Abstract
Biomarkers are the measurable characteristics of an individual that may represent risk factors for a disease or outcome, or that may be indicators of disease progression or of treatment-associated changes. In general, the process by which biomarkers, once identified, might be translated into clinical practice has received scant attention in recent psychiatric literature. A body of work in diagnostic development suggests a framework for evaluating and validating novel biomarkers, but this work may be unfamiliar to clinical and translational researchers in psychiatry. Therefore, this review focuses on the steps that might follow the identification of putative biomarkers. It first addresses standard approaches to characterizing biomarker performance, followed by demonstrations of how a putative biomarker might be shown to have clinical relevance. Finally, it addresses ways in which a biomarker-based test might be validated for clinical application in terms of efficacy and cost-effectiveness.
Keywords: biomarker, test, diagnostic, genetic, cost-effectiveness, utility
Introduction
Recent reports highlight the profound limitations of existing treatments in psychiatry, as well as the challenges faced in the development of novel treatment.1-4 As a means of speeding the development and study of new interventions, there has been growing enthusiasm for the application of biomarkers, defined as features of an individual or organism that may be measured objectively and that indicate a normal biological process, a pathogenic process or an indicator of response to an intervention.5 Biomarkers may represent risk factors for a disease or outcome, or they may represent indicators of disease progression or treatment-associated changes. Biomarkers may also be considered in the framework of mediators and moderators of treatment response.6-7 In this context, a mediator is part of the causal link between an intervention and an outcome—it explains how or why an intervention mediates an outcome. A moderator affects the relationship between intervention and outcome. Although biomarkers are usually taken to refer to explicitly ‘biological’ markers such as genomic or proteomic variation, or imaging or other physiologic measures, in principal, survey measures or rating scales could also represent candidate biomarkers.
To date, in neuropsychiatric disorders, much of the focus has been on biomarkers as surrogate outcome measures—most notably in Alzheimer’s disease, where clinical trials are typically large, long and expensive because of the modest effect sizes with current therapeutics. Rather than a 6-month study relying on batteries of cognitive tests, a shorter study might be possible that examines changes in particular cerebrospinal fluid markers of Alzheimer’s disease—if such markers could be demonstrated to be surrogates for, and potentially mediators of, clinically meaningful outcomes.8
Other potential applications of biomarkers lie in the confirmation of diagnosis and in the prediction of treatment outcomes. Such tools might reduce the uncertainty prevalent in clinical practice, ensuring that patients receive treatments that are most likely to be safe and effective for them. In doing so, they might also facilitate more efficient and reliable clinical trials and speed the development of new treatments, for example, by allowing trials to focus on patient groups that are most likely to benefit from a particular intervention. These applications for diagnosis and prediction represent the focus of this review.
Notwithstanding this recent renewal in enthusiasm, the notion of biomarkers is not new in medicine or psychiatry in particular. Experience with the dexamethasone suppression test three decades ago, however, suggests the need for a careful consideration of consequences when biomarkers are translated into practice. In brief, after initial reports that increases in cortisol levels following a dose of dexamethasone (the ‘challenge’) were associated with major depressive disorder, or with a resolution of symptoms,9 the dexamethasone suppression test rapidly became an accepted diagnostic procedure in clinical settings. Remarkably, there was little initial consideration of its reliability, validity or utility: Did it consistently measure anything? Was that measure truly associated with depression? Was its information ‘actionable’ in clinical practice? A subsequent back-lash underscored the profound limitations of this much-heralded biomarker,10 even though evidence has continued to accumulate that hypothalamic-pituitary-adrenal (HPA) axis abnormalities may well be implicated in the pathophysiology of mood disorders.11-12
In general, the process by which biomarkers, once identified, might be translated into clinical practice has received scant attention in recent psychiatric literature. Notably absent from the discussion is consideration of how a biomarker might be validated and shown to be clinically useful. Beyond the lessons of the dexamethasone suppression test, the slow pace of clinical change in disorders such as Huntington’s disease, in which robust biomarkers have already been identified, argues that translation into clinical practice is not always straightforward. On the other hand, more than 40 Food and Drug Administration (FDA)-approved medications currently include labeling that refers to biomarker testing, with many more under review.13
A body of work in diagnostic development suggests a framework for evaluating and validating novel biomarkers; however, this work may be unfamiliar to clinical and translational researchers in psychiatry. Therefore, this review focuses on the steps that might follow the identification of putative biomarkers. It first addresses standard approaches to characterizing biomarker performance. Next, it discusses how a putative biomarker might be shown to have clinical relevance. Finally, it addresses ways in which a biomarker-based test might be validated for clinical application—that is, how a tool might be shown to impact clinical outcomes—in terms of efficacy and cost effectiveness.
Approach to biomarker-based tests: discrimination and calibration
When two groups such as treatment responders and non-responders are compared, the notion of statistical significance does not necessarily imply clinical significance. The former describes differences, on average, between groups, which may be greater than that expected by chance, but reveals nothing about whether the observed differences are likely to be clinically useful. (Of course, small differences could still be scientifically useful and could lead to the development of measures or interventions that are clinically useful). A better metric familiar to clinical investigators is effect size—what is the magnitude of the difference between groups? This measure may be standardized (for example, divided by standard deviation) to indicate the size of the effect relative to the variability across a population and to facilitate comparison of studies that utilize different measures. (Another measure of effect, that is, the number needed to treat (NNT), is discussed further below). In the context of genetic studies, effects are often expressed in terms of odds ratios or population attributable risk—that is, the proportion of cases in a population that would be eliminated in the absence of a particular variant. However, these two measures of effect have important differences: the latter is dependent upon the prevalence of the risk marker in a population. Population attributable risk is commonly used to examine public health implications of a particular environmental risk.14 In complex genetic diseases, even the most successful investigations of common variance have, to date, yielded modest estimates of attributable risk, a phenomenon referred to as the problem of missing heritability.15 Finally, biomarkers may be described in terms of variance explained—the proportion of variation in a measure (for example, of disease risk) explained by a biomarker or by a model incorporating biomarkers.
Discussions about how large an effect is ‘enough’ in the abstract are unlikely to be helpful because they isolate a biomarker from its possible application. An instructive example is the test for Stevens–Johnson Syndrome risk among Asian patients treated with carbamazepine, which is referenced in the package insert for this drug.16 Among Asian individuals who test positive for the HLA-B*1502 allele, only ~10% will develop serious rash.17 However, even though the risk of rash is low in objective terms in the test-positive group, the test was still held to be valid enough in identifying individuals who can be safely treated with carbamazepine.
As the example suggests, it is often more useful to describe biomarkers in terms of their test properties, which could then be used to estimate their potential utility. When characterizing the performance of a diagnostic measure, most studies emphasize discrimination—that is, how well does a test distinguish between those with and without an outcome of interest? Even when a test yields a continuous measure, a threshold is typically applied to distinguish those in the normal versus abnormal range; although statisticians may complain about loss of precision when a continuous variable is dichotomized, medicine often requires such Boolean logic.
