Abstract
The lungs of cystic fibrosis (CF) patients are commonly colonized with Pseudomonas aeruginosa biofilms. Chronic endobronchial P. aeruginosa infections are impossible to eradicate with antibiotics, but intensive suppressive antibiotic therapy is essential to maintain the lung function of CF patients. The treatment often includes β-lactam antibiotics. How these antibiotics influence gene expression in the surviving biofilm population of P. aeruginosa is not clear. Thus, we used the microarray technology to study the effects of subinhibitory concentrations of a β-lactam antibiotic, imipenem, on gene expression in biofilm populations. Many genes showed small but statistically significant differential expression in response to imipenem. We identified 34 genes that were induced or repressed in biofilms exposed to imipenem more than fivefold compared to the levels of induction or repression for the controls. As expected, the most strongly induced gene was ampC, which codes for chromosomal β-lactamase. We also found that genes coding for alginate biosynthesis were induced by exposure to imipenem. Alginate production is correlated to the development of impaired lung function, and P. aeruginosa strains isolated from chronically colonized lungs of CF patients are nearly always mucoid due to the overproduction of alginate. Exposure to subinhibitory concentrations of imipenem caused structural changes in the biofilm, e.g., an increased biofilm volume. Increased levels of alginate production may be an unintended adverse consequence of imipenem treatment in CF patients.
Pseudomonas aeruginosa is a ubiquitous environmental bacterium and an opportunistic human pathogen, showing great adaptability and metabolic versatility (17, 36, 46). Chronic bronchopulmonary infection with P. aeruginosa is the major factor leading to the increased morbidity and premature mortality seen in patients with cystic fibrosis (CF) (26). The establishment of biofilms by alginate-producing P. aeruginosa strains is the most common mode of growth in CF patients with chronic lung infections, with the biofilms providing a protected environment against the host immune system and a number of antibiotics (1, 4, 15, 23). The presence of P. aeruginosa strains with the mucoid phenotype in CF patients with chronic lung infections is a marker of a poor prognosis (20). Endobronchial chronic infections caused by the mucoid, alginate-producing phenotype of P. aeruginosa are impossible to eradicate with antibiotics (40). The MICs of antimicrobial agents can be increased 100- to 1,000-fold when bacteria grow in biofilms (33). However, planktonic bacteria dispersed from a biofilm usually no longer demonstrate increased levels of resistance to antibiotics, suggesting that the mode of growth is of major importance (45). The switch from the nonmucoid to the mucoid phenotype may provide profound advantages to the bacteria growing in biofilms, such as increasing their levels of resistance to antibiotics (such as tobramycin) and protecting them against the host immune system (30, 35, 44, 52).
By aggressive antibacterial chemotherapy it is possible to suppress the growth of and the damage caused by P. aeruginosa in the lungs of chronically infected CF patients. Although it is not possible to eradicate P. aeruginosa, antibiotic therapy contributes to the maintenance of lung function for decades (24, 25). Therefore, the development of antibiotic resistance in P. aeruginosa is of great therapeutic concern. Antipseudomonal β-lactam antibiotics are widely used for the treatment of lung infections in CF patients. Unfortunately, the development of resistance to these drugs is a common problem (8). Resistance to β-lactam antibiotics is mainly due to stable derepression of chromosomal β-lactamase (13, 14, 31). However, the mechanisms leading to stable derepression of chromosomal β-lactamase in P. aeruginosa are not completely understood. We previously identified a new insertion sequence located in ampD, a negative regulator of ampC chromosomal β-lactamase expression, which caused stable derepressed levels of AmpC β-lactamase in clinical isolates of P. aeruginosa from patients with CF (2). Our results also suggested that multiple pathways might lead to increased levels of antimicrobial resistance due to chromosomal β-lactamase, as we registered increased levels of resistance to β-lactam antibiotics due to chromosomal β-lactamase, although we found no changes in ampD, ampC, or the transcriptional regulator ampR.
Little is known about the transcriptional regulation of AmpC in P. aeruginosa, e.g., whether the transcription of ampD is affected by the exposure to β-lactam antibiotics. In addition, it is not known whether expression of other genes (which do not affect ampC expression) is affected in a multifunctional response to β-lactam antibiotics. The responses to β-lactam antibiotics of biofilm bacteria versus those of cultured planktonic bacteria may reveal distinct differential expression profiles that are reflected by the two modes of growth. We have used the microarray technology to profile the average transcriptome of P. aeruginosa cells growing in biofilms versus that of planktonic P. aeruginosa cells exposed to imipenem.
MATERIALS AND METHODS
Strains and growth conditions.
P. aeruginosa strain PAO1 was used in this study. The growth medium (minimal medium) consisted of 0.5% NH4Cl, 0.25% NaCl, 0.025% MgSO4 · 7H2O, 0.015% KH2PO4, 1.5% morpholinepropanesulfonic acid buffer (pH 7.0), nonchelated trace elements (51), and 0.02% Casamino Acids as the carbon source.
Biofilms were grown in silicone tubes (inner diameter, 6 mm) in a once-flowthrough system at 37°C. The flow velocity was 48 ml/h and was controlled with a peristaltic pump (Watson-Marlow, Falmouth, England). The tubing was inoculated with a culture at an optical density at 600 nm (OD600) of 0.06, with the tube filled with a syringe. The flow was initiated after 1 h. For the microarray experiments biofilm material from a 10-cm tube section was harvested after 1, 2, or 3 days. The biofilm material contained from 1010 to 1012 CFU, as determined by plate counting techniques. P. aeruginosa cells growing in biofilms were exposed to imipenem (1.0 μg/ml; Tienam; Merck & Co., Inc., Rahway, N.J.) for 7 h, whereupon the bacteria were harvested. To remove the biofilm from a tube, the tube was squeezed by rolling a pencil on the outside, which easily loosened the biofilm.
Small-scale chemostats were used for planktonic chemostat cultures. The chemostats contained 100 ml of medium and a final cell density of about 108 CFU/ml. The generation time was 5 h at a flow rate of 20 ml/h, and the temperature was 37°C. The chemostats were aerated by bubbling filtered air into the system. The system was inoculated with 6 × 105 CFU, and after 1 h the flow was initiated. After 48 h the bacteria were harvested from the chemostats, which contained approximately 1010 cells, as determined by plate counting techniques and measurement of the OD600.
To study the effect of imipenem on gene expression by planktonic bacteria, imipenem was added to the chemostats at a final concentration of 0.2 μg/ml, equivalent to no more than 1/10 the MIC (imipenem MIC for strain PAO1, 2 to 4 μg/ml). Bacteria were exposed to imipenem for 7 h. The concentrations of imipenem for the experiments with planktonic cells as well as the biofilms were chosen because these levels did not affect bacterial growth.
Imipenem stability.
The chemical stability of imipenem was evaluated by measuring the concentration by a microbiological method previously described by Lautrop et al. (29) with Streptococcus sp. strain EB68 grown on Danish blood agar (State Serum Institute, Copenhagen, Denmark).
Microarray experiments and analyses.
Bacteria from tube biofilms or chemostats were immediately mixed with RNAprotect Bacteria Reagent (Qiagen), and the stabilized RNA was isolated according to the manufacturer's protocol of enzymatic lysis with mechanical disruption. Subsequently, the RNA was purified with an RNeasy Mini kit (Qiagen), including On-Column DNase digestion (Qiagen). The eluted RNA was further treated with DNase I for 1 h at 37°C (0.06 U per μg of RNA) by using the DNA-Free kit (Ambion). To optimize the final RNA purity, purification with the RNeasy Mini kit was repeated. The quality of the purified total RNA was analyzed by separating glyoxylated RNA samples by agarose gel electrophoresis by use of the Northern Max-Gly Gel system (Ambion).
cDNA synthesis was performed as described in the protocol (version 2) for the Affymetrix GeneChip P. aeruginosa Genome Array with 12 μg of total RNA as the starting material. Spiked transcripts (provided by Stephen Lory, Harvard University, Boston, Mass.) that included RNA from Saccharomyces cerevisiae, Bacillus subtilis, and Arabidopsis thalia were added for the cDNA synthesis. cDNA synthesis, fragmentation, labeling, hybridization on the GeneChip, washing, staining, and scanning were done by the Affymetrix protocol.
Affymetrix Microarray Software Suite, version 5.0 (MAS 5.0), was used for initial data analyses. To facilitate reliable comparison of the data from independent experiments, the signals on each GeneChip were scaled to an average target signal intensity of 2,500. The statistical algorithm of MAS 5.0 calculates a detection P value for each probe set on the chip, and on the basis of this P value the program assigns a call of present, marginal, or absent.
