Summary
The mammalian Target of Rapamycin (mTOR) is responsive to numerous extracellular and intracellular cues and, through the formation of two physically and functionally distinct complexes, plays a central role in the homeostatic control of cell growth, proliferation and survival. Through aberrant activation of mTOR signaling, the perception of cellular growth signals becomes disconnected from the processes promoting cell growth, and this underlies the pathophysiology of a number of genetic tumor syndromes and cancers. Here, we review the oncogenes and tumor suppressors comprising the regulatory network upstream of mTOR, highlight the human cancers in which mTOR is activated, and discuss how dysregulated mTOR signaling gives tumors a selective growth advantage. In addition, we discuss why activation of mTOR, as a consequence of distinct oncogenic events, results in diverse clinical outcomes, and how the complexity of the mTOR signaling network might dictate therapeutic approaches.
Introduction
Cell growth, division and survival are fundamental aspects of cellular physiology that are exquisitely sensitive to changes in both the extracellular and intracellular environment. In single-celled eukaryotes such as yeast, nutrient-sensing pathways are primarily responsible for the control of cell growth (an increase in cell size) and proliferation (an increase in cell number). In multi-cellular organisms, these cell autonomous processes are also regulated by growth and mitogenic factors secreted by other cells, and these signals are integrated into the ancient nutrient-sensing pathways. In metazoans, both the extracellular growth factor signaling and the intracellular nutrient-sensing cues generally activate anabolic processes, such as protein synthesis, and inhibit catabolic processes, such as autophagy. A delicate balance between these processes is necessary for normal growth control, and a disconnect between the ability of a cell to properly sense growth conditions and the pathways regulating cell growth, underlies tumorigenesis. Here, we discuss the mammalian target of rapamycin (mTOR) signaling network, which integrates signals from nutrients and growth factors and is frequently misregulated in genetic tumor syndromes and cancer.
Target of rapamycin (TOR) complexes
TOR proteins are evolutionarily conserved ser/thr kinases that belong to the phosphatidylinositol kinase-related protein kinase (PIKK) family. The first TOR proteins were identified in the budding yeast S. cerevisiae. The TOR1 and TOR2 genes were identified in a genetic screen for mutations conferring resistance to rapamycin, a naturally produced macrolide antibiotic (Heitman et al., 1991). In a complex with the intracellular cofactor FKBP12 (FK506-binding protein 12), rapamycin directly binds to and inhibits TOR proteins through an allosteric site N-terminal to its kinase domain. It is this property that allowed subsequent identification of the single TOR protein encoded by mammalian genomes (Brown et al., 1994;Chiu et al., 1994;Sabatini et al., 1994;Sabers et al., 1995). Combined with genetic evidence, the existence of two TOR proteins in yeast lead to the discovery that TOR proteins from yeast to humans exist in two physically and functionally distinct macromolecular complexes (Loewith et al., 2002;reviewed in Wullschleger et al., 2006).
In addition to the TOR kinase, several core components are also conserved within these two complexes. mTOR complex 1 (mTORC1) is composed of Raptor (Regulatory associated protein of mTOR) and mLST8 (mammalian Lethal with SEC13 protein 8), while mTORC2 consists of Rictor (Rapamycin insensitive companion of mTOR), mSIN1 (mammalian Stress-activated protein kinase interacting protein 1), and mLST8 (Frias et al., 2006;Hara et al., 2002;Jacinto et al., 2006;Jacinto et al., 2004;Kim et al., 2002;Kim et al., 2003;Loewith et al., 2002;Sarbassov et al., 2004;Yang et al., 2006). Both complexes have additional subunits that are not required for core complex function, including PRAS40 (Proline-Rich Akt Substrate of 40 kDa) in mTORC1 and PRR5 (proline-rich protein 5, also called PROTOR1) or PRR5L (PRR5-like, also called PROTOR2) in mTORC2 (Pearce et al., 2007;Sancak et al., 2007;Vander et al., 2007). Importantly, these two complexes, in yeast and humans, can be distinguished by their sensitivity to rapamycin. Rapamycin associated with FKBP12 binds to and acutely inhibits mTOR within mTORC1 but not mTORC2. However, prolonged treatment with rapamycin can block mTORC2 assembly (Sarbassov et al., 2006).
As described below, mTORC1 is controlled by cellular growth conditions and, in turn, controls cell growth in response to these stimuli. To date, only two direct downstream targets of mTORC1 have been characterized in detail, the ribosomal S6 kinases (S6K1 and S6K2) and the eukaryotic initiation factor 4E (eIF4E)-binding proteins (e.g., 4E-BP1). In response to growth stimuli, mTORC1 has been found to associate with the eukaryotic initiation factor 3 (eIF3) complex and regulate translation initiation, at least in part, through the phosphorylation of these substrates (Holz et al., 2005). Within mTORC1, Raptor appears to direct substrate binding through a small motif on the target proteins, referred to as the TOR signaling (TOS) motif (Choi et al., 2003;Holz et al., 2005;Nojima et al., 2003;Schalm and Blenis, 2002;Schalm et al., 2003). mTORC1 phosphorylates S6K1 on a residue C-terminal to its kinase domain (T389 in the 70-kD isoform of S6K1), referred to as the hydrophobic motif, which is a regulatory site conserved in many members of the protein kinase A, G, C family (AGC) that is required for full activation of these kinases. Therefore, mTORC1 activates S6K, which subsequently phosphorylates downstream targets such as the ribosomal protein S6 and eIF4B (reviewed in Ma and Blenis, 2009). mTORC1-dependent phosphorylation of 4E-BP1 at multiple sites results in its release from eIF4E at the 7-methyl-GTP cap of mRNAs, allowing subsequent assembly of an initiation complex required for cap-dependent translation. Interestingly, the ability of rapamycin to inhibit mTORC1-mediated phosphorylation events varies between these two substrates, with S6K1 being sensitive in all settings and the sites on 4E-BP1 being more resistant (e.g., (Choo et al., 2008;Wang et al., 2005)). In fact, the development of direct mTOR kinase domain inhibitors has confirmed the existence of rapamycin resistant functions for mTORC1. In addition to more completely blocking the phosphorylation of 4E-BP1, these compounds more potently inhibit protein synthesis, arrest cell proliferation, and activate autophagy, and these effects appear to be due primarily to inhibition of mTORC1 (Feldman et al., 2009;Garcia-Martinez et al., 2009;Thoreen et al., 2009).
