Abstract
The unconventional Guanine Nucleotide Exchange Factor (GEF) family comprising 11 DOCK180 related proteins is classified into four subfamilies, A through D, based on their relative GEF activity towards the closely related Rac and Cdc42 GTPases. DOCK proteins participate in the remodeling of the actin cytoskeleton and are key regulators of cell motility, phagocytosis and adhesion. Here we show that the guanine nucleotide exchange domain of DOCK7, DHR2 (for DOCK homology region 2), is a potent GEF for prenylated Cdc42 and Rac1 in a model liposome system, demonstrating that the prenylation and membrane localization of Cdc42 or Rac1 are necessary for their activation by DOCK7. Additionally, we identify DOCK7 residues that confer GTPase GEF specificity. Finally, using our liposome reconstitution assay, we show that a more narrowly defined GEF domain of DHR2 (designated DHR2s) harbors an N-terminal site distinct from the GEF active site that binds preferentially to the active, GTP-bound forms of Cdc42 and Rac1 and thereby recruits free DHR2s from solution to the membrane surface. This recruitment results in a progressive increase in the effective concentration of DHR2s at the membrane surface that in turn provides for an accelerated rate of guanine nucleotide exchange on Cdc42. The positive cooperativity observed in our reconstituted system suggests that the action of DOCK7 in vivo may involve the coordinated integration of Cdc42/Rac signaling in the context of the membrane recruitment of a DOCK7 GEF complex.
The Rho family of GTPases plays essential roles in many cellular processes, including cell polarity, motility, vesicular trafficking, cell-cycle progression, and gene expression.1–4 Their functions often depend on their cellular localization, which is mediated by a combination of a stretch of basic amino acids at the C-terminus and the post-translational modification of the C-terminal CAAX motif (where C represents cysteine, A is any aliphatic amino acid, and X is any amino acid).5,6 Prenyltransferases catalyze the modification of CAAX motifs and add a farnesyl or geranylgeranyl isoprenoid lipid tail to the cysteine residue.7 For example, both Cdc42 and Rac1 are geranylgeranylated, and this attached lipid tail of the GTPase inserts into the cellular membrane and, along with the C-terminal basic sequence, is thought to target the GTPase to the subcellular compartments where it can engage its regulators, namely GEFs (Guanine nucleotide Exchange Factors) or GAPs (GTPase Activating Proteins).8–10
Two distinct families of GEFs have been identified for Rho GTPases: the conventional Dbl family11 and the more recently identified DOCK180 family.12–14 The Dbl family consists of ~70 members, all of which contain two conserved domains, the Dbl homology (DH) and the pleckstrin homology (PH) domains.15–18 The DH domain is responsible for catalyzing guanine nucleotide exchange, while the PH domain has been demonstrated to interact with plasma membrane phosphoinositides.19,20
The mammalian DOCK180 family contains 11 members, DOCK1–11 (with DOCK180 being DOCK1).21,22 All possess two DOCK homology regions, DHR1 and DHR2, which share no sequence similarity with the DH and PH domains of the Dbl family, respectively. The DHR1 domain shares low homology with the C2 motif and associates with the cellular membrane.23 The DHR2 domain is necessary and sufficient to catalyze the exchange of GDP to GTP on Rho GTPases.22 Recently, our laboratory identified a minimal C-terminal portion of the DHR2 domain of DOCK1, designated DHR2c, which exhibited full GEF activity towards the Rac1 GTPase.24
The DOCK180 family has been shown to activate Rac1 and/or Cdc42, but not RhoA. Regarding guanine nucleotide exchange on Rac1 and Cdc42, the DOCK proteins are classified into four subfamilies according to their specificity. The DOCK-A subfamily contains DOCK1, 2, and 5, while the DOCK-B subfamily consists of DOCK3 and DOCK4, all of which are Rac1-specific GEFs. DOCK9–11 comprise the DOCK-D subfamily and are Cdc42-specific GEFs. DOCK6–8 represent the DOCK-C subfamily and are thought to be capable of activating both Rac1 and Cdc42.25 Recent studies describe two DOCK:GTPase complex structures, Cdc42 bound to the DHR2 domain of DOCK9, and Rac1 bound to the corresponding region of DOCK2. These x-ray structures have helped to elucidate the mechanism by which DOCK180 family members activate Rho GTPases, as well as shedding light on the specificity of DOCK180 proteins and their points of contact.26,27 Two residues at positions 27 and 56 of Rac1 and Cdc42 appear to be key determinants of specificity for nucleotide exchange by DOCK180 proteins. Indeed, Wu, et al. showed that residues from the α10 helix of DOCK180 proteins play a key role in the selectivity that these GEFs exhibit towards Rac1 or Cdc42.24
While DOCK-C subfamily members have been proposed to activate both Rac1 and Cdc42, traditional in vitro GEF assays have failed to show significant GEF activity for DOCK-C members, such as DOCK6 and DOCK7.27–30 In vivo, the DOCK-C members are important in the regulation of neuronal cell development. For example, DOCK6 has been reported to regulate the early development of neurite formation of neuronal cells,28,31 whereas, DOCK7 has been demonstrated to be involved in axon formation by activating Rac1 and Cdc42.29,30 DOCK7 may also play a role in growth regulation and has been shown to interact with the TSC1/TSC2 complex, which suggests that DOCK7 may contribute to the regulation of the mTOR complex.32
Here we show that DOCK7 does not exhibit GEF activity toward non-isoprenylated Rac1 or Cdc42 in solution, but was capable of a robust activation of isoprenylated Rac1 and Cdc42 in a model liposome system. In addition, we identify two key residues in DOCK7 that convey GEF specificity and, when mutated, shift the DOCK7-DHR2 activity profile. Finally, we show that a shorter DHR2s domain of DOCK7 with an N-terminal site distinct from the GEF site, binds to the active forms of Rac1 and Cdc42. We propose that this distal site helps to tethers DOCK7 to the membrane, increasing its local concentration, which results in an enhanced rate of GTPase nucleotide exchange.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
The coding region of DOCK7 (accession No. from EMBL: DQ118679) was ligated into the Bac-to-Bac baculovirus expression vector pFastBac™HT C (Invitrogen). The cDNAs encoding the DOCK truncations: DHR2 (amino acid 1433–1992), DHR2s (amino acid 1530–1970), and DHR2c (amino acid 1688–1992), were inserted into the pFastBac™HT C vector for insect cell expression. The cDNAs encoding DHR2 and DHR2s were also cloned into the ppSUMO vector, as well as being inserted into pET28b, for E. coli expression. DOCK7-DHR2c mutants were generated with the QuickChange site-directed mutagenesis kit (Stratagene). The cDNAs encoding full-length Cdc42 and Rac1 were cloned into the pFastBac™HT C, pGEX-4T-1, and pET28a vectors.
Preparation of Insect Cell-Expressed Proteins
All the target proteins cloned into pFastBac™HT C vector were expressed in Spodoptera frugiperda (Sf21) cells. The expression of Cdc42 and Rac1 was carried out at Kinnakeet Biotechnology (Midlothian, VA), and the purification of these proteins was performed as described previously.10 The DOCK7 constructs were expressed in 0.5 L Sf21 cell suspension culture for 3 days after viral infection. The cells were pelleted by centrifugation at 500g (4°C) for 5 minutes and stored at −80°C for purification. The cell pellets were suspended in 20 mL of lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 2% Triton X-100) with protease inhibitor cocktail (Roche) and disrupted by Dounce homogenization. The lysates were spun down at 9000g for 20 minutes at 4°C. The supernatants were incubated with Ni-NTA agarose beads (Qiagen) for 30 minutes at 4°C. The beads were washed with 20 volumes of washing buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% CHAPS, 30 mM imidazole), and protein was eluted with 10 mL of elution buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% CHAPS, 200 mM imidazole). The fractions containing the target protein were concentrated to a volume of approximately 3 mL.