The data processed in MAS 5.0 were used for further statistical tests in the data-processing program Data Mining Tool (Affymetrix). t tests (P < 0.05, two-tailed distribution, two-sample and unequal variances) were performed on the signal intensities obtained for each gene of the additional replicates performed for each group (control versus treated samples). Thus, for each gene the t test analysis includes the mean and the variance for the two groups and calculates the probability that they were sampled from the same distribution. The t test assumes that the variance across the replicates shows a relatively normal distribution, as we performed the t test with linear data. We have tested this, and the signal values from the chips appeared to be normally distributed. Replicates were done as biological replicates; i.e., the whole biological experiment was repeated. The fold change ratio for each gene was calculated from the average signal for the control replicates versus that for the imipenem-exposed replicates. A cutoff value of more than twofold was chosen to list the genes (loci) showing induction and repression due to exposure to imipenem. All genes showing differential expression were checked for expression levels above the lower detection level of the chips (present calls).
Real-time PCR.
The specific genes shown by the GeneChip microarray experiments to be differentially expressed due to imipenem treatment were verified by real-time PCR. For the real-time PCR, new cDNA was synthesized from total RNA as described above. The total RNA preparation was the same as that used for the microarray experiments. Thus, the verification objective was of a technical and not of a biological nature. For real-time PCR the SYBR Green PCR master mixture was used according to the protocol of the manufacturer (Applied Biosystems). Gene-specific primers for the real-time PCR were designed by using Primer Express software (Applied Biosystems). No TaqMan probes were used. The reference gene used for the real-time PCR was ampR. The real-time PCR experiments were performed in triplicate.
We used a model 7000 thermal cycler (Applied Biosystems); and the thermal cycling parameters were 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C, according to the protocol of Applied Biosystems.
To control for nonspecific amplification in the real-time PCR, dissociation profiles were determined according to the protocol of the manufacturer (Applied Biosystems). This procedure thus identified nonspecific amplification products showing differences in melting temperatures.
Chemical and enzymatic assays.
Verification of increased levels of alginate expression in treated samples was done by use of the uronic acid assay, as described previously (28). The experimental setup for these experiments included biofilms grown in silicone tubes. However, for these experiments, 1-m-long silicone tubes were used, and the treated biofilm was exposed to imipenem (1.0 μg/ml) for 24 h from day 2 to 3. The biofilms were harvested on day 3, and small aliquots were used for plate counting to determine the numbers of CFU. The cells were pelleted (15,000 × g, 15 min, 4°C); and the pellet was resuspended in 10 ml of 0.4 M NaCl, homogenized moderately for 1 min, and stirred on a magnetic stirrer for 2 h (at 4°C) to extract exopolysaccharides from the cells. Finally, the cells were pelleted (10,000 × g, 30 min, 4°C), and the levels of uronic acids in the supernatant were subsequently measured.
β-Lactamase activity was measured spectrophotometrically by using nitrocefin (51.6 μg/ml) as the substrate, as described previously (13, 39). Samples of bacteria for β-lactamase measurements were collected and were immediately prepared for the nitrocefin assay, without a time delay.
Flow-cell biofilms.
Biofilm architecture was studied by using flow cells at 30°C in a setup previously described by Christensen et al. (6). The dimensions of the flow-cell chamber were 0.3 by 4.0 by 40 mm, and minimal medium was supplied at a constant flow rate of 3 ml/h with a peristaltic pump (Watson-Marlow). The minimal medium used contained 1 mM MgCl2, 0.1 mM CaCl2, nonchelated trace elements (51), 2 g of (NH4)2SO4 per liter, 6 g of Na2HPO4 · 2H2O per liter, 3 g of KH2PO4 per liter, 3 g of NaCl per liter, and 0.01% glucose. An exponentially growing culture was diluted to an OD600 of 0.1 and was inoculated into the flow cell with a syringe. The flow was initiated after 1 h. The flow-cell channels were exposed to imipenem (0.5 μg/ml) for 0, 18, or 37 h. On day 5 the biofilms were stained with cell-permeant nucleic acid stain SYTO 62 (Molecular Probes) and examined for structural changes by scanning confocal laser microscopy (model TCS4D scanning confocal laser microscope; Leica Lasertechnik GmbH, Heidelberg, Germany), with image acquisition and quantitative image analysis performed with the computer program COMSTAT (Technical University of Denmark, Lyngby, Denmark) (22). For the visualization of polysaccharides such as alginate in the P. aeruginosa biofilms, fluorescence-labeled lectins were applied as described previously (47). The lectin concanavalin A (ConA) was conjugated with fluorescein isothiocyanate (FITC). The conjugate was prepared in phosphate buffer (1 mg/ml; pH 7.5) and was stored at −20°C. Prior to use the lectin solution was diluted 10 times in the growth medium and was used to stain the biofilm. The flow was initiated after 15 min of staining in the flow cell, and image analysis was performed by scanning confocal laser microscopy after 20 min. The P. aeruginosa strains used in the flow-cell experiments were PAO1 and mucoid strain PAO300, a mucA22 derivative of PAO1 that constitutively overproduces alginate (35).
RESULTS
Assessment of variability in microarray experiments.
To best design the protocol for our microarray analyses we studied the expression of ampC in response to imipenem. The expression of chromosomal ampC was expected to be highly induced by imipenem, as even low concentrations of this carbapenem β-lactam antibiotic cause strong induction of AmpC β-lactamase (27, 41). The maximum level of β-lactamase production in planktonic cell cultures of PAO1 was in the presence of imipenem at a concentration of 2 μg/ml. An imipenem concentration of 0.5 μg/ml affected the growth of planktonic PAO1, but no inhibition was observed at 0.2 μg/ml (data not shown).
The imipenem concentration used for the tube biofilm experiments, 1.0 μg/ml, was below the imipenem MIC and did not affect the number of CFU after 7 h of exposure in a 3-day-old biofilm (data not shown). PAO1 grown in a biofilm and exposed to 1.0 μg of imipenem/ml produced the maximum level of β-lactamase after about 10 h, followed by a decline (data not shown). At 7 h after induction, β-lactamase activity was increased about 150-fold compared to that for a noninduced 3-day-old biofilm. The decreased level of production of β-lactamase after >10 h may be due to the destabilization of imipenem at 37°C, as we found the half-life of imipenem to be approximately 16 h. Thus, the concentration of imipenem may be reduced to 0.64 μg/ml after 10 h, causing a reduced level of induction. Imipenem stability studies indicate that it has a half-life of 1.68 days at 23°C and neutral pH (49). Exposure to light does not affect the stability of imipenem (49).
In our microarray analyses, the main objective was to discover genes whose expression was affected by imipenem exposure. We would prefer false-positive to false-negative results for three reasons: (i) the results from our array experiments are intended to be a guideline for the identification of genes affected by exposure to β-lactam antibiotics, (ii) the results for particular genes should be verified by other techniques before final conclusions are made, and (iii) common trends in gene expression profiles for genes in an operon can increase the probability of true-positive results for such genes. These considerations constituted the background for performing t tests based on a P value of 0.05 in our analyses.
To assess the degree of false positivity actually present in the expression profiles, we performed analyses to estimate the biological and technical variabilities of the microarray experiments. The microarray experiments were based on two to six replicates for each condition. We performed an assessment of assay variability on the basis of two-by-two experiments, all of which were performed in the absence of imipenem, i.e., nonexposed biofilm experiments. In each case the growth and harvesting of P. aeruginosa cells growing in biofilms were followed by the independent preparation of RNA, cDNA synthesis, and hybridization with the GeneChip. The GeneChip data were grouped into those for two chips for group 1 (control group 1 [CG1]) and two chips for group 2 (control group 2 [CG2]). A preliminary sorting was carried out by sorting the genes assigned by MAS 5.0 as absent or marginal calls for more than two of the four chips. A similar stringency in the sorting process was performed for the experiments with imipenem versus the control experiments. The proportion of genes on each chip with absent and marginal calls constituted from 25 to 35% of all genes. Then, the average signal was calculated for each group, and the fold change between the two sets of data was determined for each gene. A t test was performed to estimate the probability of obtaining identical average signals for CG1 and CG2. Even before the t test, only 210 (3.8%) of the genes showed more than apparent twofold differences in levels between CG1 and CG2. However, the t test further reduced this number to only five genes, constituting only 0.09% of the genes on the chip. Thus, by using a twofold cutoff level for differential gene expression in duplicate experiments, the frequency of false-positive results was approximately 0.1%.
Differentially expressed genes in P. aeruginosa cells growing in biofilms in response to imipenem.
Microarray experiments with P. aeruginosa cells grown in biofilms in silicone tubes were carried out on days 1, 2, and 3 after inoculation of the biofilm. The biofilm contained approximately 1010 and 1012 CFU/10-cm tube on days 1 and 3, respectively. Biofilm experiments were done in duplicate on day 1, in six replicates on day 2, and in triplicate on day 3. It should be emphasized that each replicate included a new additional biofilm experiment as well as a new GeneChip assay.