Due to the later discovery of mTORC2 and its resistance to rapamycin, relative to mTORC1, knowledge of mTORC2 functions has lagged behind. By far, its best-characterized substrate is Akt (Sarbassov et al., 2005), a member of the AGC family of protein kinases. As with the phosphorylation of S6K by mTORC1, Akt is phosphorylated on its hydrophobic motif (S473) by mTORC2, and this is required to fully activate Akt (Alessi et al. 1996). In addition, mTORC2 is required for the phosphorylation of the hydrophobic motifs on other AGC kinases, including PKCα (Guertin et al., 2006;Sarbassov et al., 2004) and SGK1 (Serum Glucocorticoid-induced Kinase 1) (Garcia-Martinez and Alessi, 2008). Finally, a second conserved motif on these AGC kinases, referred to as the turn motif, is also phosphorylated in an mTORC2-dependent manner (Facchinetti et al., 2008;Ikenoue et al., 2008). Given our current void of knowledge regarding the molecular mechanisms regulating mTORC2 and the role of mTORC2 in tumorigenesis, the discussion below is focused primarily on mTORC1.
Upstream regulation of mTORC1
Eukaryotic cells dedicate a high proportion of their total nutrient and energy levels to the processes of ribosome biogenesis and mRNA translation to synthesize proteins. Increasing the protein synthetic capacity of the cell is fundamental to stimulating cell growth. Therefore, cells have evolved exquisite mechanisms to sense cellular growth conditions, in the form of nutrient and energy levels and, in multicellular eukaryotes, the presence of secreted growth factors. Signaling pathways then relay the status of these conditions to key regulators of protein synthesis, such as mTORC1. In fact, an extensive regulatory network to closely monitor a diverse array of growth cues exists to properly control mTORC1 activation.
As the essential building block of proteins, amino acid levels are closely monitored by the mTORC1 pathway, and mTORC1 signaling is shut down upon amino acid depletion. While this property is conserved back to TORC1 in yeast, the mechanisms by which amino acid levels are sensed and communicated to mTORC1 are poorly understood. A recent breakthrough in this area has come from the finding that the Rag GTPases are mTORC1-proximal components of the amino acid-sensing pathway (Kim et al., 2008;Sancak et al., 2008). This finding should facilitate future studies to elucidate the sensing and signaling mechanisms by which amino acids control mTORC1.
A complex between the products encoded by the tumor suppressor genes mutated in the tuberous sclerosis complex (TSC) disease, TSC1 and TSC2, has emerged as a master regulator of mTOR (reviewed in Huang and Manning, 2008). The TSC1-TSC2 complex negatively regulates mTORC1 through the GTPase-activating protein (GAP) activity of TSC2 toward the Ras-related small G protein Rheb (Ras homolog enriched in brain), which in its GTP-bound form is a potent and essential activator of mTORC1. Therefore, the TSC1-TSC2 complex inhibits mTORC1 activation by stimulating the intrinsic GTPase activity of Rheb, leading to conversion of active GTP-bound Rheb to the inactive GDP-bound form. Many of the cellular pathways that affect mTORC1 activity do so by affecting the ability of the TSC1-TSC2 complex to act as a GAP for Rheb. These pathways do so, in large part, through post-translational modification of TSC1 or TSC2. For instance, the ubiquitous growth factor-regulated protein kinases Akt, ERK, and RSK all directly phosphorylate TSC2 and, through unknown molecular and cellular mechanisms, inhibit the TSC1-TSC2 complex, thereby stimulating an increase in Rheb-GTP levels and activation of mTORC1 (Inoki et al., 2002;Ma et al., 2005;Manning et al., 2002;Roux et al., 2004). Under conditions of energy depletion, the highly conserved energy-sensing protein kinase AMPK is activated and phosphorylates TSC2 on additional sites that, again through an unknown mechanism, enhance the ability of the TSC1-TSC2 complex to turn off Rheb and mTORC1 (Inoki et al., 2003;Shaw et al., 2004). Therefore, growth-promoting conditions activate pathways that decrease TSC1-TSC2 complex function, while poor growth conditions activate pathways that increase TSC1-TSC2 complex function, thereby leading to the respective activation or inhibition of mTORC1.
There appears to be additional mechanisms, parallel to regulation of the TSC1-TSC2 complex, by which some of these pathways affect mTORC1 activity. For instance, mTORC1 can be further stimulated through Akt-mediated phosphorylation of PRAS40 (Sancak et al., 2007;Vander et al., 2007) and RSK-mediated phosphorylation of Raptor (Carriere et al., 2008). In addition, AMPK can further inhibit mTORC1 through phosphorylation of specific sites on Raptor (Gwinn et al., 2008). These multiple modes of regulation emphasize the importance of these pathways that sense cellular growth conditions in the control of mTORC1 activation.
Common activation of mTORC1 in human tumors
Proper regulation of mTORC1 is required to maintain homeostatic control over the anabolic processes that drive cell growth and proliferation. A ubiquitous property of tumor cells is their ability to disconnect growth-promoting processes from the perception of growth signals. This separation is achieved, in large part, through genetic events leading to aberrant activation of mTORC1. The signaling network that normally relays the presence or absence of specific growth stimuli to mTORC1 is comprised of numerous oncogenes and tumor suppressors, including those that most frequently underlie the development and progression of malignant tumors (summarized in Figure 1). Therefore, it is not surprising that elevated mTORC1 signaling has been detected in a large percentage of the most common human cancers (summarized in Table 1). Activation of mTORC1 signaling in tumors is generally scored by examining relative phosphorylation levels of its direct downstream targets, 4E-BP1 and S6K1, and/or the S6K1 substrate ribosomal S6. There are multiple phosphorylation sites on these proteins, but these are not all exclusive readouts of mTORC1 activity. In general, it is believed that phosphorylation of S6K1 on T389, S6 on S240/244, and 4E-BP1 on S65 are quite specific to mTORC1 signaling. To date, there are no phosphorylation sites on mTOR itself that are required for, and specifically indicate, the activation of mTORC1. Phosphorylation of mTOR on S2448 is often used as an indication of its activity. However, this site appears to be phosphorylated in both mTORC1 and mTORC2 (Rosner et al., 2009), and its molecular function is unknown. Therefore, in Table 1, we have excluded studies that utilized mTOR-S2448 phosphorylation as the sole readout of mTORC1 activity.
