Abstract
During the pandemic 2009 episode, we conducted laboratory-based surveillance in four countries from West Africa: Senegal, Mauritania, Cape Verde, and Guinea. Specimens were obtained from 3,155 patients: 2,264 patients from Senegal, 498 patients from Cape Verde, 227 patients from Mauritania, and 166 patients from Guinea; 911 (28.9%) patients were positive for influenza, 826 (90.7%) patients were positive for influenza A, and 85 (9.3%) patients were positive for influenza B. Among the influenza A positives, 503 (60.9%) positives were H1N1pdm09, 314 (38.0%) positives were H3N2, and 9 (1.1%) positives were seasonal H1N1. The highest detection rate for seasonal influenza viruses (17.1%) occurred in the 5–14 years age group. However, for A(H1N1)pdm09, the detection rate was highest in the 15–24 years age group (35.8%). Based on the present study data, the timeline of detection of A(H1N1)pdm09 viruses in these four countries should be Cape Verde, Guinea, Mauritania, and finally, Senegal. Genetic and antigenic analyses were performed in some isolates.
Introduction
In spring of 2009, the US Centers for Disease Control and Prevention (CDC) announced the detection of a novel A(H1N1) virus (H1N1pdm09) of influenza virus causing acute respiratory illness in humans.1 The virus spread rapidly around the world, prompting the World Health Organization (WHO) to declare on June 11, 2009 the first influenza pandemic of the 21st century.2 This new isolate was identified as a swine-origin influenza virus (S-OIV), because its RNA segments were most closely related to influenza viruses isolated from pigs in North America and Eurasia.3 By the time that the WHO declared the pandemic over in August of 2010, the virus had spread to over 215 countries, with over 80 million confirmed cases and 18,000 associated deaths reported worldwide.4
However, late detections and/or circulation of the pandemic virus were observed in the African continent, notably in West Africa. Because many West African countries lack surveillance and diagnostic capacity, it has remained unclear whether the observed late identification of the pandemic virus in the region reflected a true late introduction or poor detection capacity.
The Institute Pasteur de Dakar (IPD; Senegal) is a National Influenza Center (NIC) since 1974. Through its network, the IPD has supported regional influenza surveillance activities and provided laboratory diagnostic support to several countries in the region. In this study, we aimed to describe the epidemiology and genetic characteristics of A(H1N1)pmd09 viruses from June of 2009 to October of 2010 in four West African countries (Cape Verde, Guinea, Mauritania, and Senegal) that are part of the regional surveillance network. In addition, we report the findings for seasonal influenza to provide a complete picture of the influenza viruses that circulated in the aforementioned countries over the study period. Strengths and weaknesses of the regional surveillance network are also discussed.
Materials and Methods
Study design and setting.
Senegal is situated in the western part of Africa between the Atlantic Ocean and the Sahel. The country is bordered by Mauritania in the north and Guinea in the south. The Cape Verde Islands are a few kilometers from Dakar (the capital city of Senegal) in the Atlantic Ocean (Figure 1).
Figure 1.
Geographical proximity of Senegal, Mauritania, Guinea, and Cape Verde—four West Africa countries.
Our work was based on a prospective observational study conducted from January of 2009 to December of 2010, mainly based on Senegal. Among the four countries, only Senegal has a well-established surveillance system, with several influenza sentinel sites located in urban, suburban, and rural areas (17 sentinel sites in total). Moreover, the number of sites was expanded to other regions during the pandemic for better national coverage. For other countries (Guinea, Cape Verde, and Mauritania), samples were collected in the context of the pandemic, and most of them were collected from health centers located in capital cities.
For each country, at each sentinel site, trained physicians identified all influenza-like illness (ILI) cases presenting at the clinics from Monday to Friday. An ILI case was identified as an outpatient presenting with sudden onset of fever (≥ 38°C) and cough or sore throat accompanied or not by myalgia, prostration, headache, or malaise, with the onset of symptoms occurring within the previous 3 days.
A standardized form was used to collect demographic and clinical information from the enrolled patients.
Sample collection and laboratory procedures.
Nasal-pharyngeal and oral-pharyngeal swabs were collected from all enrolled ILI cases, placed in 2-mL cryovials containing viral transport medium (Universal Transport Medium; COPAN Diagnostics Inc., Murrieta, CA), and stored at 4°C on site. In Senegal, the specimens were transported at a controlled temperature (4°C) on a weekly basis to the Unit of Medical Virology at the Institut Pasteur de Dakar. Samples from other countries were packed using a triple packaging system and transported in refrigerated cool boxes with ice packs. On arrival at the laboratory, the specimens were processed immediately for virus isolation/detection, identification, and characterization. Aliquots of samples were also stored at −80°C for additional analysis.
RNA extraction from clinical samples.
RNA extraction was performed from 200 μL of each sample using the QIAamp Viral RNA Kit (QIAGEN, Valencia, CA) according to the manufacturer's instructions. Each RNA sample was eluted with 100 μL nuclease-free water.
One-step real-time reverse-transcription polymerase chain reaction for the detection of influenza viruses.
