Abstract
This study deals with the underlying causes of failure of chloroquine in the treatment of Plasmodium falciparum infection in some malaria-endemic regions of India. Samples were collected from 141 patients in Purulia from March of 2007 to April of 2008. In vitro drug susceptibility tests, parasitic DNA isolation followed by polymerase chain reaction, and restriction fragment-length polymorphisms of different codons of the pfcrt gene (76) and pfmdr-I genes (86, 1042, and 1246) were assessed. The responses of 141 patients to chloroquine were determined. Prevalence of double pfmdr-I (58.16%) mutation (86Y+1246Y) and some (14.89%) single pfcrt mutations with triple pfmdr-I mutation (76T+86Y+1042D+1246Y) were found. Interestingly, double pfmdr-I mutation (86Y and 1246Y codons) was observed with the early treatment failure cases. These results show, for the first time in India that in vitro chloroquine resistance and in vivo chloroquine treatment failure were caused by double pfmdr-I (P < 0.001) mutation.
Introduction
Malaria is one of the major public health problems in our country. Around 1.5 million confirmed cases are reported per annum by the National Vector Borne Disease Control Program (NVBDCP), of which 40–50% of cases are caused by Plasmodium falciparum.1 Drug resistance to P. falciparum is the major factor for death of malaria.2 The spread of multidrug-resistant P. falciparum is a serious worldwide problem considering the limited number of drugs available, the lack of vaccine, and the morbidity and mortality impact of malaria.3 In India, extensive and haphazard use of chloroquine (CQ) for more than five decades in treatment of P. falciparum malaria has resulted in the emergence of CQ- resistant P. falciparum malaria.4 Understanding the molecular mechanisms in drug-resistant malaria is of utmost importance for both designing new drugs and providing molecular markers to monitor drug activity and treatment efficacy.3 Current molecular studies of P. falciparum isolates suggest that few gene loci are associated with CQ resistance to P. falciparum. These genes have been named as pfcrt and pfmdr-I. Point mutations in the pfcrt gene were found to be associated with in vitro CQ resistance in P. falciparum isolates from Africa, South America, and South East Asia.5,6 In addition, the pfmdr-I gene located on chromosome 5 has mutations that can play a significant role in P. falciparum resistance to various antimalarials, such as mefloquine, quinine, and artemisinin derivatives.7 Specific combinations of pfcrt and pfmdr-I alleles, resulting in varying responses to CQ, seems geographically restricted,8 which may explain why some field studies reported an association between pfmdr-I polymorphisms and CQ resistance,9,10 whereas other studies did not.11 Studies from different geographical areas of the world suggest that the point mutation of asparagine to tyrosine in codon 86 (N86 to 86Y) of the pfmdr-I gene is associated with CQ resistance.9 Several other pfmdr-I polymorphisms, like 1042D and 1246Y, were implicated to varying degrees in CQ resistance.10 In India, it has been reported that the pfcrt 76T mutation of P. falciparum causes CQ resistance but not the multidrug-resistant gene of P. falciparum malaria.12,13
In India, one of the malaria-endemic zones is situated in the most remote corner of Purulia district (Bandwan Block; geographical coordinates are 22°47′0″ N and 86°30′0″ E) surrounded by deep forest, where the largest number of poor tribal people resides. CQ and sometimes quinine have been used here for more than five decades against P. falciparum malaria. Therefore, the drug pressure of CQ has gone up, and the drug efficacy has declined. Hence, the present investigation was conducted to determine the cause of in vivo CQ clinical failure and in vitro CQ resistance in this part of India.
Materials and methods
Selection of subjects.
The criteria needed to conduct the experiments included history of fever during the past 24 hours and monoinfection with P. falciparum based on the microscopic examination of Giemsa-stained thin and thick blood smears. Additionally, a rapid diagnostic test based on the detection of Plasmodium-specific lactate dehydrogenase (pLDH; OptiMAL-DT) was used with a range of parasite density of 1,000–200,000 asexual parasites/μL blood, and subjects could not have a recent history of self-medication with antimalarial drugs. Patients with signs and symptoms of severe and complicated malaria, as defined by the World Health Organization (WHO), were excluded.
Collection of sample.
The study was carried out from March of 2007 to April of 2008 before the launch of artemisinin combination therapy (ACT) by the NVBDCP. Parasite-infected blood was taken from 184 patients using ethylene diamine tetra acetic acid (EDTA)-coated vacutainer tubes; the majority of patients come from a tribal race. Giemsa-stained blood smears were examined to check for the monoinfection with P. falciparum. Finally, 141 patients were enrolled in this study. The samples were processed for in vitro assays, and aliquots were stored at −20°C before genomic DNA extraction. Informed consent was obtained from the respective patient or the patient's guardians for adult and child patients. The experimental protocol of this study was followed as per WHO guidelines and was duly approved by the Institutional Ethical Committee.
In vivo drug testing.
