kidney samples were obtained from a previously performed animal study in our laboratory. All procedures involving experimental animals adhered to the “Principles of Laboratory Animal Care” (National Institutes of Health publication no. 85-23, 1985) and were reviewed by our institution’s Institutional Animal Care and Use Committee. Male adrenalectomized (ADX) Wistar rats (Rattus rattus) weighing 225–250 g were obtained from Harlan Sprague Dawley (Indianapolis, IN). Animals were allowed to acclimate in our facility for 1 wk before study, which also ensured elimination of endogenous hormone. One day before the study, all rats were subjected to right external jugular vein cannulation under light ether anesthesia. Four animals were designated as controls (i.e., 0 time samples) and received vehicle only. The remaining 48 animals received a single 50 mg/kg dose of MPL sodium succinate (Pharmacia-Upjohn, Kalamazoo, MI) via the cannula over 30 s. Three rats were killed by exsanguination under anesthesia at each of the following time points: 0.25, 0.5, 0.75, 1, 2, 4, 5, 5.5, 6, 7, 8, 12, 18, 30, 48, and 72 h after dosing. The sampling time points were selected on the basis of previous studies describing glucocorticoid receptor (GR) dynamics and enzyme induction in liver and skeletal muscle (52, 54).
Microarrays
Both kidneys from each animal were ground into a fine powder in a mortar cooled by liquid nitrogen. Kidney powder (100 mg) from each individual animal was added to 1 ml of prechilled TRIzol reagent (Invitrogen, Carlsbad, CA), and total RNA extractions were carried out according to the manufacturer’s directions. Extracted RNAs were further purified by passage through RNAeasy minicolumns (Qiagen, Valencia, CA) according to the manufacturer’s protocols for RNA clean-up. Final RNA preparations were resuspended in RNase-free water and stored at −80°C. The RNAs were quantified spectrophotometrically, and purity and integrity were assessed by agarose gel electrophoresis. All samples exhibited A260/280 ratios of ∼2.0, and all showed intact ribosomal 28S and 18S RNA bands in an approximate ratio of 2:1 as visualized by ethidium bromide staining, demonstrating that good-quality RNA can be prepared from tissue after prolonged storage at ultralow temperatures, provided that appropriate care is taken to avoid even brief exposure of the tissue to temperature fluctuations.
Isolated RNA from each kidney sample was used to prepare the target according to the manufacturer’s protocols. The biotinylated cRNAs were hybridized to 52 individual Affymetrix GeneChips Rat Genome 230A (Affymetrix, Santa Clara, CA), which contained 15,967 probe sets. The high reproducibility of in situ synthesis of oligonucleotide chips allows accurate comparison of signals generated by samples hybridized to separate arrays. This entire dataset has been submitted to the NCBI Gene Expression Omnibus database (GSM29111-GSM29173) and is also available on line at http://pepr.cnmcresearch.org.
Initial data analysis
The Affymetrix oligonucleotide microarrays use sequence information and photolithograpy-directed combinatorial chemical synthesis to develop probe sets for the genes of interest. Each probe set consists of a series of short oligonucleotide sequences and an identical partner sequence, except for a single base mismatch in the center. The mismatch sequence provides a unique background for each sequence in the series. Affymetrix Microarray Suite 5.0 was used for initial data acquisition and basic analysis. In this first step, a “call” of “present” (P), “absent” (A), or “marginal” (M) was determined for each probe set on each chip, based on the comparison of the perfect matched and mismatched pairs for the gene sequence. The results were normalized for each chip by use of a distribution of all genes around the 50th percentile. The results from the first step were inputted to the program GeneSpring 7.2 (Silicon Genetics, Redwood City, CA) for data mining, initial clustering, and statistical analyses.
Data mining
A data mining approach previously developed by us and applied to similar datasets generated from liver and skeletal muscle from these same animals was applied to this dataset (1, 4). First, the data were transformed so that the values for all probe sets were within the same range. To accomplish this, values for each individual probe set on each chip were expressed as a ratio to the mean of the four control values for that gene, which we refer to as “normalized intensity.” Thus the average of each probe set has a value of 1 at zero time and decreases, increases, or remains not different from controls over the time series. A series of filtering steps was applied to the data in an attempt to eliminate probe sets that were not of further interest. These filters were designed to eliminate probe sets that are either not expressed in kidney tissue or are not regulated by drug from the full set of more than 15,000 probe sets on the chip, as opposed to applying statistical methods to select out probe sets that are different from controls with a certain probability. The first level of filtering was designed to eliminate probe sets not expressed in kidney and utilized the Affymetrix call feature. This first filter required that the probe set for the gene have a call of P (present) on at least 4 of the 52 chips. The second level of filtering that we applied was designed to eliminate probe sets that could not meet the basic criterion of a regulated probe. Specifically, this filtering approach was designed to eliminate probe sets whose average did not deviate from baseline by a certain value for a reasonable number of time points and employed two filters that were designed to eliminate probe sets that were neither down- nor upregulated. The first of these filters eliminated probe sets that could not meet a minimal criterion for downregulation. Starting with the 4P filtered list, we eliminated all probe sets that did not have average values below 0.65 in at least four conditions (time points). The next filter was designed to eliminate probe sets that could not meet a minimal criterion for upregulation. Starting with the 4P filtered list, we eliminated all probe sets that did not have average values above 1.5 in at least four conditions (time points). The last filter that we applied addressed the quality of the data. For this “quality control” filter we eliminated probe sets that did not meet two conditions. The first condition focused on the control chips. As indicated above, our initial operation was to divide the value of each individual probe set on each chip by the mean of the values for that probe set on the four control chips. Therefore, the quality of the control data for each particular probe set is of unique importance in defining regulation by the drug. This filter eliminated probe sets whose control values exhibited coefficients of variation (CVs) of >50%. The second condition focused on the remaining 16 time points. This filter also eliminated probe sets whose CV for more than 8 of the remaining 16 time points exceeded 50%. The final filtered dataset was analyzed by a one-way ANOVA with a Tukey post hoc test (P < 0.05).
RESULTS AND DISCUSSION
A population of ADX male Wistar rats was injected with a single bolus dose of MPL, groups of animals were killed at 16 time points over a 72-h period, and MPL-treated kidney samples were compared with vehicle-treated controls. ADX animals were used to eliminate the circadian oscillation of corticosterone and provide a stable baseline. This allowed us to identify gene transcripts that deviate from the baseline in response to MPL, and determine the duration of time it takes to return to that baseline. The times the animals were killed over the 72-h period were chosen on the basis of previous experiments indicating that the effect of the drug was most significant at the early times after dosing, but full recovery required in some cases as long as 72 h (52, 54). Affymetrix R230A chips were used to examine the temporal profile of changes in global gene expression in response to this single bolus dose of MPL. RNA samples from each individual animal were applied to a separate chip to preserve interanimal variation. Because this chip contains 15,967 probe sets, the major problem was identifying the relatively small percentage of the probe sets that are regulated by corticosteroids. To accomplish this task, we have developed an approach to data mining that is based on a series of filters designed to eliminate probe sets that do not meet certain explicit criteria, including tissue-specific expression, drug regulation, and quality control. This series of filters produces a remainder of a relatively small percentage of the total probe sets that then can become the focus of temporal and functional clustering. This approach has also been applied to similar datasets developed for skeletal muscle and liver from the same animals.