Preparation of E. coli-Expressed Proteins
The DOCK7 constructs which were cloned either into the pET28 or ppSUMO vector, and the Cdc42 and Rac1 constructs which were cloned into either the pGEX or pET28 vector, were expressed in E. coli. A single colony of E. coli BL21(DE3) or Rosetta 2 strain containing the target plasmid was inoculated in 5 mL of LB medium with 50 µg/mL kanamycin or 100 µg/mL carbenicillin (RPI) overnight at 37°C. The overnight culture was subsequently transferred to 1 L 2xYT medium with antibiotic and inoculated at 37°C until the OD600 reached 0.6, followed by induction with 0.1 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG) (RPI) at room temperature overnight. Bacteria were harvested by centrifugation at 4000g for 10 minutes at 4°C and stored at −80°C for future purification. The 6His-tagged DOCK7 pellets were suspended in lysis buffer (20 mM Tris, pH 8.0, 500 mM NaCl, 20 mM imidazole) with protease inhibitor cocktail (Roche), and lysed by sonication on ice. The lysates were centrifuged at 20000g for 30 minutes at 4°C, and the resulting supernatants were incubated with Ni-NTA agarose beads for 30 minutes at 4°C. The beads were extensively washed with washing buffer (20 mM Tris, pH 8.0, 500 mM NaCl, 40 mM imidazole), and proteins were eluted with 20 mL of elution buffer (20 mM Tris, pH 8.0, 500 mM NaCl, 200 mM imidazole).
For 6His-tagged Cdc42 and Rac1, the bacterial pellets were suspended in 25 mL of lysis buffer (20 mM Tris, pH 8.0, 500 mM NaCl, 5 mM MgCl2, 100 µM GDP, 20 mM imidazole) with protease inhibitor cocktail (Roche), and lysed by a 1 hour incubation on ice with 150 mg of lysozyme (Sigma), 45 mg of deoxycholic acid (Sigma), and DNase I (Roche). The purification procedures for the His-tagged GTPases were the same as for 6His-tagged DOCK7 proteins, except that all the buffers contained 5 mM MgCl2.
For GST-tagged Cdc42 and Rac1, bacterial pellets were suspended in 25 mL of lysis buffer (50 mM Tris, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 100 µM GDP, 1 mM DTT) with protease inhibitor cocktail, and lysed by a 1 hour incubation on ice with 150 mg of lysozyme, 45 mg of deoxycholic acid, and DNase I. The lysates were spun down and the supernatants were incubated with glutathione sepharose beads (Amersham Bioscience) at 4°C for 1 hour. The beads were washed with washing buffer (50 mM Tris, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT) and the protein was eluted with 20 mL of elution buffer (50 mM Tris, pH 8.0, 10 mM reduced glutathione, 5 mM MgCl2). 6His- and GST-tagged Cdc42 and Rac1 were stored in storage buffer (50 mM Tris, pH 7.5, 50 mM NaCl, 2 mM MgCl2, 0.5 mM DTT), concentrated to 200 µM, flash frozen by liquid nitrogen, and kept at −80°C for assays.
Preparation of Liposomes
All lipids used in this study were purchased from Avanti Polar Lipids. The model liposomes were comprised in molar percentages of 35% phosphatidylethanolamine, 35% cholesterol, 25% phosphatidylserine, and 5% phosphatidylinositol. Two approaches were used to prepare liposomes for different purposes. Rapid solvent exchange was used to make large liposomes that can be pelleted by low speed centrifugation.33 For the in vitro GEF assays, small lipid vesicles with 1 µm diameter were prepared by extrusion using Avanti mini-extruder.
Nucleotide-loading of Cdc42 and Rac1
Cdc42-Mant-GDP, Rac1-Mant-GDP, Cdc42-GTPγS, and Rac1-GTPγS were prepared by incubating insect cell-expressed isoprenylated Cdc42 or Rac1 with a 40-fold excess of Mant-GDP or a 10-fold excess of GTPγS in buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 0.1% CHAPS), together with 10 mM EDTA for 1.5 hours on ice, followed by the addition of 20 mM MgCl2 and a 0.5 hour incubation to quench the excess EDTA. Excess nucleotide was removed by Ni-column purification as described above. The eluted nucleotide-loaded Cdc42 and Rac1 were stored in 20 mM Tris, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 0.1% CHAPS and concentrated to 20 µM.
In vitro GEF Assays
Nucleotide exchange on Cdc42 or Rac1 was monitored by the fluorescence changes that accompanied the binding or dissociation of Mant nucleotide from these GTPases, by setting the excitation at 360 nm and monitoring emission at 440 nm. Using a Varian Eclipse fluorimeter, all samples were continuously stirred at 25°C in TBSM buffer (50 mM Tris, pH 7.5, 50 mM NaCl, 5 mM MgCl2) with 1 µM Mant nucleotide. For non-isoprenylated Cdc42 or Rac1, GST-Cdc42 or GST-Rac1 was added to the cuvette followed by the addition of DOCK7-DHR2s or DHR2c. For isoprenylated Cdc42 or Rac1, 6His-tagged Cdc42 or Rac1 was first incubated with liposomes for 15 minutes at room temperature, and then added to a cuvette to make a final bulk lipid concentration of 20 µM. DOCK7-DHR2s or DHR2c was added to initiate nucleotide exchange. To check whether the active form of Cdc42 or Rac1 influences the activation of Cdc42 or Rac1 by DOCK7, either DHR2s or DHR2c was incubated with Cdc42-GTPγS or Rac1-GTPγS for 15 minutes at room temperature before it was added to the reaction.
Liposome Centrifugation Assay
To assay the binding preference of DOCK7 to the three different forms of isoprenylated Cdc42 or Rac1 (nucleotide-free, GDP-bound, or GTP-bound), 0.2 nmol insect cell-expressed Cdc42 or Rac1 was incubated with 100 µL of lipids (1 µM) prepared by rapid solvent exchange for 10 minutes at room temperature, loaded with either GDP, or GTPγS, or not loaded, and then centrifuged at 15,000 ×g for 10 minutes. Supernatants were removed and lipid pellets were resuspended in 40 µL of TBSM buffer, followed by a 30 minute incubation at room temperature with 160 nmol DOCK7-DHR2s or DHR2c. The mixtures were spun down, and the supernatants and lipid pellets were examined by SDS-PAGE to examine the partitioning of DOCK7 between the liposomes versus the soluble fraction. The negative control contained lipids and DOCK7-DHR2s or DHR2c (without Cdc42 or Rac1).
GST-Cdc42/Rac1 Pull-down Assays
To check the binding of DOCK7 to non-isoprenylated Cdc42 or Rac1, E. coli-expressed GST-Cdc42 or GST-Rac1 (0.4 nmol) was prebound to 15 µL of glutathione sepharose beads in the presence of 10 mM EDTA. The negative control contained beads and DOCK7-DHR2s or DHR2c (without GST-Cdc42 or GST-Rac1). To prepare GDP- or GTPγS-preloaded GST-Cdc42 or GST-Rac1, excess GDP or GTPγS was added to samples and incubated for 15 minutes at room temperature, followed by the addition of 20 mM MgCl2 and 300 nM DOCK7-DHR2s or DHR2c. The samples were then incubated for 1 hour at 4°C. The beads were spun down, washed with TBSM (repeated twice), and examined by SDS-PAGE.