The traditional means of examining discrimination is in terms of a 2 × 2 table considering the test result (positive or negative) and true status (disease positive or disease negative). For tests that yield a continuous outcome (such as probability of an event), results are dichotomized according to a predefined threshold. For example, for hemoglobin, a particular threshold might be defined as ‘normal’ versus ‘low’ for the purpose of constructing a 2 × 2 table. This table yields four groups: the ‘true-positive’, ‘false-positive’, ‘true-negative’ and ‘false-negative’ groups; by relating the four groups, various indices of test performance can be derived. Of these, the most commonly considered are sensitivity—proportion of true positives labeled as positive—and specificity—proportion of true negatives labeled as negative. More interpretable in a clinical context is positive predictive value (PPV)—that is, the proportion of individuals with a positive test who actually have the disease—and negative predictive value—that is, the proportion of individuals with a negative test who do not have the disease. The example of carbamazepine and Stevens–Johnson syndrome reflects a PPV of 10% and a negative predictive value close to 100%; the latter characteristic, as much as the former, contributes to its clinical relevance.
However, a key distinction from sensitivity and specificity is that PPV and negative predictive value are dependent on the prevalence of the outcome being predicted. In this context, a relatively specific test for a disorder may still yield a low PPV if the disorder is rare in a particular population—for example, a problem with early human immunodeficiency virus (HIV) testing in low-risk groups. More generally, all these parameters refer to the performance of a particular test in a particular population. A test may yield poor results in an unselected population but demonstrate better discrimination in a more focused (for example, high risk) population. Notably, sensitivity and specificity also represent a trade-off. A test developer might select a threshold value on the basis of the test’s desired application—for example, focusing on sensitivity when the consequence of a false negative is particularly great, such as prediction of a serious adverse effect.
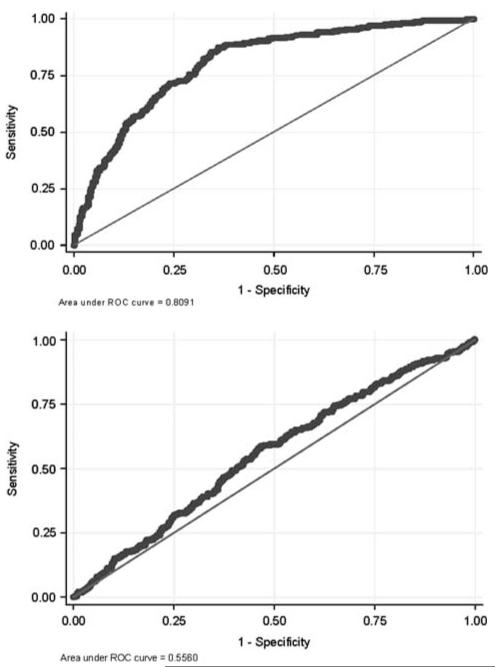
To describe the overall performance of a test across a range of cutoffs, and using a single measure, sensitivity can be plotted against 1—specificity for a range of cutoff values, generating a receiver operating characteristic (ROC) curve. That is, for every possible cutoff point of a test result, what is the resulting sensitivity and specificity? The area under the ROC curve (AUC) provides a summary measure of test discrimination, which may be interpreted as the probability that a case will be scored (ranked) higher than a control if pairs of cases and controls are picked at random. An uninformative test would have an AUC of 0.5—that is, it discriminates at the level of chance. A perfectly discriminating test would have an AUC of 1. Figure 1 illustrates two examples of ROC curves—the curve at the bottom, close to 0.5, has little discriminative ability, whereas the one at the top, close to 1, discriminates substantially better. Unfortunately, discussions of AUC often fall into the same trap as those of attributable risk or PPV: they presume that a particular value such as 0.8 is necessary for a ‘good’ test. As discussed below, in some circumstances where the clinical decision represents a ‘toss-up’, even a modest improvement in prediction may be useful. Moreover, although two markers can be directly compared in terms of AUC, in some circumstances, a test with a smaller AUC may actually be more helpful, as AUC refers to the entire curve whereas a clinician cares about a single cutoff point selected from the curve. Finally, in models predicting disease risk, the maximum AUC that may be achieved—for example, by combining across many common genetic variants to derive a polygenic risk score—depends on disease penetrance and heritability and thus varies widely (for further discussion of this emerging area, see Wray et al18). A mathematical framework for relating many of the standard measures of discrimination, based on consideration of disease probability and variance explained, has recently been described.19
Figure 1.
Example of two receiver operating characteristic (ROC) curves for alternative models to predict treatment resistance in major depression. The figure at the bottom illustrates poor discrimination (area under ROC curve of < 0.6); the one at the top illustrates improved discrimination (area under ROC curve of ~0.8).
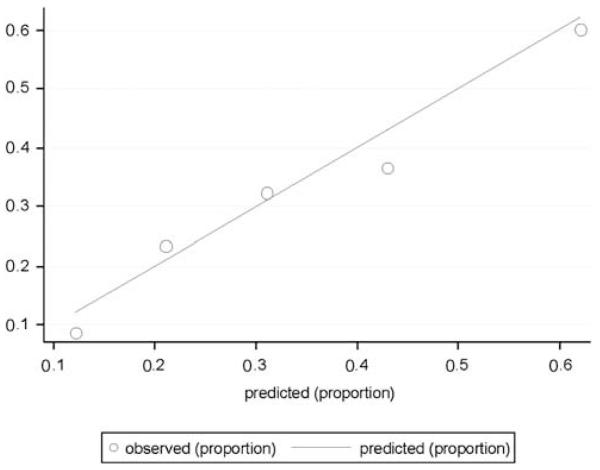
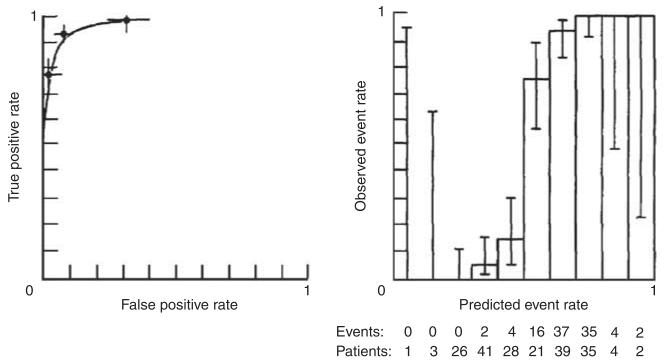
The focus on the ability of a test to correctly classify outcomes also ignores another characteristic of tests—namely, calibration—which may be particularly relevant for predicting future events.20 Calibration refers to the ability of a test to estimate risk accurately—that is, to estimate probabilities that closely match those observed in reality. For longer-term outcomes such as experiencing recurrence of mania or developing diabetes mellitus after treatment with an atypical antipsychotic, knowing that some-one’s risk is 80%, compared with 40%, may well have substantial value, even if a test cannot perfectly distinguish individuals who have a particular outcome. (In contrast, for classifying diagnosis, discrimination may be more important; stating that one’s risk of being pregnant is 80% does not seem particularly useful: one is either pregnant or not pregnant). A simple way of presenting calibration data is by plotting observed versus predicted outcomes. For example, subjects may be divided into quintiles or deciles of risk, and the proportion of outcomes in each group may be plotted against what is predicted. Figure 2 indicates the calibration of a prediction model (in this case, for likelihood of treatment resistance in antidepressant use)—the estimated number of events among those in each quintile of risk corresponds well to the actual number of events observed, indicating good calibration. Interestingly, simulation studies show that a well-calibrated test generally cannot be perfectly discriminative, indicating that a trade-off may be required in diagnostic development.21 Figure 3 shows a hypothetical test that has excellent discrimination (left panel)—that is, the AUC is very close to 1. However, calibration (right panel) is poor: there is only modest correspondence between the predicted number of events and the actual number of events, particularly in the mid-range of the test.21
Figure 2.