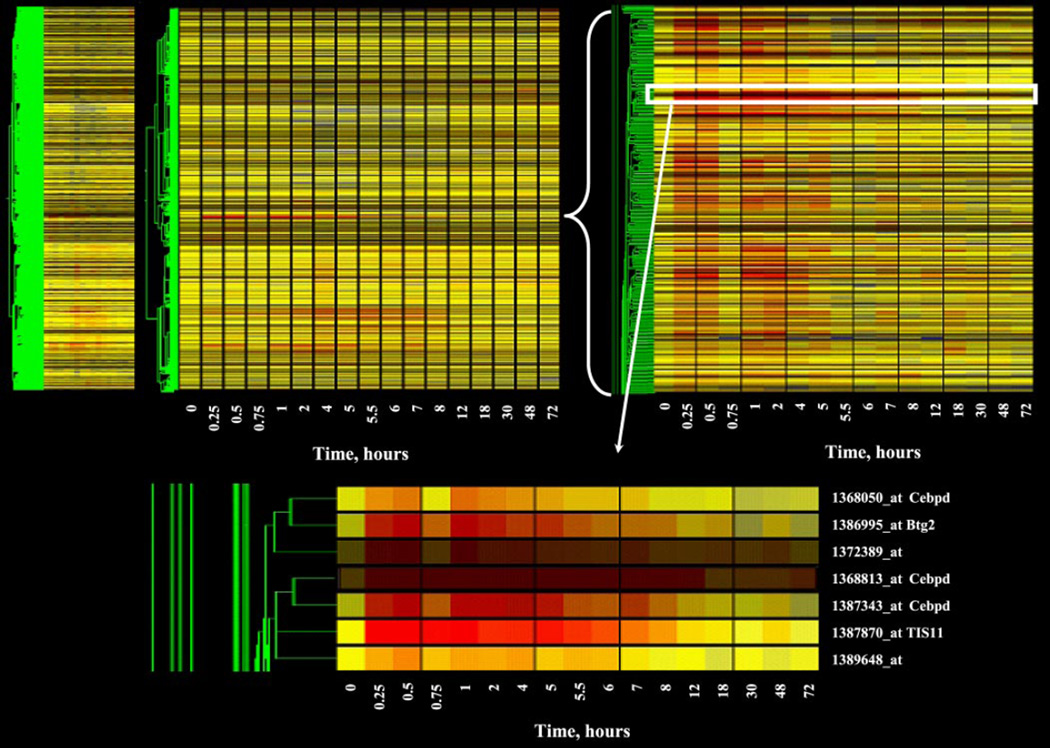
The initial step in the data mining analysis was to transform the data so that the values for all probe sets were within the same range. To accomplish this, values for each individual probe set on each chip were expressed as a ratio to the mean of the four control values for that gene, which we refer to as normalized intensity. Thus the average of each probe set has a value of 1 at zero time and decreases, increases, or remains not different from controls over the time series. To monitor the progression of the mining, we used the gene tree clustering tool developed by Eisen et al. (16). This algorithm can be used to construct a dendrogram of genes with similar patterns. A negative aspect of this tool, and most clustering algorithms when applied to time series data, is the assumption that the points in the time series are equally spaced and independent. Notwithstanding this drawback, gene trees provide an excellent method of visualizing the progression of the data analysis. Fig. 1, top left, shows the gene tree derived from the GeneSpring program for the entire dataset (15,967 probe sets at 17 time points). This tree was constructed using a Pearson correlation as the similarity index. The x-axis presents the 17 time points (including 0 time controls) studied in rank order from left to right. Vehicle controls are nominally referred to as time 0. As pointed out above, with this visualization tool each time point is equally spaced and therefore does not represent the actual temporal relationship between points. The y-axis presents the mean of the normalized value at each time point for each of the individual probe sets, represented by color and clustered by similarity. In this view, the yellow represents a value of “1,” progression toward red represents values that exceed “1,” and progression toward blue represents values that decline toward 0. The intensity of the color reflects the intensity of the original signal. To the left of the figure is a schematic tree of the relationship of all probe sets to one another based on expression pattern similarity (represented in green). Although the gene tree representation of the entire dataset is of limited value for examining individual gene patterns of regulation, it does illustrate two points. First, within the entire dataset are a vast number of genes represented by black (no expression in kidney regardless of treatment) or by yellow across the entire time frame studied. This latter group of genes exhibits no temporal regulation by the drug (i.e., their expression does not deviate from control value after drug dosing). Both represent probe sets that we wish to filter from the data set. Second, it does reflect segregation of similarly regulated genes and demonstrates that similar patterns of regulation do exist. For example, groups of intense red or blue represent clusters of genes with similar up- or downregulation, respectively. Figure 1, top right, provides a zoom-in view of one such clustering of probe sets with apparent upregulation. The location of this group of probe sets within the entire dataset (top left) is indicated by brackets. Figure 1, bottom, shows an even closer zoom-in on seven probe sets in this grouping with similar patterns.
Fig. 1.
Gene tree representation of the entire dataset of mRNA expression in kidney as a function of time after methylprednisolone (MPL) treatment. The gene tree represents the averaged normalized values of each of 15,967 probe sets at 17 different time points after MPL treatment, grouped by pattern similarities. The y-axis represents individual probe sets, with color indicating relative intensities (yellow represents no change from control; progression toward red indicates increased expression; progression toward blue represents decreased expression vs. control values). The x axis ranks samples in sequence of time after MPL treatment. Top left: the entire dataset; top right and bottom: successive zoom-in views of areas highlighted.
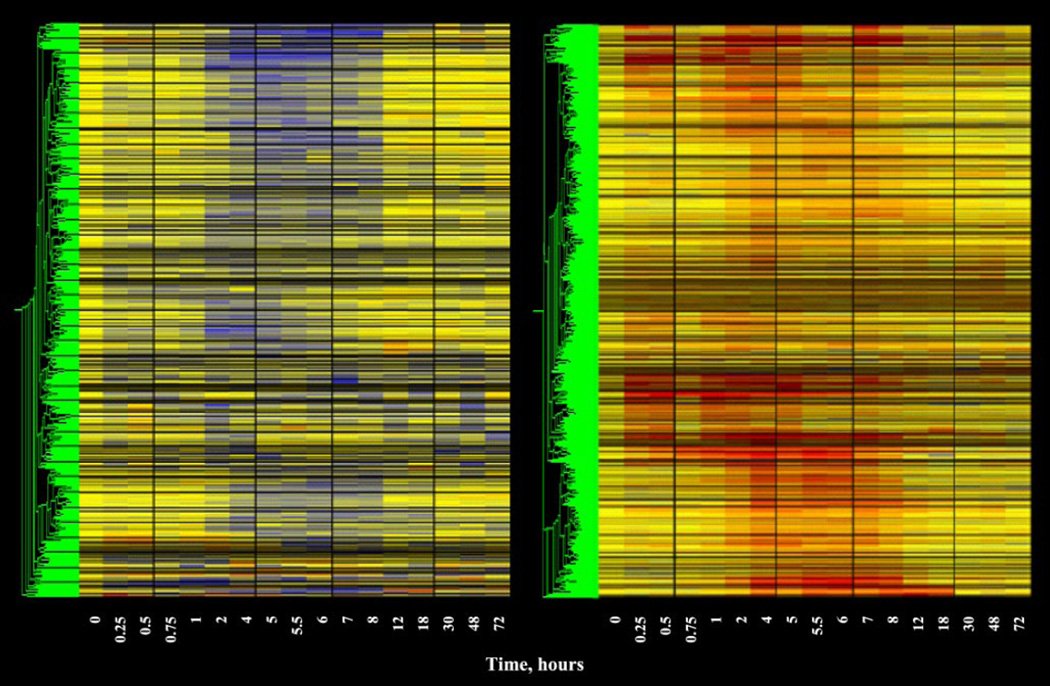
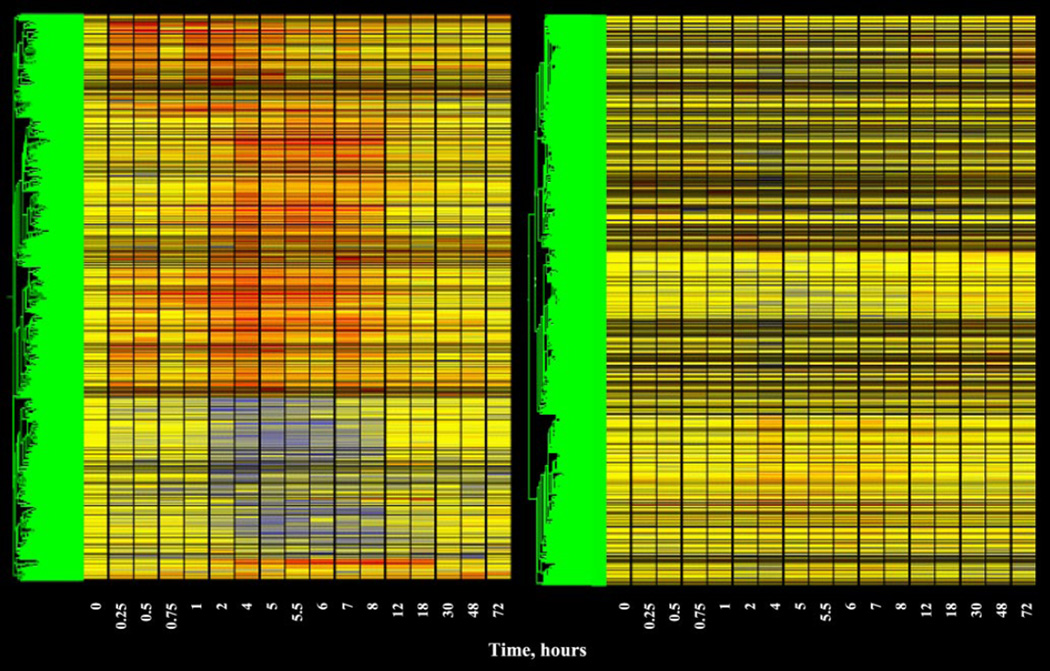
A series of filtering steps was applied to the data in an attempt eliminate probe sets that were not of further interest. The first level of filtering was designed to eliminate probe sets not expressed in kidney and utilized the Affymetrix call feature. This first filter required that the probe set for the gene have a call of P (present) on at least 4 of the 52 chips. This filter eliminated 5,555 probe sets from the dataset. The second level of filtering that we applied was designed to eliminate probe sets that could not meet the basic criterion of a regulated probe. Specifically, this filtering approach was designed to eliminate probe sets whose average did not deviate from baseline by a certain value for a reasonable number of time points, and employed two filters that were designed to eliminate probe sets that were neither down- nor upregulated. The first of these filters eliminated probe sets that could not meet a minimal criterion for downregulation. Starting with the 4P filtered list, we eliminated all probe sets that did not have average values below 0.65 in at least four conditions (time points). Figure 2, left, shows a gene tree of the 402 probe sets that were not eliminated by this filter. Most of these probe sets clearly contain a sustained run of time points represented in blue, as expected of downregulation. The next filter was designed to eliminate probe sets that could not meet a minimal criterion for upregulation. Starting with the 4P filtered list, we eliminated all probe sets that did not have average values above 1.5 in at least four conditions (time points). Figure 2, right, shows a gene tree of the 842 probe sets that were not eliminated by this filter. Most of these probe sets clearly contain a sustained run of red time points as expected of upregulation. Unlike the skeletal muscle and liver datasets, no probe sets were found in kidney that met both criteria and were biphasically regulated. Thus, using three straightforward filters, we were able to eliminate all but ∼8% of the probe sets present in the original dataset. The last filter we applied addressed the quality of the data. For this “quality control” filter we eliminated probe sets that did not meet two conditions. The first condition focused on the control chips. As indicated above, our initial operation was to divide the value of each individual probe set on each chip by the mean of the values for that probe set on the four control chips. Therefore, the quality of the control data for each particular probe set is of unique importance in defining regulation by the drug. This filter eliminated probe sets whose control values exhibited CVs of >50%. The second condition focused on the remaining 16 time points. This filter also eliminated probe sets whose CV for more than 8 of the remaining 16 time points exceeded 50%. Figure 3, left, provides a gene tree of the 977 probe sets (6%) that were not eliminated by the entire series of filters. Figure 3, right, shows the 14,990 probe sets that were filtered out by the entire set of filters. Comparing Fig. 1, top left, with Fig. 3, right, demonstrates that probes with apparent regulation are no longer present in the eliminated data set. Tables are available in supplementary online data that provide a list of the 679 probe sets of the 977 with apparent enhanced expression and a list of the 298 probe sets with apparent downregulation. Unlike the skeletal muscle and liver data sets where the number of up- and downregulated probe sets were close to equal (1, 4, 26), the number of probe sets showing enhanced regulation in the kidney is more than twice those showing downregulation.