The new standard 28-day test of therapeutic efficacy developed by the WHO was used in this study.14 Patients with positive rapid diagnostic test results and microscopically confirmed uncomplicated P. falciparum malaria were randomized with CQ (Resochin; Bayer) initially at 10 mg/kg body weight on day 0 followed by a double dose of 5 mg/kg body weight on day 1. On day 2, a single dose of 5 mg/kg body weight was administered. After pLDH confirmation at day 0, patients were given as a single dose of 10 mg/kg body weight; therefore, day 0 is day 1 for drug treatment. Additionally, day 1 can be treated as day 2, and day 2 can be treated as day 3. Large numbers of patients (31%) were below the age of 6 years, and some patients were pregnant women. Therefore, the doctors preferred a double dose of 5 mg/kg body weight instead of a single dose of 10 mg/kg body weight on day 2. The clinical conditions and parasite density were monitored on days 0, 1, 2, 3, 7, 14, 21, and 28. Blood samples were obtained by finger pricking or intravenous blood draw on enrollment and all follow-up days, including any unscheduled day to use for analysis of thick and thin blood smears. Aliquots were stored at −20°C for additional molecular analysis. The therapeutic responses were classified as adequate clinical and parasitological response (ACPR), early treatment failure (ETF), late treatment failure (LTF), and late parasitological failure (LPF) according to the criteria adopted by the WHO. The patients not responding to CQ treatment were treated with ACT (artesunate + sulfadoxine-pyrimethamine [SP]).
In vitro drug sensitivity assay.
In vitro drug sensitivity assays were performed according to the method in the work by Trager and Jensen15 on the clinical isolates, with prior adaptation to the in vitro culture conditions for 5–7 days until the parasitemia reaches > 0.8–1.0%. Infected erythrocytes were suspended in the complete folate and p-amino benzoic acid free Roswell Park Memorial Institute (RPMI) 1640 medium consisting of 0.5% Albumax II, 25 mM N-2-hydroxy ethylpiperazine-N-2 ethane sulphonic acid (HEPES), 25 mM NaHCO3, 25 μg/mL gentamicin, and 0.2% hypoxanthine at a hematocrit of 1.5% and an initial parasitemia of 0.2–1.0%. If the blood sample contained a parasitemia > 1.0%, fresh uninfected O+ erythrocytes were added to adjust the parasitemia from 0.8% to 1.0%. The one-half maximal inhibitory concentration (IC50) is a measure of the effectiveness of a compound in inhibiting biological function. Sterile RPMI 1640 was used to prepare stock solutions and dilutions of CQ (CQ phosphate; Sigma) for detecting the IC50. The final concentrations ranged from 10 to 10,000 nM for CQ. Finally, 200 μL/well suspension of parasitized erythrocytes were distributed in microculture plates (WHO plate), and 25 μL each concentration of CQ were distributed in each well. Two wells (without drug) were used as controls for the experiment, and each concentration was studied in duplicate or triplicate. Plates were incubated for 48 h at 37°C in an atmosphere of 5% O2, 5% CO2, and 90% N2 and a relative humidity of 95%. Plates were then frozen and kept at −20°C; IC50 was determined using tritiated hypoxanthine uptake assay.16 The calculation was based on non-linear regression analysis of the logarithm of concentrations plotted against the percentage of growth inhibition. Two culture-adapted cloned strains of P. falciparum (CQ-sensitive strain 3D7and CQ-resistant Dd2 strains) were used for quality controls. Reference strains were cryopreserved and thawed before each measurement.
Isolation of parasitic DNA.
Erythrocytes were separated from the patients' blood, and the parasitic DNA was extracted as described.16 The extracted DNA was air dried, resuspended in Tris EDTA (TE) buffer (10 mM Tris, 1 mM EDTA), and stored at −20°C until use. The DNA was quantified by agarose gel electrophoresis and spectrophotometrically by calculating the A260/A280 ratios and the A260 values to determine protein impurities and DNA concentrations.
Polymerase chain reaction/restriction fragment-length polymorphism analysis of pfcrt and pfmdr-I.
The regions of the pfcrt and pfmdr-I genes surrounding the polymorphisms of interest were amplified by polymerase chain reaction (PCR) using the Eppendorf thermal cycler under the following conditions: approximately 200 ng genomic DNA, 15 pmol primers, reaction buffer (10 mM Tris, 50 mM KCl, pH 8.3), 2.5 mM MgCl2, 250 μM 2′-deoxynucleoside 5′-triphosphate (dNTP), and 1 unit Taq DNA polymerase (Roche Applied Science) in a 25-μL reaction mixture at different reaction conditions for different genes. Primers were designed (Table 1) on the basis of the complete P. falciparum Dd2/Indochina strain sequence (accession number AF030694) available in GenBank.16,17 Single nucleotide polymorphisms of the pfcrt and pfmdr-I genes at their specific codons were determined by enzymatic digestion of specific restriction enzymes. In the pfcrt gene, the ApoI (New England Biolabs) enzyme digested the amplicon (10 μL) at 50°C for 1 hour and identified the wild-type 76 lysine. Digestion with AflIII enzyme at 37°C for 1 hour recognized the mutant tyrosine amino acid at codon 86 of the pfmdr-I gene, whereas the AsnI (New England Biolabs) enzyme identified the asparagine amino acid at codon 1042 of the pfmdr-I gene. The EcoRV (New England Biolabs) enzyme detected the mutant tyrosine at 1246 codon after 1 hour of digestion at 37°C.17 DNA fragments were separated by 1.2% agarose gel electrophoresis and visualized under an ultraviolet transilluminator after staining with ethidium bromide. In this experiment, 3D7 and Dd2 served as control strains.
Table 1.