One-step real-time reverse-transcription polymerase chain reaction (RT-PCR) was performed using the ABI 7500 platform according to the CDC protocol for the identification of influenza A(H1 and H3) and B viruses (courtesy of the Centers for Disease Control, Atlanta, GA) during the pandemic episode. The reagents and positive control material were kindly provided by the CDC. Total reaction volumes were 25 μL, containing 0.5 μL Superscript III/Platinum enzyme mix (Invitrogen, Paisley, United Kingdom), 5.5 μL H2O, 12.5 μL 2× buffer, 0.5 μL each primer and probe (0.8 μM forward primer [40 μM], 0.8 μM reverse primer [40 μM], and 0.2 μM probe [10 μM]), and 5 μL purified RNA. Each RNA sample was tested for the presence of RNAs derived from three genes: RNAse P was targeted as a measure of the extraction procedure to assess the specimen content of human RNA and lack of reaction inhibitors, the matrix gene segment was for the identification of influenza A viruses, and the nucleoprotein gene was for the identification of influenza B viruses. A second real-time RT-PCR, using the same reagent proportions, was performed for the subtyping of influenza A viruses using primers targeting hemagglutinin genes of seasonal (H1 and H3) viruses and A(H1N1)pdm09. Reverse transcription was performed at 50°C for 3 minutes and terminated at 95°C for 2 minutes. PCR was assessed after 45 cycles of 95°C for 15 seconds and 55°C for 30 seconds.5
Virus isolation and sequencing.
A selection of the influenza-positive specimens was chosen for virus isolation attempts based on the real-time RT-PCR positive (lowest Ct values), timing (taking into account periods of detection for a better coverage), and country of sampling. Samples were inoculated onto monolayers of Madin–Darby canine kidney (MDCK) cells with a maintenance media supplemented with 2 μg/mL trypsin (TPCK; reference 93630; Sigma-Aldrich) as described previously6 and incubated at 35°C for 3–7 days with daily monitoring of cytopathic effects (CPEs) by microscopy. After this step, tissue-culture fluids were harvested and screened for influenza virus by hemagglutination (HA) titration. Influenza A viruses in positive cultures were subtyped (H1 or H3) by the HA inhibition (HI) using specific antisera, which is recommended in WHO standard protocols.7
A subset of selected viruses was sent to the WHO Collaborating Center for Influenza at the National Institute for Medical Research in London, Unite Kingdom, for antigenic characterization by HA inhibition assays (HIAs) to assess the antigenic property of circulating strains and for genetic characterizations. HA and neuraminidase (NA) genes of selected viruses were sequenced using ABI Prism BigDye terminator cycle sequencing kits and an ABI-3730XL DNA analyzer. Data in fasta format were sent to the laboratory for analysis.
Data management and analysis.
Case reports were entered into an Epi Info database (Centers for Disease Control and Prevention, Atlanta, GA) and merged with laboratory data, and frequencies were analyzed using Epi Info.
Sequence alignments were performed using the BioEdit Sequence alignment Editor.8 For phylogenetic analysis, HA1 complete segments of isolates from Senegal, Mauritania, and Cape Verde were compared with 37 HA1 sequences of diverse isolates in the world downloaded from GenBank and GISAID EpiFlu databases: http://www.ncbi.nlm.nih.gov and http://platform.gisaid.org/epi3/frontend. MEGA version 5 was used for constructing a Maximum Likelihood Tree using the Tamura-nei evolutionary model with 100 bootstrap replicates, and only bootstrap values over 70 are shown.9
Statistical analysis.
We compared the distribution of pandemic cases into the different age groups to verify whether the associated rates were statistically supported. The Fisher's exact test was used, and a P value < 0.05 was considered statistically significant. We used the 0–4 year group as the reference group. The R.15.1 tool was used to perform the analyses.
Ethical considerations.
The surveillance protocol was approved as less than minimal risk research by the Senegalese National Ethical Committee of the Ministry of Health, and written consent forms were not required. Throughout the study, the database was shared with the Epidemiology Department at the Senegalese Ministry of Health and Prevention for appropriate public health actions.
Results
Demographic and clinical characteristics of ILI patients.
A total of 3,155 samples from patients presenting with ILI at the different sentinel sites in the four countries were received and analyzed in the Senegal NIC during the study period (2009 to October of 2010): 2,264 (71.8%) were from Senegal, 498 (15.8%) were from Cape Verde, 227 (7.2%) were from Mauritania, and 166 (5.3%) were from Guinea. Patient ages ranged from 1 month to 96 years (median = 3 years), with 52% of all ILI patients being less than 5 years. The male to female ratio was 1.1 (1,476 [46.8%] females and 1,666 [52.8%] males) (Table 1).
Table 1.