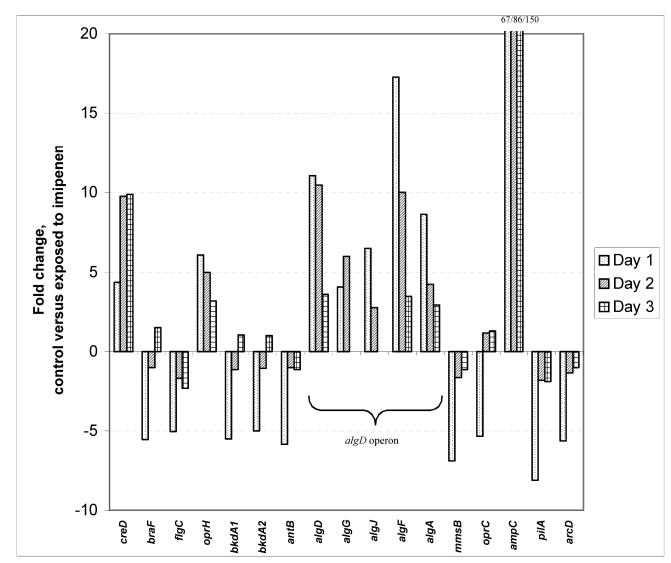
For P. aeruginosa cells growing in biofilms and exposed to imipenem, 336 genes were induced or repressed at least twofold, and the P value from the t test was less than 0.05 on at least 1 of the 3 days. A complete table including all 336 genes is available at http://www.immi.ku.dk/bagge-arrays2003. Table 1 includes 76 differentially expressed genes organized in known or putative operons, genes induced or repressed more than twofold, or genes of special interest. The gene showing the strongest differential expression on days 1, 2, and 3 was ampC. On day 3, imipenem exposure for 7 h induced ampC expression 150-fold. Of the 336 genes that were induced or repressed at least twofold due to imipenem exposure, only 34 genes were induced or repressed more than fivefold (Table 1). The algD to algA gene cluster was induced more than 10-fold, although this was less distinctive on day 3 than on days 1 and 2 (Table 1). On day 3, the hybridization signals of several alginate genes were absent or marginal. However, no significant differences in the levels of expression of the alginate genes were found in the noninduced biofilms when the levels of expression on days 1, 2, and 3 were compared. Genes of assigned function that were induced or repressed more than fivefold are illustrated in Fig. 1. As supported by the data in Table 1, Fig. 1 illustrates that the individual genes present in an operon, such as the alginate operon, in most cases revealed similar responses, although differences in the magnitudes of differential expression within the operon were seen.
TABLE 1.
Effects of imipenem on genes of P. aeruginosa growing in biofilmsa
| Gene no.b | Description | Fold change in expression level (P value)c
|
||
|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | ||
| 0102 | Carbonic anhydrase | 5.9 (1.2E-2) | 2.5 (6.7E-3) | 1.4 (2.1E-1) |
| 0320 | Hypothetical protein | 24.4 (7.2E-2) | 8.8 (6.1E-3) | 8.4 (9.6E-2) |
| 0327 | Hypothetical protein | 5.3 (2.2E-2) | 3.3 (3.7E-2) | 2.3 (1.5E-1) |
| 0376 | rpoH, sigma factor | 2.1 (2.9E-2) | 1.1 (3.1E-1) | 1.3 (1.5E-2) |
| 0464 | creC, two-component sensor | 2.2 (1.3E-2) | Δd | 1.0 (9.7E-1) |
| 0465 | creD, inner membrane protein | 4.4 (1.0E-1) | 9.8 (1.2E-2) | 9.9 (9.8E-2) |
| 0466 | Hypothetical protein | 5.0 (8.6E-2) | 7.0 (3.5E-2) | 8.7 (8.6E-2) |
| 0755 | Probable porin | 1.2 (5.3E-1) | 6.6 (2.1E-3) | 5.5 (8.4E-2) |
| 0762e | algU, sigma factor | 2.0 (1.6E-2) | 1.7 (1.1E-2) | 1.8 (4.4E-2) |
| 0763 | mucA, anti-sigma factor | 2.0 (4.9E-3) | 1.7 (1.4E-3) | 1.8 (1.1E-1) |
| 0764 | mucB, regulator, alginate biosynthesis | 2.2 (1.8E-2) | 1.5 (3.1E-1) | 1.3 (2.3E-1) |
| 0765 | mucC, regulator, alginate biosynthesis | 2.4 (1.9E-3) | 1.1 (8.0E-1) | 1.2 (3.7E-1) |
| 1071 | braF, amino acid transport protein | −5.5 (1.0E-1) | −1.1 (6.0E-1) | 1.5 (4.2E-2) |
| 1078 | flgC, flagellar basal-body rod protein | −5.0 (8.4E-3) | −1.7 (1.8E-2) | −2.3 (2.2E-1) |
| 1079 | flgD, flagellar basal-body rod protein | −3.9 (2.2E-2) | −1.7 (7.8E-3) | −2.6 (2.2E-1) |
| 1080 | flgE, flagellar hook protein | −4.1 (6.2E-4) | −1.5 (1.8E-1) | −2.4 (3.0E-1) |
| 1081 | flgF, flagellar basal-body rod | −2.8 (3.6E-2) | −1.6 (3.1E-2) | −2.6 (2.2E-1) |
| 1084 | flgI, flagellar P-ring protein | −2.2 (1.2E-2) | −1.4 (1.6E-1) | −2.2 (1.6E-1) |
| 1178 | oprH, outer membrane protein H1 precursor | 6.1 (1.2E-3) | 5.0 (4.1E-4) | 3.2 (3.5E-2) |
| 1179 | phoP, two-component regulator | 5.1 (1.3E-1) | 2.1 (1.1E-3) | 2.3 (1.5E-2) |
| 1180 | phoQ, two-component sensor | 4.1 (4.6E-2) | 2.5 (1.0E-2) | 1.2 (4.4E-1) |
| 1673 | Hypothetical protein | −5.5 (2.0E-2) | −1.3 (2.2E-1) | 1.0 (9.8E-1) |
| 2247 | bkdA1, 2-oxoisovalerate dehydrogenase | −5.5 (3.6E-2) | −1.2 (3.3E-1) | 1.0 (8.6E-1) |
| 2248 | bkdA2, 2-oxoisovalerate dehydrogenase | −5.0 (1.8E-2) | −1.1 (5.1E-1) | 1.0 (9.1E-1) |
| 2367 | Hypothetical protein | −5.3 (5.2E-4) | −1.1 (7.8E-1) | 1.2 (6.4E-1) |
| 2441 | Hypothetical protein | 1.5 (1.8E-1) | 2.4 (2.3E-2) | 5.6 (4.0E-3) |
| 2513 | antB, anthranilate dioxygenase subunit | −5.8 (1.8E-2) | 1.0 (9.8E-1) | −1.2 (5.0E-1) |
| 2658 | Hypothetical protein | 5.0 (1.7E-2) | 2.4 (1.5E-3) | 2.2 (8.0E-3) |
| 2659 | Hypothetical protein | 7.2 (7.5E-3) | 3.4 (2.9E-4) | 3.3 (4.0E-2) |
| 2760 | Probable outer membrane protein | −5.0 (4.6E-2) | −1.1 (5.6E-1) | 1.4 (3.0E-2) |
| 3540 | algD, GDP-mannose 6-dehydrogenase | 11.1 (1.1E-1) | 10.5 (5.7E-3) | 3.6 (1.3E-1) |
| 3541 | Alginate biosynthesis protein | 2.8 (2.2E-2) | 4.8 (2.4E-2) | Δ |
| 3542 | Alginate biosynthesis protein | 4.9 (8.6E-2) | 3.8 (2.5E-4) | Δ |
| 3543 | algK, alginate biosynthetic protein | 13.6 (2.0E-1) | Δ | Δ |
| 3544 | algE, outer membrane protein | 3.3 (5.3E-2) | 3.7 (1.6E-3) | Δ |
| 3545 | algG, alginate-c5-mannuronan-epimerase | 4.1 (7.3E-2) | 6.0 (3.4E-4) | Δ |
| 3546 | algX, alginate biosynthesis protein | 4.2 (1.8E-2) | 3.3 (3.8E-3) | Δ |
| 3547 | algL, poly(beta-d-mannuronate) lyase | 5.6 (9.6E-2) | 3.0 (1.5E-3) | 1.3 (4.1E-1) |
| 3548 | algI, alginate O-acetyltransferase | 2.8 (8.2E-2) | 2.8 (6.3E-3) | Δ |
| 3549 | algJ, alginate O-acetyltransferase | 6.5 (2.6E-2) | 2.8 (6.2E-4) | Δ |
| 3550 | algF, alginate O-acetyltransferase | 17.3 (4.1E-2) | 10.0 (3.8E-3) | 3.5 (2.1E-2) |
| 3551 | algA, phosphomannose isomerase 5-diphospho-d-mannose pyrophosphorylase | 8.6 (5.0E-2) | 4.2 (3.6E-5) | 2.9 (9.9E-4) |
| 3554 | Hypothetical protein | 5.1 (4.9E-2) | 1.5 (9.0E-2) | 1.4 (3.3E-1) |
| 3559 | Probable nucleotide sugar dehydrogenase | 5.9 (1.7E-2) | 1.9 (3.8E-2) | 2.0 (1.1E-2) |
| 3569 | mmsB, dehydrogenase | −6.9 (9.6E-3) | −1.7 (1.2E-2) | −1.1 (5.0E-1) |
| 3790 | oprC, outer membrane protein | −5.3 (1.3E-2) | −1.2 (6.7E-1) | 1.3 (1.9E-1) |
| 4110 | ampC, β-lactamase precursor | 67.0 (9.5E-3) | 86.0 (5.5E-4) | 150.5 (3.3E-3) |