PCR primer sequences and reaction conditions were used for the amplification of sequences encoding P. falciparum pfcrt and pfmdr-I gene
| Primer name | Primer sequence | PCR product size (bp) | PCR cycling conditions |
|---|---|---|---|
| pfcrt | |||
| K76T F (sense) | 5′-TGTGCTCATGTGTTTAAACTT-3′ | 134 | 95°C for 5 minutes; 30 cycles of 95°C for 30 seconds, 50°C for 30 seconds, and 72°C for 1 minute; 72°C for 4 minutes |
| K76T R (antisense) | 5′-CAAAACTATAGTTACCAATTTTG-3′ | 134 | |
| pfmdr-I | |||
| N86Y F (sense) | 5′-TTTACCGTTTAAATGTTTACCTGC-3′ | 310 | 95°C for 5 minutes; 40 cycles of 95°C for 30 seconds, 56°C for 35 seconds, and 72°C for 1 minute; 72°C for 5 minutes |
| N86Y R (antisense) | 5′-CCATCTTGATAAAAAACACTTCTT-3′ | 310 | |
| N1042D F (sense) | 5′-TATGTCAAGCGGAGTTTTTGC-3′ | 337 | 95°C for 5 minutes; 42 cycles of 95°C for 30 seconds, 56°C for 40 seconds, and 72°C for 1 minute; 72°C for 5 minutes |
| N1042D R (antisense) | 5′-TCTGAATCTCCTTTTAAGGAC-3′ | 337 | |
| D1246Y F (sense) | 5′-GTGGAAAATCAACTTTTATGA-3′ | 499 | 95°C for 5 minutes; 44 cycles of 95°C for 30 seconds, 58°C for 40 seconds, and 72°C for 1 minute; 72°C for 8 minutes |
| D1246Y R (antisense) | 5′-TTAGGTTCTCTTAATAATGCT-3′ | 499 | |
Multiplicity of infection and prevalence of monoclonal infections.
The multiplicity of infection, defined as the highest number of alleles detected at either of the two loci, was estimated by using an allelic family-specific nested PCR (MAD20 and K1 for pfmsp-1 and 3D7 Africa and FC27 for pfmsp-2).18 Clonality was defined as the highest number of alleles detected at either of the two loci, and it used to classify isolates as monoclonal or polyclonal infection and distinguish recrudescence from new infection for all patients failing therapy after the seventh day (isolates from day 0 and day of recurrence). All PCR amplifications contained a positive control (genomic DNA from strains FCM29 and 3D7 Africa) and a negative control (no target DNA).
Assessment of antimalarial drug pressure and population mobility.
Cross-sectional surveys were carried out from May of 2007 to July of 2007 in Purulia. Thirty households from the four blocks were randomly selected. Three randomly selected individuals in each household were interviewed about their recent travel and antimalarial drug consumption as previously described.19 Data collected from the individuals and the household were secondarily aggregated to characterize each site.
Statistical analysis.
The data were expressed as mean ± SEM. Fisher exact and Mann–Whitney U tests were used to study the relation between IC50 values and genotypes. The relationship between in vivo and in vitro phenotypes with molecular genotypes was studied by Fisher exact tests. All the statistical analyses were performed using a statistical package (Origin 6.1; Northampton, MA) with Fisher exact or Mann–Whitney U test; P < 0.05 was the limit of significance (GraphPad InStat software 3.0).
Results
Clinical feature of CQ.
The clinical data were available for 184 suspected cases with high fever, headache, and other symptoms compatible with this disease. A total of 141 (76.63%) isolates from 184 suspected cases were enrolled, and they were carried with P. falciparum monoinfection. Monoclonal P. falciparum isolates were confirmed by using allelic family-specific nested PCR. This group consisted of 41% females and 31% children below the age of 6 years. CQ treatment exerted ACPR in 54 subjects (38.29%). ETF was found in 76 (53.90%) isolates, with a median of day 2 (range = 1–3 days), whereas LTF was observed in 11 (7.80%) isolates, with a median of day 9 (range = 4–14 days).
In vitro susceptibility to CQ.
In vitro drug susceptibility tests were performed after clonal selection of P. falciparum isolates. In vitro assay for CQ yielded interpretable results on all 141 monoclonal P. falciparum isolates. It was found, through in vitro testing, that only 12 (8.51%) isolates were CQ-sensitive (geometric mean IC50 = 45 nM, range = 15–60 nM), 17 (12.06%) of 141 isolates were intermediately susceptible to CQ (mean IC50 = 90 nM, range = 61–100 nM) and 112 (79.43%) isolates were highly resistant to CQ (mean IC50 = 521.22 nM, range = 110–2,250 nM. 98 (87.5%) of 112 resistant isolates were found with double pfmdr-I mutation (86Y+1246Y). Mixed isolates were taken as mutant isolates.
PCR/restriction fragment-length polymorphism: test for pfcrt and pfmdr-I genes.
The region of pfcrt and pfmdr-I genes flanking the polymorphism of interest was amplified by PCR (multiplex PCR) followed by digestion with specific restriction enzymes to detect each variant. The total numbers of pfcrt mutant isolates at codon 76 were 31.91%. We found that 5.67% of isolates contained mixed K76T allele and 62.41% of isolates were wild K76 allele. The two alternative forms of codon 76 (i.e., threonine, and lysine) were discriminated by digestion with ApoI enzyme. The frequencies composed of pfmdr-I mutant isolates at codons 86, 1042, and 1246 were, respectively, 87.23%, 17.02%, and 67.38% (Table 2). Double pfmdr-I (86Y+1246Y) mutation was predominantly found in 58.16% of isolates, and 14.89% of isolates were observed with single pfcrt mutation and triple pfmdr-I (76T+86Y+1042D+1246Y) mutation. Additionally, some triple mutations were found with single pfcrt mutations and double pfmdr-I mutations (76T+86Y+1246Y). Some triple pfmdr-I mutations (86Y+1042D+1246Y) were also found here.
Table 2.