RESULTS
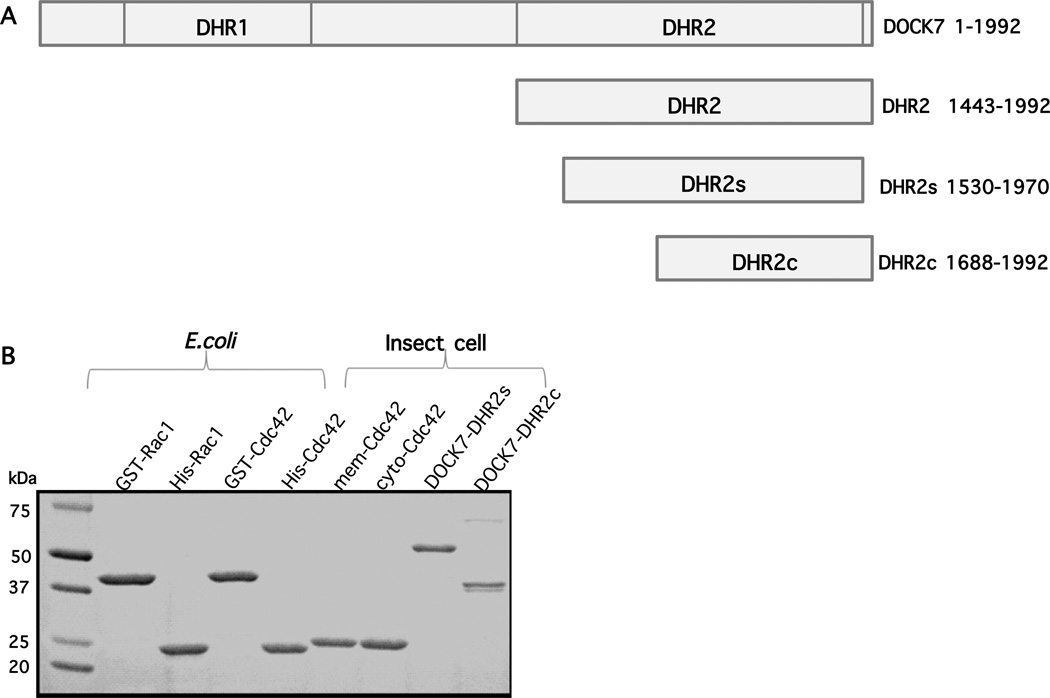
All DOCK180 family members have a C-terminal domain of approximately 500 amino acids, termed the DHR2 domain, which is necessary and sufficient for GEF activity.22 Previous studies have demonstrated that the E. coli recombinant full-length DHR2 domain of DOCK180 or DOCK9 was not stable in solution, whereas a slightly shorter version of DHR2 was sufficiently stable for GEF assays and crystallization.24,26 Therefore, we designed four different constructs for expression in insect cells to characterize the biochemical features of DOCK7 (Figure 1A). The full-length DOCK7 protein contains 1992 residues including 22 residues that are downstream from the DHR2 domain but aggregated and was difficult to express in usable quantities. The construct designated as DHR2s lacked 87 residues from the N-terminus of DHR2 and 22 amino acids from the C-terminus. DHR2c represents the C-terminal two-thirds of DHR2, which was designed to test whether DOCK7 possessed a similar domain capable of GEF activity as was earlier shown for DOCK180.24 In fact, we were able to express and purify DOCK7-DHR2s and DHR2c from insect cells (Figure 1B). As previously described, GST-tagged Rac1 and Cdc42 recombinant proteins were used for the GEF assays (Figure 1B). We also expressed 6His-tagged Rac1 and Cdc42 in E. coli as the counterparts of the same His-tagged constructs expressed in insect cells (Figure 1B). The Cdc42 and Rac1 proteins present in the membrane fractions of insect cells are isoprenylated, while the cytosolic fractions contain non-isoprenylated forms of these GTPases (Figure 1B).10 The isoprenylated, recombinant Cdc42 has been shown to bind to both insect cell membranes and reconstituted model liposomes.10
Figure 1.
Protein expression (A) Schematic representation of the DOCK7 and DHR2 constructs that were examined in this study. (B) SDS-PAGE and Colloidal Blue-staining of wild-type proteins used in this study.
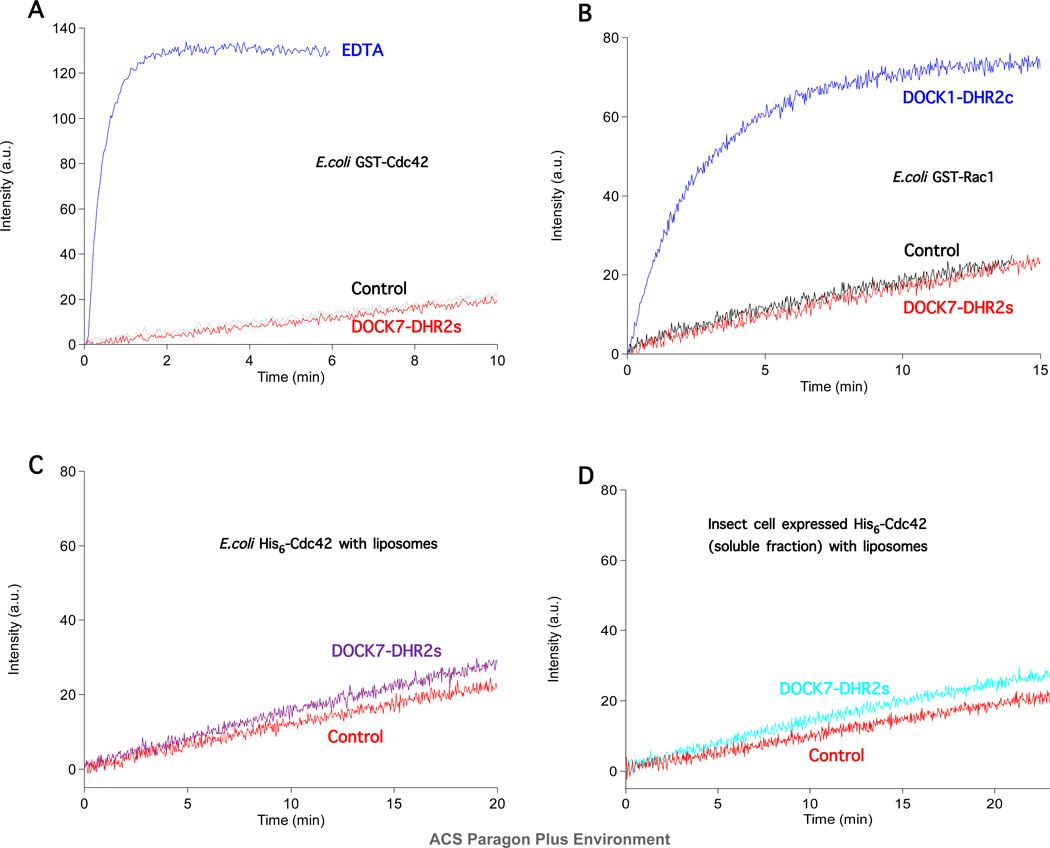
We first tested whether DOCK7-DHR2s or DOCK7-DHR2c could activate Cdc42 or Rac1. By monitoring the fluorescence changes accompanying the binding of Mant-GDP to these GTPases, we observed no detectable activation of either GST-Cdc42 or GST-Rac1 when assayed for Mant-GDP/GDP exchange in the presence of DOCK7-DHR2s in solution, as indicated by the data in Figures 2A–2D. Likewise, DOCK7-DHR2c did not initiate a noticeable activation of either GST-Cdc42 or GST-Rac1 (data not shown). However, the addition of EDTA caused a significant exchange of Mant-GDP for GDP on E. coli-expressed GST-Cdc42 (Figure 2A), as did the Rac1-specific GEF, DHR2c of DOCK1, when assaying E. coli-expressed GST-Rac1 (Figure 2B). To rule out that the GST tag might present an obstacle for the interaction between DOCK7-DHR2s or DOCK7-DHR2c with Cdc42 or Rac1, the replacement of GST-Cdc42 and GST-Rac1 by E. coli-expressed 6His-Cdc42 (Figure 2C) and 6His-Rac1, respectively, still resulted in no detectable activation by these domains. These data were consistent with the findings of another DOCK-C member, the DHR2 domain of DOCK6, which showed no activation of Cdc42 or Rac1 using the Mant-labeled nucleotide fluorescence assay.27 Although a weak activation of Rac1 and Cdc42 by the DHR2 domain of DOCK7 was detected by using a more sensitive GEF assay that monitored nucleotide exchange using radio-labeled GDP,29,30 these observations, when taken together with our findings, made it difficult to arrive at conclusions regarding the GTPase specificity of DOCK-C members as well as the conditions under which they are fully functional GEFs.
Figure 2.
DHR2s and DHR2c of DOCK7 do not catalyze the nucleotide exchange activity of non-isoprenylated Cdc42 or Rac1. (A) DOCK7-DHR2s (160 nM) was added at t=0 with Mant-GDP with or without bacterially expressed GST-Cdc42 (200 nM). The same concentration of Cdc42 without DHR2s was EDTA treated (10 mM) to demonstrate maximum Mant-GDP binding. (B) As in (A), DOCK7-DHR2s (160 nM) was added at t=0 with Mant-GDP (1 µM) with or without E. coli-expressed GST-Rac (200 nM). To test the exchange reactivity of Rac, 200 nM (final concentration) of E. coli-expressed GST-Rac was mixed with 200 nM (final concentration) of the DOCK1 exchange domain, DHR2c.24 (C) 20 µM liposomes (bulk concentration) were mixed with or without DOCK7-DHR2s (160 nM) and 1 mM Mant-GDP, with 200 nM E. coli-expressed 6His-Cdc42. (D) Insect cells expressing 6His-Cdc42 were separated into soluble and particulate fractions and Cdc42 purified from the soluble fraction (200 nM Cdc42) was tested for reactivity with E. coli-expressed DHR2s (200 nM).