Fig. 2.
Gene tree of probe sets remaining after filtering for non-drug-regulated probe sets. Left: the 402 probe sets remaining after filtering out of probe sets not exhibiting repressed expression as described in text. Right: the 842 probe sets remaining after filtering out of probe sets not exhibiting enhanced expression as described in the text. A description of the gene tree is provided in Fig. 1.
Fig. 3.
Gene trees of the 979 probe sets remaining after completion of all filtering steps described in text (left) and of the 14,988 eliminated probe sets (right). Gene trees are as described in Fig. 1.
The aforesaid set of filters were designed to eliminate probe sets that are not regulated from the full set of >15,000 probe sets on the chip as opposed to applying statistical methods for selecting out probe sets that are different from controls with a certain probability. As a final statistical evaluation of the results, we performed a one-way ANOVA with a Tukey post hoc test (P < 0.05) on both lists. From the enhanced regulated list, 543 of 679 probe sets passed this test. From the downregulated list, 230 of 298 probe sets passed this test. Those probe sets that failed this test are marked by an asterisk in the supplementary tables (supplementary materials may be found at http://ajpendo.physiology.org/cqi/content/full/0196.2005/DC1).
The purpose of this experiment was to use gene array technology as a method of high-throughput data collection to obtain the data necessary for developing mechanism-based PK/PD models of the response of the kidney to MPL. Similar models for selected genes have been developed for both liver and skeletal muscle from these animals (26, 43, 52–54). Data for the development of those models were obtained using both methods that measured individual genes and gene arrays. The use of a rich time-series design for the examination of drug effects using high-throughput methods such as gene arrays provides the advantages that multiple independent measurements taken over relatively short time intervals abrogate the need for independent confirmation of gene regulation necessary with single-point studies, and provides a definitive analysis of the pattern of regulation that is not available or may even be obscured by single point studies. However, the extremely large datasets generated by such a design provides challenges in clustering analysis.
Clustering genes into groups with similar mechanisms of regulation is a complex process that, to be adequately accomplished, should involve not only clustering algorithms but also available biological data. Initially with the liver dataset, we treated data mining and clustering as a single process and used clustering algorithms based on similarity indexes of Euclidian distance and correlation coefficients (26). However, neither of these methods incorporated true time intervals, in that they treated all time domains as equal in magnitude and all time points as independent. In our time series design, 9 of the 16 points are within the first 6 h and 12 of the 16 points are within the first 12 h after drug dosing. The interval between time domains ranged from 0.25 h in the beginning to 24 h at the end. The assumption that these time domains are equal and that all time observations are independent greatly impairs the effectiveness of most current mathematical tools for mining and clustering biologically relevant time-series data. A pharmacogenomic time series is uniquely different from other types of time series that have employed gene arrays. Unlike time series that analyze processes such as biological development, the drug can be viewed as a stimulus that simply perturbs a system that is otherwise in homeostasis or stable balance. An acute stimulus such as we have applied here will allow the system to return to the original state over time. In this perspective, pharmacogenomic time series consists of interdependent observations displaced along time.
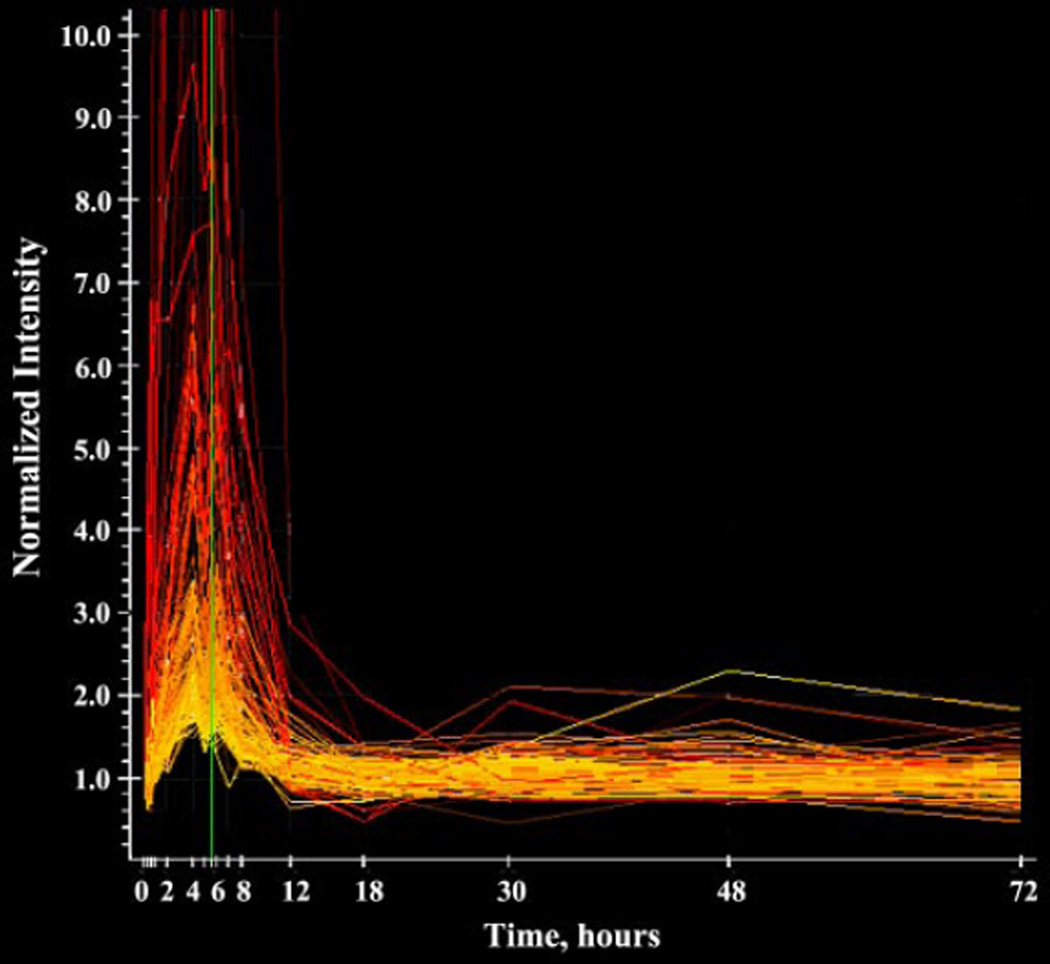
Due to the limitations in applying currently available clustering algorithms to a rich time-series dataset such as this, we approached data mining and clustering as distinct processes. We have previously observed that clustering algorithms based on a single similarity measurement, such as correlation (K-means) or geometric distance (SOM), provide different results for the same data set. This result supports the proposition that purely computational approaches to grouping genes into clusters with common mechanisms of regulation should not be relied on as the sole criterion. The unique nature of this pharmacogenomic time series coupled with the wealth of data provided by the gene arrays provides an approach to exploring ways of introducing biological information into the clustering process. The liganded glucocorticoid receptor serves as a transcription factor that binds to a cis-acting sequence in the 5′ region of responsive genes. From the upregulated group we used the complex correlation feature of GeneSpring to identify a group of 114 probe sets with a 0.9 Pearson correlation with each other (Fig. 4). This group contained several genes [serine dehydratase, lipocalin 7, insulin-like growth factor-binding protein-1 (IGFBP-1), and connective tissue growth factor] with well-documented enhanced expression by corticosteroids (13, 33, 37, 59). This group of genes also had response profiles very similar to two well-characterized glucocorticoid-responsive genes: tyrosine aminotransferase (TAT) in liver and glutamine synthetase (GS) in muscle. Our previous work characterized TAT and GS response profiles in tissues from these same animals by using both Northern hybridization and gene array techniques (4, 26, 52, 54).
Fig. 4.
Temporal profiles extracted directly from the GeneSpring program of a group of 114 probe sets exhibiting regulation patterns similar to prototype glucocorticoid-enhanced genes. The x-axis represents time after drug dosing; the y-axis represents normalized intensities.