Distribution of pfcrt and pfmdr-I genotypes in 141 blood samples from Purulia with P. falciparum infection
| Codons | n | Wild | Mutant | Mixed |
|---|---|---|---|---|
| pfcrt 76 | 141 | 88 (62.41%) | 45 (31.91%) | 8 (5.67%) |
| pfmdr-I 86 | 141 | 12 (8.51%) | 123 (87.23%) | 6 (4.26%) |
| pfmdr-I 1042 | 141 | 108 (76.60%) | 24 (17.02%) | 9 (6.38%) |
| pfmdr-I 1246 | 141 | 24 (17.02%) | 95 (67.38%) | 22 (15.60%) |
Relationship between pfcrt and pfmdr-I genotypes in in vitro and in vivo data.
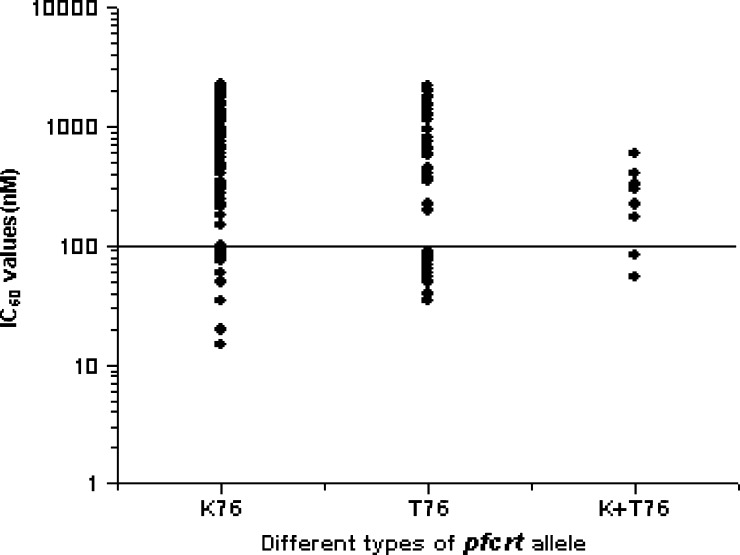
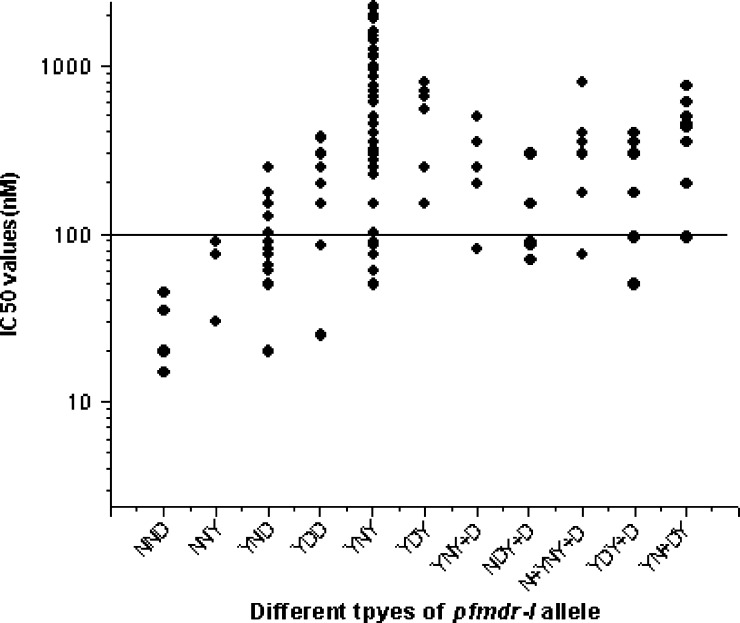
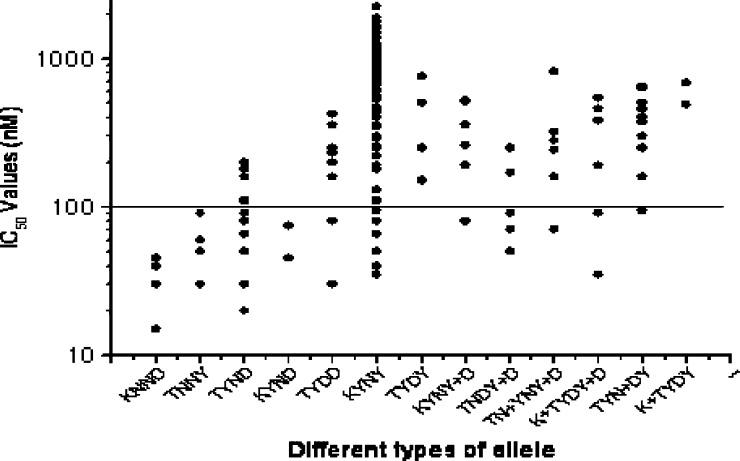
It was observed that the presence of pfmdr-I point mutations was linked to in vitro resistance to CQ, whereas there was no such correlation observed with pfcrt mutation, because there was no single 76T mutation. Here, the 76T mutation was always associated with other pfmdr-I mutations like 86Y, 1246Y, or both (86Y+1246Y). All of these mutant alleles possessed very high IC50 values for CQ and showed in vitro resistance to CQ (Fisher test: CQ, P < 0.05 for 76T+86Y; CQ, P < 0.05 for 76T+86Y+1246Y). The phenotype of in vitro susceptibility to CQ was associated with pfmdr-I genotypes at positions 86 and 1246 but not 1042 (Fisher test: CQ, P < 0.001 for codon 86 and P < 0.01 for codon 1246; P was not significant at the level of 0.05 for the 1042 codon) (Table 3). Double pfmdr-I (86Y+1246Y) mutations were found in 82 isolates; 75 (91.46%) of 82 isolates were in vitro CQ-resistant. Very high IC50 values were observed with wild-type pfcrt genotypes (more specifically with K76 alleles than mutant 76T alleles, which was very unlikely) (Figure 1 ). Double pfmdr-I (86Y+1246Y) mutations were also associated with very high IC50 values for CQ and resulted in in vitro resistance to CQ (Figure 2 ) (Fisher test: P < 0.001). Therefore, we compared the whole haplotype sequence of pfcrt and pfmdr-I genes with IC50 values. Most interestingly, it was observed that 82 (93.18%) of 88 wild-type K76 alleles were found with double pfmdr-I (86Y+1246Y) mutations (Figure 3 ). Because the wild-type K76 allele was found with the double pfmdr-I (86Y+1246Y) mutation, it possessed higher IC50 values for CQ. From Figure 3, it was observed that the wild-type K76+N86+N1042+D1246 (KNND) haplotype possessed lower IC50 values for CQ.