Demographical characteristics and symptoms
| Characteristics | Countries | Total (N = 3,155) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Senegal | Cape Verde | Mauritania | Guinea | ||||||
| 2009 (N = 936) | 2010 (N = 1,328) | 2009 (N = 463) | 2010 (N = 35) | 2009 (N = 108) | 2010 (N = 119) | 2009 (N = 48) | 2010 (N = 118) | ||
| Sex no. (%) | |||||||||
| Male | 512 (54.7) | 705 (53.1) | 199 (43.0) | 25 (71.4) | 73 (67.6) | 75 (63.0) | 18 (37.5) | 59 (50.0) | 1,666 (52.8) |
| Female | 424 (45.3) | 622 (46.8) | 263 (56.8) | 10 (28.6) | 32 (29.6) | 38 (32.0) | 30 (62.5) | 57 (48.3) | 1,476 (46.8) |
| Missing | 0 | 1 (0.1) | 1 (0.2) | 0 | 3 (2.8) | 6 (5.0) | 0 | 2 (1.7) | 13 (0.4) |
| Age (years) no. (%) | |||||||||
| 0–4 | 662 (70.7) | 656 (49.4) | 69 (14.9) | 19 (54.3) | 84 (77.8) | 71 (59.7) | 15 (31.3) | 66 (55.9) | 1,642 (52.0) |
| 5–14 | 91 (9.7) | 234 (17.6) | 57 (12.3) | 2 (5.7) | 9 (8.3) | 14 (11.8) | 2 (4.2) | 13 (11.0) | 422 (13.4) |
| 15–24 | 43 (4.6) | 148 (11.1) | 108 (23.3) | 5 (14.3) | 1 (0.9) | 0 | 10 (20.8) | 9 (7.6) | 324 (10.3) |
| 25–44 | 76 (30.89) | 157 (87.7) | 154 (62.6) | 3 (1.7) | 3 (1.2) | 0 | 13 (5.3) | 19 (10.6) | 351 (11.1) |
| 45–64 | 42 (40.0) | 52 (84.6) | 52 (49.5) | 3 (4.6) | 7 (6.7) | 0 | 4 (3.8) | 7 (10.8) | 244 (7.7) |
| 65+ | 5 (0.5) | 9 (0.7) | 8 (1.7) | 2 (5.7) | 0 | 0 | 3 (6.3) | 3 (2.5) | 30 (0.9) |
| Missing | 17 (1.8) | 69 (5.2) | 14 (3.0) | 1 (2.8) | 4 (3.7) | 34 (28.6) | 1 (2.1) | 1 (0.8) | 142 (4.5) |
| Clinical signs no. (%) | |||||||||
| Myalgia | 83 (8.9) | 264 (19.9) | 37 (8.0) | 0 | 1 (0.9) | 0 | 6 (12.5) | 7 (5.9) | 398 (12.6) |
| Fever | 835 (89.2) | 1,298 (97.7) | 412 (89.0) | 35 (100.0) | 97 (89.8) | 116 (97.5) | 34 (70.8) | 96 (81.3) | 2,923 (92.6) |
| Cough | 832 (88.9) | 1,212 (91.3) | 67 (14.5) | 6 (17.1) | 69 (63.9) | 84 (70.6) | 42 (87.5) | 114 (96.6) | 2,426 (76.9) |
| Diarrhea | 50 (5.3) | 86 (6.5) | 15 (3.2) | 0 | 3 (2.8) | 7 (5.9) | 0 | 1 (0.8) | 162 (5.1) |
| Sore throat | 468 (50.0) | 512 (38.5) | 207 (44.7) | 17 (48.6) | 47 (43.5) | 49 (41.2) | 17 (35.4) | 62 (52.5) | 1,379 (43.7) |
| Headache | 55 (5.9) | 442 (33.3) | 104 (22.5) | 2 (5.7) | 15 (13.9) | 1 (0.8) | 18 (37.5) | 46 (39.0) | 683 (21.6) |
| Nausea | 65 (6.9) | 108 (8.1) | 22 (4.7) | 0 | 0 | 4 (3.4) | 1 (2.1) | 1 (0.8) | 201 (6.4) |
| Rhinitis | 836 (89.3) | 1,118 (84.2) | 112 (24.2) | 6 (17.1) | 79 (73.1) | 72 (60.5) | 29 (60.4) | 104 (88.1) | 2,356 (74.7) |
| Laryngitis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 15 (12.7) | 15 (0.4) |
| Pharyngitis | 135 (14.4) | 226 (17.0) | 2 (0.4) | 0 | 36 (33.3) | 32 (26.9) | 22 (45.8) | 100 (84.7) | 553 (17.5) |
All enrolled patients presented with fever, because fever was the prime inclusion criteria, although it was not always reported in the data. Cough (76.9%), rhinitis (74.4%), sore throat (43.7%), headache (21.6%), pharyngitis (17.5%), and myalgia (12.6%) were also reported in enrolled patients. The comparison of symptoms between patients infected by the pandemic virus and patients infected by seasonal influenza viruses (H1N1, H3N2, and influenza B) showed that cases of myalgia are significantly higher (P < 0.001) in patients infected by the pandemic virus. However, other symptoms did not show significant difference between these two groups.
Influenza detection.
Of 3,155 samples tested, 911 (28.9%) were positive for influenza virus as assessed by the real-time RT-PCR method (Table 2). Of these samples, 826 (90.7%) were influenza A, and 85 (9.3%) were influenza B. Among the influenza A-positives, 503 (60.9%) were H1N1pdm09, 314 (38.0%) were H3N2, and 9 (1.1%) were seasonal H1N1. The detection rate for influenza over the study period was highest in Senegal (717/2264; 31.7%) followed by Cape Verde (132/498; 26.5%), Mauritania (42/227; 18.5%), and Guinea (20/166; 12%). In Cape Verde, the majority of H1N1pdm09 viruses (131/132; 99.2%) was detected in 2009. Conversely, most H1N1pdm09 viruses in Senegal (344/344; 100%), Mauritania (24/26; 92.3%), and Guinea (7/9; 77.8%) were detected in 2010.
Table 2.