Rho GTPases consist of the classical 5 α-helices and 6 β-pleated sheet architecture characteristic of all small GTPases as well as the α subunits of large heterotrimeric G-proteins. The C-termini of Rho GTPases contain a polybasic region (PBR) followed by the CAAX motif. Both the PBR and CAAX motif are important for the cellular functions of Rho GTPases.5,6 The positively charged Arg and Lys residues of the PBR are thought to facilitate the binding of Rho GTPases to cellular membranes, particularly those containing phosphoinositol headgroups. The attached isoprenyl tail, linked to the cysteine residue of the CAAX motif, can insert into membranes where the GTPases are able to engage their regulators and effectors. Since DOCK7-DHR2s or DOCK7-DHR2c could not promote significant activation of either Cdc42 or Rac1 in solution, we further investigated the isoprenylated forms Cdc42 and Rac1 in a model liposome system.
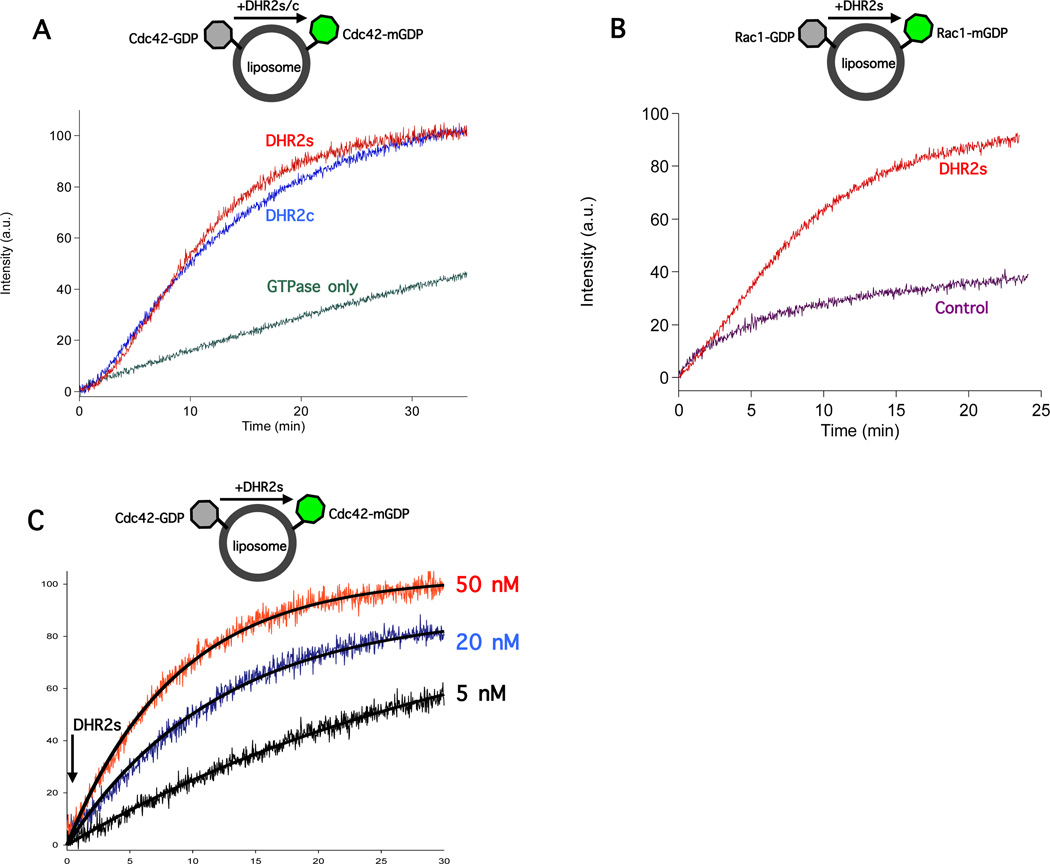
The insect cell-expressed geranylgeranylated Cdc42 was incubated with liposomes at room temperature to allow for membrane binding by the GTPase. Cdc42 was then able to bind Mant-GDP upon the addition of DOCK7 DHR2s or DHR2c (Figure 3A). Similarly, insect cell-expressed Rac1 was activated upon the addition of DOCK7-DHR2s (Figure 3B). The kinetics of exchange by DOCK7-DHR2s were dose-dependent and well described by a single pseudo-first order rate constant as shown for Cdc42 in Figure 3C. Although at 30 minutes the reactions at 20 nM and 5 nM DHR2s are incomplete, the fluorescence levels are eventually comparable to that for 50 nM DHR2s and this is borne out by the curve fitting results tabulated in the figure legend to Figure 3C. Regarding the inability of non-membrane bound GTPases to participate in DOCK-7 mediated nucleotide exchange, non-isoprenylated (i.e., E. coli-expressed) Cdc42, when suspended in the same liposome mixture, did not undergo nucleotide exchange in the presence of DOCK7-DHR2s (Figure 2C). Furthermore, the cytosolic (non-isoprenylated) fraction of insect cell-expressed Cdc42 did not exhibit any significant nucleotide exchange when assayed in the presence of DOCK7-DHR2s (Figure 2D). These data demonstrate that the isoprenylation and membrane localization of Cdc42 and Rac1 were essential for activation by DOCK7. Additionally, DOCK7-DHR2c was able to activate Cdc42 to an extent similar to the activation by DOCK7-DHR2s, indicating that the DHR2 domain of DOCK7 contains a C-terminal domain which is necessary and sufficient for activating Rho GTPases as observed for the DHR2 domain of DOCK180.24
Figure 3.
DOCK7-DHR2s and DOCK7-DHR2c activate isoprenylated Cdc42 and Rac1 in the presence of liposomes. 20 µM liposomes (bulk lipid concentration) pre-incubated with either added DOCK7-DHR2s (50 nM) (red) or DOCK7-DHR2c (50 nM) (blue) at the zero-time point to initiate the GDP to Mant-GDP exchange reaction with insect cell-expressed, isoprenylated Cdc42 (200 nM) (A) or Rac1 (200 nM) (B). No GEF was added in the control curves. (C) The activation of Cdc42 (200 nM) by DHR2s of DOCK7 was dose-dependent with the DHR2s concentrations indicated. Similar Ymax values for each condition were attained after sufficient reaction time. The exchange data were fit to a single exponential (Ft=Fmax[1−e(−k’t)] where Ft is the observed fluorescence at time t, Fmax is the fluorescence observed at the completion of exchange, and k’ is apparent first order rate constant). This yielded values for {F(max), k’(s−1)} of {100.8+/−2.5, 0.028+/−0.001)}, {89.4+/−0.3, 0.083+/−0.001} and {102.9+/−0.3, 0.115+/−0.001} for the 5 nM, 20 nM and 50 nM curves, respectively.