| 4111 | Hypothetical protein | 18.8 (6.4E-4) | 13.0 (1.1E-3) | 14.8 (1.9E-2) |
| 4112 | Probable sensor-response regulator | 2.3 (6.9E-3) | 1.2 (9.7E-2) | 1.4 (2.6E-1) |
| 4306 | Hypothetical protein | −6.5 (1.9E-2) | −1.1 (7.9E-1) | −1.1 (8.7E-1) |
| 4407 | ftsZ, cell division protein | 1.3 (1.9E-1) | 2.5 (7.3E-3) | 2.7 (1.9E-4) |
| 4408 | ftsA, cell division protein | 1.7 (3.4E-2) | 2.1 (8.1E-5) | 2.2 (2.3E-2) |
| 4409 | ftsQ, cell division protein | 1.3 (1.6E-1) | 2.3 (1.2E-5) | 1.9 (9.3E-3) |
| 4410 | ddlB, d-alanine-d-alanine ligase | 1.3 (3.0E-1 | 2.3 (7.0E-3) | 2.2 (4.5E-2) |
| 4411 | murC, UDP-N-acetylmuramate-alanine ligase | 1.1 (2.3E-1) | 2.5 (3.5E-3) | 2.0 (1.6E-2) |
| 4412 | murG, transferase | 1.5 (2.1E-1) | 2.4 (1.9E-3) | 2.6 (1.2E-2) |
| 4413 | ftsW, cell division protein | 1.2 (2.6E-1) | 2.0 (3.9E-3) | 2.5 (4.8E-4) |
| 4414 | murD, UDP-N-acetylmuramate-d-glutamate ligase | 1.1 (1.5E-1) | 2.1 (4.5E-4) | 2.4 (1.8E-3) |
| 4415 | mraY, phospho-N-acetylmuramate-peptidyl transferase | 1.2 (2.9E-1) | 2.1 (1.3E-4) | 2.1 (3.1E-3) |
| 4416 | murF, UDP-N-acetylmuramate-d-glutamyl- diaminopimelate-d-alanyl-d-alanyl ligase | 1.2 (5.8E-2) | 2.6 (2.3E-4) | 2.1 (4.1E-3) |
| 4417 | murE, UDP-N-acetylmuramoylalanyl-d-glutamate- diaminopimelate ligase | 1.1 (3.2E-1) | 2.3 (2.7E-3) | 2.1 (7.7E-3) |
| 4419 | ftsL, cell division protein | 1.0 (8.9E-1) | 2.4 (1.9E-3) | 2.6 (7.3E-3) |
| 4420 | Hypothetical protein | 1.1 (5.2E-1) | 2.1 (1.6E-2) | 2.1 (4.7E-3) |
| 4421 | Hypothetical protein | −1.1 (3.6E-1) | 2.2 (9.9E-4) | 1.9 (8.4E-3) |
| 4507 | Hypothetical protein | −6.9 (1.7E-2) | −2.1 (1.9E-2) | −1.6 (1.1E-1) |
| 4525 | pilA, type 4 fimbrial precursor | −8.1 (3.5E-2) | −1.8 (1.1E-1) | −1.9 (2.9E-1) |
| 4526 | pilB, type 4 fimbrial biogenesis protein | −3.0 (2.7E-2) | −1.3 (2.2E-1) | −1.5 (1.8E-1) |
| 5040 | pilQ, type 4 fimbrial biogenesis protein | −2.5 (1.7E-2) | −1.2 (3.3E-1) | 1.1 (5.9E-1) |
| 5041 | pilP, type 4 fimbrial biogenesis protein | −3.1 (1.6E-2) | −1.1 (5.5E-1) | −1.0 (8.7E-1) |
| 5042 | pilO, type 4 fimbrial biogenesis protein | −2.3 (6.0E-3) | −1.2 (1.8E-1) | 1.1 (4.9E-1) |
| 5044 | pilM, type 4 fimbrial biogenesis protein | −2.1 (4.7E-2) | −1.1 (4.9E-1) | 1.2 (4.1E-1) |
| 5170 | arcD, arginine-ornithine antiporter | −5.7 (2.3E-3) | −1.4 (1.2E-1) | −1.0 (9.4E-1) |
| 5182 | Hypothetical protein | 6.0 (2.2E-2) | 3.5 (9.9E-4) | 2.7 (2.2E-3) |
| 5261e | algR, alginate biosynthesis regulatory protein | 1.9 (1.2E-1) | 1.3 (1.9E-1) | 1.4 (2.0E-1) |
| 5262e | algZ, alginate biosynthesis protein | 1.5 (2.8E-1) | 1.2 (4.3E-2) | 1.3 (2.0E-1) |
| 5322e | algC, phosphomannomutase | 1.8 (1.3E-2) | 1.4 (4.6E-2) | 1.4 (2.9E-2) |
The table includes only a fraction (76 of 336) of the genes differentially expressed in response to imipenem. The complete list of all 336 differentially expressed genes is available at http://www.immi.ku.dk/bagge-arrays2003.
Gene number from the P. aeruginosa genome project (http://www.pseudomonas.com). Genes indicated in boldface are mentioned in the text. Arrows indicate the transcriptional orientations of the operons of interest.
E, × 10 (e.g., 1.2E−2 = 1.2 × 10−2).
Δ, MAS 5.0 indicated that the signal was absent; i.e., the levels of expression were below the lower detection level of the chip.
The gene was not significantly differentially expressed more than twofold on day 1, 2, or 3; but the trend for the gene may be of interest.
FIG. 1.
Genes of P. aeruginosa growing in biofilms which were up- or downregulated more than fivefold in response to imipenem (1.0 μg/ml). Only genes of assigned function are shown.
The algU-mucABC gene cluster (PA0762 to PA0765), which encodes a sigma factor, an anti-sigma factor, and regulators of the algD to algA gene cluster, was slightly induced by imipenem (Table 1) (42, 43). The AlgU polypeptide is a member of the sigma E-like family of stress sigma factors (42). AlgU activates transcription of rpoH (PA0376), which encodes a sigma factor, RpoH, that affects the expression of other systems participating in defense against temperature and oxidative stresses (16). Exposure to imipenem caused a slightly increased level of expression of rpoH (just above twofold) on day 2.
The expression of flagellum-encoding genes flgC to flgI (PA1078 to PA1084) was repressed up to fivefold due to exposure to imipenem, but the repression was the most significant in the younger biofilm. Also, the pilus-encoding genes pilA, pilB, and pilM to pilQ (PA5044 to PA5040) were downregulated; again, this was most significant in the younger biofilm. Genes involved in cell division and the peptidoglycan biosynthetic pathway (PA4421 to PA4407) were induced by imipenem by approximately twofold in the 2- and 3-day-old biofilms. Genes other than ampC previously reported to encode genes influencing β-lactamase production, i.e., ampR, ampD, ampG, and, possibly, ampE, were found to be constantly expressed in the presence and absence of imipenem.
Verification of microarray results by real-time PCR.
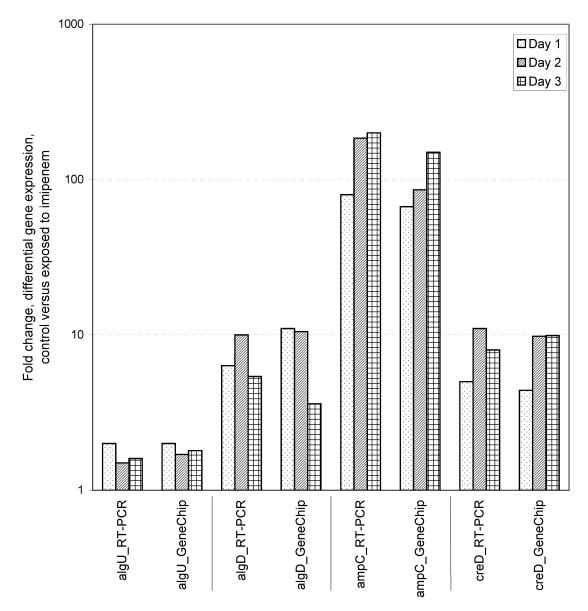
Real-time PCR was used to verify the gene expression results for selected genes obtained by the microarray experiments. Four genes were selected for real-time PCR: algU, algD, ampC, and creD. The result for each sample of total RNA (from all replicates) used for GeneChip microarray analyses was verified by real-time PCR. The results correlated with the gene expression results from the microarray experiments (Fig. 2). Repeat of the whole real-time PCR procedure, i.e., reverse transcription and the real-time PCR assay, showed assay-to-assay variations in the range of 2 to 25%. The ampR gene was chosen as a reference gene for real-time PCR. The average signal intensity of ampR was 1,319 ± 81 (P = 0.05).