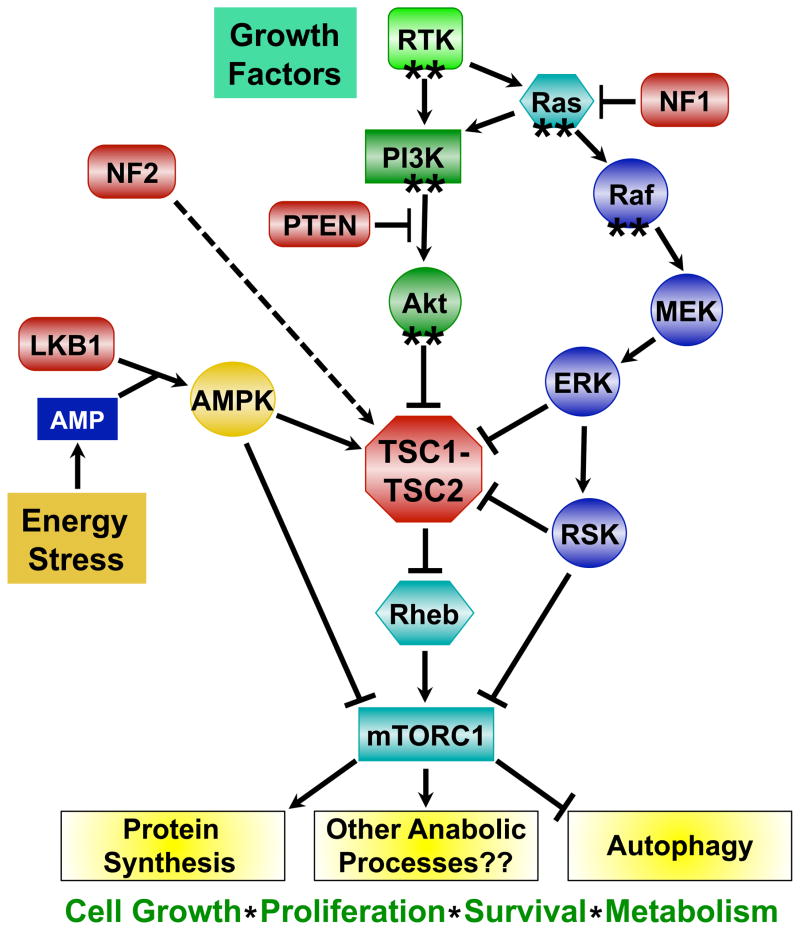
Figure 1. Oncogenes and tumor suppressors converge on the regulation of mTORC1.
Model of the common oncogenic signaling pathways regulating mTORC1 activation. Proteins encoded by oncogenes are indicated with asterisks (**), and those encoded by tumor suppressor genes are depicted in red. See text for details.
Table 1.
Activation of mTORC1 signaling in human cancers
| Cancer | p4E-BP1 | pS6K (T389) | pS6 | pmTOR (S2448)* | N | References |
|---|---|---|---|---|---|---|
| Breast | 71.9% | 58.5% (S235/236) | 44.9% | 89 | (Lin et al., 2005) | |
| 41.2% (S65) | 42.4% | 165 | (Zhou et al., 2004) | |||
| 87.3% (T70) | 77.7% | 77.7% (S240/244) | 103 | (Rojo et al., 2007) | ||
| Colorectal | 82.1% (T37/46) | 66.1% | 60.7% | 56 | (Zhang et al., 2009) | |
| 40% | 69 | (Nozawa et al., 2007) | ||||
| Endometrial | 61% (S235/236) | 75 | (Lu et al., 2008) | |||
| Glioblastoma | 56% | 56 | (Chakravarti et al., 2004) | |||
| 94% | 75% | 268 | (Pelloski et al., 2006) | |||
| Hepatocellular Carcinoma | 47.7% (S240/244) | 86 | (Villanueva et al., 2008) | |||
| 88.3% (S235/236) | 528 | (Zhou et al., 2009) | ||||
| Lung Adenocarcinoma | 84% (S235/236) | 77 | (McDonald et al., 2008) | |||
| 100% 54% high (S235/236) |
37 | (Conde et al., 2006) | ||||
| Lymphoma | 66% (T70) | 66% | 66% (S240/244) | 66% | 29 | (Vega et al., 2006) |
| Melanoma | 73% (S235/236) | 107 | (Karbowniczek et al., 2008) | |||
| Ovarian | 41.1% (T70) | 26.4% | 15.5% (S240/244) | 129 | (Castellvi et al., 2006) | |
| Prostate | 90.6% (T70) | 71.7% (S235/236) | 96.2% | 84 | (Kremer et al., 2006) | |
| Renal Cell Carcinoma | 59% (S235/236) | 29 | (Robb et al., 2007) | |||
| 85% (S235/236) | 375 | (Pantuck et al., 2007) |
Studies using P-mTOR (S2448) alone were not included, as the functional relevance of this site is unknown.