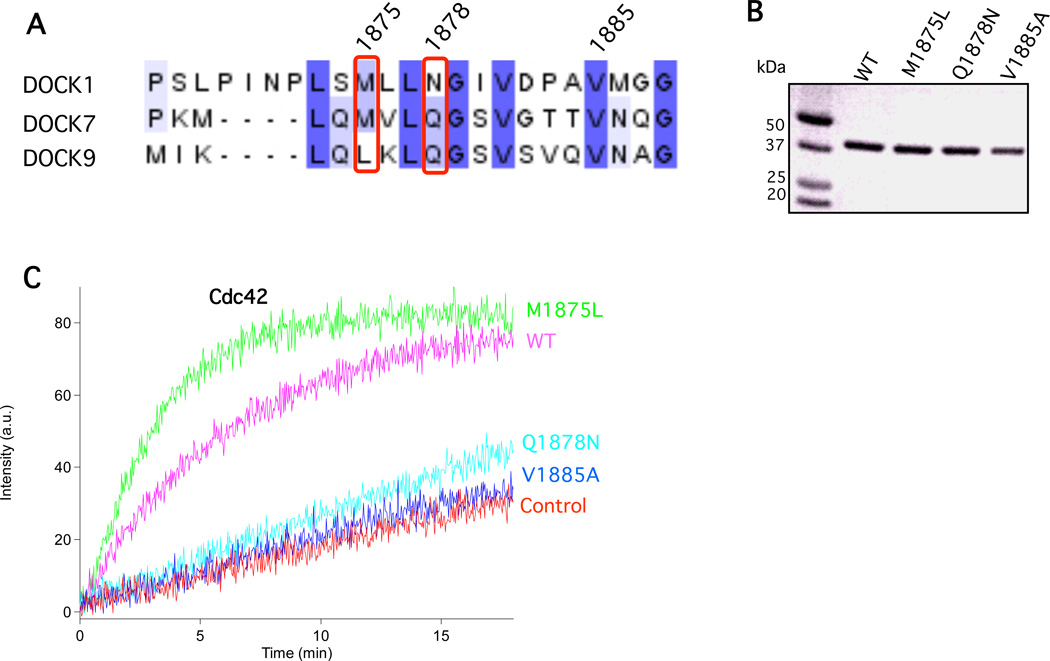
The recently solved x-ray structures for the Cdc42-DOCK9 and Rac1-DOCK2 complexes shed some light regarding the DOCK-GTPase specificity.26,27 Phe56 of Cdc42, and Trp56 of Rac1, are key specificity residues for the recognition by DOCK proteins. Substitution of Phe56 of Cdc42 with tryptophan yielded a dramatic decrease in DOCK9 GEF activity, while the W56F mutant of Rac1 showed a much lower degree of activation by DOCK180. Phe56 of Cdc42 interacts with residues Leu1955, Gln1958, and Gly1959 of DOCK9 (Figure 4A). Among them, Leu1955 is conserved in the DOCK-D (Cdc42-specific) subfamily, Gln1958 is invariant in both the DOCK-C and DOCK-D subfamilies, while Gly1959 is conserved in all the DOCK180 family members. Methionine is the corresponding residue in the DOCK-A/B (Rac-specific) and DOCK-C subfamilies to residue Leu1955 in DOCK9; whereas asparagine in the DOCK-A/B (Rac-specific) subfamily corresponds to residue Gln1958 in DOCK9. These alignments collectively suggest that the combination of leucine and glutamine in the DOCK-D subfamily encode Cdc42 specificity, and that the combination of methionine and asparagine in the DOCK-A/B subfamily determine Rac specificity. Therefore, the combination of methionine and asparagine in DOCK7 offered an explanation for its dual GEF function for both Cdc42 and Rac1. The M1875L mutant of DOCK7 was expected to convert it to a Cdc42-specific GEF while the Q1878N mutant should shift DOCK7 to a Rac-specific GEF. The DOCK7-DHR2c M1875L mutant, as well as the Q1878N mutant, were expressed and purified (Figure 4B). Confirming the importance of this residue for Cdc42-specific exchange, the DOCK7-M1875L mutant exhibited a more robust GEF activity toward Cdc42 than wild-type DOCK7-DHR2c (Figure 4C). Conversely, the DOCK7-DHR2c Q1878N mutant showed no significant GEF activity toward Cdc42 (Figure 4C) but rather an increased level of GEF activity towards Rac1 (data not shown). The DOCK7 GEF-defective V1885A mutant, as predicted, showed no activity (Figure 4C).
Figure 4.
Residues specific for Cdc42 and Rac nucleotide exchange activity. (A) Sequence comparison in the specificity encoding region for DOCK subfamilies A, C, and D members: DOCK1, DOCK7, and DOCK9, respectively. Numbered residues shown refer to the human DOCK7 sequence (Acc # AAZ38451). (B) SDS-PAGE and Colloidal Blue-staining of wild-type DOCK7-DHR2c and DOCK7-DHR2c mutants tested in this figure. (C) Substitution of methionine 1875 of DHR2c to leucine increases its GEF activity toward Cdc42; while mutation of glutamine 1878 of DHR2c to asparagine decreases its GEF activity toward Cdc42. The DHR2c protein concentration for each mutant was added to a final concentration of 40 nM at t=0. The concentration of insect cell-expressed isoprenylated Cdc42 was 200 nM for each trace with Mant-GDP present at 1 µM. The exchange-deficient DOCK7 mutant V1885A serves as a negative control as does liposome-bound Cdc42 alone. All traces are representative of 4 replicates of each condition and were repeated with at least two different protein purifications.
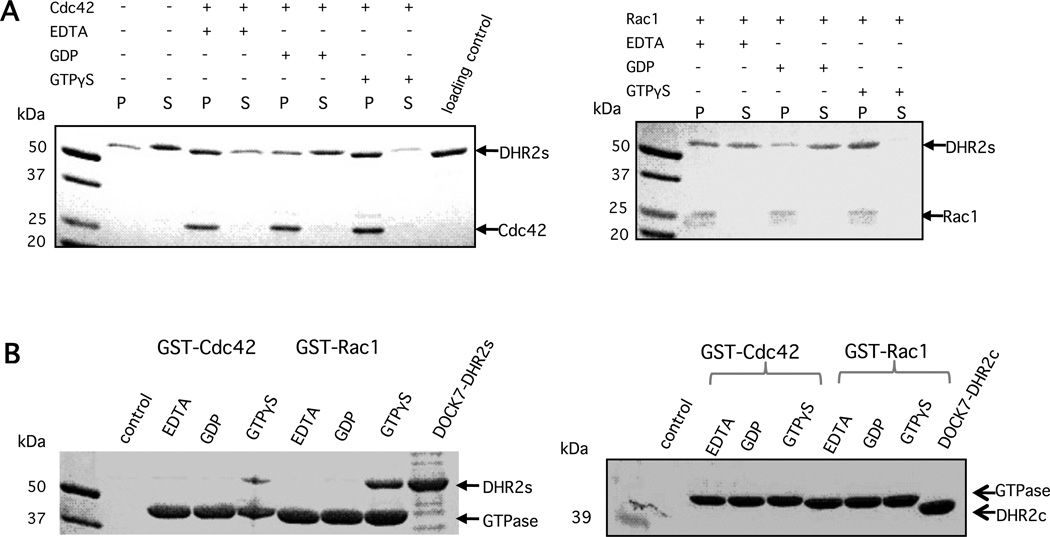
The DHR2s and DHR2c domains of DOCK7 showed similar GEF activity, but differed in their ability to interact with distinct nucleotide-bound states of the GTPases. When isoprenylated Cdc42 was bound to liposomes which were pelleted at a low speed, and then treated with either EDTA, GDP, or GTPγS, DOCK7-DHR2s appeared to bind to nucleotide-free and GTPγS-bound forms of Cdc42 with higher affinity than to Cdc42-GDP, upon comparing the relative amounts of DHR2s in the supernatant (unbound) and pellet (bound) fractions (Figure 5A, left panel). Additionally, when compared to nucleotide-free Cdc42, DHR2s appears to exhibit a slight preference for GTPγS-bound Cdc42 (Figure 5A, left panel). A similar behavior was observed with geranylgeranylated Rac1 (Figure 5A, right panel). However, DOCK7-DHR2c acted as a typical GEF and bound preferentially to liposomes containing the nucleotide-free forms of its cognate GTPases, again when comparing the relative amounts of DOCK7-DHR2s in the supernatant and pellet functions (data not shown). To test whether the DHR2c domain is responsible for binding the inactive forms of GTPases in order to catalyze nucleotide exchange, while the extra N-terminal portion of DHR2s is involved in the binding and recruitment of the active forms of the GTPases, we performed pulldown assays to examine the interactions of DOCK7-DHR2s and DOCK7-DHR2c, with GST-Cdc42 and GST-Rac1 (Figure 5B). When non-isoprenylated E. coli-expressed GST-Cdc42/Rac1 was bound to glutathione beads and subsequently treated with EDTA, GDP, or GTPγS, we only observed stable complexes between the DHR2s domain and the GTPγS-bound forms of Cdc42 or Rac1 (Figure 5B, left panel). We were unable to detect affinity precipitated complexes between DHR2c and any form of GST-Cdc42 or Rac1 (Figure 5B, right panel).
Figure 5.
DOCK7-DHR2s, but not DOCK7-DHR2c, preferentially binds to the active (GTP-bound) forms of Cdc42 and Rac1. (A) DOCK7-DHR2s was incubated with isoprenylated Cdc42 (left panel) or Rac1 (right panel) pre-bound to liposomes. EDTA-treated (nucleotide-free), and GDP- and GTPγS-bound forms of the GTPases are indicated. (B) DOCK7-DHR2s or DOCK7-DHR2c at 300 nM was incubated with either E. coli-expressed GST-Cdc42 or GST-Rac1 (0.4 nmol=20 µg) and affinity precipitated with glutathione-conjugated beads after loading the respective GTPases with GDP, GTPγS, or leaving them nucleotide-free.