Detection rates of influenza virus by type and subtype in Senegal, Cape Verde, Mauritania, and Guinea from 2009 to 2010
| Virus no. (%) | Senegal | Cape Verde | Mauritania | Guinea | ||||
|---|---|---|---|---|---|---|---|---|
| 2009 (N = 936) | 2010 (N = 1,328) | 2009 (N = 463) | 2010 (N = 35) | 2009 (N = 108) | 2010 (N = 119) | 2009 (N = 48) | 2010 (N = 118) | |
| Total | 274 (29.3) | 443 (33.4) | 131 (28.3) | 1 (2.8) | 18 (16.7) | 24 (20.2) | 10 (20.8) | 10 (8.5) |
| Influenza B | 0 | 76 (5.7) | 1 (0.2) | 0 | 3 (2.8) | 0 | 3 (6.2) | 2 (1.7) |
| Influenza A | 274 (29.3) | 367 (27.6) | 130 (28.1) | 1 (2.8) | 15 (13.9) | 24 (20.2) | 7 (14.6) | 8 (6.8) |
| H3N2 | 271 (28.9) | 22 (1.7) | 8 (1.7) | 0 | 7 (6.5) | 0 | 5 (10.4) | 1 (0.8) |
| HINI | 3 (0.3) | 0 | 0 | 0 | 6 (5.5) | 0 | 0 | 0 |
| A(H1N1)pdm09 | 0 | 345 (26.0) | 122 (26.3) | 1 (2.8) | 2 (1.8) | 24 (20.2) | 2 (4.2) | 7 (5.9) |
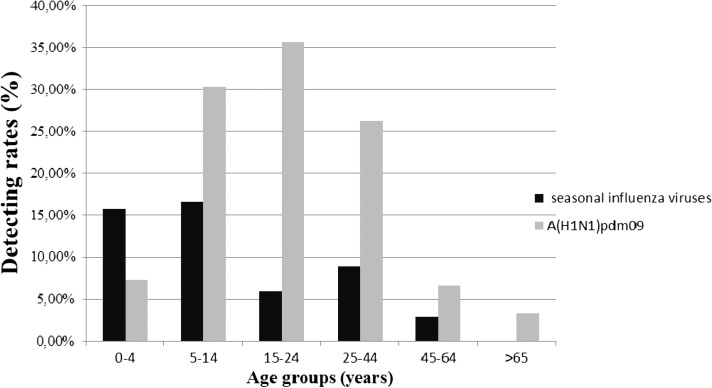
The highest detection rate for seasonal influenza viruses (72/422; 17.1%) occurred in the 5–14 years age group (Figure 2) followed by the < 5 years group (262/1642; 16.2%), and the rate was less than 10% in each of the remaining age groups. However, for A(H1N1)pdm09, the detection rate was highest in the 15–24 years age group (116/324; 35.8%; P < 0.001) (Table 3) followed by the 5–14 years age group (128/422; 30.2%; P < 0.001) and the 25–44 years age group (92/351; 26.2%; P < 0.001), with only 7.2% in the < 5 years age group (119/1,642) used as reference group in the Fisher's exact test. In terms of the comparative distribution of the pandemic and seasonal viruses in each age group, we note that the seasonal viruses were predominant only among children under 5 years of age in contrast to the other age groups in which detection rates for the A(H1N1)pdm09 virus were significantly higher than the rates for seasonal viruses.
Figure 2.
Infection rates of influenza viruses [seasonal and A(H1N1)pmd09] among different age groups during the pandemic period in Senegal, Cape Verde, Mauritania, and Guinea (data gathered).
Table 3.
Comparison of the distribution of pandemic-positive cases into the different age groups
| Viral type | Age groups (years) n (%) | |||||
|---|---|---|---|---|---|---|
| 0–4 | 5–14 | 15–24 | 25–44 | 45–64 | > 64 | |
| A(H1N1)pdm09 | 262 (15.9%) | 128 (30.1%) | 116 (35.6%) | 92 (26.2%) | 21 (8.6%) | 2 (6.4%) |
| P value | Reference group | P < 0.001 | P < 0.001 | P < 0.001 | P < 0.001 | P = 0.76 |
Values in bold are considered statistically significant.
A(H1N1)pdm09 viruses were first detected in three specimens from Cape Verde, sent to Dakar in week 25/2009 (June 15–21), with sample collection dates of June 18.10 Pandemic viruses were subsequently confirmed at the end of 2009 in Guinea (collection date of December 12 for the first case), Mauritania at week 51 in 2009 (collection date of December 14 for the first case), and finally, Senegal in week 1 in 2010 (collection date of January 5 for the first case), not in September of 2009 as stated elsewhere.11
Phylogenetic analysis of the HA1 of A(H1N1)pdm09 virus isolates.