Table 3.
Distribution of pfcrt and pfmdr-I genotypes in relation to CQ treatment efficacy and in vitro susceptibility to CQ in Purulia
| Number | pfcrt genotype 76 | pfmdr-I genotype | In vivo response to CQ | In vitro susceptibility to CQ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 86 | 1042 | 1246 | ACPR | ETF | LTF | S | I | R | ||
| 4 | K | N | N | D | 4/4 | – | – | 4 | – | – |
| 3 | T | N | N | Y | 3/3 | – | – | 1 | 2 | – |
| 11 | T | Y | N | D | 10/11 | – | 1/11 | 3 | 3 | 5 |
| 2 | K | Y | N | D | 2/2 | – | – | – | 1 | 1 |
| 7 | T | Y | D | D | 5/7 | 1/7 | 1/7 | 1 | 1 | 5 |
| 77 | K | Y | N | Y | 13/77 | 61/77 | 3/77 | 2 | 4 | 71 |
| 4 | T | Y | D | Y | 1/4 | 2/4 | 1/4 | – | – | 4 |
| 5 | K | Y | N | Y+D | 2/5 | 3/5 | – | – | 1 | 4 |
| 5 | T | N | D | Y+D | 5/5 | – | – | 1 | 2 | 2 |
| 6 | T | N+Y | N | Y+D | 4/6 | 1/6 | 1/6 | – | 1 | 5 |
| 6 | K+T | Y | D | Y+D | 3/6 | 3/6 | – | 1 | 1 | 4 |
| 9 | T | Y | N+D | Y | 2/9 | 3/9 | 4/9 | – | 1 | 8 |
| 2 | K+T | Y | D | Y | – | 2/2 | – | – | – | 2 |
CQ treatment responses are classified as ACPR, ETF, and LTF. Here, in vitro test responses are classified as susceptible (S), intermediate (I), and resistant (R).
Figure 1.
Relationship between the phenotype determined by in vitro drug sensitivity assays, expressed as IC50 of CQ, and the pfcrt genotype, defined by either the absence of mutations (wild type) or presence of point mutations. Here, different genotypes are classified as K76, T76, and K+T76. The solid line (corresponding to 100 nM CQ; highly resistant) hypothetically shows the resistance levels for in vitro CQ resistance.
Figure 2.
Relationship between in vitro IC50 values of CQ and pfmdr-I genotypes, defined by either the absence of mutations (wild type) or presence of point mutations. Here, different genotype are classified as NND (N86, N1042, D1246), NNY (N86, N1042, Y1246), YND (Y86, N1042, D1246), YDD (Y86, D1042, D1246), YNY (Y86, N1042, Y1246), YDY (Y86, D1042, Y1246), YNY+D (Y86, N1042, Y+D1246), NDY+D (N86, D1042, Y+D1246), N+YNY+D (N+Y86, N1042, Y+D1246), YDY+D (Y86, D1042, Y+D1246), and YN+DY (Y86, N+D1042, Y1246). The solid line (corresponding to 100 nM; highly resistant) hypothetically shows the resistance levels for in vitro CQ resistance.
Figure 3.
Relationship between the phenotype determined by in vitro drug sensitivity assays, expressed as IC50 of CQ, and the pfcrt and pfmdr-I genotypes, defined by either the absence of mutations (wild type) or presence of point mutations. Here, different genotype are classified as KNND (K76, N86, N1042, D1246), TNNY (T76, N86, N1042, Y1246), TYND (T76, Y86, N1042, D1246), KYND (K76, Y86, N1042, D1246), TYDD (T76, Y86, D1042, D1246), KYNY (K76, Y86, N1042, Y1246), TYDY (T76, Y86, D1042, Y1246), KYNY+D (K76, Y86, N1042, Y+D1246), TNDY+D (T76, N86, D1042, Y+D1246), TN+YNY+D (T76, N+Y86, N1042, Y+D1246), K+TYDY+D (K+T76, Y86, D1042, Y+D1246), TYN+DY (T76, Y86, N+D1042, Y1246), and K+TYDY (K+T76, Y86, D1042, Y1246). The solid line (corresponding to 100 nM; highly resistant) hypothetically shows the resistance levels for in vitro CQ resistance.
In Table 3, different genotypes of all isolates with complete genotyping in relation to in vivo and in vitro results were described. Because mixed and mutant genotypes showed similar in vitro phenotypes, mixed genotypes were considered as mutant for this analysis. Most of the samples (87.5%) exhibited in vitro resistance to CQ, and they were mainly associated with double pfmdr-I (86Y+1246Y), triple pfmdr-I (86Y+1042D+1246Y), or single pfcrt and triple pfmdr-I (76T+86Y+1042D+1246Y) mutations (Table 3).