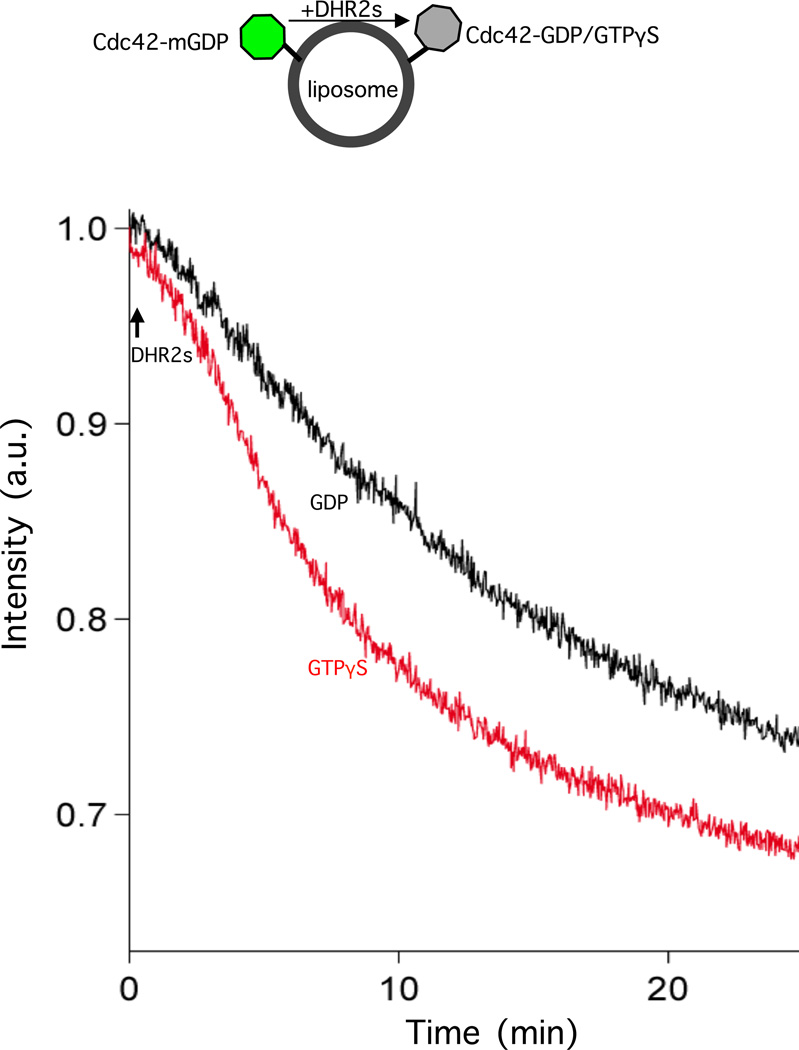
Sos, a Ras-specific GEF, has been shown to possess a second distal GTPase-binding site, which was distinct from the catalytic site. This distal site is specifically recognized by GTP-bound Ras, with the binding of activated Ras regulating the rate of GEF-catalyzed nucleotide exchange that results in a positive feedback activation of Ras.34 Likewise, we reported an interaction site for activated Cdc42 within the amino-terminal portion of DOCK11, distinct from the DHR2 domain.35 The interaction of GTP-bound Cdc42/Rac1 with DOCK7-DHR2s implied that DHR2s might harbor a distal site which interacts with Cdc42/Rac1-GTP, providing for a potential positive feedback mechanism in the activation of Rac1 and Cdc42 by DOCK7. In fact, we have found that Cdc42 pre-loaded with Mant-GDP (i.e. Cdc42-mGDP) shows a modest but consistent acceleration in Mant-GDP dissociation when exchanged with excess GTPγS, compared to GDP, upon the addition of DOCK7-DHR2s (Figure 6).
Figure 6.
DOCK7-DHR2s activates Cdc42 with a nucleotide-dependent exchange rate that is accelerated by GTPγS. Cdc42 (200 nM final concentration) pre-loaded with Mant-GDP, and then separated from excess Mant-GDP, was mixed with 50 µM unlabeled GDP or GTPγS prior to the addition of DHR2s (160 nM final concentration; at t=0 indicated by arrow).
DISCUSSION
The DOCK180 family members are important regulators which have been demonstrated to impact multiple cellular processes, including cell migration, cell phagocytosis, cytoskeleton reorganization,36–41 and neuronal differentiation.29–31 They function as upstream activators (GEFs) of Cdc42 and/or Rac1, two GTPases that are critical players in actin dynamics as well as in cell growth regulation. The recent structural and functional studies of the DOCK9-Cdc42 and DOCK2-Rac1 complexes have provided insights regarding the mechanisms used by DOCK180 members to activate their cognate GTPases, and how these GEFs recognize their specific GTPase targets. The Rac1-specific DOCK-A member DOCK2, and the Cdc42-specific DOCK-D member DOCK9/Zizimin, make similar contacts and presumably use the same mechanism to activate Rac1 and Cdc42, respectively. DOCK-C subfamily members make similar, though less discriminating contacts, with their GTPase targets, given that replacing the strictly conserved catalytic residue Val1885 of DHR2c from DOCK7 with alanine abolished its GEF activity (Figure 4C).
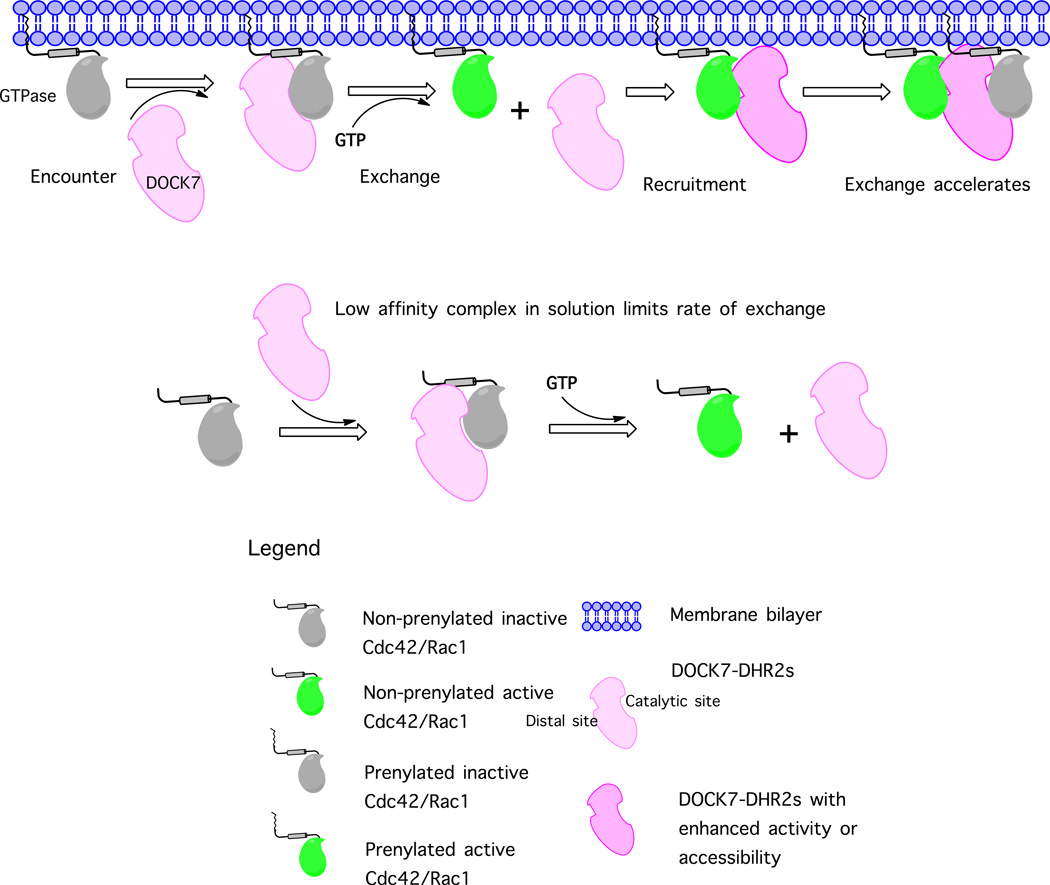
The GEF activity and specificity of the DOCK-A and the DOCK-D subfamilies have been well studied.22,42 However, the biochemical characteristics of the DOCK-C subfamily members, such as DOCK7, are less well understood, primarily because Dock-C members fail to display significant GEF activity toward Cdc42 or Rac1 in solution using in vitro GEF assays. In this study, we now show that both the DHR2s and DHR2c domains of DOCK7 exhibit robust GEF activity towards prenylated Cdc42 or Rac1, but only in the presence of membranes (Figure 2). DOCK7 was able to stimulate the activation of GTPases at nanomolar concentrations in the presence of liposomes, whereas it was unable to do so even at micromolar concentrations in solution. This suggests that DOCK7 is active only at the membrane surface, where the local concentrations of GTPase and GEF are sufficiently high to permit appreciable amounts of the GEF-GTPase complex to form (Figure 7). These observations are reminiscent of studies of the GEF activity catalyzed by the Ras-GEF Sos, which demonstrated the necessity for the farnesylation of Ras to facilitate Sos-mediated nucleotide exchange.43,44
Figure 7.