It has been proposed that one approach to validation of purely analytic methods of clustering is to show that coex-pressed genes share more transcription factor binding sites (TFBS) in upstream noncoding regions than among genes that are not co-expressed (36). However, TFBS motifs, such GREs, are short (5–9 bp) and fairly degenerate, so most putative TFBS matches occur by chance alone and are not functional. An approach to distinguishing bonafide functional sites from those that occur by random chance is to ascertain which are evolutionarily conserved (22). The gene array temporal profiles provide a unique opportunity to test this approach using a group of genes that should, based on their response signature, contain GREs. Current thought is that GREs are constructed of two hexamers with a three-nucleotide random-hinge region in between. However, a good consensus is only available for one hexamer, TGTTCT. Using the Rat and Mouse Genome9999 programs of GeneSpring, we searched the 9,999 nucleotides in the 5′ region of TAT. For our search we used the TGTTCT half-site. Table 1 shows that, although seven TGTTCT sites were found 5′ to TAT in both rat and mouse, only four showed conservation in flanking regions between the two species. The 114 probe sets identified by correlation to be similar to documented glucocorticoid-responsive genes contained 43 probe sets corresponding to 41 identified genes. The remainder of the probe sets in this group corresponded to ESTs. Table 2 shows those 41 identified genes. Of the 41 genes, we were able to obtain both rat and mouse 5′ sequences for 18. Table 3 shows a similar analysis of GRE half-sites in these 18 genes. Those results demonstrate that in all cases the 5′-flanking regions of at least one of the putative GREs is conserved between rat and mouse. At the bottom of Table 3 are also listed two genes whose expression is well documented to be enhanced by glucocorticoids but whose 5’-flanking regions are not available in the rat. As a further test of the conservation hypothesis, we searched these two genes in mouse and human. In both cases, there is conservation of at least one site in the 5’-flanking regions between mouse and human. These data, together with the temporal profiles, provide strong evidence for the inclusion of at least these 20 genes and probably all 41 genes in a cluster whose expression is directly enhanced by corticosteroids through ligand-bound receptor interacting with GRE regulatory sites.
Table 1.
Comparison of GRE-half sites in 5’-regulatory regions of rat and mouse TAT
| Species | Position | Sequence | |
|---|---|---|---|
| Not conserved | Rat | −3553 | ctgtacaggaTGTTCTagctacttta |
| Rat | −5534 | cgcgcacatcTGTTCTtcttggaggg | |
| Rat | −9484 | ggtttggttcTGTTCTttagtagctg | |
| Mouse | −1807 | tattattagtTGTTCTtattagtgtg | |
| Mouse | −5005 | tggagtctagTGTTCTctacccatac | |
| Mouse | −8757 | gataaataaaTGTTCTgctctcgtgg | |
| Conserved pairs | Rat | −7847 | CTTGTGACTGTGTTCTAGCAGGAtTT |
| Mouse | −7591 | CTTGTGACTGTGTTCTAGCAGGAcTT | |
| Rat | −4601 | TGtAAGCAGCTGTTCTaGAGCTtTGC | |
| Mouse | −3753 | TGcAAGCAGCTGTTCTgGAGCTcTGC | |
| Rat | −3664 | CTCTTCTCAGTGTTCTCTATCaCAGG | |
| Mouse | −2604 | CTCTTCTCAGTGTTCTCTATCgCAGG | |
| Rat | −1798 | CTgCCCAgTTTGTTCTAAcTGGAGTC | |
| Mouse | −760 | CTtCCCAtTTTGTTCTAAaTGGAGTC |
Underlined regions indicate the GRE half-site consensus sequence. Non-conserved nucleotides are presented in lowercase letters.
Table 2.
Probable corticosteroid-enhanced genes
| Probe Set ID | Gene Name | Abbreviation |
|---|---|---|
| 1367960_at | ADP-ribosylation-like 4 | Arl4 |
| 1369870_at | Adrenomedullin receptor | Admr |
| 1368272_at | Aspartate aminotransferase cytosolic (cAspAT) | Got1 |
| 1368563_at | Aspartoacylase | Aspa |
| 1372051_at | Brain-enriched SH3-domain protein | Besh3 |
| 1387389_at | Calcitonin receptor activity modifying protein 3 | Ramp3 |
| 1368614_at | Carbohydrate responsive element-binding protein | ChREBP |
| 1371027_at | Casitas B-lineage lymphoma b | Cblb |
| 1387005_at | Cathepsin S | Ctss |
| 1370215_at | Complement component 1, q subcomponent β | C1qb |
| 1367631_at | Connective tissue growth factor | Ctgf |
| 1367771_at | Glucocorticoid-induced leucine zipper | Gilz |
| 1367894_at | Growth response protein | LOC64194 |
| 1368025_at | HIF-1 responsive RTP801 | Rtp801 |
| 1374117_at | Insulin receptor substrate p53 | IRSp53 |
| 1368160_at | Insulin-like growth factor-binding protein-1 | IGFBP1 |
| 1369956_at | Interferon-γ receptor | Ifngr |
| 1368249_at | Kruppel-like factor 15 | Klf15 |
| 1370209_at | Kruppel-like factor 9 | Klf9 |
| 1367650_at | Lipocalin 7; glucocorticoid-inducible protein | gis5 |
| 1368172_a_at | Lysyl oxidase | Lox |
| 1368171_at | Lysyl oxidase | Lox |
| 1367568_a_at | Matrix Gla protein | Mgp |
| 1367858_at | Matrix metalloproteinase 11 | Mmp11 |
| 1367669_a_at | Microtubule-associated protein-1 light chain 3 | MPL3 |
| 1368436_at | Nuclear distribution gene C | Nudc |
| 1367847_at | Nuclear protein 1 | Nupr1 |
| 1387109_at | P-450 (cytochrome) oxidoreductase | Por |
| 1370445_at | Phosphatidylserine-specific phospholipase A1 | Pspla1 |
| 1387074_at | Regulator of G protein signaling 2 | Rgs2 |
| 1368877_at | Renal transcription factor; kidney 1; zinc finger protein 354A | Znf354a |
| 1368080_at | Response gene to complement 32 | Rgc32 |
| 1367562_at | Secreted acidic cystein-rich glycoprotein (osteonectin) | Sparc |
| 1369864_a_at | Serine dehydratase | Sds |
| 1367802_at | Serum/glucocorticoid regulated kinase | Sgk |
| 1369412_a_at | Solute carrier family 19, member 1 (Na/H exchanger) | Slc19a1 |
| 1387585_at | Solute carrier family 29, member 2 (nucleoside transporters) | Slc29a2 |
| 1368043_at | Sorting nexin 1 | Snx1 |
| 1370195_at | Synaptosomal-associated protein 23 | Snap23 |
| 1370194_at | Synaptosomal-associated protein 23 | Snap23 |
| 1387703_a_at | Ubiquitin-specific, cysteine protease | Usp2 |
| 1367574_at | Vimentin (Vim) | Vim |
| 1388130_at | Zyxin | Zyx |
Table 3.
Cross-species identification of GRE half-sites in 5’-regulatory regions of corticosteroid-enhanced genes
| Gene Name | Species | Position | Sequence |
|---|---|---|---|
| Insulin-like growth factor-binding protein-1 (IGFBP1) | Rat | −356 | TGGTCTAATCTGTTCTTgCTTTtcct |
| (6, 6) | Mouse | −315 | TGGTCTAATCTGTTCTTtCTTTagtc |
| Solute carrier family 19, member 1 | Rat | −8139 | AGgCCCGTctTGTTCTTTCTCTTCCg |
| (6, 2) | Mouse | −8313 | AGtCCCGTgcTGTTCTTTCTCTTCCt |
| Rat | −8099 | cagatTCAAGTGTTCTCAAGTGAGCC | |
| Mouse | −8274 | gcagcTCAAGTGTTCTCAAGTGAGCC | |
| Solute carrier family 29, member 2 | Rat | −8252 | AtTTTGGATGTGTTCTAACCTCAgCA |
| (6, 3) | Mouse | −6778 | AgTTTGGATGTGTTCTAACCTCAaCA |
| Calcitonin; receptor activity modifying protein 3 | Rat | −2761 | CCaagccTgCTGTTCTGCCAGGACAA |
| (3, 1) | Mouse | −2527 | CCtcaggTcCTGTTCTGCCAGGACAA |
| Cathepsin S | Rat | −3800 | CagattgAaATGTTCTgTTTCAaCAT |
| (3, 6) | Mouse | −1027 | CctggccAgATGTTCTaTTTCAgCAT |
| Rat | −8469 | CAGGGAAGTCTGTTCTGttgaGtaaA | |
| Mouse | −4642 | CAGGGAAGTCTGTTCTGcattGaggA | |
| Complement component 1, q subcomponent β | Rat | −9034 | caGGGTCTaTTGTTCTGTtTTGGTCA |
| (5, 5) | Mouse | −9282 | agGGGTCTgTTGTTCTGTgTTGGTCA |
| Interferon-γ receptor | Rat | −1793 | AActgTGTCCTGTTCTaGCTTGTATT |
| (5, 8) | Mouse | −1754 | AAtcaTGTCCTGTTCTgGCTTGTATT |
| Aspartoacylase | Rat | −9804 | CAGTATTCTATGTTCTGAGAAAGAAA |
| (3, 4) | Mouse | −9062 | CAGTATTCTATGTTCTGAGAAAGAAA |
| Rat | −3324 | acCCcaGCTCTGTTCTGGTTATCGTT | |
| Mouse | −3194 | taCCtgGCTCTGTTCTGGTTATCGTT | |
| Nuclear distribution gene C | Rat | −432 | TTTGTAAAgCTGTTCTAtaCAAACTA |
| (4, 2) | Mouse | −388 | TTTGTAAAaCTGTTCTActCAAACTA |
| Aspartate aminotransferase cytosolic | Rat | −1792 | AATATTTATTTGTTCTTAGCCAAACt |
| (10, 2) | Mouse | −1771 | AATATTTATTTGTTCTTAGCCAAACc |
| Lysyl oxidase | Rat | −199 | TGTTCCGTTTTGTTCTTTCCCCTTAA |
| (9, 1) | Mouse | −213 | TGTTCCGTTTTGTTCTTTCCCCTTAA |
| Kruppel-like factor 15 | Rat | −3174 | TGACAAGCCATGTTCTACTGCAcAGt |
| (2, 3) | Mouse | −8369 | TGACAAGCCATGTTCTACTGCAtAGa |
| HIF-1 responsive RTP801 | Rat | −1164 | GGCCCTGCAGTGTTCTGTCCCtaagg |