No such correlation was observed with CQ treatment outcome and pfcrt mutation. Here, 76 (53.90%) ETF and 11 (7.80%) LTF cases were found. Whatever might be the other mutation in the pfcrt and pfmdr-I genes, most interestingly, 72 of 76 ETF isolates were found with double pfmdr-I (86Y+1246Y) mutation (P < 0.001). In concise view, however, 64 (84.21%) of 76 ETF cases were predominantly found with the 86Y+1246Y mutation (KYNY haplotype). Three (2.13%) LTF cases were also found with this double pfmdr-I mutation, and 18 (12.77%) in vitro CQ-resistant isolates and 10 (7.09%) ETF cases were found with single pfcrt and triple pfmdr-I (76T+86Y+1042D+1246Y) mutations. Five (3.55%) LTF cases were also observed with this mutation. One (0.71%) LTF case was associated with a single 76T mutation and a single 86Y mutation (76T+86Y), whereas another LTF was found because of an isolate presenting with a single pfcrt mutation and a double pfmdr-I (76T+86Y+1246Y) mutation. One LTF case was associated with a single pfcrt mutation and a double pfmdr-I mutation (76T+86Y+1042D) (Table 3).
Prevalence of monoclonal infections.
Multiplicity of infection was analyzed for a subset of 184 isolates, and it showed that the proportion of monoclonal infections was very high. The results of multilocus genotype analysis of 141 isolates had a single allelic form and were considered as monoclonal infection. These monoclonal infection isolates were considered in the study.
Antimalarial drug pressure and population mobility.
Three hundred sixty individuals were interviewed (mean age = 22.17 years, age range = 3–78 years), and 24% declared that they had been prescribed an antimalarial treatment in the previous 30 days at the health center or hospital. Surprisingly, CQ was the most commonly prescribed antimalarial drug by clinicians (73%) followed by SP (18%) and quinine (QU; 9%). The combination of artesunate plus amodiaquine or artesunate plus SP was not prescribed. Among the 38% of individuals who had taken an antimalarial drug at home, 62% declared that they had used CQ, 28% declared that they had used QU, and 10% declared that they had used SP. A population mobility survey showed that one-seventh of the individuals lived at another site 1 year before the interview. The proportion of individuals who traveled outside the site in the previous 30 days was estimated as 3.14%.
Discussion
The present study deals with the association of pfmdr-I genotype with in vivo CQ clinical treatment failure and in vitro CQ resistance in Purulia, West Bengal, India. CQ was efficient in treating malaria attacks in different parts of India, although in recent times, most of the isolates were in vitro-resistant to CQ. The NVBDCP has introduced ACT as the first-line option to keep abreast of treating all P. falciparum cases in the country.1 This study was carried out to evaluate the efficacy of CQ before the launch of ACT by the NVBDCP in eastern India.
Several factors may explain the dissimilarity between in vitro- and in vivo-resistant patterns of isolates. Sometimes, this dissimilarity pattern was increased. Treatment failures reflect a combination of several parameters, including parasite resistance to the drug, drug level achieved in the host, and action of the host immune response. The increasing failure rate of several antimalarial drugs in the majority of the malaria-affected areas indicates the essentiality of close monitoring of the epidemiology and dynamics of drug resistance. The identification and validation of easy and rapid molecular markers of drug resistance would greatly facilitate this process and allow us to overcome difficulties in using of traditional methods for assaying drug sensitivity.
The former accepted view revealed that the major role of CQ resistance was determined by polymorphisms in the pfcrt gene that encoded a transporter, which promoted drug efflux from the parasite digestive vacuole in its mutated forms,8 whereas pfmdr-I mutation modulates the level of in vitro CQ resistance.20
In our study, we combined in vitro and in vivo tests as well as molecular genotyping at pfcrt and pfmdr-I loci. CQ treatments failed in 87 (61.70%) patients, of which 76 (53.90%) patients were ETFs and 11 (7.80%) patients were LTFs. 112 (79.43%) in vitro CQ-resistant isolates and 12 (8.57%) CQ-sensitive isolates were also found here. 58.16% of isolates presented the double pfmdr-I (86Y+1246Y) mutation, and 14.89% of isolates showed the single pfcrt and triple pfmdr-I (76T+86Y+1042D+1246Y) mutation. However, this result indicated a very high rate of in vitro CQ resistance as well as enormous drug pressure of CQ over this population, because more than 60% of cases belonged to in vivo CQ treatment failure cases. This enormous drug pressure of CQ might be because of the extensive and haphazard use of CQ for more than five decades. This molecular genotype and in vitro resistance as well as in vivo CQ treatment failure were strongly related.