Model for Cdc42/Rac1 activation by DOCK7 via a recruitment feed-back loop. In solution, DHR2s of DOCK7 could not stimulate the activation of Cdc42 or Rac1, whereas in liposomes, DHR2s initiated nucleotide exchange on Cdc42 or Rac1. Addition of Cdc42-GTPγS accelerated the nucleotide exchange reaction.
Amino acid 56 of Cdc42 and Rac1 (phenylalanine and tryptophan, respectively) has been identified as the key residue responsible for the specific recognition by DOCK180 proteins.24,27 The side-chain of residue 56 of Cdc42 or Rac1 inserts into a binding pocket of the specific DOCK180 family member and interacts with the side-chains of residues in the pocket, including Leu1955 and Gln1958 of DOCK9 for Cdc42, or Met1529 and Asn1532 of DOCK2 for Rac1. Supporting the critical role for the Met(DOCK9):Trp(Rac) or the Leu(DOCK2):Phe(Cdc42) interaction specificity in Rac-specific nucleotide exchange, the replacement of Met1524 of DOCK1 can restore about 25% of the full activation of the Rac1 W56F mutant.24 The smaller side-chain of Phe56 of Cdc42 favors the bulkier side-chains of Leu1955 and Gln1958 of DOCK9, whereas the bulkier side-chain Trp56 of Rac1 is complementary to the smaller side-chains of Met1529 and Asn1532 of DOCK2 (or Met1524 and Asn1527 of DOCK1). Therefore, substitution of Phe56 of Cdc42 with tryptophan results in steric hindrance in the pocket of DOCK9, whereas substitution of Trp56 of Rac1 with phenylalanine is unable to make sufficient contact with DOCK2 or DOCK1. How then does DOCK7 recognize both Cdc42 and Rac1? It appears that DOCK7 combines Met1875 from the DOCK-A subfamily and Gln1878 from the DOCK-D subfamily at the corresponding positions, which is sufficient to make contact with Phe56 of Cdc42, but without the steric hindrance with Trp56 of Rac1. By virtue of this hybrid design, DOCK7 is capable of the dual recognition of both Cdc42 and Rac1 at the cost of binding affinity for either GTPase. This point is further underscored by the effect of the M1875L and Q1878N mutants of DOCK7, as these substitutions result in a respective increase or decrease in nucleotide exchange activity towards Cdc42 when compared to wild-type DHR2s (Figure 4C).
Previously, we identified a C-terminal region (designated DHR2c) of DOCK1, which exhibited comparable GEF activity to the full-length DHR2.24 From the data presented here, it is clear that DOCK7 possesses a similar DHR2c nucleotide exchange domain, which exhibits comparable catalytic activity as DHR2s (Figure 3A). The N-terminus of DOCK9-DHR2 has been shown to contribute slightly to GEF activity,26 although it is not clear that all the DOCK180-related members require this smaller domain in order to fully activate their cognate GTPase. However, in contrast to DHR2c, the longer DHR2s of DOCK7 stably binds to the active forms of both Rac and Cdc42, which suggests the potential for a membrane translocation-based positive cooperativity resulting from the progressive recruitment of DHR2s to the liposome surface (Figure 7). The ability of GTP-bound Cdc42 to interact with DOCK7 is somewhat reminiscent of our earlier work with DOCK11.35 In that case, the active form of Cdc42 was shown to form a stable complex with the N-terminal portion of DOCK11. This in turn enabled an additional interaction to occur between GTP-bound Cdc42 and the DHR2 domain of DOCK11, thereby leading to a positive-feedback activation of Cdc42. A similar set of interactions may be occurring between GTP-bound Cdc42 and DOCK7. While the amino-terminal portion of the DHR2s domain of DOCK7 is capable of forming a stable complex with activated Cdc42, a secondary interaction between GTP-bound Cdc42 and the smaller DHR2c might also occur, as we have observed some degree of positive cooperativity when assaying the ability of this more limited domain to stimulate nucleotide exchange on Cdc42. A more detailed picture of how GTP-bound Cdc42 associates with these DOCK7 domains awaits high resolution structural information.
Some form of positive feedback mechanism may also underlie the activities of Dbl-family GEFs, whereby, for example, the PH domain of PDZ-RhoGEF has been demonstrated to bind the active form of RhoA at a proposed distal, effector-binding site.45 Specifically, the DH-PH domain of PDZ-RhoGEF can form a ternary complex with the active form of RhoA bound to the PH domain, and with the inactive form of RhoA bound to the DH domain. However, the binding by activated RhoA does not affect the GEF activity of PDZ-RhoGEF in vitro. It is possible that, in order to enhance the accessibility of GEFs by concentrating them at the membrane surface, cells use this spatial regulatory strategy to recruit GEFs to membranes through their ability to bind to the active forms of isoprenylated Rho GTPases. It therefore remains of fundamental importance to delineate the events leading to full DOCK GEF activity as evidence accumulates that DOCK family members are recruited by the active forms of their cognate GTPases to the membrane surface, providing for the spatial and temporal regulation of their abilities to catalyze guanine nucleotide exchange.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM040654 and GM047458).