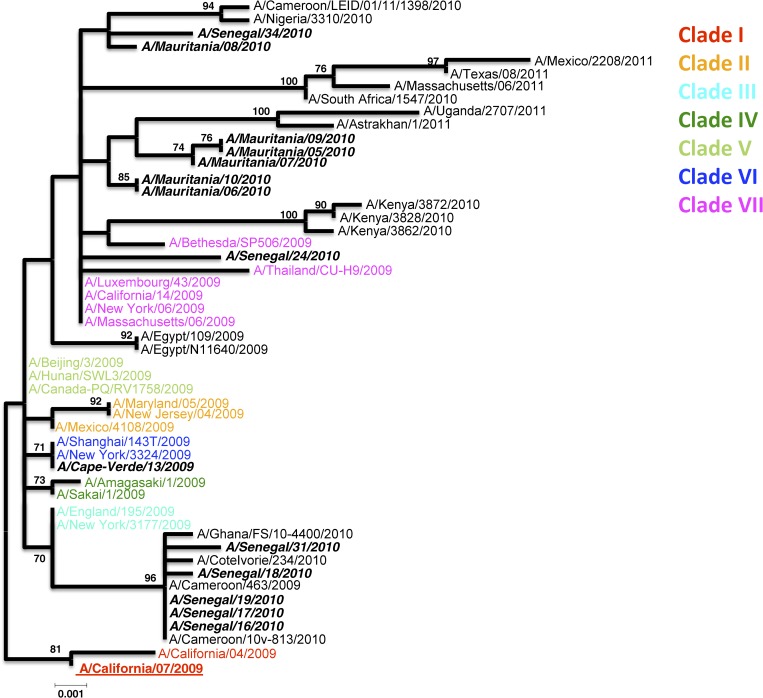
A total of 14 HA1 A(H1N1)pmd09 sequences were obtained: 7 from Senegal, 6 from Mauritania, and 1 from Cape Verde. Unfortunately, no isolate sequence was obtained from Guinea because of the poor quality of the clinical specimens for viral isolation.
To evaluate the degree of genetic variability of strains originating from Senegal, Cape Verde, and Mauritania, strains belonging to the seven clades previously defined for the phylogeny of A(H1N1)pdm09 viruses were added (Figure 3).12
Figure 3.
Maximum-likelihood phylogenetic tree analysis of HA1 genes of H1N1pdm strains circulating in Senegal, Cape Verde, and Mauritania. Strains from the previously determined seven clades are included. Senegalese, Cape Verdian, and Mauritanian strains are shown by names in bold and italics.
Therefore, the phylogeny based on the HA1 gene sequences (first 1,200 bp) shows an interesting grouping of five isolates from Senegal (A/Senegal/16/2010, A/Senegal/17/2010, A/Senegal/18/2010, A/Senegal/19/2010, and A/Senegal/31/2010) with isolates from other West African countries (Ivory Coast, Ghana, and Cameroon). This cluster seems genetically closer to the reference strains A/England/195/2009 and A/New York/3177/2009. These two strains belong to clade 3. This proximity is in accordance with the antigenic test results that showed isolates A/Senegal/16/2010, A/Senegal/17/2010, A/Senegal/18/2010, and A/Senegal/19/2010 reacting well with the A/England/195/2009 antiserum (Table 4) and also, most other reference sera (A/California/07/2009, A/Auckland/3/2009, and A/Lviv/N6/2009). A/Senegal/24/2010 is closer with isolates from clade 7 (A/Luxembourg/43/2009, A/Thailand/CU-H9/2009, A/New-York/06/2009, etc…) at the genetic level. The remaining isolate from Senegal, A/Senegal/34/2010, forms a cluster with three other West African strains (A/Cameroon/LEID/01/11/1398/2010, A/Nigeria/33/2010, and A/Mauritania/08/2010). Antigenetically, A/Senegal/24/2010 and A/Senegal/34/2010 isolates reacted better with A/England/195/2009 and A/Auckland/3/2009 reference antisera. The five remaining isolates from Mauritania are together in a cluster in which we find two other isolates from 2011: A/Uganda/2707/2011 and A/Astrakhan/1/2011. These clusters likely symbolized evolved strains from clade 7. Antigenically, isolates from Mauritania reacted well with antiserum against the reference strains A/Auckland/3/2009 and A/Lviv/N6/2009. In contrast, the values obtained with the antisera against the Californian isolates are very low. The single sequenced isolate from Cape Verde (A/Cape-Verde/13/2009) seems genetically closer to isolates belonging to clade 6 (A/New York/3324/2009 and A/Shanghai/143T/2009).
Table 4.
HIA titers of isolates from Senegal and Mauritania tested with reference antisera
| Viruses | Isolation history | A/Cal 4/09 C4/F14/09 | Post-infection ferret sera | ||||
|---|---|---|---|---|---|---|---|
| A/Cal 7/09 C4/31/09 | A/Eng 195/09 NIBSC F17/09 | A/Auck 3/09 C4/17/09 | A/Bayern 69/09 C4/33/09 | A/Lviv N6/2009 C4/34/09 | |||
| Reference viruses | |||||||
| A/California/4/2009 | C1, E2 | 2,560 | 2,560 | 2,560 | 5,120 | 2,560 | 5,120 |
| A/California/7/2009 | E6 | 640 | 2,560 | 640 | 2,560 | 1,280 | 5,120 |
| A/England/195/2009 | MDCK3/SIAT1 | 1,280 | 1,280 | 1,280 | 2,560 | 1,280 | 2,560 |
| A/Auckland/3/2009 | Ex+3 | 1,280 | 2,560 | 1,280 | 2,560 | 1,280 | 2,560 |
| A/Bayern/69/2009 | MDCK4/SIAT1 | 80 | 320 | 40 | 80 | 320 | 320 |
| A/Lviv/N6/2009 | MDCK4/SIAT1 | 320 | 640 | 80 | 160 | 1,280 | 1,280 |
| Test viruses | |||||||
| A/Mauritania/05/2010 | SIAT3 | 1,280 | 1,280 | 1,280 | 2,560 | 640 | 1,280 |
| A/Mauritania/06/2010 | SIAT3 | 1,280 | 1,280 | 1,280 | 2,560 | 640 | 1,280 |
| A/Mauritania/07/2010 | SIAT3 | 320 | 1,280 | 160 | 640 | 640 | 1,280 |
| A/Mauritania/08/2010 | SIAT3 | 320 | 1,280 | 640 | 1,280 | 640 | 640 |
| A/Senegal/016/2010 | MDCK2/SIAT1 | 2,560 | 2,560 | 2,560 | 2,560 | 1,280 | 2,560 |
| A/Senegal/017/2010 | MDCK2/SIAT1 | 2,560 | 2,560 | 2,560 | 5,120 | 1,280 | 2,560 |
| A/Senegal/018/2010 | MDCK2/SIAT1 | 2,560 | 2,560 | 2,560 | 2,560 | 1,280 | 2,560 |
| A/Senegal/019/2010 | MDCK2/SIAT1 | 1,280 | 1,280 | 1,280 | 2560 | 640 | 1,280 |
| A/Senegal/024/2010 | MDCK2/MDCK1 | 1,280 | 1,280 | 2,560 | 2,560 | 1,280 | 2,560 |
| A/Senegal/031/2010 | MDCK2/MDCK1 | 2,560 | 5,120 | 2,560 | 2,560 | 1,280 | 2,560 |
| A/Senegal/034/2010 | MDCK2/MDCK1 | 1,280 | 1,280 | 2,560 | 2,560 | 1,280 | 2,560 |
Amino acid substitutions in the HA and NA proteins.