Example of a calibration curve for a model of treatment resistance in major depression. The curve plots observed outcomes against expected (predicted) outcomes across five quintiles of risk. In a perfectly calibrated test, all groups would lie on the diagonal from 0,0 to 1,1.
Figure 3.
An example of a classification model with high discrimination (left panel) but poor calibration (right panel)—reprinted from Diamond.21
The role of reclassification
The test parameters addressed so far assume that the alternative is no test at all—that is, treatment as usual. However, in some cases, useful biomarkers already exist, even if they are not considered as such. One example of such a marker in psychiatry might be anxious depression: the presence or absence of anxiety appears to be a predictor of differential antidepressant response.22 Outside of psychiatry, the use of risk stratification models in clinical practice is well established. An example is the venerable Framingham Risk Score, which predicts cardiovascular outcomes on the basis of sociodemographic and laboratory studies.23 Therefore, rather than attempting to replace these prediction models, biomarker studies try to improve upon them—that is, the value of a new marker is considered in terms of its improvement in test performance and not on the basis of its performance in isolation (that is, in a univariate model). An example of such model building is the Reynolds Risk Score,24 which attempts to improve upon the Framingham score in predicting cardiovascular risk in women rather than simply reporting individual predictor variables.
One approach to examining a new biomarker would be to consider it solely in terms of improvement in discrimination, typically by an increase in AUC; two AUC measures can simply be tested for statistically significant differences. However, a single marker with strong evidence of association with an outcome in univariate analysis, even with a large odds ratio, may have very modest effects on AUC.25 Nevertheless, in many cases, a new marker may substantially improve test accuracy without changing AUC through an improvement in calibration, as noted above.
An alternative means of examining the value of a new marker is to explicitly consider its impact on existing classifications—that is, the extent to which it leads subjects to be reclassified more accurately. For defined categories, this is the net reclassification index, which reflects higher-risk subjects correctly moving to higher-risk categories and vice versa; an extension of this measure that does not require predefined categories is the integrated discrimination improvement.26 An example of the utility of considering the net reclassification index comes from a report by Kathiresan et al.27 examining the addition of genetic markers of risk for cardiovascular events to clinical predictors. In that study, individual single nucleotide polymorphism (SNPs) were associated with lipid levels with P-values as low as 3 × 10−21, and a risk score derived by simply counting the number of risk alleles was strongly associated with cardiovascular outcomes. However, the addition of this risk score to a clinical prediction model yielded no change in discrimination—that is, the AUC was 0.80 with or without genetic predictors. Notably, consideration of the net reclassification index indicated that the genetic predictors did significantly improve risk classification. Subjects who subsequently developed cardiovascular disease were more likely to be moved into higher-risk categories when genetic risk was included, whereas subjects who did not develop disease were more likely to be moved into lower-risk categories.
Unfortunately, to date, very few risk models have been validated in psychiatry;28 therefore, there is little to improve upon. Nevertheless, a notable finding in the risk models reported to date is the value of combining novel biomarkers with existing clinical predictors (for examples outside of psychiatry, see Pencina et al.,26 Kathiresan et al.27 and Seddon et al.29) rather than expecting a biomarker to simply replace clinical assessment. The example of cardiovascular disease also suggests that relying on discrimination alone, as indicated by AUC, could lead investigators to overlook clinically meaningful additions to risk models.
Pitfalls in describing test performance
The metrics of performance of a test rely on some comparison between the observed outcome and the predicted outcome, such as disease or treatment response. However, if the outcome cannot be accurately measured—that is, if the gold standard is imperfect—this will yield a ceiling effect in test performance. As an example, the ability of a test to distinguish bipolar disorder from major depressive disorder cannot exceed the ability of the structured interview used to confirm diagnosis in order to make this distinction.
Another limitation in interpreting test performance is the aptly named problem of optimism. When a model is fit to a particular data set, it is likely to incorporate the characteristics of the data set that actually represent chance variation and that are unique to those data. For example, if all patients born in January happened to have poorer outcomes in a given data set, the month of birth might be incorporated in a model, improving the fit of the multi-variable model for the data. This aspect of modeling is referred to as overfitting and leads to predictions about model performance that are overly optimistic—that is, inflated.30 The tendency of a model to be overfit depends on the manner in which that model has been created—different approaches (for example, logistic regression, neural networks and support vector machines) have different strengths and weaknesses in this regard. It also reflects a phenomenon referred to as the ‘curse of dimensionality’—the increasing risk of false-positive results as more tests for associations are conducted, whether because of multiple phenotypes or as a result of multiple putative predictors.
The problem of overfitting has been a particular problem in psychiatric biomarker studies in which cohort collection is labor intensive. After identifying univariate associations, the temptation to estimate their ‘real-world’ prospects by model building in the same data set is high. An example of this is a genome-wide association study of antidepressant response among hospitalized patients with major depressive disorders.31 After identifying the top markers associated with outcome, the investigators showed that, when those markers were combined and examined in the same data set, they were highly predictive of outcome, yielding P-values on the order of 1 × 10 −19. However, the strength of association was far weaker when an independent cohort of patients was added, likely illustrating overfitting in the original model.
To obtain more accurate measures of model performance, a standard approach is cross-validation—partitioning a model into multiple subsets, building models in subsets and examining performance in the other subsets. Particularly for smaller data sets, such estimates will continue to be optimistic. A far better alternative is to build a model on a ‘training’ subset of data (typically, although not necessarily, randomly selected two-thirds of the data) and estimate model performance in the remaining, independent ‘testing’ subset. Note that cross-validation can still be usefully applied for model development within the original training subset. The disadvantage of this alternative, of course, is that it is less efficient, requiring some data to be set aside for validation. Moreover, because both cohorts are drawn from the same population, this approach does not fully protect against overfitting. The optimal measure of performance entails the use of a fully independent data set—for example, a different population of patients32—although such independent data sets may be challenging to identify.
A related challenge in understanding test performance comes from the recognition that tests may perform differently depending upon the population investigated. Biomarkers are often identified initially in highly selected groups such as research patient populations, which may be poorly representative of general clinical populations. The problem is analogous to that of efficacy versus effectiveness: How well does an intervention work in randomized controlled trials (RCTs), versus ‘the real world’? In a large effectiveness study of major depressive disorder (MDD), which did not utilize advertising or traditional inclusion criteria, only 22% of participants would have been eligible for a typical randomized trial.33 In general, both treatments and tests often fare more poorly in the wilds of the clinic, where there are far more co-occurring medical and psychiatric conditions, medication interactions and other obstacles to treatment adherence. A test designed to predict antidepressant efficacy based on a randomized trial will perform poorly in clinical populations in which ~50% of patients do not consistently ingest the antidepressant,34 unless it incorporates predictors of adherence as well.