Oncogenic events leading to misregulation of the PI3K-Akt and/or ERK signaling pathways are very common in malignant tumors (reviewed in Engelman et al., 2006;Roberts and Der, 2007;Shaw and Cantley, 2006). This often occurs through activating mutations and amplifications leading to ligand-independent signaling from upstream receptor tyrosine kinases (e.g., EGFR, Her-2/Neu, MET) or scaffolding adaptors (e.g., BCR-Abl). In addition, oncogenic Ras activates both the PI3K-Akt and Erk pathways and is among the most common oncogenes in human cancers. Furthermore, Ras is negatively regulated by the NF1 tumor suppressor (also referred to as neurofibromin), which acts as a GAP for Ras. Activating mutations in B-RAF and, more commonly, PI3K are also found in a large variety of cancers and lead to growth factor-independent activation of ERK and Akt, respectively. Finally, among tumor suppressor genes, loss of PTEN appears to occur at a rate second to only p53 in malignant tumors, and this leads to aberrant activation of Akt. Therefore, these two critical pathways through which growth factors signal to mTORC1 are constitutively activated in most cancers, rendering mTORC1 active even under conditions of growth factor withdrawal.
Other tumor suppressor and oncogene pathways also converge on mTORC1 regulation. For instance, the tumor suppressor kinase LKB1 is a critical upstream activator of AMPK and is required for mTORC1 inhibition under conditions of energy stress (reviewed in Shaw, 2009). Therefore, in tumor cells with loss of LKB1, which occurs frequently in non-small cell lung cancer, mTORC1 signaling is elevated and no longer sensitive to perturbation in intracellular ATP levels. Furthermore, both the Wnt and TNFα pathways, which contribute to the development and progression of some malignancies, have inputs into the TSC1-TSC2 complex that could result in mTORC1 activation in tumors (Inoki et al., 2006;Lee et al., 2007).
An important shared property of these oncogenic pathways converging on mTORC1 is that they are negatively regulated by tumor suppressors that are mutated in genetic tumor syndromes. Germline mutations in the gene encoding the Ras-GAP NF1 give rise to neurofibromatosis type 1, which is the most prevalent of the tumor syndromes classified as neurocutaneous disorders, or phakomatoses (McClatchey, 2007). Individuals heterozygous for mutations in PTEN are more rare, but develop a variety of phenotypically distinct disorders (e.g., Cowden syndrome and Bannayan-Riley Ruvalcaba syndrome), collectively referred to as PTEN hamartoma tumor syndrome (reviewed in Orloff and Eng, 2008). Homozygous loss of either NF1 or PTEN results in growth factor-independent Akt-mediated phosphorylation and inhibition of TSC2, leading to aberrant activation of mTORC1 (Johannessen et al., 2005;Manning et al., 2002). Interestingly, these two tumor suppressors are also frequently mutated in sporadic glioblastoma multiforme (Cancer Genome Atlas Research Network, 2008), in which mTORC1 signaling has been found to be commonly activated (Chakravarti et al., 2004;Pelloski et al., 2006); Table 1). While the molecular mechanism remains unknown, loss of the NF2/merlin tumor suppressor, which is mutated in neurofibromatosis type 2, has also recently been found to result in growth factor-independent activation of mTORC1 signaling (James et al., 2009;Lopez-Lago et al., 2009). In addition to the frequent occurrence in sporadic lung cancer, mutations in the LKB1 tumor suppressor are the underlying cause of Peutz-Jeghers syndrome, and the colon polyps universal to patients with this disease display elevated mTORC1 signaling (Shackelford et al., 2009). Finally, as the convergence point of these upstream pathways, it is not surprising that germline mutations affecting the function of the TSC1 or TSC2 tumor suppressors also give rise to a genetic tumor syndrome, TSC, and mutations in TSC2 can also give rise to sporadic lymphangioleiomyomatosis (LAM). Homozygous loss of the TSC genes results in constitutive activation of mTORC1, and tumors arising in TSC and LAM patients invariably display elevated levels of mTORC1 signaling (reviewed in Crino et al., 2006;Huang et al., 2008).
As aberrant mTORC1 activation is a common molecular event in both cancer and genetic tumor syndromes, there is much interest in developing and testing mTORC1 inhibitors as anti-tumor agents. To date, there are completed or ongoing clinical trials testing the efficacy of rapamycin (sirolimus) and/or its analogs (e.g., CCI-779/temsirolimus, RAD001/everolimus) against nearly all major forms of cancer, as well as TSC and LAM (clinicaltrials.gov; (reviewed in Dowling et al., 2009)). However, since recent studies have found that these allosteric inhibitors only block a subset of mTORC1 functions, it will be important in the future to test the anti-tumor activities of mTOR kinase domain inhibitors in preclinical and clinical studies (Feldman et al., 2009;Garcia-Martinez et al., 2009;Thoreen et al., 2009).
Below, we address two major questions related to the pathological and therapeutic implications of the common mTORC1 activation in tumors. First, what is gained by the tumor in its activation of mTORC1? Second, if mTORC1 activation is a major molecular event shared in cancer and tumor syndromes, why are the pathological consequences of the genetic events discussed above so distinct?
Selective advantage gained by mTORC1 activation in tumors
In the now classic review from Hanahan and Weinberg (Hanahan and Weinberg, 2000), malignant transformation is proposed to be dictated by six essential alterations in cell physiology: self-sufficiency in growth signals, insensitivity to growth-inhibitory signals, evasion of apoptosis, limitless replicative potential, sustained angiogenesis, and tissue invasion and metastasis. To varying degrees, the uncontrolled activation of mTORC1 might contribute to all of these areas and provide a substantial selective advantage to tumor cells.