FIG. 2.
Verification of GeneChip microarray results by real-time PCR (RT-PCR). The average fold changes in gene expression due to exposure to imipenem on days 1, 2, and 3 are shown.
Gene expression versus concentration of imipenem.
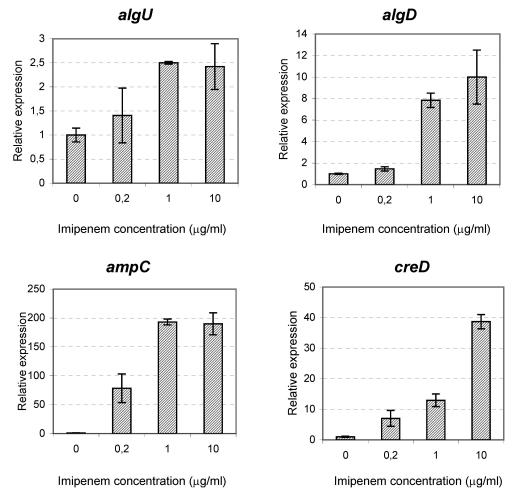
A subinhibitory concentration of imipenem (1.0 μg/ml) was chosen for use in the microarray experiments to avoid any growth-related effects on gene expression (MICs, 2 to 4 μg/ml). However, clinically, we intend to expose P. aeruginosa to higher concentrations of imipenem. Genes that were marginally affected by imipenem in our microarray experiments could actually be significantly affected by higher concentrations of imipenem. Figure 3 shows the real-time PCR results for P. aeruginosa cells growing in biofilms exposed to imipenem at 0, 0.2, 1.0, or 10.0 μg/ml. Imipenem was administered for 7 h at day 2. Besides algU, algD, and ampC, the experiments also included the gene for an inner membrane protein, creD. The results for algU, algD, and ampC showed that the level of gene expression in the presence of the highest imipenem concentration, 10 μg/ml, was not significantly higher than the level at 1.0 μg/ml. For creD, the level of expression in the presence of imipenem at 10 μg/ml was threefold higher than that in the presence of the drug at 1.0 μg/ml. Comparison of the levels of expression in the presence of imipenem at 0, 0.2, and 1.0 μg/ml showed a correlation between the level of imipenem and the level of gene expression.
FIG. 3.
Expression of algU, algD, ampC, and creD genes in the presence of increasing concentrations of imipenem measured by real-time PCR. Each experiment was done in duplicate. The data are presented as means, and error bars represent 95% confidence intervals. In all four charts, the expression levels are normalized to the level of expression of the noninduced biofilms.
Imipenem increases the level of alginate production.
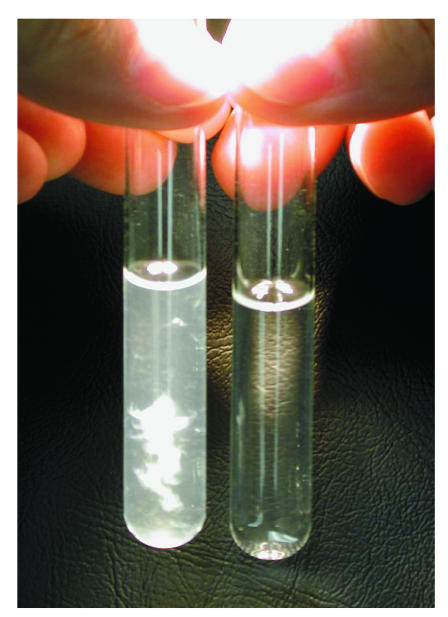
The increased levels of expression of alginate genes in the microarray experiments indicated that PAO1 cells growing in biofilms might have increased levels of alginate production in the presence of imipenem. To quantify the amount of alginate in P. aeruginosa cells growing in biofilms, we performed an uronic acid assay with cells from biofilms exposed to imipenem (1.0 μg/ml) for 24 h and noninduced biofilms. The 24-h induction was intended to maximize the production of alginate in response to imipenem exposure. The results showed a 20-fold higher level of alginate in the biofilm matrix of the imipenem-exposed PAO1 biofilms than in the control biofilms. Precipitates of the extracted exopolysaccharide matrix showed a clear difference between the noninduced and the imipenem-induced biofilms (Fig. 4). However, P. aeruginosa from imipenem-exposed biofilms subsequently cultured on nonselective Luria-Bertani agar plates did not demonstrate a mucoid phenotype.
FIG. 4.
Ethanol precipitation of exopolysaccharides from a P. aeruginosa biofilm exposed to imipenem (1.0 μg/ml) (left) compared to that for a nonexposed biofilm (right). The biofilm was exposed to imipenem for 24 h. Uronic acid assay measurements verified the increased levels of production of uronic acids from the imipenem-exposed P. aeruginosa biofilm, showing an approximately 20-fold higher level of uronic acids.
Differentially expressed genes in planktonic P. aeruginosa in response to imipenem.
Planktonic P. aeruginosa cells for microarray experiments were cultured in chemostats. The gene expression results for cells exposed to imipenem (0.2 μg/ml) for 7 h compared to those for cells that were not exposed from experiments performed in quadruplicate are presented in Table 2. The ampC gene and PA2659 were the only genes that were significantly induced (i.e., more than twofold; P < 0.05) in response to imipenem. We also included in Table 2 the results for a number of genes, even though they did not fulfill these criteria, as we wanted to observe the trends in differential gene expression of those genes from experiments with planktonic cells compared with the results obtained for the P. aeruginosa cells growing in biofilms and exposed to different concentrations of imipenem (cf. the results in Table 2 and those in Table 1).
TABLE 2.
Effects of imipenem on planktonic P. aeruginosa genes
| Gene no.a | Description | Fold change in expression level (P value)b |
|---|---|---|
| 0376 | rpoH, sigma factor | −1.1 (3.3E-1) |
| 0464 | creC, two-component sensor | −1.3 (7.0E-1) |
| 0465 | creD, inner membrane protein | −1.2 (8.4E-1) |
| 0762 | algU, sigma factor | 1.2 (5.5E-1) |
| 0763 | mucA, anti-sigma factor | 1.1 (7.0E-1) |
| 0764 | mucB, regulator for alginate biosynthesis | 1.1 (9.3E-1) |
| 0765 | mucC, regulator for alginate biosynthesis | −1.0 (9.6E-1) |
| 1078 | flgC, flagellar basal-body rod protein | −1.2 (3.7E-1) |
| 1079 | flgD, flagellar basal-body rod protein | −1.3 (3.7E-1) |
| 1080 | flgE, flagellar hook protein | −1.1 (8.6E-1) |
| 1081 | flgF, flagellar basal-body rod protein | −1.1 (5.3E-1) |
| 1084 | flgI, flagellar P-ring protein precursor | −1.2 (4.7E-1) |
| 1178 | oprH, outer membrane protein H1 precursor | 1.6 (3.8E-2) |
| 2659 | Hypothetical protein | 2.1 (3.3E-2) |
| 3540 | algD, GDP-mannose 6-dehydrogenase | 2.9 (1.4E-1) |
| 3541 | Alginate biosynthesis protein | 1.9 (2.1E-1) |
| 3542 | Alginate biosynthesis protein | 1.5 (1.2E-1) |
| 3543 | algK, alginate biosynthetic protein | 4.8 (2.3E-1) |
| 3544 | algE, outer membrane protein | 1.7 (2.0E-1) |
| 3545 | algG, alginate-c5-mannuronan-epimerase | 1.8 (3.0E-1) |
| 3546 | algX, alginate biosynthesis protein | 1.6 (3.3E-1) |
| 3547 | algL, poly(beta-d-mannuronate) lyase precursor | 1.4 (2.5E-1) |
| 3548 | algI, alginate O-acetyltransferase | 1.2 (6.5E-1) |
| 3549 | algJ, alginate O-acetyltransferase | 1.3 (4.3E-1) |
| 3550 | algF, alginate O-acetyltransferase | 3.0 (1.7E-1) |
| 3551 | algA, phosphomannose isomerase/guanosine | 1.9 (2.7E-1) |
| 4110 | ampC, β-lactamase precursor | 19.1 (4.2E-2) |
| 4407 | ftsZ, cell division protein | 1.1 (4.6E-1) |
| 4408 | ftsA, cell division protein | 1.2 (1.3E-1) |
| 4409 | ftsQ, cell division protein | 1.3 (9.4E-2) |
| 4410 | ddlB, d-alanine-d-alanine ligase | 1.2 (2.0E-1) |
| 4411 | murC, UDP-N-acetylmuramate-alanine ligase | 1.1 (4.7E-1) |
| 4412 | murG, transferase | 1.3 (8.6E-2) |
| 4413 | ftsW, cell division protein | 1.1 (5.0E-1) |
| 4414 | murD, UDP-N-acetylmuramate-d-glutamate ligase | 1.4 (8.4E-2) |
| 4415 | mraY, phospho-N-acetylmuramate-peptidylpept.-transferase | 1.2 (1.2E-1) |
| 4416 | murF, UDP-N-acetylmuramate-d-glutamyl-diaminopimelate-d-alanyl-d-alanyl ligase | 1.3 (9.9E-2) |
| 4417 | murE, UDP-N-acetylmuramoylalanyl-d-glutamate-diaminopimelate ligase | 1.1 (3.6E-1) |
| 4419 | ftsL, cell division protein | 1.2 (7.2E-1) |
| 4420 | Hypothetical protein | 1.4 (4.4E-1) |
| 4421 | Hypothetical protein | 1.2 (5.8E-1) |
| 4525 | pilA, type 4 fimbrial precursor | −1.3 (5.7E-1) |
| 4526 | pilB, type 4 fimbrial biogenesis protein | −1.2 (5.6E-1) |
| 5040 | pilQ, type 4 fimbrial biogenesis protein | −1.3 (2.2E-1) |
| 5041 | pilP, type 4 fimbrial biogenesis protein | −1.2 (2.8E-1) |
| 5042 | pilO, type 4 fimbrial biogenesis protein | −1.2 (2.8E-1) |
| 5044 | pilM, type 4 fimbrial biogenesis protein | −1.1 (5.6E-1) |
| 5261 | algR, alginate biosynthesis regulatory protein | 1.3 (4.9E-1) |
| 5262 | algZ, alginate biosynthesis protein | 1.3 (5.0E-1) |
| 5322 | algC, Phosphomannomutase | 1.1 (1.0E-1) |
Gene number from the P. aeruginosa genome project (http://www.pseudomonas.com). Genes indicated in boldface passed the t test (P < 0.05). Arrows indicate the transcriptional orientations of the operons of interest.