At the amino acid level, compared with the sequences of the vaccine strain A/California/7/09 (H1N1), which was recommended for the southern hemisphere since the 2010 influenza season, a number of variations were detected in both the HA and NA sequences (Table 5).
Table 5.
Major mutations in HA and NA genes in pandemic strains isolated from Dakar in 2010 compared with the vaccine strain amino acid sequence
| Strains | Positions in HA | Positions in NA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 49 | 100 | 101 | 212 | 220 | 222 | 239 | 338 | 391 | 106 | 248 | |
| A/California/7/2009 | L | P | S | A | S | R | D | I | E | V | N |
| A/Mauritania/05/2010 | – | S | – | – | T | K | – | V | – | I | D |
| A/Mauritania/06/2010 | – | S | N | – | T | – | – | V | K | I | D |
| A/Mauritania/07/2010 | – | S | – | – | T | K | – | V | K | I | D |
| A/Mauritania/08/2010 | – | S | – | – | T | – | – | V | – | I | D |
| A/Mauritania/09/2010 | – | S | – | – | T | K | – | V | – | I | D |
| A/Mauritania/10/2010 | – | S | N | – | T | – | – | V | K | – | D |
| A/Senegal/16/2010 | I | S | – | – | – | – | – | V | – | – | D |
| A/Senegal/17/2010 | I | S | – | – | – | – | – | V | – | – | D |
| A/Senegal/18/2010 | I | S | – | – | – | – | – | V | – | – | D |
| A/Senegal/19/2010 | I | S | – | – | – | – | – | V | – | – | D |
| A/Senegal/24/2010 | – | S | – | – | T | – | – | V | – | I | D |
| A/Senegal/31/2010 | I | S | – | V | – | – | – | V | – | – | D |
| A/Senegal/34/2010 | – | S | – | – | T | – | E | V | – | I | D |
Regarding the HA sequence, amino acid substitutions defining clades were found in our isolate A(H1N1)pdm09 strains (P83S, S203T, and I321V).12,13 The P83S (P100S, signal peptide included) and I321V (I338V) were mapped in all isolates, and the S203T (S220T) was mapped in 8 of 13 sequenced isolates. All isolates possessed D204 in the receptor binding site. The isolate from Senegal A/Senegal/34/2010 had a D222E (D239E) substitution in the receptor binding domain (RBD). In three isolates from Mauritania (A/Mauritania/05/2010, A/Mauritania/07/2010, and A/Mauritania/09/2010), we observed an R222K mutation. The substitution E391K exists in three isolates sequences (A/Mauritania/06/2010/H1N1, A/Mauritania/007/2010/H1N1, and A/Mauritania/10/2010/H1N1) from Mauritania, and lastly, five isolates from Senegal (A/Senegal/16/2010/H1N1, A/Senegal/17/2010/H1N1, A/Senegal/018/2010/H1N1, A/Senegal/19/2010/H1N1, and A/Senegal/31/2010/H1N1) exhibited an L49I mutation.
For NA amino acid sequence, the well-associated Oseltamivir resistance H275Y mutation was not observed. However, all isolates were N248D variants, and the V106I substitution was characterized in 7 of 14 isolates sequenced (A/Mauritania/05/2010, A/Mauritania/06/2010, A/Mauritania/07/2010, A/Mauritania/08/2010, A/Mauritania/09/2010, A/Senegal/24/2010, and A/Senegal/34/2010). These mutations have only a phylogenetic value and are not so far associated with resistance to neuraminidase inhibitors. The Mauritanian isolates A/Mauritania/06/2010 and A/Mauritania/10/2010 showed an N221Y mutation in the neuraminidase protein sequence.