| (6, 9) | Mouse | −906 | GGCCCTGCAGTGTTCTGTCCCcctaa |
| Rat | −1844 | AAGACCTGGCTGTTCTAAAAaTCAGG | |
| Mouse | −1608 | AAGACCTGGCTGTTCTAAAAgTCAGG | |
| Rat | −6010 | GGGTGCACcCTGTTCTcTAGCCGCCt | |
| Mouse | −6086 | GGGTGCACtCTGTTCTtTAGCCGCCc | |
| Connective tissue growth factor | Rat | −3424 | gGGGtaAaAATGTTCTCTTGAAGTCA |
| (6, 6) | Mouse | −3184 | tGGGgtAgAATGTTCTCTTGAAGTCA |
| Vimentin (VIM) | Rat | −5475 | CCACCTAAgTTGTTCTGCtTtGAATA |
| (19, 8) | Mouse | −4962 | CCACCTAAATTGTTCTGCcTaGAATA |
| Osteonectin; secreted acidic cystein-rich glycoprotein | Rat | −300 | AGGCCTGtcCTGTTCTAGGTGATTCC |
| (3, 5) | Mouse | −291 | AGGCCTGgCCTGTTCTAGGTGATTCC |
| Rat | −8667 | tcGgtTtcaCTGTTCTGGAATCaTAG | |
| Mouse | −8405 | agGccTggcCTGTTCTGGAATCtTAG | |
| ADP-ribosylation-like 4 | Rat | −1975 | ACCCTCCAGATGTTCTCtAGGgCACC |
| (6, 6) | Mouse | −2831 | ACCCTCCAGATGTTCTCcAGGaCACC |
| Regulator of G protein signaling 2 | Rat | −8277 | AGtATAAAACTGTTCTTATTTGATcg |
| (3, 2) | Mouse | −7555 | AGcATAAAACTGTTCTTATTTGATgt |
| Lipocalin 7; glucocorticoid-inducible protein | Mouse | −2553 | tacCCAGGtCTGTTCTGTTaAAGCCt |
| (6, 6) | Human | −1502 | gtaCCAGGcCTGTTCTGTTgAAGCCa |
| Serum/glucocorticoid regulated kinase | Mouse | −972 | AaGaACAgAATGTTCTCgGAGATTAg |
| (10, 6) | Human | −1164 | AcGgACAaAATGTTCTCaGAGATTAc |
| Mouse | −1034 | CCTCCTCACGTGTTCTTGGCATGGCt | |
| Human | −1226 | CCTCCTCACGTGTTCTTGGCATGGCa |
Numbers in parentheses under Gene Name indicate numbers of GRE half-sites identified in rat and mouse respectively. Sequence column presents regions containing half-sites with conservation in flanking regions. GRE half-sites are underlined, and nonconserved nucleotides are presented in lowercase letters.
The corticosteroid-enhanced expression of 7 of the genes in this group of 20 also provides insight into kidney pathology associated with prolonged corticosteroid treatment. Prolonged use of corticosteroids causes kidney hypertrophy and fibrosis. Although corticosteroids downregulate IGF-I expression in the liver and in peripheral tissues, increased amounts of this protein are found in the kidney within the context of reduced amounts of IGF-I mRNA (29). It has been proposed that the enhanced expression of IGFBP-1 serves to trap circulating IGF-I in the kidney, causing an increase in its hypertrophying influence. Similarly, the enhanced expression of connective tissue growth factor (CTGF) by corticosteroids may play an important role in the pathogenesis of fibrotic disease (21, 25, 60). CTGF has potent effects on fibroblast proliferation and extracellular matrix deposition. Consistent with the effects on extracellular matrix are the enhanced expression of cathepsin S, vimentin, lipocalin 7, osteonectin, and lysyl oxidase (10, 12, 17, 42, 46).
To further explore the evolutionary conservation hypothesis, we selected a group of probes whose expression was reduced after MPL treatment. Because we did not have an exemplar, we used K-means with a Pearson correlation to divide the list of downregulated probes into five groups with similar temporal profiles. We then selected a few probe sets from each of the five K-mean clusters for which we could obtain rat and mouse 5’-flanking regions and looked for conserved GRE half-sites. The result was that 8 of 18 selected genes did have conserved half-sites (Table 4). There are several possible explanations for the presence of conserved GRE half-sites in the 5′ region of these eight genes. One is that the half-site mediates the down-regulation of the probe. However, the eight genes were distributed across all five clusters obtained using K-means, indicating a lack of similar temporal profiles. The second possibility is that, like TAT in liver, the gene is upregulated by corticosteroids in another tissue but not in kidney. Therefore, we searched the literature for each of the eight genes to ascertain whether corticosteroid-enhanced regulation had been observed. The first gene is urokinase-type plasminogen activator (uPA). We found such a report of the transcriptional upregulation of uPA in mammary epithelial cells by hydrocortisone and dexamethasone (47). The literature similarly confirms that corticosteroids reduce the amount of uPA mRNA in many tissues including kidney (23,41). The downregulation of this gene that is involved in fibrinolytic activity, along with the systemic hypercholesterolemia, probably contributes to the kidney damage associated with chronic corticosteroid treatment. Because this reduction requires protein synthesis, it is postulated that corticosteroids enhance the expression of a factor that either increases the degradation of uPA mRNA or reduces its transcription in the kidney (5). Another gene that was downregulated in the kidney but contained a conserved GRE half-site was 17β-hydroxysteroid dehydrogenase type 2 (17β-HSD2). This enzyme converts sex steroids to their less active forms: 17(3-estradiol into estrone and testosterone into androstenedione. The conserved half-site in the 5′ region of this gene may not be related to the effects of corticosteroids but rather to the effects of progesterone. The sequence TGTTCT is shared by DNA-binding sites for glucocorticoids, progestins, mineralo-corticoids, and androgens. Progesterone has been shown to enhance the expression of 17β-HSD2 in endometrial tissue under some conditions, possibly explaining the presence of this conserved sequence in the 5′ region. Another gene in this group with a conserved half-site is ferroportin 1 (Slc39a1), which is essential for iron efflux from cells. The expression of this gene impacts both local and systemic iron homeostasis. Few data are available on its regulation other than that its 5′ region contains an iron-responsive element and its expression is enhanced locally by inflammation (14, 35). Another gene in this group with a conserved half-site is monocarboxylic acid transporter (Slc16a1, MCT1) (57). MCT1 is widely distributed and was originally cloned from kidney. It is thought that MCT1 transports lactic acid into cells. No data are available concerning its regulation by any of the four steroids that share the TGTTCT-binding site. Plasmolipin, whose 5′ region contains two conserved sites, was initially isolated from kidney plasma membranes but also is present in brain myelin tracts (9). In kidney, it is restricted to the apical surface of tubular epithelial cells and is a transmembrane protein involved in ion channel formation. The downregulation in kidney by corticosteroids is a novel observation as is the presence of the conserved DNA-binding site. Another downregulated gene with a conserved half-site is angiopoietin-like 2 (Angptl2) (45). Tyrosine kinase, which contains immunoglobulin-like loops 2 (Tie2), is an endothelial receptor tyrosine kinase that is activated by Angptl1. Angptl2 is a ligand that blocks the activation of Tie2 by Angptl1. Tie2 activation promotes endothelial cell proliferation. Therefore, the downregulation of Angptl2 after corticosteroid treatment is consistent with the development of nephropathy. The presence of a conserved half-site can be explained by the observation that progestins enhance the expression of Angptl2 in the uterus. The last of the downregulated genes with a conserved half-site in its 5′ region is kynurenine 3-hydroxylase (Kmo). Kmo is a mitochondrial enzyme that is expressed in many tissues, including kidney (55). It converts kynurenine to 3-hydroxykynurenine in the pathway that converts tryptophan to quinolinic acid. The kynurenine pathway is important to many biological processes ranging from antioxidant status to neuronal and immunological function. The observation that corticosteroids downregulate the expression of a key enzyme in the kynurenine pathway in kidney is novel. However, our previous work (1, 4) showed that it was also downregulated in liver but not in skeletal muscle. Interestingly, pharmacological manipulation of Kmo has been proposed for a variety of disease processes ranging from cancer to cataracts. The remaining 10 genes in Table 4 did not contain conserved half-sites.
Table 4.