E, × 10 (e.g., 3.3E−1 = 3.3 × 10−1).
The entire alginate operon (PA3540 to PA3551) was slightly induced (approximately 1.5- to 2-fold) due to imipenem exposure (Table 2). The expression of alginate regulatory genes also showed a slight induction due to imipenem. Flagellum-encoding genes flgB to flgL, the pilus-encoding genes, and the genes involved in the peptidoglycan biosynthetic pathway showed differential gene expression patterns comparable to those of the samples from the biofilms.
Effect of imipenem exposure on biofilm structure.
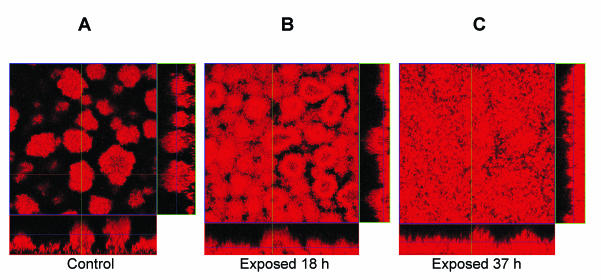
To study the influence of imipenem on the structure of the biofilm, we performed experiments in flow cells. On day 5 we compared the results for PAO1 from nontreated biofilms to those for PAO1 from biofilms exposed to imipenem (0.5 μg/ml) for 18 or 37 h (Fig. 5 and Table 3). The biofilms were inoculated and cultured simultaneously in parallel flow-cell channels and supplied with the same medium. The results of quantitative image analyses revealed by the COMSTAT computer program showed significant differences in the biofilm architectures due to imipenem (Fig. 5 and Table 3). Biofilms exposed to imipenem developed more biomass per substratum area and an increased average thickness. The control (noninduced) biofilms formed numerous towers and pillars of microcolonies separated by areas devoid of cells, thereby revealing a higher roughness coefficient (Fig. 5 and Table 3). The smoother biofilm structure of the imipenem-exposed biofilms also resulted in a lower surface-to-volume ratio, and this ratio was significantly lower in the biofilm exposed to imipenem for 37 h, in which few towers and pillars were observed.
FIG. 5.
Scanning confocal photomicrographs of P. aeruginosa PAO1 biofilms established in flow cells. The effects of exposure to imipenem (0.5 μg/ml) for 37 h (C) or 18 h (B) or no exposure (A) were examined on day 5. For each condition (A to C) a top view and two side views of the biofilms are shown. Biofilms were stained with the cell-permeant nucleic acid stain SYTO 62.
TABLE 3.
Data from quantitative image analysis of P. aeruginosa PAO1 biofilmsa
| Duration of imipenem exposure | Biomass (μm3/μm2) | Avg thickness (μm) | Surface-to-biovolume ratio (μm2/μm3) |
|---|---|---|---|
| Control | 10.8 ± 1.2 | 15.2 ± 2.4 | 1.7 ± 0.1 |
| 18 h | 17.2 ± 1.7 | 22.0 ± 2.5 | 1.5 ± 0.3 |
| 37 h | 18.1 ± 1.5 | 21.0 ± 1.8 | 1.2 ± 0.1 |
Quantitative image analyses were performed to compare the architectures of imipenem (0.5 μg/ml)-exposed biofilms and unexposed biofilms (control) on day 5. The values are the averages of 10-image stacks from each flow cell channel acquired in one experiment. The biomass is calculated as the biomass volume (in cubic micrometers) per substratum surface area (in square micrometers). The thickness is the average depth of the biofilm. The surface-to-biovolume ratio describes the surface area of the biofilm (in square micrometers) per biomass volume of the biofilm community (in cubic micrometers). The data are presented as means ± 95% confidence intervals. The data were quantified with the computer program COMSTAT.
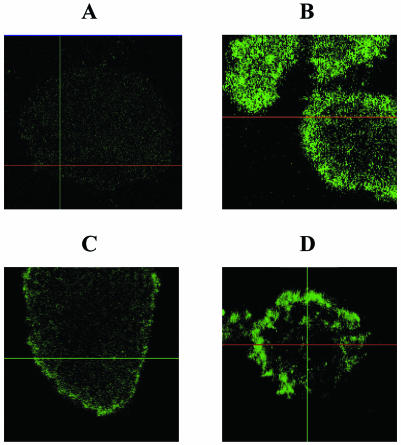
To visualize whether imipenem exposure caused increased levels of polysaccharides such as alginate in the biofilms, the biofilms were treated with a fluorescent lectin (ConA-FITC). PAO1 biofilms were treated on day 5 after inoculation, and PDO300 biofilms were treated on day 3 after inoculation. Mucoid strain PDO300 showed a higher level of staining (Fig. 6B) compared to that for PAO1 not exposed to imipenem (Fig. 6A). However, exposure of PAO1 to imipenem for 18 h (Fig. 6C) or 37 h (Fig. 6D) caused increased levels of staining due to ConA-FITC binding to alginate.
FIG. 6.
Scanning confocal photomicrographs of P. aeruginosa biofilms established in flow cells. Select microcolonies stained with ConA-FITC, which reveals polysaccharides such as alginate, are shown from the top. (A) PAO1 biofilm not exposed to antibiotics; (B) PDO300 (a derivate of PAO1 that constitutively overproduces alginate) not exposed to antibiotics; (C) PAO1 exposed to imipenem for 18 h; (D) PAO1 exposed to imipenem for 37 h.
DISCUSSION
We have used the GeneChip microarray technology to identify changes in the global gene expression pattern of P. aeruginosa exposed to subinhibitory concentrations of the β-lactam antibiotic imipenem. It is well established that chromosomal AmpC β-lactamase often compromises antipseudomonal β-lactam chemotherapy in patients with chronic P. aeruginosa lung infections (13, 14). The results of this study are consistent with the hypothesis that AmpC is an important part of the response to imipenem. The biofilm microarray experiments showed that the level of induction of ampC increased 67- to 150-fold after exposure to imipenem, which correlated with the level of induction measured by the β-lactamase assay. Owing to the strong induction potential of imipenem, the high level of expression of ampC was expected (27, 32).
We used a low concentration of imipenem for our biofilm experiments, which corresponded to 0.5 the MIC for planktonic cell cultures. The intention of antipseudomonal treatment of patients with CF is to administer the antibiotics at concentrations that exceed the MIC. However, as the antimicrobial treatments do not lead to clearance of the P. aeruginosa cells growing in biofilms from the lungs of CF patients, we would expect that persistent bacteria are exposed to subinhibitory concentrations (or at least sub-minimal bactericidal concentrations). The scope of the present study was to reveal the specific effects of imipenem treatment on gene expression, i.e., the consequence of treatment on the average expression profile among all cells constituting the bacterial communities studied. Subinhibitory concentrations of imipenem were applied to avoid obtaining a gene expression pattern that reflected both the specific effects of imipenem and the general growth inhibition effects. However, this experimental scenario is a difficult balancing act, as the super-MIC actions of β-lactam antibiotics actually affect growth by blocking peptidoglycan synthesis.
Among the genes that responded to imipenem exposure were the alginate biosynthesis genes (algD to algA), which, in particular, were induced 5- to 10-fold on days 1 and 2. The increased level of expression of the alginate genes was supported by the increased level of production of alginate, as measured by the uronic acid assay. Our results are in accordance with the previous observations of Takahashi et al. (48) and Majtán and Hybenová (34), which showed the induction of alginate production by planktonic P. aeruginosa cells exposed to β-lactam antibiotics, although they did not study the response to imipenem of P. aeruginosa cells growing in biofilms.