Discussion
We described the epidemiology of influenza A(H1N1)pdm09 in four West African countries during the pandemic period. Among these countries, only Senegal had a functional surveillance system at the time of the emergence of the new reassortant influenza virus. However, Senegal was the last country where the pandemic virus was detected. Among the four countries surveyed in this study, the first cases of A(H1N1)pdm09 were detected in Cape Verde in June of 2009. Therefore, it is important to clarify that the flu monitoring in these countries began months and even years (for Mauritania) before the 2009 pandemic episode. Indeed, we started receiving samples from Cape Verde in the beginning of 2009 (week 2); for Mauritania, we were already implicated in the flu monitoring (since 2004), and for Guinea, the first samples were received at week 25 of 2009.
This finding is consistent with the detection time of A(H1N1)pdm09 in other parts of Africa. The first cases of influenza A(H1N1)pdm09 in Africa were detected in Egypt, Ivory Coast, South Africa, and Ethiopia in June of 2009. However, despite the detection of the first cases of A(H1N1)pdm09 around June and August of 2009 in several countries in Africa, delayed community transmission of A(H1N1)2009 (toward the end of 2009 or beginning of 2010) was observed in several West and Central Africa countries (e.g., Ghana, Ivory Coast, Nigeria, Niger, Cameroon, and Mali).14 All these countries had functional influenza surveillance system that documented community transmission 3–5 months after the detection of the first A(H1N1)pdm09 case in the country. This situation is similar to what we have observed in Senegal, Mauritania, and Guinea.
The reasons for the delayed circulation of A(H1N1)pdm09 in these countries are not clear, but unfavorable temperature and humidity conditions from June to November could explain some of the observed patterns.15,16 Interestingly, a recent work in Niger investigated the visibility as an additional significant parameter in the virus propagation.17
We compared the distribution of the 2009–2010 cumulative cases in age groups between the pandemic and seasonal influenza (Figure 2). A significant difference was observed between the age distribution of positive cases infected with seasonal and pandemic influenza. Indeed, seasonal viruses show a strong prevalence only in children less than 5 years. In other age groups, the pandemic is clearly the predominant virus. This finding is consistent (excepted for elders groups) with other studies, in which the greatest increase in influenza cases during 2009 (H1N1) pandemic influenza period was seen for school-aged children, adolescents, and younger adults.18,19 Considering the elderly, the A(H1N1)pdm09 detection rate (2/30; 6.6%; P value = 0.76) (Table 3) is because of the very small number of elderly people in consultation for ILI in these countries during the 2009–2010 period (0.9%). In fact, previous studies have shown that the elderly have better protective immunity acquired during previous contact with the virus.20
For Senegal, the seasonality is well-defined after years of regular flu monitoring.21 We have a regular circulation of influenza virus during the year with a peak in August (in the middle of the rainy season) in Senegal. The pandemic A(H1N1) 2009 virus that has globally emerged during the second quarter of 2009 was not detected in Senegal until 2010; interestingly, it occurred with a shift on seasonality that was reflected by a peak of infection that occurred in the first quarter of that year. For other countries, the seasonality is not well-defined because of the absence of data from a system of regular monitoring. Therefore, it was in the panic of the pandemic that Cape Verde and Guinea tried to send an exhaustive number of samples from patients with ILI to the Senegal NIC. For Mauritania, based on the results in the earlier years of collaboration, it is tempting to speculate that a peak of influenza occurs at the beginning of each year (January–February).
Regarding the HA and NA amino acid sequences, amino acid substitutions defining clades were found in our isolate H1N1 pdm09 strains. Therefore, the amino acid changes mapped in the NA (V106I and N248D) and the S203T (S220T) mapped in the HA indicated that the majority of isolates from the three countries seems to belong to clades 3, 6, and 7, which is in agreement with recent results.13 The P83S (P100S) and I321V (I338V) were mapped in all isolates, and the S203T (S220T) was in 8 of 14 sequenced isolates. It is important to specify that, in a recent study conducted in Brazil,22 the S220T mutation was identified in 78.6% of fatal pandemic influenza cases.
All isolates possessed D204 in the receptor binding site, which is known to confer binding of H1 viruses to human receptors,23 supporting efficient transmission of these viruses in humans.24 One isolate from Senegal, A/Senegal/34/2010, had a D239E that represents both a receptor binding residue and a Ca antigenic site.25 The substitution D239G is clinically very interesting; this substitution was detected more frequently in viruses isolated from patients with fatal outcomes and isolates from lungs.12,26 This strain was isolated from sample from a 20-year-old girl who came for consultation in a hospital in Dakar (Hôpital principal de Dakar). The symptoms were severe, with fever, intense coughing, myalgia, rhinitis, vomiting, and abdominal pain. The patient had undergone a rapid test for malaria diagnosis, but the test was negative. The substitution D239G/E/N has also been reported very recently. Globally, it should be noted that there is an expansion of mutant viruses with HA mutations at the position D239 (D239E; D239G and D238N mutations) that characterizes viruses of the second wave of the pandemic (H1N1) 2009. Residue Q310 is well-conserved in Senegal, Mauritania, and Cape Verde strains sequences. Indeed, the Q310H mutation is not observed. Q310H mutant influenza viruses were recently isolated from post-mortem blood and lung tissue samples of patients with clinically lethal forms of influenza pandemic (H1N1) 2009.26 Therefore, other mutations in HA (R222K, E391K, and L49I) are mapped in or around known antigenic sites and may have antigenic importance and implications in vaccine efficacy. Hence, it will be very interesting to perform additional antigenic and phenotypic tests. The H275Y mutation, a known marker for sensitivity to the neuraminidase inhibitor Oseltamivir, is not observed,27 which correlates with the phenotypic test results that revealed that all pandemic viruses isolated in IPD are sensitive to this antiviral drug.