Abbreviations
- DH
Dbl homology
- DHR2
DOCK homology region 2
- GAP
GTPase activating protein
- GEF
guanine nucleotide exchange factor
- PBR
polybasic region
- PH
pleckstron homology
- Sf21
Spodoptera frugiperda 21 cells
- Sos
Son-of-sevenless
REFERENCES
- 1.Cerione RA. Cdc42: new roads to travel. Trends Cell Biol. 2004;14:127–132. doi: 10.1016/j.tcb.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Chardin P. The ras superfamily proteins. Biochimie. 1998;70:865–868. doi: 10.1016/0300-9084(88)90226-x. [DOI] [PubMed] [Google Scholar]
- 3.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 4.Tapon N, Hall A. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr. Opin. Cell Biol. 1997;9:86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- 5.Williams CL. The polybasic region of Ras and Rho family small GTPases: a regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell. Signal. 2003;15:1071–1080. doi: 10.1016/s0898-6568(03)00098-6. [DOI] [PubMed] [Google Scholar]
- 6.Roberts PJ, Mitin N, Keller PJ, Chenette EJ, Madigan JP, Currin RO, Cox AD, Wilson O, Kirschmeier P, Der CJ. Rho family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J. Biol. Chem. 2008;283:25150–25163. doi: 10.1074/jbc.M800882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sebti SM, Der CJ. Opinion: Searching for the elusive targets of farnesyltransferase inhibitors. Nat. Rev. Cancer. 2003;3:945–951. doi: 10.1038/nrc1234. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman GR, Cerione RA. Signaling to the Rho GTPases: networking with the DH domain. FEBS Lett. 2002;513:85–91. doi: 10.1016/s0014-5793(01)03310-5. [DOI] [PubMed] [Google Scholar]
- 9.Nassar N, Hoffman GR, Manor D, Clardy JC, Cerione RA. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat. Struct. Biol. 1998;5:1047–1052. doi: 10.1038/4156. [DOI] [PubMed] [Google Scholar]
- 10.Johnson JL, Erickson JW, Cerione RA. New insights into how the Rho guanine nucleotide dissociation inhibitor regulates the interaction of Cdc42 with membranes. J. Biol. Chem. 2009;284:23860–23871. doi: 10.1074/jbc.M109.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hart MJ, Eva A, Evans T, Aaronson SA, Cerione RA. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the dbl oncogene product. Nature. 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- 12.Takai S, Hasegawa H, Kiyokawa E, Yamada K, Kurata T, Matsuda M. Chromosomal mapping of the gene encoding DOCK180, a major Crk-binding protein, to 10q26.13-q26.3 by fluorescence in situ hybridization. Genomics. 1996;35:403–404. doi: 10.1006/geno.1996.0378. [DOI] [PubMed] [Google Scholar]
- 13.Hasegawa H, Kiyokawa E, Tanaka S, Nagashima K, Gotoh N, Shibuya M, Kurata T, Matsuda M. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol. Cell. Biol. 1996;16:1770–1776. doi: 10.1128/mcb.16.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson JW, Cerione RA. Structural elements, mechanism, and evolutionary convergence of Rho protein-guanine nucleotide exchange factor complexes. Biochemistry. 2004;43:837–842. doi: 10.1021/bi036026v. [DOI] [PubMed] [Google Scholar]
- 15.Worthylake DK, Rossman KL, Sondek J. Crystal structure of the DH/PH fragment of Dbs without bound GTPase. Structure. 2004;12:1078–1086. doi: 10.1016/j.str.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Feng Q, Albeck JG, Cerione RA, Yang W. Regulation of the Cool/Pix proteins: key binding partners of the Cdc42/Rac targets, the p21-activated kinases. J. Biol. Chem. 2002;277:5644–5650. doi: 10.1074/jbc.M107704200. [DOI] [PubMed] [Google Scholar]
- 17.Feng Q, Baird D, Cerione RA. Novel regulatory mechanisms for the Dbl family guanine nucleotide exchange factor Cool-2/alpha-Pix. EMBO J. 2004;23:3492–3504. doi: 10.1038/sj.emboj.7600331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baird D, Feng Q, Cerione RA. The Cool-2/alpha-Pix protein mediates a Cdc42-Rac signaling cascade. Curr. Biol. 2005;15:1–10. doi: 10.1016/j.cub.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 19.Lemmon MA, Ferguson KM. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- 20.Snyder JT, Rossman KL, Baumeister MA, Pruitt WM, Siderovski DP, Der CJ, Lemmon MA, Sondek J. Quantitative analysis of the effect of phosphoinositide interactions on the function of Dbl family proteins. J. Biol. Chem. 2001;276:45868–45875. doi: 10.1074/jbc.M106731200. [DOI] [PubMed] [Google Scholar]
- 21.Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, Macara IG, Madhani H, Fink GR, Ravichandran KS. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO compelx. Nat. Cell Biol. 2002;4:574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 22.Cote JF, Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 2002;115:4901–4913. doi: 10.1242/jcs.00219. [DOI] [PubMed] [Google Scholar]
- 23.Cote JF, Motoyama AB, Bush JA, Vuori K. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 2005;7:797–807. doi: 10.1038/ncb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, Ramachandran S, Lin MC, Cerione RA, Erickson JW. A minimal Rac activation domain in the unconventional guanine nucleotide exchange factor Dock180. Biochemistry. 2011;50:1070–1080. doi: 10.1021/bi100971y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cote JF, Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 2007;17:383–393. doi: 10.1016/j.tcb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Zhang Z, Roe SM, Marshall CJ, Barford D. Activation of Rho GTPases by DOCK exchange factors is mediated by a nucleotide sensor. Science. 2009;325:1398–1402. doi: 10.1126/science.1174468. [DOI] [PubMed] [Google Scholar]
- 27.Kulkarni K, Yang J, Zhang Z, Barford D. Multiple factors confer specific Cdc42 and Rac protein activation by dedicator of cytokinesis (DOCK) nucleotide exchange factors. J. Biol. Chem. 2011;286:25341–25351. doi: 10.1074/jbc.M111.236455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyamoto Y, Yamauchi J, Sanbe A, Tanoue A. Dock6, a Dock-C subfamily guanine nucleotide exchanger, has the dual specificity for Rac1 and Cdc42 and regulates neurite outgrowth. Exp. Cell Res. 2007;313:791–804. doi: 10.1016/j.yexcr.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 29.Watabe-Uchida M, John KA, Janas JA, Newey SE, Van Aelst L. The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of stathmin/Op18. Neuron. 2006;51:727–739. doi: 10.1016/j.neuron.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 30.Yamauchi J, Miyamoto Y, Chan JR, Tanoue A. ErbB2 directly activates the exchange factor Dock7 to promote Schwann cell migration. J. Cell. Biol. 2008;181:351–365. doi: 10.1083/jcb.200709033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyamoto Y, Yamauchi J. Cellular signaling of Dock family proteins in neural function. Cell Signal. 2010;22:175–182. doi: 10.1016/j.cellsig.2009.09.036. [DOI] [PubMed] [Google Scholar]
- 32.Nellist M, Burgers PC, van den Ouweland AM, Halley DJ, Luider TM. Phosphorylation and binding partner analysis of the TSC1-TSC2 complex. Biochem. Biophys. Res. Commun. 2005;333:818–826. doi: 10.1016/j.bbrc.2005.05.175. [DOI] [PubMed] [Google Scholar]
- 33.Buboltz JT, Feigenson GW. A novel strategy for the preparation of liposomes: rapid solvent exchange. Biochim. Biophys. Acta. 1999;1417:232–245. doi: 10.1016/s0005-2736(99)00006-1. [DOI] [PubMed] [Google Scholar]
- 34.Margarit SM, Sondermann H, Hall BE, Nagar B, Hoelz A, Pirruccello M, Bar-Sagi D, Kuriyan J. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112:685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 35.Lin Q, Yang W, Baird D, Feng Q, Cerione RA. Identification of a DOCK180-related guanine nucleotide exchange factor that is capable of mediating a positive feedback activation of Cdc42. J. Biol. Chem. 2006;281:35253–35262. doi: 10.1074/jbc.M606248200. [DOI] [PubMed] [Google Scholar]
- 36.Wu YC, Horvitz HR. C. elegans phagocytosis and cell-migration protein CED5 is similar to human DOCK180. Nature. 1998;392:501–504. doi: 10.1038/33163. [DOI] [PubMed] [Google Scholar]
- 37.Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, Klibanov AL, Mandell JW, Ravichandran KS. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Wu YC, Fadok VA, Lee MC, Gengyo-Ando K, Cheng LC, Ledwich D, Hsu PK, Chen JY, Chou BK, Henson P, Mitani S, Xue D. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science. 2003;302:1563–1566. doi: 10.1126/science.1087641. [DOI] [PubMed] [Google Scholar]
- 39.Wu YC, Tsai MC, Cheng LC, Chou CJ, Weng NY. C. elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev. Cell. 2001;1:491–502. doi: 10.1016/s1534-5807(01)00056-9. [DOI] [PubMed] [Google Scholar]
- 40.Henson PM. Engulfment: ingestion and migration with Rac, Rho and TRIO. Curr. Biol. 2005;15:R29–R30. doi: 10.1016/j.cub.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 41.Yajnik V, Paulding C, Sordella R, McClatchey AI, Saito M, Wahrer DC, Reynolds P, Bell DW, Lake R, van den Heuvel S, Settleman J, Haber DA. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112:673–684. doi: 10.1016/s0092-8674(03)00155-7. [DOI] [PubMed] [Google Scholar]
- 42.Kwofie MA, Skowronski J. Specific recognition of Rac2 and Cdc42 by DOCK2 and DOCK9 guanine nucleotide exchange factors. J. Biol. Chem. 2008;283:3088–3096. doi: 10.1074/jbc.M705170200. [DOI] [PubMed] [Google Scholar]
- 43.Porfiri E, Evans T, Chardin P, Hancock JF. Prenylation of Ras proteins is required for efficient hSOS1-promoted guanine nucleotide exchange. J. Biol. Chem. 1994;269:22672–22677. [PubMed] [Google Scholar]
- 44.Pechlivanis M, Ringel R, Popkirova B, Kuhlmann J. Prenylation of Ras facilitates hSOS1-promoted nucleotide exchange, upon Ras binding to the regulatory site. Biochemistry. 2007;46:5341–5348. doi: 10.1021/bi602353k. [DOI] [PubMed] [Google Scholar]
- 45.Chen Z, Medina F, Liu MY, Thomas C, Sprang SR, Sternweis PC. Activated RhoA binds to the pleckstrin homology (PH) domain of PDZ-RhoGEF, a potential site for autoregulation. J. Biol. Chem. 2010;285:21070–21081. doi: 10.1074/jbc.M110.122549. [DOI] [PMC free article] [PubMed] [Google Scholar]