The oncogenic signaling events detailed above give rise to mTORC1 activation that is independent of growth factors and/or insensitive to perturbations in cellular growth conditions. This aberrant activation of mTORC1 has the potential to be a major contributing factor to the uncontrolled cell growth, proliferation, and survival of tumor cells. While it is likely that mTORC1 controls multiple anabolic processes contributing to cell growth, its best-characterized function is in driving protein synthesis. As described above, mTORC1 stimulates protein synthesis both acutely, through specific effects on cap-dependent translation initiation, and in a more sustained manner, through less well-defined mechanisms promoting ribosome biogenesis. Importantly, while mRNAs are generally translated in a cap-dependent fashion, it appears that a subset of mRNAs, such as those with extensive secondary structure, are particularly sensitive to mTORC1 signaling for their efficient translation (reviewed in Ma et al., 2009). These mRNAs include those encoding Cyclin D1 and c-Myc, which are important regulators of cell-cycle entry (Gera et al., 2004). Interestingly, in addition to decreasing cell size, rapamycin treatment generally causes a G1 phase cell-cycle arrest, demonstrating a role for mTORC1 signaling in promoting cell proliferation. Through the phosphorylation and inhibition of 4E-BP1, mTORC1 signaling increases eIF4E-mediated translation initiation. In many studies, eIF4E has been found to have oncogenic properties and to promote aspects of cell proliferation and survival (e.g., (Lazaris-Karatzas et al., 1990;Ruggero et al., 2004;Wendel et al., 2004).
The mechanisms by which mTORC1 signaling blocks apoptosis are not fully understood and may vary significantly in different settings. Translation of the pro-survival Bcl-2 family member MCL-1 can be stimulated by mTORC1 activity, and this mechanism has been shown to contribute to cell survival in a mouse lymphoma model (Mills et al., 2008). In addition, in a study of evasion from TRAIL-induced apoptosis in glioblastoma multiforme, mTORC1 signaling was found to induce translation of the FLIPS protein, which blocks caspase-8 activation downstream of TRAIL (Panner et al., 2005). Finally, the poorly defined role of mTORC1 as a negative regulator of autophagy could be both detrimental and beneficial to the tumor, as autophagy acts as a mechanism of both cell survival and death that is dependent on the metabolic status of the cell and its nutrient environment (Mathew et al., 2007).
Uncontrolled mTORC1 signaling is also likely to contribute to the metabolic reprogramming common to tumor cells. TORC1 plays a clear role in nutrient uptake in yeast cells (reviewed in Wullschleger et al., 2006). While mTORC1 has been suggested to stimulate nutrient uptake in mammalian cells (Edinger and Thompson, 2002), the molecular mechanism for such an effect is currently unknown. Interestingly, mTORC1 signaling increases the levels of the hypoxia-inducible factor-1α (HIF-1α) transcription factor, and this occurs, at least in part, through increased translation (e.g., (Hudson et al., 2002;Thomas et al., 2006;Zhong et al., 2000)). Through its transcriptional targets, HIF-1α enhances glucose uptake and its glycolytic conversion to lactate (Denko, 2008), a property common to cells under hypoxic growth conditions. However, cancer cells exhibit this switch to glycolytic metabolism even under normoxic conditions, often referred to as the Warburg effect (Vander Heiden et al., 2009), and aberrant upregulation of HIF-1α is likely to contribute to this metabolic switch. In both genetic and xenograft tumor models in mice, HIF-1α has been implicated in the mTORC1-dependent (i.e., rapamycin sensitive) increase in glucose uptake into tumors (Majumder et al., 2004;Shackelford et al., 2009;Thomas et al., 2006). Along with the hypoxic tumor microenvironment, uncontrolled mTORC1 signaling is likely to contribute to tumor angiogenesis and, perhaps, metastasis via further up-regulation of both HIF-1α and HIF-2α. Among the many HIF1α/2α targets, the gene encoding vascular endothelial growth factor A (VEGF-A) is likely to contribute most significantly to a role for mTORC1 in promoting tumor angiogenesis. However, observed anti-angiogenic effects of rapamycin might also be attributed to inhibition of mTORC2 signaling within vascular endothelial cells, as the mTORC2-mediated phosphorylation of Akt appears to be particularly sensitive to rapamycin in these cells (Phung et al., 2006).
Complexity of mTOR signaling and its implication for targeted therapeutics
On the surface, the molecular wiring of the oncogenic pathways converging on mTORC1 suggests that rapamycin and its analogs (collectively referred to as rapamycin here) should be effective therapeutics for the treatment of both genetic tumor syndromes and sporadic cancers. However, while many trials are still underway, thus far, rapamycin has had somewhat limited success in the clinic (Chiang and Abraham, 2007). Tumor shrinkage (40-50%) has been reported in trials for specific manifestations of TSC, but upon withdrawal from rapamycin, these normally slow growing tumors rapidly regain their original size (Bissler et al., 2008;Davies et al., 2008;Franz et al., 2006). This finding is consistent with the cells in the tumor getting smaller upon rapamycin treatment, without significant loss of cell number. Thus, upon removal of rapamycin, the tumor can rapidly regrow just by adding mass to the existing cells, and this is likely stimulated by a restoration of uncontrolled mTORC1 signaling. With very few exceptions, rapamycin elicits a cytostatic response on eukaryotic cells, from yeast to human, generally resulting in a G1 phase cell-cycle arrest. This is likely the reason that, in its more successful clinical trials in cancer, rapamycin treatment results in stable disease with little effect on tumor volume (Easton and Houghton, 2006;Faivre et al., 2006). One important outstanding question is whether the many catalytic domain inhibitors of mTOR that are in development, which have uncovered rapamycin-resistant functions of mTORC1 (Feldman et al., 2009;Garcia-Martinez et al., 2009;Thoreen et al., 2009), will elicit cytotoxic responses and have stronger effects on tumor regression.
In addition to the varying effects of rapamycin on mTORC2 assembly and stability (Sarbassov et al., 2006), other complex effects on the signaling network are elicited by specific inhibition of mTORC1. This is due to the fact that multiple mTORC1-dependent negative feedback mechanisms exist to dampen the activation of upstream components of the network. The best characterized of these is the mTORC1-and S6K1-mediated phosphorylation of the insulin receptor substrate (IRS-1), leading to a block in insulin signaling to PI3K and other downstream pathways (reviewed in Harrington et al., 2005). This and other mechanisms of feedback regulation inhibiting Akt activation are relieved by rapamycin and, under most growth conditions, results in increased Akt phosphorylation. Signs of Akt activation by rapamycin have also been detected in tumor biopsies from clinical trials (e.g., (Cloughesy et al., 2008;O’Reilly et al., 2006;Tabernero et al., 2008). This suggests that rapamycin, while inhibiting growth and proliferation in many tumor cells, might actually inhibit tumor cell apoptosis by activating the pro-survival kinase Akt. These feedback mechanisms are a major motivation behind the development and testing of mTOR catalytic domain inhibitors to hit both mTORC1 and mTORC2, inhibitors that block both mTOR and PI3K, or combination therapies of rapamycin plus inhibitors of upstream RTKs (Guertin and Sabatini, 2009).