A particular concern about generalizability relates to ethnic differences. To minimize the risk of spurious results due to population admixture (that is, confounding), many genetic association studies have focused solely on a single population, typically but not always comprising Caucasians. Thus, it is likely that the informativeness of tests developed for this population may be substantially lower in non-Caucasians, which could further exacerbate disparities in care if, for example, tests are found to be cost-effective only in particular groups.35
Determining test utility
An adage of clinical practice holds that it is not worth ordering a test if the results will not influence patient management—that is, if the test is not actionable. A biomarker may be shown to be extremely precise in its prediction, but if it does not lead to a change in the clinician’s or patient’s behavior its clinical value is dubious. In contrast, there are many circumstances in medicine in which two treatment strategies have similar utility—the decision is a toss-up.36 In some scenarios, even modestly discriminative tests may be useful,32 shifting behavior from one strategy to another. A commonly applied risk model for invasive breast cancer, for example, has an AUC of < 0.6 in some studies.37-38 This is the reason that trying to specify an arbitrary threshold for clinical utility—say, 95% sensitivity or specificity39—oversimplifies the case.
Of course, biomarkers have an abundance of other potential applications—for example, in guiding investigations of pathophysiology in vitro or in drug development; however, for clinical application, they must potentially move the posterior probability across a threshold at which behavior would change. For example, suppose a health-care system decides that any depressed patient with at least a 70% chance of being treatment resistant will be referred for cognitive-behavior therapy. Unless a test has the possibility to move (that is, reclassify) some patients into or out of this high-risk group, it will not be useful, at least in the context of referral for cognitive therapy. Many current examples of the importance of reclassification relate to cardiology or oncology, where a specific threshold for risk is often applied to determine which patients should receive more intensive follow-up or particular interventions. Therefore, a diagnostic that reclassifies subjects across this threshold more accurately (that is, one in which the net reclassification index is high) would be likely to be valuable.
In considering the potential application of a biomarker, the distinction first drawn in oncology between ‘prognostic’ and ‘predictive’ tests is useful. Prognostic tests are those that yield information about outcomes such as recurrence independent of treatment, whereas predictive tests yield information that may influence treatment selection itself.40 Tests that lack treatment specificity have been criticized as unlikely to be helpful clinically, giving patients and clinicians more reason to feel pessimistic about the course of illness.41 For example, pharmacogenetic studies that include only a single medication or medication class may identify moderators associated with poor outcome in general, rather than moderators of outcome specific to a treatment. On the other hand, in oncology, prognostic tests may still be useful in determining the intensity of treatment for the prevention of recurrence, even if they do not predict the specific chemotherapy regimen required. By analogy, a predictor of high risk of subsequent manic episodes might not dictate a specific pharmacotherapy but might dictate closer monitoring of symptoms, addition of structured psychosocial treatment or willingness to consider combination medication treatments.
Notably, although behavior is most often considered in terms of the clinician—that is, does a test change the clinician’s behavior—this need not be the case. A test that improves the patient’s adherence—even if it does not change treatment strategy—might still have value, because it changes the patient’s behavior. An ongoing investigation of statin use (AKROBATS; http://www.clinicaltrials.gov) addresses this possibility, hypothesizing that individuals who are aware that they carry cardiovascular risk variants will be more likely to adhere to treatment. Likewise, a biomarker might also have value in helping individuals plan for the future, although the thresholds for action here are more difficult to define and characterize. Testing for BRCA1 and 2, or for expanded Huntingtin alleles, might fall into this latter category. It is noteworthy that, in the absence of education about how to interpret test results, it is also possible that testing may adversely affect outcomes. Anecdotally, for example, many clinicians believe that patients with CYP450 2D6 alleles that cause them to be poor metabolizers must not be treated with medications that are 2D6 substrates, rather than simply adjusting the dose42—which might lead them to overlook potentially efficacious interventions.
Two key tools in considering the potential usefulness of a novel diagnostic are decision analysis and cost-effectiveness analysis (CEA). In the former, by estimating outcome probabilities and assigning value (utility) to particular outcomes (remitted depression, depressed mood and death), different testing and treatment strategies can be compared. For example, a strategy in which all schizophrenia patients receive the same antipsychotic treatment might be compared with one in which they are tested and then assigned to treatment on the basis of test results.43 It might seem that any test with some degree of accuracy would be useful; however, it is to be noted that tests may have substantial financial costs as well as consequences in terms of utility—for example, if more patients undergo painful, risky or expensive procedures as a result. Indeed, this is a major consideration in, for example, debates on the frequency and mode of screening for prostate or breast cancer.44-47
CEA is an important extension of such models that incorporates not just utility but also explicit monetary costs while comparing strategies.48 After constructing a decision tree including alternate strategies and outcomes, the cost of each strategy can be calculated and compared. For example, to model the potential cost-effectiveness of a test for selective serotonin reuptake inhibitor (SSRI) responsiveness, a decision tree compared testing before treatment with testing after an initial treatment failure and with not testing at all.49 This set of tools is extremely useful to policymakers in cost-constrained environments and also allows questions like ‘how big an effect size matters’ to be answered in concrete terms. Key to such analyses is the availability of reliable cost data and the ability to estimate other parameters (for example, risk of 1-year recurrence in a general population of individuals with major depression). In large health-care systems, it may be possible to estimate the value of an intervention in that system by incorporating known parameters derived from billing data. Nevertheless, given the difficulty in obtaining precise estimates of parameters, CEA typically presents a ‘base case’ (that is, the results with the initial model), as well as a sensitivity analysis that examines the effects of varying multiple parameters. Often, the sensitivity analysis is the most valuable portion of the analysis as it identifies the key features that will determine the value of a biomarker in a given context. Numerous tools are available to facilitate decision analysis and CEA.50
An often-overlooked aspect of CEA is the fact that it may help in identifying alternative strategies that may be ‘dominating’—that is, more cost-effective. For example, the application of CYP450 testing while prescribing antidepressants has received considerable attention.51-52 Rather than trying to identify non-wild-type drug metabolizers, however, it may be more cost-effective simply to treat with antidepressants for which common CYP450 variation has little or no effect. (The notion of a dominating choice also affects the potential usefulness of test validation study designs; see below.)
In lieu of CEA, an increasingly popular strategy is to report the effects of an intervention in terms of NNT—that is, how many individuals need to be treated to prevent a single outcome—which is another form of effect size. For example, the UK’s National Institute for Health and Clinical Excellence has suggested that an NNT threshold of 10 or less is required for clinical significance. The same concept can be applied to examine a biomarker, yielding a ‘number needed to test’ in order to achieve or prevent a particular outcome. NNT is straightforward to calculate from case-control data, as it represents 1 over the difference in risk for an outcome between two strategies. If the risk of antidepressant-associated mania was 10% at baseline, and 5% following a test, NNT would be 1/(0.1–0.05) or 20—that is, it would be necessary to test 20 patients to prevent one instance of antidepressant-associated mania. However, the implications of a given NNT still depend on the context, particularly in terms of the utility/expense of the outcome and the expense of the test. NNT may therefore be most useful for comparing similar strategies (for example, two alternative genetic tests for antipsychotic-associated weight gain).