PCR-based methods did not detect minor clones in a mixed population; although a wild-type clone might remain undetected, this finding was unlikely for in vitro susceptibility, because IC50 mainly reflects the susceptibility of the major clones present in the blood sample. The correlation between pfmdr-I genotypes and quinoline resistance has often generated conflicting results. It has been suggested that pfmdr-I 86Y can be correlated with increased CQ resistance in parasites that originated from different areas of the world,7,8 but in India, field studies had not corroborated these findings, and the results of the P. falciparum genetic cross indicated that CQ resistance did not depend on the pfmdr-I gene.12,21 Different groups of workers showed that, in India, CQ-resistant P. falciparum and the CQ treatment failure mainly occurred because of the mutation in the pfcrt and not the pfmdr-I gene.13
Our present findings implicated that double pfmdr-I mutations (86Y+1246Y) were highly correlated (P < 0.001) with in vitro CQ resistance in Purulia. Because the presence of both N86Y and D1246Y in our samples was largely dependent on their CQ response, it indicated that CQ seemed to exert a selective pressure on this area, which was at odds with previous findings from India.13
Our results confirmed that pfcrt 76T was a key mutation; it can cause in vitro resistance to CQ, with association with 86Y or 86Y+1246Y mutation (P < 0.05), but it cannot cause early treatment failure. No single 76T mutation was found here; instead, of the 14 isolates, isolates were found either with the 76T+86Y or 76T+1246Y mutation. They showed moderate to high IC50 values for in vitro CQ response but exerted only a single in vivo LTF case. In six isolates, 76T mutation was found along with double pfmdr-I mutation (86Y+1246Y), and five isolates showed very high IC50 values for CQ but possessed only one ETF case and one LTF case. Similarly, very high IC50 value was also found with single pfcrt and triple pfmdr-I mutation (76T+86Y+1042D+1246Y). Importantly, 10 ETF and 5 LTF cases were found with these mutations. The impact of the pfcrt/pfmdr-I combination mutation depends on the genetic background of the strain, which was shown by studies with genetically manipulated lines or recombinant progeny of experimental crosses,22 and the history of using antimalarial drugs (CQ and QU being the two mains drugs used in West Bengal, India.).
Finally, these findings aroused some most important questions. (1) What was the cause of this large number of ETF cases? (2) What would be the possible cause of the less prevalent pfcrt mutant parasite population despite huge drug pressure of CQ, which was largely different from other parts of India? (3) Was monitoring of isolates with pfcrt mutant alleles the best way to assess the incidence of CQ resistance in West Bengal, India, as previously described?12,13,21
Our results provided the answer for the question of the large number of treatment failures. Prevalence of double pfmdr-I mutation (86Y+1246Y) was highly associated (P < 0.001) with in vitro CQ resistance as well as ETF (P < 0.001). All the ETF isolates were found with this double pfmdr-I mutation, whereas the 76T and 1042D mutations increased the degree of resistance indicated by very high IC50 values.
Here, we observed that CQ was the most frequently prescribed drug in the public sector for nearly five decades (starting in late 1950s), and this use might be the cause of the specific selective pressures exerted on the parasite population over CQ drug in Purulia, West Bengal, India. Additional factors contributing to the uncommon drug resistance situation in West Bengal might be its geographical position, the broad range of malaria epidemiological strata, in which the three Plasmodium species (P. falciparum, P. vivax, and P. malariae) were present, and the admixture of inhabitants with multiple tribal ethnic origins. We also confirmed from our study that monitoring only the pfcrt mutant allele was not the best way to assess the incidence of CQ resistance in this part of India. Monitoring of pfmdr-I genotype was also very much essential for detecting the CQ-resistant pattern.
Similar types of results were found in Madagascar as previously described.15 In Madagascar, single nucleotide polymorphisms at codons 86, 184, and 1246 in pfmdr-I were highly prevalent, likely reflecting the widespread use of quinolines, such as CQ and QU. The elevated frequency of pfmdr-I mutant alleles (YYD, NFD, YFD, and YFY) associated with CQ resistance and the low frequency of pfcrt mutant genotypes might account for the high rate of late clinical failure of CQ treatment.8,23
pfcrt mutations were known to have an effect on CQ resistance, which was proven by genetic cross-breeding between sensitive and resistant parasites.13 However, the unbalanced numbers of genotypes here did not allow us to draw any conclusion on the impact of pfcrt mutations.
It was concluded that in vitro CQ resistance and failure of CQ treatment in this area of India was caused mainly by the combination of two pfmdr-I (86Y and 1246Y) mutations but not the pfcrt (76T) mutation, which was a very new finding in the field of drug-resistant P. falciparum in India. The increase in the number of pfmdr-I mutations was strongly correlated to CQ resistance. Additional studies are needed to determine the precise incidence of the combination of pfcrt and pfmdr-I gene mutations and the role of double pfmdr-I mutation on CQ and other antimalarial drug efficacy. New cheap antimalarial combinations (except ACTs) should be tested for treatment of the drug-resistant P. falciparum.
ACKNOWLEDGMENTS
The authors thank Vidyasagar University, Midnapore, for providing the facilities to execute these studies. The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
Disclaimer: We declare that we have no conflict of interest.
Footnotes
Authors' addresses: Sabyasachi Das, Subhankari Prasad Chakraborty, and Somenath Roy, Immunology and Microbiology Laboratory, Department of Human Physiology with Community Health, Vidyasagar University, Midnapore, West Bengal, India, E-mails: sdas.vu@gmail.com, subhankariprasad@gmail.com, and roysomenath@hotmail.com. Amiya Kumar Hati, Department of Medical Entomology, Calcutta School of Tropical Medicine, Kolkata, West Bengal, India, E-mail: amiya_hati@rediffmail.com.