While mTORC1 activation is a shared consequence of the many oncogenic events described above, the pathological outcome appears to vary greatly between different tumor syndromes and cancers. Based solely on our knowledge of the wiring of the upstream regulatory network, there are several possible explanations for these differences. The most obvious is the extensive branching of the upstream pathways, such that aberrant mTORC1 activation is only one of many downstream effects of pathway misregulation. In fact, some of the most integrated nodes of cell signaling (e.g., Ras, PI3K, Akt, ERK, and AMPK) comprise this upstream network. Another likely underlying cause of clinical diversity is that the degree to which mTORC1 signaling becomes constitutively activated is highly dependent on the genetic event leading to pathway misregulation. For example, loss of PTEN will render mTORC1 signaling resistant to growth factor withdrawal, but it will remain sensitive to cellular energy levels. Likewise, loss of LKB1 leads to mTORC1 activation that is insensitive to energy stress, but it remains dependent on growth factors. Related to this point is that the ultimate level of aberrant mTORC1 signaling is dictated by which oncogenes, tumor suppressors, or combinations thereof are affected in a given tumor cell. The levels of mTORC1 activation become important in the context of the feedback mechanisms discussed above, which can affect tumor progression. For instance, among the upstream tumor suppressors, loss of TSC1 or TSC2 results in the highest levels of mTORC1 signaling, but results in a disease with the lowest malignancy potential. In cells lacking the TSC genes, the mTORC1-driven feedback mechanisms affecting the IRS-1 protein render PI3K unresponsive to insulin and insulin-like growth factor (IGF1) (Harrington et al., 2004;Shah et al., 2004). In addition, the TSC1-TSC2 complex normally promotes mTORC2 activation, and mTORC2 signaling to Akt, PKCα, and SGK1 is strongly attenuated in TSC gene-deficient cells and tumors (Huang J, 2009;Huang et al., 2008). Therefore, together with the mTORC1-dependent feedback mechanisms, Akt activation and signaling to its other downstream targets is lost upon disruption of these particular tumor suppressors. Using mouse genetic approaches to bypass the feedback effects on PI3K activation, it has been found that this loss of Akt signaling in TSC-deficient tumors can account for their very slow growing nature, at least in some tumor types (Manning et al., 2005). Therefore, while mTORC1 activation contributes to tumor development and progression, it can also have tumor suppressive effects due to feedback mechanisms. Hence, the positioning of oncogenic perturbation(s) within the upstream regulatory network will dictate the pathological consequences of dysregulated mTOR signaling.
Conclusions
As cancer researchers, these are exciting times in the mTOR field. While the increasing complexity of the mTOR signaling network can be daunting at times, given its widespread activation in tumors, it is essential that we delineate the mechanistic details of how it is truly wired. Only then will we understand its role in tumorigenesis, how best to perturb the network for therapeutic intervention, and how to interpret the clinical outcomes of these interventions. To maximize progress, we must embrace the continuum of basic, preclinical, and clinical studies. Substantial voids in our knowledge currently exist in all three of these areas. For the basic science, further elucidation of the upstream regulatory mechanisms, including spatial and structural insights, and the downstream functions of the mTOR complexes is needed. In preclinical studies, we must understand the contributions of mTOR signaling to tumor development and progression and test targeted therapeutics in relevant genetic mouse tumor models, rather than xenograft models where anti-angiogenic effects can dominate. In the clinic, it is essential to learn from the basic and preclinical studies in choosing which compounds and combinations to test and which biomarkers to monitor. Currently, there is a paucity of clinically validated predictive markers for response to mTOR inhibition, and studies to reveal such markers will greatly enhance patient selection. Finally, to come full circle, emerging clinical data should yield novel mechanistic hypotheses and lead to the development of new preclinical models to better understand and target mTOR signaling in tumors.