Cross-species identification of GRE half-sites in 5’-regulatory regions of downregulated genes
| Gene Name | Species | Position | Sequence |
|---|---|---|---|
| Urinary plasminogen activator; urokinase (uPA) | Rat | −3158 | ATCTgAGAtgTGTTCTTTGCCcCCcA |
| (5, 6) | Mouse | −2608 | ATCTaAGAccTGTTCTTTGCCtCCaA |
| Rat | −3248 | TcCTgTTCTCTGTTCTGCATTCACTG | |
| Mouse | −2698 | TaCTtTTCTCTGTTCTGCATTCACTG | |
| Rat | −5592 | CCATACTGAATGTTCTTAGTATAGCA | |
| Mouse | −4620 | CCATACTGAATGTTCTTAGTATAGCA | |
| n-acetyltransferase; Camello 4 (Cml4) | Rat | −7848 | ACAtTgAAAGTGTTCTTCAGAGgCCA |
| (5, 10) | Mouse | −7258 | ACAcTtAAAGTGTTCTTCAGAGaCCA |
| 17β-Hydroxysteroid dehydrogenase type 2 (HSD17β2) | Rat | −4817 | gaAAGGCACaTGTTCTCAGCAACgTT |
| (7, 6) | Mouse | −4877 | agAAGGCACgTGTTCTCAGCAACaTT |
| Ferroportin 1; iron exporter (Slc39a1) | Rat | −1064 | aGctCCCACCTGTTCTTAcCcCTCag |
| (6, 4) | Mouse | −1661 | gGacCCCACCTGTTCTTAgCaCTCtc |
| Monocarboxylic acid transporter (Slc16a1) | Rat | −4034 | cGTAAGATTATGTTCTCTTTcTAAAA |
| (6, 8) | Mouse | −4120 | tGTAAGATTATGTTCTCTTTtTAAAA |
| Rat | −9123 | TGAGACCATGTGTTCTAAGACAcCAA | |
| Mouse | −8740 | TGAGACCATGTGTTCTAAGACAtCAA | |
| Plasmolipin | Rat | −3656 | gCctCccTCTTGTTCTTTGTTTTTTG |
| (6, 3) | Mouse | −2900 | cCtcCtgTCTTGTTCTTTGTTTTTTG |
| Rat | −3968 | AgCCCcGCTTTGTTCTGAGCTTGGaC | |
| Mouse | −3128 | AcCCCaGCTTTGTTCTGAGCTTGGtC | |
| Angiopoietin-like 2 (Angptl2) | Rat | −1825 | AtTAACtACCTGTTCTtTGGCtaTtG |
| (4, 8) | Mouse | −1848 | AgTAACcACCTGTTCTcTGGCctTgG |
| Rat | −2194 | ACCAcATAgCTGTTCTTGCCCCtGTc | |
| Mouse | −2224 | ACCAtATAaCTGTTCTTGCCCCaGTg | |
| Kynurenine 3-hydroxylase (Kmo) | Rat | −1043 | ccttCcagTGTGTTCTCATCAGAAAT |
| (4, 4) | Mouse | −819 | atgcCattTGTGTTCTCATCAGAAAT |
| Rat | −2948 | cTTTGCACTCTGTTCTTTAtGGGATG | |
| Mouse | −2867 | aTTTGCACTCTGTTCTTTAgGGGATG | |
| Rat | −3001 | GgACCTGCTTTGTTCTTGTTGATGCC | |
| Mouse | −2918 | GaACCTGCTTTGTTCTTGTTGATGCC | |
| Glucocorticoid receptor (GCR) | NM | ||
| Calcium-dependent, elastin-binding protein (Fbln5) | NM | ||
| Insulin-like growth factor 2 receptor (IGF2R) | NM | ||
| Uterine sensitization-associated gene-1 (Usag1) | NM | ||
| Pyruvate dehydrogenase phosphatase isoenzyme 2 (PDP2) | NM | ||
| Potassium inwardly-rectifying channel, subfamily J, member 16 | NM | ||
| Thyroid hormone receptor-β (Thrb) | NM | ||
| Phosphotriesterase related (Pter) | NM | ||
| Histamine n-methyltransferase (Hnmt) | NM | ||
| es-Type nucleoside transporter (Slc29a1) | NM |
Numbers in parentheses under Gene Name indicate numbers of GRE half-sites identified in rat and mouse, respectively. Sequence column presents regions containing half-sites with conservation in flanking regions. GRE half-sites are underlined, and nonconserved nucleotides are presented in lowercase letters. NM indicates no matches.
To further extend the test of the hypothesis, we filtered out a group of genes that were expressed in the kidney but were unaffected by MPL. The constraints of this filter were that the probe set must have a Affymetrix call of P on all 52 chips and an average no less than 0.95 or greater than 1.05 in any of the 17 conditions. Of the 11 genes selected, four contained conserved 5′ half-sites. One gene that contained a conserved half-site whose expression was unchanged by MPL in the kidney is granzyme K (Gzmk). Granzymes (Gzms) are granule-stored lymphocyte serine proteases that are implicated in T- and natural killer cell-mediated cytotoxicity. In general, corticosteroids appear to reduce the expression of Gzms in these cells. However, in one report on leukemic cells, cortico-steroids where shown to enhance the expression of granzyme A through an identified GRE (56). A second gene that was not affected in kidney but contained a conserved half-site was pleiotropic regulator 1 (Plrg1). Plrg1 is a component of a multiprotein complex that is a subunit of the spliceosome. Our own data show that this gene is upregulated by MPL in liver but not skeletal muscle (1, 4). The third gene that is not regulated by MPL in the kidney but has a conserved half-site is cis-Golgi matrix protein (GM130). GM130 appears to be important for endoplasmic reticulum-Golgi traffic, and for Golgi reassembly after cytokinesis (20). No data are available concerning the effects of corticosteroids on its expression. The last gene in this group is a component of the exocytotic complex, sec6. This protein is part of a multiprotein complex essential for targeting exocytic vesicles to specific docking sites on the plasma membrane (24). No data are available concerning its regulation by corticosteroids.
The analysis of the 5′ region of genes for the cross-species conservation of a TFBS seems to provide an approach to augmenting clustering based solely on analytic methods. In addition, the results of the analysis demonstrate that the number of conserved half-sites ranges from as few as one to as many as four. What is most interesting is that, in no case, including TAT, was a “textbook” full GRE found. However, even the textbook GRE does not seem to have a consensus for the 5′ hexamer. For example, in Lewin’s Genes V, published in 1994, the 5′ hexamer was described as TGGTCA (30), whereas in the most recent addition, Genes VIII, the 5′ hexamer is described as a reverse repeat of the 3′ hexamer TCTTGT (31), and others have described it as GGTACA (7). Including TAT, we have 21 exemplars of genes with very similar temporal response profiles and conservation of the sequence TGTTCT, which does seem to represent a true consensus. In addition, we have identified eight genes that were downregulated and four genes that were expressed in kidney but not regulated by MPL that also contained conserved sites. In many cases, the effect of steroids on these genes in other tissues supported the presence of the conserved site. The significance of variable numbers, locations, and structure of 5′ GREs is at present not clear. However, as additional sequence data become available, being able to identify the structure, number, and location of GREs in the 5′ region of a large number of genes with a defined response signature to a specific dosing regimen should provide insight into the significance of the organization of this enhancer.
The approach we used in the current analysis employed the Genome9999 program, a component of the GeneSpring software package. An obvious limitation of that software is that it searches only the 9,999 nucleotides 5′ to the start site of a gene. Although in many cases GREs have been found within this somewhat limited region, it is certainly possible that additional GREs may be present at locations more distant from the core promoter. A second limitation to our analysis was the current lack of availability of sequence information for the 5′ of many of the genes of interest in the rat. A third limitation is that we focused only on GREs, and it is highly likely that analysis of other transcription factor-binding sites may provide additional insight into understanding patterns of regulation by the drug. As sequence information for the rat genome becomes more complete, and with the development of more sophisticated tools for searching such sequences, the analysis of TFBS will likely become an important adjunct to analytic clustering methods for exploring such datasets.
This report describes the mining of the third of three Affymetrix gene array datasets developed using tissue from a group of animals subjected to a single dose of MPL. This treatment yielded a rich time series for developing this and the two parallel datasets (liver and skeletal muscle) to obtain the data necessary for beginning to understand the broad multiorgan origins of the adverse systemic effects of corticosteroids. A single bolus dose of MPL was used so that we could define the temporal cascade of events that, if perpetuated by repeated dosing, causes systemic pathologies. The intent is to use these data to develop quantitative hypotheses in the form of mechanism-based PK/PD models. All three data sets are available online in a user-friendly format that requires no specialized knowledge or software normally necessary for examining gene array data (http://pepr.cnmcresearch.org) (3, 11). This Oracle web database includes a novel time series query analysis tool, enabling live generation of graphs and spreadsheets showing the action of any transcript of interest over time. It allows workable access to all of our data to anyone desiring to query any gene(s) and allows one to download several types of raw data for independent analysis.
Table 5.
Cross-species identification ofGRE half-sites in 5’-regulatory regions of nonregulated genes
| Gene Name | Species | Position | Sequence |
|---|---|---|---|
| Granzyme K (Gzmk) | Rat | −5939 | AcCATgTGGTTGTTCTCTGaATTTGA |
| (2, 7) | Mouse | −5340 | AgCATaTGGTTGTTCTCTGcATTTGA |
| Pleiotropic regulator 1 (Plrg1) | Rat | −1978 | TGATTTAAAATGTTCTTTTCTAgGtt |
| (6, 1) | Mouse | −1037 | TGATTTAAAATGTTCTTTTCTAtGgc |
| cis-Golgi matrix protein | Rat | −5686 | ctgtgGtTCCTGTTCTTAGGcCTCTG |
| (4, 4) | Mouse | −5831 | gctgtGgTCCTGTTCTTAGGtCTCTG |
| Exocytotic complex (sec6) | Rat | −2485 | TTACCTAcAtTGTTCTTTCATTggCT |
| (7, 8) | Mouse | −834 | TTACCTAtAcTGTTCTTTCATTtaCT |
| Rat | −8035 | ACCATAGGATTGTTCTTTTAGTTggA | |
| Mouse | −6306 | ACCATAGGATTGTTCTTTTAGTTtaA | |
| Uroporphyrinogen decarboxylase (Urod) | NM | ||
| MAP kinase kinase (Map2k1) | NM | ||
| Calcineurin subunit A α (Calna1, Ppp3ca) | NM | ||
| 17β-Hydroxysteroid dehydrogenase 10 (HSD17B10) | NM | ||
| Peroxiredoxin 1 (Prdx1) | NM | ||
| BCL2/1.9 kDa-interacting protein 3-like (Bnip31) | NM | ||
| 5’-AMP-activated protein kinase, γ-1 (Prkga1) | NM |
Numbers in parentheses under Gene Name indicate numbers of GRE half-sites identified in rat and mouse, respectively. Sequence column presents regions containing half-sites with conservation in flanking regions. GRE half-sites are underlined, and nonconserved nucleotides are presented in lowercase letters. NM, no matches.