References
- 1.Government of India . Guidelines for Diagnosis and Treatment of Malaria in India. Government of India: NIMR/TRS-06/APR-2009; 2009. [Google Scholar]
- 2.Peters W. Chemotherapy and Drug Resistance in Malaria. 2nd Ed. London, United Kingdom: Academic Press; 1987. Resistance of human malaria I, III and IV. [Google Scholar]
- 3.Aubouy A, Jafari S, Huart V, Florence M, Mayombo J, Durand R, Mohamed B, Jacques Le B, Deloron P. DHFR and DHPS genotypes of Plasmodium falciparum isolates from Gabon correlate with in vitro activity of pyrimethamine and cycloguanil, but not with sulfadoxine–pyrimethamine treatment efficacy. J Antimicrob Chemother. 2003;52:43–49. doi: 10.1093/jac/dkg294. [DOI] [PubMed] [Google Scholar]
- 4.Sehgal PN, Sharma MID, Sharma Sl, Gopal S. Resistance to chloroquine in falciparum malaria in Assam state, India. J Commun Dis. 1973;5:175–180. [Google Scholar]
- 5.Durand R, Jafari S, Vauzelle J, Delabre JF, Jesic ZZ, Le Bras J. Analysis of pfcrt point mutations and chloroquine susceptibility in isolates of Plasmodium falciparum. Mol Biochem Parasitol. 2001;114:95–102. doi: 10.1016/s0166-6851(01)00247-x. [DOI] [PubMed] [Google Scholar]
- 6.Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV, Coulibaly D. A molecular marker for chloroquine resistant falciparum malaria. N Engl J Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 7.Duraisingh MT, Cowman AF. Contribution of the pfmdr-1 gene to antimalarial drug-resistance. Acta Trop. 2005;94:181–190. doi: 10.1016/j.actatropica.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Andriantsoanirina V, Ratsimbasoa A, Bouchier C, Tichit M, Jahevitra M, Rabearimanana S, Raherinjafy R, Mercereau-Puijalon O, Durand R, Menard D. Chloroquine clinical failures in P. falciparum malaria are associated with mutant Pfmdr-1, not Pfcrt in Madagascar. PLoS One. 2010;5:e13281. doi: 10.1371/journal.pone.0013281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma YD. Genetic alteration in drug resistance markers of Plasmodium falciparum. Indian J Med Res. 2005;121:13–22. [PubMed] [Google Scholar]
- 10.Than N, Manoj D, Michael R, Alan CF. Analysis of Pfcrt, pfmdr-1, dhfr and dhps mutation and drug sensitivities in Plasmodiuml falciparum isolates from patients in Vietnam before and after treatment with artemisinin. Am J Trop Med Hyg. 2003;68:350–356. [PubMed] [Google Scholar]
- 11.Mehlotra RK, Mattera G, Bockarie MJ, Maguire JD, Baird JK. Discordant patterns of genetic variation at two chloroquine resistance loci in worldwide populations of the malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother. 2008;52:2212–2222. doi: 10.1128/AAC.00089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vathsala PG, Pramanik A, Dhanasekaran S, Rangarajan PN, Padmanaban G. Widespread occurrence of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) gene haplotype SVMNT in P. falciparum malaria in India. Am J Trop Med Hyg. 2004;70:256–259. [PubMed] [Google Scholar]
- 13.Valecha N, Joshi H, Mallick PK, Sharma SK, Kumar A, Tyagi PK, Shahi B, Das MK, Nagpal BN, Dash AP. Low efficacy of chloroquine: time to switchover to artemisinin-based combination therapy for falciparum malaria in India. Acta Trop. 2009;111:21–28. doi: 10.1016/j.actatropica.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization (WHO) Assessment and Monitoring of Antimalarial Drug Efficacy for the Treatment of Uncomplicated Falciparum Malaria. Geneva, Switzerland: WHO; 2003. [Google Scholar]
- 15.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 16.Basco KL, Ringwald P. Molecular epidemiology of malaria in Yaounde, Cameroon VI. Sequence variations in the Plasmodium falciparum dihydrofolate reductase-thymidylate synthase gene and in vitro resistance to pyrimethamine and cycloguanil. Am J Trop Med Hyg. 2000;62:271–276. doi: 10.4269/ajtmh.2000.62.271. [DOI] [PubMed] [Google Scholar]
- 17.Lopes D, Rungsihirunrat K, Nogueira F, Seugorn A, Gil JP, do Rosário VE, Cravo P. Molecular characterizations of drug-resistant Plasmodium falciparum from Thailand. Malar J. 2002;1:12. doi: 10.1186/1475-2875-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93:369–374. doi: 10.1016/s0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- 19.Gardella F, Assi S, Simon F, Bogreau H, Eggelte T, Ba F, Foumane V, Henry MC, Kientega PT, Basco L, Trape JF, Lalou R, Martelloni M, Desbordes M, Baragatti M, Briolant S, Almeras L, Pradines B, Fusai T, Rogier C. Antimalarial drug use in general populations of tropical Africa. Malar J. 2008;7:124. doi: 10.1186/1475-2875-7-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinayak S, Biswas S, Dev V, Kumar A, Ansari MA, Sharma YD. Prevalence of the K76T mutation in the pfcrt gene of Plasmodium falciparum among chloroquine responders in India. Acta Trop. 2003;87:287–293. doi: 10.1016/s0001-706x(03)00021-4. [DOI] [PubMed] [Google Scholar]
- 22.Sa JM, Twu O, Hayton K, Reyes S, Fay MP. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci USA. 2009;106:18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valerie A, Arsene R, Christiane B, Martial J, Stephane R, Rogelin R, Voahangy A, Tantely R, Ange RM, Magali T, Leon RP, Odile M-P, Remy D, Didier M. Plasmodium falciparum drug resistance in Madagascar: facing the spread of unusual pfdhfr and pfmdr-1 haplotypes and the decrease of dihydroartemisinin susceptibility. Antimicrob Agents Chemother. 2009;53:4588–4597. doi: 10.1128/AAC.00610-09. [DOI] [PMC free article] [PubMed] [Google Scholar]