In Azotobacter vinelandii, a gram-negative aerobic soil bacterium, inactivation of the ampDE operon caused a significantly increased level of algD expression (38). A. vinelandii does not contain an inducible ampC β-lactamase gene, suggesting that the function of AmpD is solely related to recycling of muropeptides in A. vinelandii. The transcriptional regulators of alginate and the alginate biosynthetic operon show high degrees of homology in A. vinelandii and P. aeruginosa, as is also the case for the ampDE operon. Núñez et al. (38) presented evidence suggesting that peptidoglycan recycling participates in the regulation of alginate production in A. vinelandii. They suggested that muropeptides might affect an AmpR-like regulator of the alginate regulatory circuit (38).
The mechanism of alginate expression in P. aeruginosa is complex and involves several regulatory genes affecting the transcription of the alginate biosynthetic genes (algD to algA) (16). Transcription of the alginate operon is dependent on the sigma factor AlgU, but the response regulator AlgR and the DNA-binding protein AlgZ are also essential for alginate synthesis and transcriptional activation (3). All the details of the regulatory circuitry controlling the expression of alginate are not completely clarified. However, it may be hypothesized that the recycling of muropeptides is linked to the regulation of alginate expression, e.g., by affecting the properties of AlgZ or AlgR.
The results of our microarray experiments showed increased levels of expression of almost all genes involved in the alginate circuit. Some regulatory genes, e.g., algZ, algR, and algC, revealed only slight tendencies to have increased levels of expression. AlgU has been reported to induce the transcription of rpoH, which causes the expression of systems participating in different stress responses (16). AlgU has also been associated with the negative control of flagellum synthesis (10). This correlates with our microarray results, as all of the genes of the flagellum biosynthetic operon, i.e., flgC to flgI (PA1078 to PA1084), were repressed in response to imipenem.
It is tempting to agree with the hypothesis that there is a link between recycling of muropeptides and alginate expression (38). On the other hand, expression of alginate is also induced in the presence of ofloxacin or norfloxacin. In this case the target of these quinolone antibiotics is DNA gyrase, which does not affect the peptidoglycan layer (48). Therefore, multiple antibiotic-induced pathways leading to alginate induction may exist. An algU-independent pathway has been proposed, in which alginate production and transcription of the critical algD promoter depend on another alternative sigma factor, RpoN (sigma 54) (5). However, this gene was not induced in our experiments.
To examine whether the transcriptional response to imipenem was affected by the mode of growth in the biofilm, we performed experiments with planktonic cells in chemostats. The trends were comparable to the results obtained with the biofilms, but as we could use only low levels of imipenem in the experiments with planktonic cells, the responses were muted in comparison to the biofilm responses. Thus, our experiments did not reveal a mode-of-growth-dependent response to imipenem exposure that could account for the differences in the levels of resistance of P. aeruginosa cells growing in biofilms and those of planktonic cells.
The increased level of expression of alginate in response to imipenem exposure may be an unintended adverse consequence of imipenem treatment of chronic P. aeruginosa lung infections in CF patients, as alginate is known to provide a protected environment for biofilm bacteria. Increased production of alginate decreases the susceptibilities of bacteria growing in biofilms to numerous antibiotics. The penetration of antibiotics through the biofilm may be reduced due to binding to alginate, as has been reported for aminoglycosides, e.g., tobramycin (1, 15, 21, 23). However, exposure to tobramycin does not affect the expression of alginate in P. aeruginosa biofilms (PAO1), as reported by Whiteley et al. (50). The expression profile effected by imipenem versus that effected by tobramycin showed only common dominant genes, hypothetical genes PA2703 and PA3819, which were slightly upregulated (twofold) with imipenem exposure (cf. http://www.immi.ku.dk/bagge-arrays2003) (50). Alginate is not believed to act as a barrier to β-lactam antibiotics, as these are relatively uncharged agents (11, 37). However, if P. aeruginosa produces high levels of β-lactamase, the enzyme may accumulate in the extracellular biofilm matrix and hydrolyze β-lactam antibiotics as they penetrate the biofilm matrix (9). Imipenem-induced β-lactamase production has previously been shown in P. aeruginosa biofilms, and P. aeruginosa secretes chromosomal AmpC β-lactamase packaged in membrane vesicles, which may accumulate in the biofilm matrix (7, 12). This may explain previous observations that a major part of the β-lactamase in biofilms is located extracellularly (7, 14). The prolonged imipenem exposure resulted in structural changes of the P. aeruginosa biofilms, causing the biofilm to have an increased thickness and biomass (Fig. 5 and Table 3). This may increase the potential of β-lactamase accumulation in the biofilm, thereby reducing the effect of β-lactam treatment on P. aeruginosa cells growing in biofilms. If the increased biomass and lower surface-to-biovolume ratio of the biofilm were due to the increased level of production of alginate, this may reduce the accessibility of antibiotics such as aminoglycosides to the bacteria growing in biofilms, thereby reducing the efficacies of the antimicrobial treatments. The results of our experiments showing increased levels of lectin binding to the imipenem-exposed biofilms point to the presence of increased levels of alginate in the biofilms. Increased levels of alginate in biofilms have been reported to contribute to oxygen limitations in the biofilm (18). If compartments of the biofilm become anaerobic, the efficacies of aminoglycosides are significantly reduced or abolished (19). Reduced accessibility to oxygen and nutrients may reduce the growth rate of the bacteria in the biofilm, thereby reducing the antimicrobial actions of β-lactam antibiotics, as the action of transpeptidase inhibition is related to the growth rate.
Mucoid isolates embedded in alginate, which constitutes a protected environment, may be less affected than nonmucoid isolates by a transient induction by a β-lactam antibiotic. The effect of induction may be overshadowed by the constitutive production of alginate. However, the relative level of alginate present in the biofilm matrices of nonmucoid isolates may be significantly increased due to the inductive effect of imipenem, as alginate is not a significant constituent of the extracellular polymeric material of nonmucoid P. aeruginosa cells involved in the initial colonization of the lungs of CF patients (53). Thus, the induction of alginate may compromise the efficacy of antimicrobial treatment, increasing the risk that nonmucoid colonizers will survive in the lungs of CF patients.
Acknowledgments
We thank Jette Teglhus Møller, Lora Huang, and Jessica Anna Linton for excellent technical assistance. We also thank Stephen Lory for supplying the control spiked transcripts for GeneChip microarray analysis. We thank Pradeep K. Singh for helpful discussions and assistance with digital imaging.
This work was supported by grants from H:S (Hovedstadens Sygehusfællesskab, Copenhagen, Denmark), The Danish Medical Research Council, The Danish Technical Research Council, The Villum Kann Rasmussen Foundation (Denmark), and the National Institute of General Medicine (grant GM59026). Support was also provided through grants from the American Cystic Fibrosis Foundation.