ACKNOWLEDGMENTS
We acknowledge the expert assistance of Ms. Suzette Mis in preparation of this manuscript.
GRANTS
This work was supported by Grants GM-24211 and GM-67650 from the National Institute of General Medical Sciences, National Institutes of Health (NIH). This dataset was developed under the auspices of a grant from the National Heart, Lung, and Blood Institute/NIH Programs in Genomic Applications HL-66614.
REFERENCES
- 1.Almon R, DuBois D, Jin J, Jusko W. Pharmacogenomic responses of rat liver to methylprednisolone: an approach to mining a rich microarray time series. AAPS J. 2005;7:E156–E194. doi: 10.1208/aapsj070117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almon R, DuBois DC, Pearson K, Stephan D, Jusko WJ. Gene arrays and temporal patterns of drug response: corticosteroid effects on rat liver. Funct Integr Genomics. 2003;3:171–179. doi: 10.1007/s10142-003-0090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almon RR, Chen J, Snyder G, DuBois D, Jusko W, Hoffman E. In vivo multi-tissue corticosteroid microarray time series available online at Public Expression Profile Resource (PEPR) Pharmacogenomics. 2003;4:791–799. doi: 10.1517/phgs.4.6.791.22816. [DOI] [PubMed] [Google Scholar]
- 4.Almon RR, DuBois DC, Piel W, Jusko WJ. The genomic response of skeletal muscle to methylprednisolone using microarrays: tailoring data mining to the structure of the pharmacogenomic time series. Pharmaco-genomics. 2004;5:525–552. doi: 10.1517/14622416.5.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Althus M, Nagamine Y. Protein synthesis inhibition stabalizes urokinase-type plasminogen activator mRNA. J Biol Chem. 1991;266:21190–21196. [PubMed] [Google Scholar]
- 6.Baxter J. Advances in glucocorticoid therapy. Adv Intern Med. 2000:317–349. [PubMed] [Google Scholar]
- 7.Beato M, Chalepakis G, Schauer M, Slater E. DNA regulatory elements for steroid hormones. J Steroid Biochem. 1989;32:737–748. doi: 10.1016/0022-4731(89)90521-9. [DOI] [PubMed] [Google Scholar]
- 8.Bialis M, Routledge P. Adverse effects of corticosteroids. Adverse Drug React Toxicol Rev. 1998;17:227–235. [PubMed] [Google Scholar]
- 9.Bosse F, Hasse B, Pippirs U, Greiner-Petter K, Muller H. Prote-olipid plasmolipin: localization in polarized cells, regulated expression, and lipid raft association in CNS and PNS myelin. J Neurochem. 2003;86:508–518. doi: 10.1046/j.1471-4159.2003.01870.x. [DOI] [PubMed] [Google Scholar]
- 10.Burns-Kurtis CL, Olzinski AR, Needle S, Fox JH, Capper EA, Kelly FM, McQueney MS, Romanic AM. Cathepsin S expression is up-regulated following balloon angioplasty in the hypercholesterolemic rabbit. Cardiovasc Res. 2004;62:610–620. doi: 10.1016/j.cardiores.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Zhao P, Massaro D, Clerch L, Almon RR, DuBois DC, Jusko WJ, Hoffman E. The PEPR GeneChip Data Warehouse, and implementation of a dynamic time series query tool (SGQT) with graphical interface. Nucleic Acids Res. 2004;32:578–581. doi: 10.1093/nar/gkh003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Contente S, Csiszar K, Kenyon K, Friedman R. Structure of the mouse lysyl oxidase gene. Genomics. 1993;16:395–400. doi: 10.1006/geno.1993.1202. [DOI] [PubMed] [Google Scholar]
- 13.Dammeier J, Beer HD, Brauchle M, Werner S. Dexamethasone is a novel potent inducer of connective tissue growth factor expression. Implications for glucocorticoid therapy. J Biol Chem. 1998;273:18185–18190. doi: 10.1074/jbc.273.29.18185. [DOI] [PubMed] [Google Scholar]
- 14.di Patti MC, Persichini T, Mazzone V, Polticelli F, Colasanti M, Musci G. Interleukin-1beta up-regulates iron efflux in rat C6 glioma cells through modulation of ceruloplasmin and ferroportin-1 synthesis. Neuro-sci Lett. 2004;363:182–186. doi: 10.1016/j.neulet.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Edwards C, Stewart P, Burt D, Brett L, McIntyre M, Sutanto W, deKloet E, Monder C. Localization of 11B-hydroxysteroid dehy-drogenase-tissue specific promotor of the mineralocorticoid receptor. Lancet. 1988:986–989. doi: 10.1016/s0140-6736(88)90742-8. [DOI] [PubMed] [Google Scholar]
- 16.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Favre CJ, Mancuso M, Maas K, McLean JW, Baluk P, McDonald DM. Expression of genes involved in vascular development and angio-genesis in endothelial cells of adult lung. Am J Physiol Heart Circ Physiol. 2003;285:H1917–H1938. doi: 10.1152/ajpheart.00983.2002. [DOI] [PubMed] [Google Scholar]
- 18.Frauman A. An overview of the adverse reactions to adrenal corticosteroids. Adverse Drug React Toxicol Rev. 1996;15:203–206. [PubMed] [Google Scholar]
- 19.Funder J, Pearce P, Smith R, Smith A. Mineralocorticoid action: target tissue specificity is enzyme, not receptor, mediated. Science. 1988;242:583–585. doi: 10.1126/science.2845584. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Mata R, Sztul E. The membrane-tethering protein p115 interacts with GBF1, an ARF guanine-nucletide-exchange factor. EMBO Reports. 2003;4:320–325. doi: 10.1038/sj.embor.embor762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gore-Hyer E, Shegogue D, Markiewicz M, Lo S, Hazen-Martin D, Greene EL, Grotendorst G, Trojanowska M. TGF-fj and CTGF have overlapping and distinct fibrogenic effects on human renal cells. Am J Physiol Renal Physiol. 2002;283:F707–F716. doi: 10.1152/ajprenal.00007.2002. [DOI] [PubMed] [Google Scholar]
- 22.Hardison R. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;26:225–228. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- 23.Henderson B, Kefford R. Dexamethasone decreases urokinase plasminogen activator mRNA stability in MAT 13762 mammary carcinoma cells. Br J Cancer. 1993;67:99–101. doi: 10.1038/bjc.1993.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu SC, Hazuka CD, Foletti DL, Scheller RH. Targeting vesicles to specific sites on the plasma membrane: the role of the sec6/8 complex. Trends Cell Biol. 1999;9:150–153. doi: 10.1016/s0962-8924(99)01516-0. [DOI] [PubMed] [Google Scholar]
- 25.Inoue T, Okada H, Kobayashi T, Watanabe Y, Kanno Y, Kopp JB, Nishida T, Takigawa M, Ueno M, Nakamura T, Suzuki H. Hepatocyte growth factor counteracts transforming growth factor-beta1, through attenuation of connective tissue growth factor induction, and prevents renal fibrogenesis in 5/6 nephrectomized mice. FASEB J. 2003;17:268–270. doi: 10.1096/fj.02-0442fje. [DOI] [PubMed] [Google Scholar]
- 26.Jin JY, Almon RR, DuBois DC, Jusko WJ. Modeling of corticosteroid pharmacogenomics in rat liver using gene microarrays. J Pharmacol Exp Ther. 2003;307:93–109. doi: 10.1124/jpet.103.053256. [DOI] [PubMed] [Google Scholar]
- 27.Jusko WJ, Jin JY, DuBois DC, Almon RR. Pharmacodynamics and pharmacogenomics of corticosteroids: microarray studies. In: D’ Argenio D, editor. Advanced Methods of Pharmacokinetic and Pharmacodynamics Systems Analysis. Boston, MA: Kluwer Academic; 2004. [Google Scholar]
- 28.Kennedy B, Elayan H, Ziegler MG. Glucocorticoid hypertension and nonadrenal phenylethanolamine N-methyltransferase. Hypertension. 1993;21:415–419. doi: 10.1161/01.hyp.21.4.415. [DOI] [PubMed] [Google Scholar]
- 29.Lang C, Nystrom G, Frost R. Burn-induced changes in IGF-I and IGF-binding proteins are partially glucocorticoid dependent. Am J Physiol Regul Integr Comp Physiol. 2002;282:R207–R215. doi: 10.1152/ajpregu.00319.2001. [DOI] [PubMed] [Google Scholar]
- 30.Lewin B. Genes V. New York: Oxford Univ. Press; 1994. [Google Scholar]
- 31.Lewin B. Genes VIII. Pearson-Prentice Hall; 2004. [Google Scholar]
- 32.Locsey L, Asztalos L, Kincses Z, Gyorfi F, Berczi C. Dyslipidaemia and hyperlipidaemia following renal transplantation. Int Urol Nephrol. 1996;28:419–430. doi: 10.1007/BF02550506. [DOI] [PubMed] [Google Scholar]