Prospective studies of test utility
The path to demonstrating that an intervention has efficacy in medicine is well trodden and usually culminates in RCTs. Although there is a prelude—smaller, ‘proof-of concept’ studies—and a concluding act—real-world-effectiveness or cost-effectiveness studies, or ‘predictor’ studies to determine whether subgroups respond preferentially—the RCT is the main attraction.
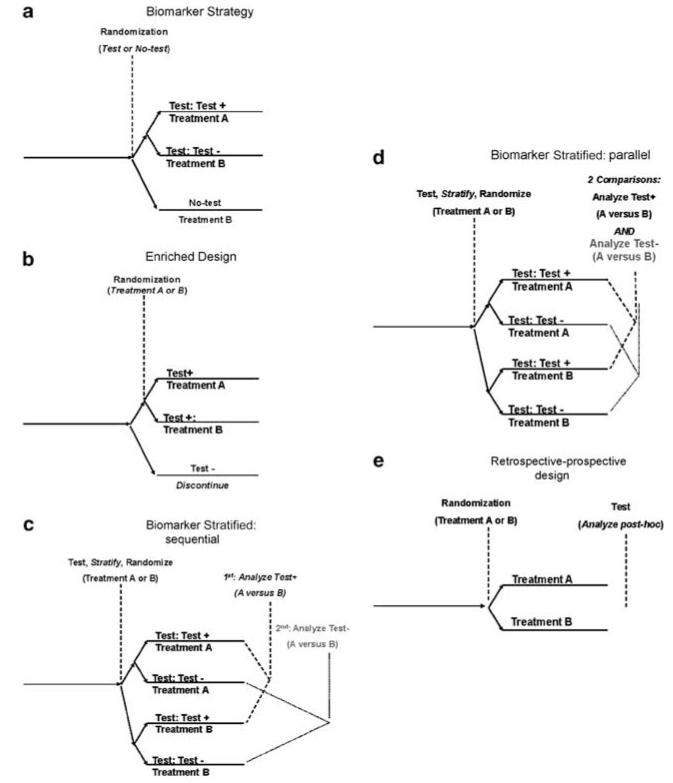
For diagnostic tests, the path to validation is less clear. Initial confirmation of association between the biomarker and the outcome of interest, such as diagnosis or treatment response, may be a relatively simple matter of collecting a replication cohort. However, simply confirming that a test measures what it purports to measure is only the initial step in the validation process; obtaining a more precise estimate of test performance and showing that the test changes behavior in a meaningful way are also required. That is, the focus shifts from ‘is it real’ to ‘is it useful’. Figure 4 depicts multiple strategies that may be considered for validating a biomarker for clinical application. The most straightforward approach is an RCT in which one test might be compared with another test, with a clinical ‘gold standard’, or with usual clinical practice. This approach is typically referred to as the ‘biomarker-strategy’ design (Figure 4a) as it compares two (or more) strategies, at least one of which does not utilize the biomarker. For example, a marker for antidepressant response might be compared with clinical judgment alone, with individuals at risk for SSRI non-response receiving a non-SSRI antidepressant as first-line treatment. However, studying biomarkers prospectively with RCTs poses certain scientific risks, most notably when the field itself is advancing at a rapid pace. For example, a study of warfarin metabolism that used a single marker (CYP2C19) to determine optimal warfarin dose for anticoagulation would become essentially obsolete as soon as an additional marker useful for prediction (VKORC1) is identified. Similarly, studies using the serotonin transporter polymorphism for antidepressant selection became potentially less relevant as the value of this biomarker came into question.53
Figure 4.
Alternative designs for clinical investigation of biomarkers. (a) Biomarker strategy, (b) enriched design, (c) biomarker strategy: sequential, (d) biomarker strategy: parallel and (e) retrospective–prospective design.
A second challenge in the biomarker-strategy approach is the fact that larger sample sizes will often be required to achieve adequate statistical power. The key problem here is that, in many cases, the biomarker-driven strategy and the non-marker-driven (default) strategy will be the same, rendering these subjects essentially uninformative. (So, for example, only those for whom a test dictates not receiving SSRI treatment would contribute to detection of a difference from treatment as usual in which all subjects receive SSRI treatment.) As a rough estimate, a comparison of sample size requirements in oncology studies found that sample size requirements were 2–3 times greater in the biomarker-strategy approach, in which 30–50% of the cohort carried the marker of interest.54 To minimize the diluting effect of test results that do not change behavior while retaining the biomarker-strategy design, it might be possible to contrast outcomes only among subjects in whom the test is, or would have been, informative—that is, to contrast test-positive subjects who did or did not receive assay-guided treatment.
Another concern with this approach is that it may not be possible to distinguish an effective biomarker from a more effective treatment strategy, as the comparison includes differences in both diagnostic and treatment. That is, if the biomarker-strategy approach includes a treatment that is more effective, regardless of biomarker status, it may indicate greater efficacy for that strategy even if the biomarker is actually uninformative. This problem is equivalent to the dominating treatment option already described. Consider, for example, a biomarker-strategy study that compares valproate treatment (for all patients) with a gene-guided test that triages a subset of patients to lithium in lieu of valproate. If lithium is more efficacious than valproate, this study would show the test-guided strategy to be superior even if the marker itself is not, as it leads to more patients receiving the more efficacious option.
Importantly, rather than traditional efficacy measures (or in addition to such measures), a biomarker trial might focus on the clinician’s behavior. For example, a less resource-intensive strategy is to examine changes in the physician’s behavior as a short-term proxy measure. That is, one might ask, ‘Did the availability of the biomarker cause physicians to behave differently than they otherwise would have?’ This information may be obtained simply by asking clinicians to report their treatment choice before receiving results, and then after receiving results. Ideally, such studies would be randomized at the level of the patient, physician or clinic. Although influencing behavior might be a prerequisite for a useful test, a matter of concern about this approach is that it makes the (large) assumption that the physician’s behavior changes in a manner that improves patient outcomes.
A key question to be answered is whether randomization is necessary and feasible in all cases. In lieu of an RCT, it may sometimes be reasonable to consider historical controls—as was done, for example, in the ‘D-04’ component of the FDA registration trial of vagus nerve stimulation for major depressive disorders,55 where outcomes were collected for a comparable cohort of treatment-resistant subjects who did not undergo the procedure. Similarly, in another study, outcomes were compared between subjects who received warfarin doses according to the results of a genetic test for warfarin metabolism and a matched group of patients who received treatment the previous year but who were otherwise similar in terms of risk; outcomes were markedly improved among the test-guided group.56
Some authors question whether a prospective trial is necessary at all to validate a biomarker. Where large prospectively studied cohorts exist, an alternative is the ‘retrospective–prospective design’57 (Figure 4e). In this approach, a putative biomarker is examined retrospectively in terms of prospectively measured outcomes. The utility of a novel predictor of recurrence risk might be examined in a completed study in which biomaterials such as DNA have been banked. It should be emphasized that this is fundamentally different from the exploratory approach typical in such studies. Here, a predefined biomarker is examined in an independent data set. Otherwise, where a marker is derived and validated in the same data set, the risk for overly optimistic estimates of test performance is high (see overfitting, above). The retrospective–prospective approach depends on the availability of these large cohorts, with treatment trials at least somewhat representative of real-world strategies. The recent emphasis on real-world ‘effectiveness’ studies has yielded multiple psychiatric cohorts that may be amenable to this approach. Ongoing efforts to create repositories for biomaterials in psychiatry will also facilitate these analyses.