REFERENCES
- 1.Allison, D. G., and M. J. Matthews. 1992. Effect of polysaccharide interactions on antibiotic susceptibility of Pseudomonas aeruginosa. J. Appl. Bacteriol. 73:484-488. [DOI] [PubMed] [Google Scholar]
- 2.Bagge, N., O. Ciofu, M. Hentzer, J. I. A. Campbell, M. Givskov, and N. Høiby. 2002. Constitutive high expression of chromosomal beta-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob. Agents Chemother. 46:3406-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baynham, P. J., A. L. Brown, L. L. Hall, and D. J. Wozniak. 1999. Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA-binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol. Microbiol. 33:1069-1080. [DOI] [PubMed] [Google Scholar]
- 4.Bolister, N., M. Basker, N. A. Hodges, and C. Marriott. 1991. The diffusion of beta-lactam antibiotics through mixed gels of cystic fibrosis-derived mucin and Pseudomonas aeruginosa alginate. J. Antimicrob. Chemother. 27:285-293. [DOI] [PubMed] [Google Scholar]
- 5.Boucher, J. C., M. J. Schurr, and V. Deretic. 2000. Dual regulation of mucoidy in Pseudomonas aeruginosa and sigma factor antagonism. Mol. Microbiol. 36:341-351. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools in physiological studies of microbial biofilms. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 7.Ciofu, O., T. J. Beveridge, J. Kadurugamuwa, J. Walther-Rasmussen, and N. Høiby. 2000. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 45:9-13. [DOI] [PubMed] [Google Scholar]
- 8.Ciofu, O., B. Giwercman, S. S. Pedersen, and N. Høiby. 1994. Development of antibiotic resistance in Pseudomonas aeruginosa during two decades of antipseudomonal treatment at the Danish CF Center. APMIS 102:674-680. [PubMed] [Google Scholar]
- 9.Dibdin, G. H., S. J. Assinder, W. W. Nichols, and P. A. Lambert. 1996. Mathematical model of beta-lactam penetration into a biofilm of Pseudomonas aeruginosa while undergoing simultaneous inactivation by released beta-lactamase. J. Antimicrob. Chemother. 38:757-769. [DOI] [PubMed] [Google Scholar]
- 10.Garrett, E. S., D. Perlegas, and D. J. Wozniak. 1999. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU). J. Bacteriol. 181:7401-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert, P., and M. R. W. Brown. 1998. Biofilms and beta-lactam activity. J. Antimicrob. Chemother. 41:571-578. [DOI] [PubMed] [Google Scholar]
- 12.Giwercman, B., E. T. Jensen, N. Høiby, A. Kharazmi, and J. W. Costerton. 1991. Induction of beta-lactamase production in Pseudomonas aeruginosa biofilm. Antimicrob. Agents Chemother. 35:1008-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giwercman, B., P. A. Lambert, V. T. Rosdahl, G. H. Shand, and N. Høiby. 1990. Rapid emergence of resistance in Pseudomonas aeruginosa in cystic fibrosis due to in vivo selection of stable derepressed beta-lactamase producing strains. J. Antimicrob. Chemother. 26:247-259. [DOI] [PubMed] [Google Scholar]
- 14.Giwercman, B., C. Meyer, P. A. Lambert, C. Reinert, and N. Høiby. 1992. High-level beta-lactamase activity in sputum samples from cystic fibrosis patients during antipseudomonal treatment. Antimicrob. Agents Chemother. 36:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon, C. A., N. A. Hodges, and C. Marriott. 1988. Antibiotic interaction and diffusion through alginate and exopolysaccharide of cystic fibrosis-derived Pseudomonas aeruginosa. J. Antimicrob. Chemother. 22:667-674. [DOI] [PubMed] [Google Scholar]
- 16.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg, E. P. 2000. Pump up the versatility. Nature 406:947-948. [DOI] [PubMed] [Google Scholar]
- 18.Hassett, D. J. 1996. Anaerobic production of alginate by Pseudomonas aeruginosa: alginate restricts diffusion of oxygen. J. Bacteriol. 178:7322-7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassett, D. J., J. Cuppoletti, B. Trapnell, S. V. Lymar, J. J. Rowe, S. S. Yoon, G. M. Hilliard, K. Parvatiyar, M. C. Kamani, D. J. Wozniak, S.-H. Hwang, T. R. McDermott, and U. A. Ochsner. 2002. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Deliv. Rev. 54:1425-1443. [DOI] [PubMed] [Google Scholar]
- 20.Henry, R. L., C. M. Mellis, and L. Petrovic. 1992. Mucoid Pseudomonas aeruginosa is a marker of poor survival in cystic fibrosis. Pediatr. Pulmonol. 12:158-161. [DOI] [PubMed] [Google Scholar]
- 21.Hentzer, M., G. M. Teitzel, G. J. Baltzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersbøll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program Comstat. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 23.Hodges, N. A., and C. A. Gordon. 1991. Protection of Pseudomonas aeruginosa against ciprofloxacin and beta-lactams by homologous alginate. Antimicrob. Agents Chemother. 35:2450-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Høiby, N. 2000. Prospects for the prevention and control of pseudomonal infection in children with cystic fibrosis. Paediatr. Drugs 2:451-463. [DOI] [PubMed] [Google Scholar]
- 25.Høiby, N. 2002. New antimicrobials in the management of cystic fibrosis. J. Antimicrob. Chemother. 49:235-238. [DOI] [PubMed] [Google Scholar]
- 26.Høiby, N., G. Döring, and P. O. Schiøtz. 1987. Pathogenic mechanisms of chronic Pseudomonas aeruginosa infections in cystic fibrosis patients. Antibiot. Chemother. 39:60-76. [PubMed] [Google Scholar]
- 27.Jones, R. N. 1998. Important and emerging beta-lactamase-mediated resistances in hospital-based pathogens: the AmpC enzymes. Diagn. Microbiol. Infect. Dis. 31:461-466. [DOI] [PubMed] [Google Scholar]
- 28.Kintner, P. K., and J. P. Van Buren. 1982. Carbohydrate interference and its correction in pectin analysis using the m-hydroxydiphenyl method. J. Food Sci. 47:756-764. [Google Scholar]
- 29.Lautrop, H., N. Høiby, A. Bremmelgaard, and B. Korsager. 1979. Bakteriologiske undersøgelsesmetoder. FADL, Copenhagen, Denmark.
- 30.Learn, D. B., E. P. Brestel, and S. Seetharama. 1987. Hypochlorite scavenging by Pseudomonas aeruginosa alginate. Infect. Immun. 55:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livermore, D. M. 1987. Clinical significance of beta-lactamase induction and stable derepression in gram-negative rods. Eur. J. Clin. Microbiol. 6:439-445. [DOI] [PubMed] [Google Scholar]
- 32.Livermore, D. M., and Y.-J. Yang. 1987. Beta-lactamase lability and inducer power of newer beta-lactam antibiotics in relation to their activity against beta-lactamase-inducibility mutants of Pseudomonas aeruginosa. J. Infect. Dis. 155:775-782. [DOI] [PubMed] [Google Scholar]
- 33.Mah, T.-F. C., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 34.Majtán, V., and D. Hybenová. 1996. Inhibition of Pseudomonas aeruginosa alginate expression by subinhibitory concentrations of antibiotics. Folia. Microbiol. (Prague) 41:61-64. [DOI] [PubMed] [Google Scholar]
- 35.Mathee, K., O. Ciofu, C. Sternberg, P. W. Lindum, J. I. A. Campbell, P. Jensen, A. H. Johnsen, M. Givskov, D. E. Ohman, S. Molin, N. Høiby, and A. Kharazmi. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145:1349-1357. [DOI] [PubMed] [Google Scholar]
- 36.McCarthy, M. 2000. Pseudomonas genome reveals a formidable foe. Lancet 356:918. [DOI] [PubMed] [Google Scholar]
- 37.Nichols, W. W., M. J. Evans, M. P. E. Slack, and H. L. Walmsley. 1989. The penetration of antibiotics into aggregates of mucoid and non-mucoid Pseudomonas aeruginosa. J. Gen. Microbiol. 135:1291-1303. [DOI] [PubMed] [Google Scholar]
- 38.Núñez, C., S. Moreno, L. Cárdenas, G. Soberón-Chávez, and G. Espín. 2000. Inactivation of the ampDE operon increases transcription of algD and affects morphology and encystment of Azotobacter vinelandii. J. Bacteriol. 182:4829-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Callaghan, C. H., A. Morris, S. M. Kirby, and A. H. Shingler. 1972. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob. Agents Chemother. 1:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedersen, S. S., C. Koch, and N. Høiby. 1992. Prevention and early treatment of Pseudomonas aeruginosa infection, p. 135-144. In Cystic fibrosis, basic and clinical research. Elsevier, Amsterdam, The Netherlands.
- 41.Sanders, C. C. 1987. Chromosomal cephalosporinases responsible for multiple resistance to newer beta-lactam antibiotics. Annu. Rev. Microbiol. 41:573-593. [DOI] [PubMed] [Google Scholar]
- 42.Schurr, M. J., H. Yu, J. Martinez-Salazar, J. C. Boucher, and V. Deretic. 1996. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J. Bacteriol. 178:4997-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schurr, M. J., H. Yu, J. C. Boucher, N. S. Hibler, and V. Deretic. 1995. Multiple promoters and induction by heat shock of the gene encoding the alternative sigma factor AlgU (sigma E) which controls mucoidy in cystic fibrosis isolates of Pseudomonas aeruginosa. J. Bacteriol. 177:5670-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson, J. A., S. E. Smith, and R. T. Dean. 1989. Scavenging by alginate of free radicals released by macrophages. Free. Radic. Biol. Med. 6:347-353. [DOI] [PubMed] [Google Scholar]
- 45.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 46.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Logrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 47.Strathmann, M., J. Wingender, and H.-C. Flemming. 2002. Application of fluorescently labelled lectins for the visualization and biochemical characterization of polysaccharides in biofilms of Pseudomonas aeruginosa. J. Microbiol. Methods 50:237-248. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi, A., S. Yomoda, Y. Ushijima, I. Kobayashi, and M. Inoue. 1995. Ofloxacin, norfloxacin and ceftazidime increase the production of alginate and promote the formation of biofilm of Pseudomonas aeruginosa in vitro. J. Antimicrob. Chemother. 36:743-745. [DOI] [PubMed] [Google Scholar]
- 49.Walker, S. E., P. R. Walshaw, and H. Grad. 1997. Imipenem stability and staining of teeth. Can. J. Hosp. Pharm. 50:61-67. [Google Scholar]
- 50.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 51.Widdel, F., and F. Bak. 1992. The procaryotes. Springer-Verlag, New York, N.Y.
- 52.Worlitzsch, D., G. Herberth, M. Ulrich, and G. Döring. 1998. Catalase, myeloperoxidase and hydrogen peroxide in cystic fibrosis. Eur. Respir. J. 11:377-383. [DOI] [PubMed] [Google Scholar]
- 53.Wozniak, D. J., T. J. O. Wyckoff, M. Starkey, R. Keyser, P. Azadi, G. A. O'Toole, and M. R. Parsek. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 100:7907-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]