- 33.Matsuda K, Fukushima C. Identification of glucocorticoid and cyclic AMP responsive elements of the rat serine dehydratase gene: difference in responses of the transfected and chromosomal genes. Biochem Biophys Res Commun. 1991;176:385–391. doi: 10.1016/0006-291x(91)90936-2. [DOI] [PubMed] [Google Scholar]
- 34.McKay LI, DuBois DC, Sun YN, Almon RR, Jusko WJ. Cortico-steroid effects in skeletal muscle: gene induction/receptor autoregulation. Muscle Nerve. 1997;20:1318–1320. doi: 10.1002/(sici)1097-4598(199710)20:10<1318::aid-mus17>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 35.Mok H, Jelinek J, Pai S, Cattanach BM, Prchal JT, Youssoufian H, Schumacher A. Disruption of ferroportin 1 regulation causes dynamic alterations in iron homeostasis and erythropoiesis in polycythaemia mice. Development. 2004;131:1859–1868. doi: 10.1242/dev.01081. [DOI] [PubMed] [Google Scholar]
- 36.Moses A, Chiang D, Pollard D, Iyer V, Eisen M. MONKEY: identifying conserved transcription-factor binding sites in multiple alignments using a binding site-specific evolutionary model. Genome Biol. 2004;5:R98. doi: 10.1186/gb-2004-5-12-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukai K, Mitani F, Nagasawa H, Suzuki R, Suzuki T, Suematsu M, Ishimura Y. An inverse correlation between expression of a prepro-cathepsin B-related protein with cysteine-rich sequences and steroid 11be-ta-hydroxylase in adrenocortical cells. J Biol Chem. 2003;278:17084–17092. doi: 10.1074/jbc.M301477200. [DOI] [PubMed] [Google Scholar]
- 38.Mune T, Rogerson F, Nikkila J, Agarwal A, White P. Human hypertension caused by mutations in the kidney isozyme of 11B-hydrox-ysteroid dehydrogenase. Nat Genet. 1995;10:394–399. doi: 10.1038/ng0895-394. [DOI] [PubMed] [Google Scholar]
- 39.Newton R. Molecular mechanisms of glucocorticoid action: what is important? Thorax. 2000;55:603–613. doi: 10.1136/thorax.55.7.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nichols AI, Boudinot FD, Jusko WJ. Second generation model for prednisolone pharmacodynamics in the rat. J Pharmacokinet Biopharm. 1989;17:209–227. doi: 10.1007/BF01059029. [DOI] [PubMed] [Google Scholar]
- 41.Pearson D, Altus M, Horiechi A, Nagamine Y. Dexamethasone coordinately inhibits plasminogen activator gene expression and enzyme activity in porcine kidney cells. Biochem Biophys Res Commun. 1987;143:329–336. doi: 10.1016/0006-291x(87)90669-3. [DOI] [PubMed] [Google Scholar]
- 42.Pichler RH, Hugo C, Shankland SJ, Reed MJ, Bassuk JA, Andoh TF, Lombardi DM, Schwartz SM, Bennett WM, Alpers CE, Sage EH, Johnson RJ, Couser WG. SPARC is expressed in renal interstitial fibrosis and in renal vascular injury. Kidney Int. 1978;50:1978–1989. doi: 10.1038/ki.1996.520. [DOI] [PubMed] [Google Scholar]
- 43.Ramakrishnan R, DuBois DC, Almon RR, Pyszczynski NA, Jusko WJ. Fifth-generation model for corticosteroid pharmacodynamics: application to steady-state receptor down-regulation and enzyme induction patterns during seven-day continuous infusion of methylprednisolone in rats. J Pharmacokinet Pharmacodyn. 2002;29:1–24. doi: 10.1023/a:1015765201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynolds RM, Walker BR. Human insulin resistance: the role of glucocorticoids. Diabet Obes Metab. 2003;5:5–12. doi: 10.1046/j.1463-1326.2003.00221.x. [DOI] [PubMed] [Google Scholar]
- 45.Rizkalla B, Forbes JM, Cooper ME, Cao Z. Increased renal vascular endothelial growth factor and angiopoietins by angiotensin II infusion is mediated by both AT1 and AT2 receptors. J Am Soc Nephrol. 2003;14:3061–3071. doi: 10.1097/01.asn.0000099374.58607.c9. [DOI] [PubMed] [Google Scholar]
- 46.Sanai T, Sobka T, Johnson T, el-Essawy M, Muchaneta-Kubara EC, Ben Gharbia O, el Oldroyd S, Nahas AM. Expression of cytoskel-etal proteins during the course of experimental diabetic nephropathy. Diabetologia. 2000;43:91–100. doi: 10.1007/s001250050012. [DOI] [PubMed] [Google Scholar]
- 47.Sasaki CY, Lin H, Passaniti A. Regulation of urokinase plasmino-gen activator (uPA) activity by E-cadherin and hormones in mammary epithelial cells. J Cell Physiol. 1999;181:1–13. doi: 10.1002/(SICI)1097-4652(199910)181:1<1::AID-JCP1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 48.Schaaf M, Cidlowski J. Molecular mechanisms of glucocorti-coid action and resistance. J Steroid Biochem Mol Biol. 2003;83:37–48. doi: 10.1016/s0960-0760(02)00263-7. [DOI] [PubMed] [Google Scholar]
- 49.Schacke H, Docke WD, Asadullah K. Mechanisms involved in the side effects of glucocorticoids. Pharmacol Ther. 2002;96:23–43. doi: 10.1016/s0163-7258(02)00297-8. [DOI] [PubMed] [Google Scholar]
- 50.Schimmer B, Parker K. Adrenocorticotrophic hormones, adrenocor-tical steroids and their synthetic analogs, inhibitors of synthesis and actions of adrenocortical hormones. In: Hardman J, Limbird L, Molinoff B, Ruddon R, Goodman-Gilman A, editors. Goodman & Gilman’s The Pharmacological Basis of Therapeutics. New York: McGraw Hill; 1996. pp. 1459–1486. [Google Scholar]
- 51.Stewart P, Corrie J, Schackleton C, Edwards C. Syndrome of apparent mineralocorticoid excess: a defect in the cortisol-cortisone shuttle. J Clin Invest. 1988;82:340–349. doi: 10.1172/JCI113592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun YN, DuBois DC, Almon RR, Jusko WJ. Fourth-generation model for corticosteroid pharmacodynamics: a model for methylpred-nisolone effects on receptor/gene-mediated glucocorticoid receptor down-regulation and tyrosine aminotransferase induction in rat liver. J Phar-macokinet Biopharm. 1998;26:289–317. doi: 10.1023/a:1023233409550. [DOI] [PubMed] [Google Scholar]
- 53.Sun YN, DuBois DC, Almon RR, Pyszczynski NA, Jusko WJ. Dose-dependence and repeated-dose studies for receptor/gene-mediated pharmacodynamics of methylprednisolone on glucocorticoid receptor down-regulation and tyrosine aminotransferase induction in rat liver. J Pharmacokinet Biopharm. 1998;26:619–648. doi: 10.1023/a:1020746822634. [DOI] [PubMed] [Google Scholar]
- 54.Sun YN, McKay LI, DuBois DC, Jusko WJ, Almon RR. Phar-macokinetic/Pharmacodynamic models for corticosteroid receptor down-regulation and glutamine synthetase induction in rat skeletal muscle by a receptor/gene-mediated mechanism. J Pharmacol Exp Ther. 1999;288:720–728. [PubMed] [Google Scholar]
- 55.Tankiewicz A, Pawlak D, Topczewska-Bruns J, Buczko W. Kidney and liver kynurenine pathway enzymes in chronic renal failure. Adv Exp Med Biol. 2003;527:409–414. doi: 10.1007/978-1-4615-0135-0_48. [DOI] [PubMed] [Google Scholar]
- 56.UM Shen L, Oshida T, Miyauchi J, Yamada M, Miyashita T. Identification of novel direct transcriptional targets of glucocorticoid receptor. Leukemia. 2004;18:1850–1856. doi: 10.1038/sj.leu.2403516. [DOI] [PubMed] [Google Scholar]
- 57.Vannucci S, Simpson I. Developmental switch in brain nutrient transporter expression in the rat. Am J Physiol Endocrinol Metab. 2003;285:E1127–E1134. doi: 10.1152/ajpendo.00187.2003. [DOI] [PubMed] [Google Scholar]
- 58.Xu ZX, Sun YN, DuBois DC, Almon RR, Jusko WJ. Third-generation model for corticosteroid pharmacodynamics: roles of glucocor-ticoid receptor mRNA and tyrosine aminotransferase mRNA in rat liver. J Pharmacokinet Biopharm. 1995;23:163–181. doi: 10.1007/BF02354270. [DOI] [PubMed] [Google Scholar]
- 59.Yeagley D, Guo S, Unterman T, Quinn P. Gene- and activation-specific mechanisms for insulin inhibition of basal and glucocorticoid-induced insulin-like growth factor binding protein-I and phosphoenolpyru-vate carboxykinase transcription. J Biol Chem. 2001;36:33705–33710. doi: 10.1074/jbc.M101215200. [DOI] [PubMed] [Google Scholar]
- 60.Yokoi H, Sugawara A, Mukoyama M, Mori K, Makino H, Suganami T, Nagae T, Yahata K, Fujinaga Y, Tanaka I, Nakao K. Role of connective tissue growth factor in profibrotic action of transforming growth factor-beta: a potential target for preventing renal fibrosis. Am J Kidney Dis. 2001;38:S134–S138. doi: 10.1053/ajkd.2001.27422. [DOI] [PubMed] [Google Scholar]