The retrospective–prospective approach could be considered particularly in early intervention or in ‘at risk’ populations, where the goal is to identify individuals at high risk for a given disorder and potentially intervene for primary prevention. A major logistical challenge in these studies is the long follow-up period. At present, these studies tend to focus on individuals ‘at risk’ based not only on family history but also on the emergence of some symptoms. Unfortunately, this approach does not require individuals to be truly presymptomatic or predisease at entry—they may no longer be at risk but may actually be in the early stages of illness. For example, a sleep disturbance may represent the initial symptoms of a major depressive episode rather than be a true predictor of subsequent episodes.
Validating drug–diagnostic combinations
A special case of biomarker development arises when a diagnostic is developed in parallel with a novel treatment, with the intention of marketing the two together. To date, these circumstances often arise when a proof-of-concept or phase 2 study fails to achieve its primary goal, but post hoc analysis suggests a subgroup with a particularly good (or poor) response. For example, in a phase 2 study of bapineuzumab for mild-to-moderate Alzheimer’s disease in which the drug was not superior to placebo, an exploratory analysis suggested benefit in the subset of patients who were APOE4 epsilon-4 non-carriers.58 Two design possibilities for follow-on investigations include biomarker-enriched (Figure 4b) and biomarker-stratified (Figures 4c and d) approaches.54,59,60
Clinical investigators describe a trial with an enriched design as one in which the study population is selected on the basis of a particular feature, such as responsiveness to medication. Thus, studies in which all subjects first receive open treatment with an atypical antipsychotic, and are then randomized to remain on treatment or discontinue it, would be said to be enriched for acute treatment tolerability and response—subjects unable to tolerate and remain stable on acute treatment would not enter the subsequent trial. The biomarker-enriched design makes a strong assumption about biomarker effects: only subjects who test positive for the marker are enrolled and randomized. In psychiatry, one of the first suggestions of this approach was to use variation in the serotonin transporter (SLC6A4) promoter region to identify likely placebo non-responders in antidepressant treatment studies suffering from major depressive disorders and in this way increase drug–placebo differences. Preliminary data indicated that enriching for subjects not carrying the ‘short’ allele of SLC6A4 in antidepressant trials would better separate drug and placebo,61 although these data were neither published nor replicated.
A recent example of this approach was an investigation of buspirone and melatonin in anxious depression,62 in which the presence of anxiety assessed on a rating scale can be considered as a simple (if low tech) biomarker. Anxiety was anticipated to be a moderator of the association between treatment and outcome. That is, the study made the assumption that placebo-like response was likely to be lower in individuals with anxiety and therefore enriching for this group would maximize drug–comparator separation—an assumption that proved to be correct.
An advantage of this strategy is its efficiency—subjects unlikely to contribute to drug–comparator separation are not enrolled, although they must still be screened.63 If the putative marker is informative, a trial should have greater power to show statistical significance or should have a smaller sample size, although if it does not, the trial will likely be slower to complete, and more costly, compared with the ‘all-comers’ approach. A notable disadvantage of this design is that it does not demonstrate, in and of itself, the utility of the biomarker. The buspirone/melatonin combination might be equally efficacious in non-anxious patients, so further study would be required to determine the treatment specificity of the biomarker.
An alternative design allows the treatment specificity to be determined directly, although at the cost of requiring a more complex design and larger sample sizes. This biomarker-stratified approach is also useful when investigators are less confident in the utility of a biomarker and more interested in knowing the overall efficacy of an intervention. In this study design (Figures 4c and d), randomization to two or more interventions proceeds as in any other controlled trial, but randomization is stratified by a biomarker to ensure a balanced distribution of biomarker status across groups. An investigation of the serotonin transporter for prediction of response to ondansetron in alcohol use disorders utilized this approach.64 These designs have become particularly common in oncology; see, for example, the MARVEL study of erlotinib in lung cancer.65 For a reasonably large study, such stratification is in fact unnecessary (randomization should ensure balanced distribution); however, for smaller studies, in which power is more affected by small deviations, it may be useful. This approach may be considered as a form of ‘match-mismatch’ design: subjects are randomized to treatment that either does, or does not, ‘match’ that specified by a biomarker.
The analysis of such studies offers a range of possibilities. The simplest is to examine overall drug effects compared with a comparator, then to look for interactions with the biomarker, although this assumes that the study is powered to show a main effect for the drug even after ‘dilution’ by the biomarker-negative group. An alternative strategy receiving increasing attention is the ‘step-up’ or sequential design, which proceeds sequentially through a set of comparisons. This analytic approach has been specifically cited by FDA officials as being well suited to the study of drug–diagnostic combinations. First, the ‘test-positive’ group is examined to determine whether the drug is superior to placebo. If so, the ‘test-negative’ group is examined in the same way. The drug–diagnostic combination is validated only if the former is true. If both are true, the drug is confirmed as validated, but not the diagnostic. The advantage of such a sequential approach is that the control of type I error is straightforward. Moreover, it does not require a second trial to confirm that a biomarker is treatment-specific, as would be the case with the biomarker-enriched design. The trade-off here is the need for larger cohorts, as each of the groups (biomarker positive and biomarker negative) must be adequately powered to stand on its own. In essence, two adequately powered studies are conducted in parallel. When the biomarker prevalence is further from 50% in either direction, problems in feasibility may arise. A variant of this method is the parallel design, in which both the biomarker-positive and biomarker-negative groups are examined with P < 0.025 in order to control overall study α at 0.05.
Hybrid strategies may also be considered for all the designs noted here. For example, one might consider an adaptive design in which the trial proceeds initially without stratification by biomarker, but an interim analysis examines putative biomarkers defined a priori. If this interim analysis identifies greater benefit in the biomarker-positive group, the study is then altered (in a prespecified manner) to enroll only biomarker-positive subjects. Study designs may also be combined. For example, a cohort may be enriched for one biomarker, then randomized with stratification by another (for a real-world example, see the SLCG0601 lung cancer study described further in Freidlin et al.54).
Conclusion
As greater consistency emerges in biomarker studies in the field of psychiatry, pressure to begin using such markers to reduce uncertainty in clinical practice will increase. Translating such findings to practice will require careful and systematic development of diagnostic tools, characterization of their performance and examination of their utility. Particularly useful concepts here include consideration of test calibration in addition to discrimination, attention to overfitting and consideration of the clinical context in which a diagnostic tool may be used. Standard metrics exist for all these processes. Ultimately, as with nearly any intervention in medicine, randomized investigations will likely be required to demonstrate the efficacy of a biomarker as diagnostic; however, a range of designs merit consideration. As they are validated, biomarkers should begin to deliver on their often-cited, rarely-studied potential, with opportunities for more personalized treatments, focused clinical trials and primary or secondary prevention.
Acknowledgments
I thank Shaun Purcell, PhD, and Pamela Sklar, MD, PhD, for helpful discussion. This work was supported by R01 MH086026 and by the Stanley Center for Psychiatric Research.
Footnotes
Conflict of interest
Dr Perlis has received consulting fees and royalties from Concordant Rater Systems and consulting fees from Proteus Biomedical, RIDVentures and Geno-mind.