References
- Bissler JJ, McCormack FX, Young LR, Elwing JM, Chuck G, Leonard JM, et al. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N Engl J Med. 2008;358:140–151. doi: 10.1056/NEJMoa063564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriere A, Cargnello M, Julien LA, Gao H, Bonneil E, Thibault P, et al. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr Biol. 2008;18:1269–1277. doi: 10.1016/j.cub.2008.07.078. [DOI] [PubMed] [Google Scholar]
- Castellvi J, Garcia A, Rojo F, Ruiz-Marcellan C, Gil A, Baselga J, et al. Phosphorylated 4E binding protein 1: a hallmark of cell signaling that correlates with survival in ovarian cancer. Cancer. 2006;107:1801–1811. doi: 10.1002/cncr.22195. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Zhai G, Suzuki Y, Sarkesh S, Black PM, Muzikansky A, et al. The prognostic significance of phosphatidylinositol 3-kinase pathway activation in human gliomas. J Clin Oncol. 2004;22:1926–1933. doi: 10.1200/JCO.2004.07.193. [DOI] [PubMed] [Google Scholar]
- Chiang GG, Abraham RT. Targeting the mTOR signaling network in cancer. Trends Mol Med. 2007;13:433–442. doi: 10.1016/j.molmed.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Chiu MI, Katz H, Berlin V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc Natl Acad Sci U S A. 1994;91:12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KM, McMahon LP, Lawrence JC., Jr Two motifs in the translational repressor PHAS-I required for efficient phosphorylation by mammalian target of rapamycin and for recognition by raptor. J Biol Chem. 2003;278:19667–19673. doi: 10.1074/jbc.M301142200. [DOI] [PubMed] [Google Scholar]
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloughesy TF, Yoshimoto K, Nghiemphu P, Brown K, Dang J, Zhu S, et al. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008;5:e8. doi: 10.1371/journal.pmed.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde E, Angulo B, Tang M, Morente M, Torres-Lanzas J, Lopez-Encuentra A, et al. Molecular context of the EGFR mutations: evidence for the activation of mTOR/S6K signaling. Clin Cancer Res. 2006;12:710–717. doi: 10.1158/1078-0432.CCR-05-1362. [DOI] [PubMed] [Google Scholar]
- Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355:1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- Davies DM, Johnson SR, Tattersfield AE, Kingswood JC, Cox JA, McCartney DL, et al. Sirolimus therapy in tuberous sclerosis or sporadic lymphangioleiomyomatosis. N Engl J Med. 2008;358:200–203. doi: 10.1056/NEJMc072500. [DOI] [PubMed] [Google Scholar]
- Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- Dowling RJ, Pollak M, Sonenberg N. Current status and challenges associated with targeting mTOR for cancer therapy. BioDrugs. 2009;23:77–91. doi: 10.2165/00063030-200923020-00002. [DOI] [PubMed] [Google Scholar]
- Easton JB, Houghton PJ. mTOR and cancer therapy. Oncogene. 2006;25:6436–6446. doi: 10.1038/sj.onc.1209886. [DOI] [PubMed] [Google Scholar]
- Edinger AL, Thompson CB. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell. 2002;13:2276–2288. doi: 10.1091/mbc.01-12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Facchinetti V, Ouyang W, Wei H, Soto N, Lazorchak A, Gould C, et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27:1932–1943. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov. 2006;5:671–688. doi: 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz DN, Leonard J, Tudor C, Chuck G, Care M, Sethuraman G, et al. Rapamycin causes regression of astrocytomas in tuberous sclerosis complex. Ann Neurol. 2006;59:490–498. doi: 10.1002/ana.20784. [DOI] [PubMed] [Google Scholar]
- Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16:1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum-and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, et al. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009;421:29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gera JF, Mellinghoff IK, Shi Y, Rettig MB, Tran C, Hsu JH, et al. AKT activity determines sensitivity to mammalian target of rapamycin (mTOR) inhibitors by regulating cyclin D1 and c-myc expression. J Biol Chem. 2004;279:2737–2746. doi: 10.1074/jbc.M309999200. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009;2:e24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LS, Findlay GM, Lamb RF. Restraining PI3K: mTOR signalling goes back to the membrane. Trends Biochem Sci. 2005;30:35–42. doi: 10.1016/j.tibs.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Wu CL, Manning BD. Signaling events downstream of mammalian target of rapamycin complex 2 are attenuated in cells and tumors deficient for the tuberous sclerosis complex tumor suppressors. Cancer Res. 2009;69:6107–6114. doi: 10.1158/0008-5472.CAN-09-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, et al. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol. 2002;22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 2008;27:1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- James MF, Han S, Polizzano C, Plotkin SR, Manning BD, Stemmer-Rachamimov AO, et al. NF2/Merlin is a Novel Negative Regulator of mTOR Complex 1 and Activation of mTORC1 is Associated with Meningioma and Schwannoma Growth. Mol Cell Biol. 2009;29:4250–4261. doi: 10.1128/MCB.01581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowniczek M, Spittle CS, Morrison T, Wu H, Henske EP. mTOR is activated in the majority of malignant melanomas. J Invest Dermatol. 2008;128:980–987. doi: 10.1038/sj.jid.5701074. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer CL, Klein RR, Mendelson J, Browne W, Samadzedeh LK, Vanpatten K, et al. Expression of mTOR signaling pathway markers in prostate cancer progression. Prostate. 2006;66:1203–1212. doi: 10.1002/pros.20410. [DOI] [PubMed] [Google Scholar]
- Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK, Wei Y, et al. IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell. 2007;130:440–455. doi: 10.1016/j.cell.2007.05.058. [DOI] [PubMed] [Google Scholar]
- Lin HJ, Hsieh FC, Song H, Lin J. Elevated phosphorylation and activation of PDK-1/AKT pathway in human breast cancer. Br J Cancer. 2005;93:1372–1381. doi: 10.1038/sj.bjc.6602862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Lopez-Lago MA, Okada T, Murillo MM, Socci N, Giancotti FG. Loss of the Tumor Suppressor NF2/Merlin Constitutively Activates Integrin-dependent mTORC1 Signaling. Mol Cell Biol. 2009;29:4235–4249. doi: 10.1128/MCB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KH, Wu W, Dave B, Slomovitz BM, Burke TW, Munsell MF, et al. Loss of tuberous sclerosis complex-2 function and activation of mammalian target of rapamycin signaling in endometrial carcinoma. Clin Cancer Res. 2008;14:2543–2550. doi: 10.1158/1078-0432.CCR-07-0321. [DOI] [PubMed] [Google Scholar]
- Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- Manning BD, Logsdon MN, Lipovsky AI, Abbott D, Kwiatkowski DJ, Cantley LC. Feedback inhibition of Akt signaling limits the growth of tumors lacking Tsc2. Genes Dev. 2005;19:1773–1778. doi: 10.1101/gad.1314605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey AI. Neurofibromatosis. Annu Rev Pathol. 2007;2:191–216. doi: 10.1146/annurev.pathol.2.010506.091940. [DOI] [PubMed] [Google Scholar]
- McDonald JM, Pelloski CE, Ledoux A, Sun M, Raso G, Komaki R, et al. Elevated phospho-S6 expression is associated with metastasis in adenocarcinoma of the lung. Clin Cancer Res. 2008;14:7832–7837. doi: 10.1158/1078-0432.CCR-08-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ, et al. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci U S A. 2008;105:10853–10858. doi: 10.1073/pnas.0804821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, et al. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- Nozawa H, Watanabe T, Nagawa H. Phosphorylation of ribosomal p70 S6 kinase and rapamycin sensitivity in human colorectal cancer. Cancer Lett. 2007;251:105–113. doi: 10.1016/j.canlet.2006.11.008. [DOI] [PubMed] [Google Scholar]
- O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orloff MS, Eng C. Genetic and phenotypic heterogeneity in the PTEN hamartoma tumour syndrome. Oncogene. 2008;27:5387–5397. doi: 10.1038/onc.2008.237. [DOI] [PubMed] [Google Scholar]
- Panner A, James CD, Berger MS, Pieper RO. mTOR controls FLIPS translation and TRAIL sensitivity in glioblastoma multiforme cells. Mol Cell Biol. 2005;25:8809–8823. doi: 10.1128/MCB.25.20.8809-8823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantuck AJ, Seligson DB, Klatte T, Yu H, Leppert JT, Moore L, et al. Prognostic relevance of the mTOR pathway in renal cell carcinoma: implications for molecular patient selection for targeted therapy. Cancer. 2007;109:2257–2267. doi: 10.1002/cncr.22677. [DOI] [PubMed] [Google Scholar]
- Pearce LR, Huang X, Boudeau J, Pawlowski R, Wullschleger S, Deak M, et al. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007;405:513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloski CE, Lin E, Zhang L, Yung WK, Colman H, Liu JL, et al. Prognostic associations of activated mitogen-activated protein kinase and Akt pathways in glioblastoma. Clin Cancer Res. 2006;12:3935–3941. doi: 10.1158/1078-0432.CCR-05-2202. [DOI] [PubMed] [Google Scholar]
- Phung TL, Ziv K, Dabydeen D, Eyiah-Mensah G, Riveros M, Perruzzi C, et al. Pathological angiogenesis is induced by sustained Akt signaling and inhibited by rapamycin. Cancer Cell. 2006;10:159–170. doi: 10.1016/j.ccr.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb VA, Karbowniczek M, Klein-Szanto AJ, Henske EP. Activation of the mTOR signaling pathway in renal clear cell carcinoma. J Urol. 2007;177:346–352. doi: 10.1016/j.juro.2006.08.076. [DOI] [PubMed] [Google Scholar]
- Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- Rojo F, Najera L, Lirola J, Jimenez J, Guzman M, Sabadell MD, et al. 4E-binding protein 1, a cell signaling hallmark in breast cancer that correlates with pathologic grade and prognosis. Clin Cancer Res. 2007;13:81–89. doi: 10.1158/1078-0432.CCR-06-1560. [DOI] [PubMed] [Google Scholar]
- Rosner M, Siegel N, Valli A, Fuchs C, Hengstschlager M. mTOR phosphorylated at S2448 binds to raptor and rictor. Amino Acids. 2009 doi: 10.1007/s00726-008-0230-7. [DOI] [PubMed] [Google Scholar]
- Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci U S A. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, et al. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Schalm SS, Blenis J. Identification of a conserved motif required for mTOR signaling. Curr Biol. 2002;12:632–639. doi: 10.1016/s0960-9822(02)00762-5. [DOI] [PubMed] [Google Scholar]
- Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- Shackelford DB, Vasquez DS, Corbeil J, Wu S, Leblanc M, Wu CL, et al. mTOR and HIF-1{alpha}-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proc Natl Acad Sci U S A. 2009;106:11137–11142. doi: 10.1073/pnas.0900465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Shaw RJ. LKB1 and AMP-activated protein kinase control of mTOR signalling and growth. Acta Physiol (Oxf) 2009;196:65–80. doi: 10.1111/j.1748-1716.2009.01972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- Tabernero J, Rojo F, Calvo E, Burris H, Judson I, Hazell K, et al. Dose- and schedule-dependent inhibition of the mammalian target of rapamycin pathway with everolimus: a phase I tumor pharmacodynamic study in patients with advanced solid tumors. J Clin Oncol. 2008;26:1603–1610. doi: 10.1200/JCO.2007.14.5482. [DOI] [PubMed] [Google Scholar]
- Thomas GV, Tran C, Mellinghoff IK, Welsbie DS, Chan E, Fueger B, et al. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat Med. 2006;12:122–127. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander HE, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Vega F, Medeiros LJ, Leventaki V, Atwell C, Cho-Vega JH, Tian L, et al. Activation of mammalian target of rapamycin signaling pathway contributes to tumor cell survival in anaplastic lymphoma kinase-positive anaplastic large cell lymphoma. Cancer Res. 2006;66:6589–6597. doi: 10.1158/0008-5472.CAN-05-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–1983. doi: 10.1053/j.gastro.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Beugnet A, Murakami M, Yamanaka S, Proud CG. Distinct signaling events downstream of mTOR cooperate to mediate the effects of amino acids and insulin on initiation factor 4E-binding proteins. Mol Cell Biol. 2005;25:2558–2572. doi: 10.1128/MCB.25.7.2558-2572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel HG, de SE, Fridman JS, Malina A, Ray S, Kogan S, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Dai Q, Sun DF, Xiong H, Tian XQ, Gao FH, et al. mTOR Signaling Pathway Is a Target for the Treatment of Colorectal Cancer. Ann Surg Oncol. 2009;16:2617–2628. doi: 10.1245/s10434-009-0555-9. [DOI] [PubMed] [Google Scholar]
- Zhong H, Chiles K, Feldser D, Laughner E, Hanrahan C, Georgescu MM, et al. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 2000;60:1541–1545. [PubMed] [Google Scholar]
- Zhou L, Huang Y, Li J, Wang Z. The mTOR pathway is associated with the poor prognosis of human hepatocellular carcinoma. Med Oncol. 2009 doi: 10.1007/s12032-009-9201-4. [DOI] [PubMed] [Google Scholar]
- Zhou X, Tan M, Stone HV, Klos KS, Lan KH, Yang Y. Activation of the Akt/mammalian target of rapamycin/4E-BP1 pathway by ErbB2 overexpression predicts tumor progression in breast cancers. Clin Cancer Res. 2004;10:6779–6788. doi: 10.1158/1078-0432.CCR-04-0112. [DOI] [PubMed] [Google Scholar]