References
- 1.Rush AJ, Trivedi MH, Wisniewski SR, Nierenberg AA, Stewart JW, Warden D, et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am J Psychiatry. 2006;163:1905–1917. doi: 10.1176/ajp.2006.163.11.1905. [DOI] [PubMed] [Google Scholar]
- 2.Kaitin KI. Deconstructing the drug development process: the new face of innovation. Clin Pharmacol Ther. 2010;87:356–361. doi: 10.1038/clpt.2009.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perlis RH, Ostacher MJ, Patel JK, Marangell LB, Zhang H, Wisniewski SR, et al. Predictors of recurrence in bipolar disorder: primary outcomes from the systematic treatment enhancement program for bipolar disorder (STEP-BD) Am J Psychiatry. 2006;163:217–224. doi: 10.1176/appi.ajp.163.2.217. [DOI] [PubMed] [Google Scholar]
- 4.Keefe RS, Bilder RM, Davis SM, Harvey PD, Palmer BW, Gold JM, et al. Neurocognitive effects of antipsychotic medications in patients with chronic schizophrenia in the CATIE Trial. Arch Gen Psychiatry. 2007;64:633–647. doi: 10.1001/archpsyc.64.6.633. [DOI] [PubMed] [Google Scholar]
- 5.Biomarkers Definitions Working Group Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 6.Kraemer HC, Stice E, Kazdin A, Offord D, Kupfer D. How do risk factors work together? Mediators, moderators, and independent, overlapping, and proxy risk factors. Am J Psychiatry. 2001;158:848–856. doi: 10.1176/appi.ajp.158.6.848. [DOI] [PubMed] [Google Scholar]
- 7.Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51:1173–1182. doi: 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- 8.Hampel H, Frank R, Broich K, Teipel SJ, Katz RG, Hardy J, et al. Biomarkers for Alzheimer’s disease: academic, industry and regulatory perspectives. Nat Rev Drug Discov. 2010;9:560–574. doi: 10.1038/nrd3115. [DOI] [PubMed] [Google Scholar]
- 9.Greden JF, Albala AA, Haskett RF, James NM, Goodman L, Steiner M, et al. Normalization of dexamethasone suppression test: a laboratory index of recovery from endogenous depression. Biol Psychiatry. 1980;15:449–458. [PubMed] [Google Scholar]
- 10.Arana GW, Baldessarini RJ, Ornsteen M. The dexamethasone suppression test for diagnosis and prognosis in psychiatry. Commentary and review. Arch Gen Psychiatry. 1985;42:1193–1204. doi: 10.1001/archpsyc.1985.01790350067012. [DOI] [PubMed] [Google Scholar]
- 11.Young E, Korszun A. Sex, trauma, stress hormones and depression. Mol Psychiatry. 2010;15:23–28. doi: 10.1038/mp.2009.94. [DOI] [PubMed] [Google Scholar]
- 12.Heim C, Newport DJ, Mletzko T, Miller AH, Nemeroff CB. The link between childhood trauma and depression: insights from HPA axis studies in humans. Psychoneuroendocrinology. 2008;33:693–710. doi: 10.1016/j.psyneuen.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Anonymous Table of Valid Genomic Biomarkers in the Context of Approved Drug Labels. 2010 Available from: http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm.
- 14.Northridge ME. Public health methods—attributable risk as a link between causality and public health action. Am J Public Health. 1995;85:1202–1204. doi: 10.2105/ajph.85.9.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomson Reuters Clinical Editorial Staff . Carbamazepine. 2010. Physician’s Desk Reference [serial on the Internet] [Google Scholar]
- 17.Chung WH, Hung SI, Hong HS, Hsih MS, Yang LC, Ho HC, et al. Medical genetics: a marker for Stevens-Johnson syndrome. Nature. 2004;428:486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]
- 18.Wray NR, Yang J, Goddard ME, Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 2010;6:e1000864. doi: 10.1371/journal.pgen.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.So HC, Sham PC. A unifying framework for evaluating the predictive power of genetic variants based on the level of heritability explained. PLoS Genet. 2010;6:e1001230. doi: 10.1371/journal.pgen.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook NR. Statistical evaluation of prognostic versus diagnostic models: beyond the ROC curve. Clin Chem. 2008;54:17–23. doi: 10.1373/clinchem.2007.096529. [DOI] [PubMed] [Google Scholar]
- 21.Diamond GA. What price perfection? Calibration and discrimination of clinical prediction models. J Clin Epidemiol. 1992;45:85–89. doi: 10.1016/0895-4356(92)90192-p. [DOI] [PubMed] [Google Scholar]
- 22.Fava M, Rush AJ, Alpert JE, Balasubramani GK, Wisniewski SR, Carmin CN, et al. Difference in treatment outcome in outpatients with anxious versus nonanxious depression: a STAR*D report. Am J Psychiatry. 2008;165:342–351. doi: 10.1176/appi.ajp.2007.06111868. [DOI] [PubMed] [Google Scholar]
- 23.Eichler K, Puhan MA, Steurer J, Bachmann LM. Prediction of first coronary events with the Framingham score: a systematic review. Am Heart J. 2007;153:722–731. 31, e1–8. doi: 10.1016/j.ahj.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 24.Ridker PM, Buring JE, Rifai N, Cook NR. Development and validation of improved algorithms for the assessment of global cardiovascular risk in women: the Reynolds Risk Score. JAMA. 2007;297:611–619. doi: 10.1001/jama.297.6.611. [DOI] [PubMed] [Google Scholar]
- 25.Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007;115:928–935. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
- 26.Pencina MJ, D’Agostino RB, Sr, D’Agostino RB, Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–172. doi: 10.1002/sim.2929. discussion 207-12. [DOI] [PubMed] [Google Scholar]
- 27.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 28.Perlis RH, Ostacher MJ, Miklowitz DJ, Hay A, Nierenberg AA, Thase ME, et al. Clinical features associated with poor pharmacologic adherence in bipolar disorder: results from the STEP-BD study. J Clin Psychiatry. 2010;71:296–303. doi: 10.4088/JCP.09m05514yel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seddon JM, Reynolds R, Maller J, Fagerness JA, Daly MJ, Rosner B. Prediction model for prevalence and incidence of advanced age-related macular degeneration based on genetic, demographic, and environmental variables. Invest Ophthalmol Vis Sci. 2009;50:2044–2053. doi: 10.1167/iovs.08-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ioannidis JP. Why most discovered true associations are inflated. Epidemiology. 2008;19:640–648. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- 31.Ising M, Lucae S, Binder EB, Bettecken T, Uhr M, Ripke S, et al. A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch Gen Psychiatry. 2009;66:966–975. doi: 10.1001/archgenpsychiatry.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wisniewski SR, Rush AJ, Nierenberg AA, Gaynes BN, Warden D, Luther JF, et al. Can phase III trial results of antidepressant medications be generalized to clinical practice? A STAR*D report. Am J Psychiatry. 2009;166:599–607. doi: 10.1176/appi.ajp.2008.08071027. [DOI] [PubMed] [Google Scholar]
- 34.Serna MC, Cruz I, Real J, Gasco E, Galvan L. Duration and adherence of antidepressant treatment (2003 to 2007) based on prescription database. Eur Psychiatry. 2010;25:206–213. doi: 10.1016/j.eurpsy.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 35.Haga SB. Impact of limited population diversity of genome-wide association studies. Genet Med. 2010;12:81–84. doi: 10.1097/GIM.0b013e3181ca2bbf. [DOI] [PubMed] [Google Scholar]
- 36.Kassirer JP, Pauker SG. The toss-up. N Engl J Med. 1981;305:1467–1469. doi: 10.1056/NEJM198112103052409. [DOI] [PubMed] [Google Scholar]
- 37.Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Schairer C, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 38.Mealiffe ME, Stokowski RP, Rhees BK, Prentice RL, Pettinger M, Hinds DA. Assessment of clinical validity of a breast cancer risk model combining genetic and clinical information. J Natl Cancer Inst. 2010;102:1618–1627. doi: 10.1093/jnci/djq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braff DL, Freedman R. Clinically responsible genetic testing in neuropsychiatric patients: a bridge too far and too soon. Am J Psychiatry. 2008;165:952–955. doi: 10.1176/appi.ajp.2008.08050717. [DOI] [PubMed] [Google Scholar]
- 40.Driessen E, Hollon SD. Cognitive behavioral therapy for mood disorders: efficacy, moderators and mediators. Psychiatr Clin North Am. 2010;33:537–555. doi: 10.1016/j.psc.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon GE, Perlis RH. Personalized medicine for depression: can we match patients with treatments? Am J Psychiatry. 2010;167:1445–1455. doi: 10.1176/appi.ajp.2010.09111680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirchheiner J, Brockmoller J. Clinical consequences of cytochrome P450 2C9 polymorphisms. Clin Pharmacol Ther. 2005;77:1–16. doi: 10.1016/j.clpt.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 43.Perlis RH, Ganz DA, Avorn J, Schneeweiss S, Glynn RJ, Smoller JW, et al. Pharmacogenetic testing in the clinical management of schizophrenia: a decision-analytic model. J Clin Psychopharmacol. 2005;25:427–434. doi: 10.1097/01.jcp.0000177553.59455.24. [DOI] [PubMed] [Google Scholar]
- 44.Lebovic GS, Hollingsworth A, Feig SA. Risk assessment, screening and prevention of breast cancer: a look at cost-effectiveness. Breast. 2010;19:260–267. doi: 10.1016/j.breast.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Kopans DB. The 2009 U.S. Preventive Services Task Force guidelines ignore important scientific evidence and should be revised or withdrawn. Radiology. 2010;256:15–20. doi: 10.1148/radiol.10100057. [DOI] [PubMed] [Google Scholar]
- 46.Thrall JH. US Preventive Services Task Force recommendations for screening mammography: evidence-based medicine or the death of science? J Am Coll Radiol. 2010;7:2–4. doi: 10.1016/j.jacr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 47.DeAngelis CD, Fontanarosa PB. US Preventive Services Task Force and breast cancer screening. JAMA. 2010;303:172–173. doi: 10.1001/jama.2009.1990. [DOI] [PubMed] [Google Scholar]
- 48.Rutigliano MJ. Cost effectiveness analysis: a review. Neurosurgery. 1995;37:436–43. doi: 10.1227/00006123-199509000-00011. discussion 43-4. [DOI] [PubMed] [Google Scholar]
- 49.Perlis RH, Patrick A, Smoller JW, Wang PS. When is pharmacogenetic testing for antidepressant response ready for the clinic? A cost-effectiveness analysis based on data from the STAR*D study. Neuropsychopharmacology. 2009;34:2227–2236. doi: 10.1038/npp.2009.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Introduction to cost-effectiveness analysis (CEA) [database on the Internet] Available from: http://wwwherc.research.va.gov/methods/cea.asp.
- 51.Kirchheiner J, Rodriguez-Antona C. Cytochrome P450 2D6 genotyping: potential role in improving treatment outcomes in psychiatric disorders. CNS Drugs. 2009;23:181–191. doi: 10.2165/00023210-200923030-00001. [DOI] [PubMed] [Google Scholar]
- 52.Perlis RH. Cytochrome P450 genotyping and antidepressants. Bmj. 2007;334:759. doi: 10.1136/bmj.39169.547512.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kraft JB, Peters EJ, Slager SL, Jenkins GD, Reinalda MS, McGrath PJ, et al. Analysis of association between the serotonin transporter and antidepressant response in a large clinical sample. Biol Psychiatry. 2007;61:734–742. doi: 10.1016/j.biopsych.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 54.Freidlin B, McShane LM, Korn EL. Randomized clinical trials with biomarkers: design issues. J Natl Cancer Inst. 2010;102:152–160. doi: 10.1093/jnci/djp477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daban C, Martinez-Aran A, Cruz N, Vieta E. Safety and efficacy of Vagus Nerve Stimulation in treatment-resistant depression. A systematic review. J Affect Disord. 2008;110:1–15. doi: 10.1016/j.jad.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 56.Epstein RS, Moyer TP, Aubert RE, O Kane DJ, Xia F, et al. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study) J Am Coll Cardiol. 2010;55:2804–2812. doi: 10.1016/j.jacc.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 57.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101:1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salloway S, Sperling R, Gilman S, Fox NC, Blennow K, Raskind M, et al. A phase 2 multiple ascending dose trial of bapineuzumab in mild to moderate Alzheimer disease. Neurology. 2009;73:2061–2070. doi: 10.1212/WNL.0b013e3181c67808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simon R. The use of genomics in clinical trial design. Clin Cancer Res. 2008;14:5984–5993. doi: 10.1158/1078-0432.CCR-07-4531. [DOI] [PubMed] [Google Scholar]
- 60.Mandrekar SJ, Sargent DJ. Clinical trial designs for predictive biomarker validation: theoretical considerations and practical challenges. J Clin Oncol. 2009;27:4027–4034. doi: 10.1200/JCO.2009.22.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manji H. NIMH New Clinical Drug Evaluation Unit Meeting. NCDEU; Phoenix, AZ: 2001. 2001. Serotonin Transporter Promoter Polymorphisms and Placebo Response. [Google Scholar]
- 62.Fava M. A randomized, controlled trial of buspirone, melatonin, or the combination for anxious major depressive disorder. American Psychiatric Association; San Francisco, CA: 2009. [Google Scholar]
- 63.Simon R, Maitournam A. Evaluating the efficiency of targeted designs for randomized clinical trials. Clin Cancer Res. 2004;10:6759–6763. doi: 10.1158/1078-0432.CCR-04-0496. [DOI] [PubMed] [Google Scholar]
- 64.Johnson BA, Ait-Daoud N, Chamindi S, Roache JD, Javors MA, Wang X, et al. Pharmacogenetic approach at the serotonin transporter gene as a method to reduce the severity of drinking alcohol. Am J Psychiatry. 2011;168:265–275. doi: 10.1176/appi.ajp.2010.10050755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wakelee H, Kernstine K, Vokes E, Schiller J, Baas P, Saijo N, et al. Cooperative group research efforts in lung cancer 2008: focus on advanced-stage non-small-cell lung cancer. Clin Lung Cancer. 2008;9:346–351. doi: 10.3816/CLC.2008.n.050. [DOI] [PubMed] [Google Scholar]