However, our study suffers from some limitations. The limited standardization of procedures across countries hindered the possibility to implement more robust comparison. In addition, the lack of surveillance systems in Mauritania, Guinea, and Cape Verde before the pandemic period does not allow us to compare the time of circulation of influenza A(H1N1)pdm09 with the influenza season. This limitation is also reflected in the limited number of samples obtained from some countries, which hindered the possibility of observing well-defined seasonal patterns as well as the quality of samples that did not allow for sufficient virus isolation.
Despite the aforementioned limitations, our study remains important, because it documents, for the first time, the circulation of seasonal and pandemic influenza in four West African countries during the pandemic period.
In conclusion, delayed A(H1N1)pdm09 transmission occurred in June of 2009 (week 25) in Cape Verde, late 2009 in Mauritania and Guinea (weeks 51 and 52), and at the beginning of 2010 (week 1) in Senegal. Therefore, A(H1N1)pdm09 became the dominant influenza subtype in 2010 in these countries (Senegal, Mauritania, and Guinea). The results of this study show the importance of the IPD Regional Influenza Surveillance Network that provides a valuable platform to monitor influenza activities in a region that has limited influenza surveillance and diagnostic capacity. However, it also highlights its weaknesses and the need for improvement in terms of standardization of procedures and development of timely and efficient systems for shipment of samples.
ACKNOWLEDGMENTS
The authors thank Dr. Stefanot Tempia for his great help in writing the draft, data analysis, and revisions of the paper. We thank Rod Daniels and Vicki Gregory from WHO Collaborating Centre (WHO CC) for Influenza, National Institute for Medical Research (NIMR), London, for their contributions in antigenic and genetic characterization of the majority of the viruses isolated in Dakar, Senegal. The authors thank CDC Atlanta for providing kit and detection protocols. The authors thank all the staff of the Epidemiology Unit of Pasteur Institute of Dakar for their collaboration. The authors thank Oumy Niass (PhD student at the Immunology unit, Institut Pasteur de Dakar, IPD) for her great help for the statistical analyses.
Footnotes
Financial support: This work was funded by Institut Pasteur de Dakar (Senegal), the WHO, the French Ministry of Health/Établissement de Préparation et de Réponse aux Urgences Sanitaires (EPRUS), and the US Department of Health and Human Services.
Authors' addresses: Ndongo Dia, Mbayame Niang Ndiaye, and Ousmane M. Diop, Institut Pasteur de Dakar, Dakar, Senegal, E-mails: ndia@pasteur.sn, niang@pasteur.sn, and diopo@who.int. Maria de Lourdes Monteiro, Ministry of Health, Praia, Cape Verde, E-mail: marial.Monteiro@ms.gov.cv. Lamine Koivogui, Laboratoire de Virologie, Hôpital Donka, Conackry, Guinea, E-mail: koivogui@biasy.net. Mohamed Ould Bara, Institut National de Recherches en Santé Publique (INRSP), Laboratoire de Virologie, Nouakchott, Mauritanie, E-mail: elbaraahmed@yahoo.fr.
Reprint requests: Mbayame Niang Ndiaye, Institut Pasteur de Dakar, 36 Avenue Pasteur, Dakar, Senegal, BP 220, E-mail: niang@pasteur.sn.
References
- 1.Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 2.Enserink M, Cohen J. Virus of the year. The novel H1N1 influenza. Science. 2009;326:1607. doi: 10.1126/science.326.5960.1607. [DOI] [PubMed] [Google Scholar]
- 3.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, López-Gatell H, Olivera H, López I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization Pandemic (H1N1) 2009—Update 112. 2010. http://www.who.int/csr/don/2010_08_06/en/index.html Available at.
- 5.CDC . 2009 H1N1 Flu: International Situation Update. Atlanta, GA: Centers for Disease Control and Prevention; 2009. [Google Scholar]
- 6.Dosseh A, Ndiaye K, Spiegel A, Sagna M, Mathiot C. Epidemiological and virological influenza survey in Dakar, Senegal: 1996–1998. Am J Trop Med Hyg. 2000;62:639–643. doi: 10.4269/ajtmh.2000.62.639. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization . WHO Manual on Animal influenza Diagnosis and Surveillance. Geneva: World Health Organization; 2002. [Google Scholar]
- 8.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 9.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization . Global Alert and Response (GAR). Situation Updates—Pandemic (H1N1) 2009. Geneva: World Health Organization; 2009. [Google Scholar]
- 11.World Health Organization Recommended Viruses for Influenza Vaccines for Use in the 2010–2011 Northern Hemisphere Influenza Season, 2010. http://www.who.int/influenza/vaccines/virus/recommendations/20100_Recommendation.pdf Available at. [PubMed]
- 12.Potdar VA, Chadha MS, Jadhav SM, Mullick J, Cherian SS, Mishra AC. Genetic characterization of the influenza A pandemic (H1N1) 2009 virus isolates from India. PLoS One. 2010;5:e9693. doi: 10.1371/journal.pone.0009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson M, Spiro D, Wentworth D, Beck E, Fan J, Ghedin E, Halpin R, Bera J, Hine E, Proudfoot K, Stockwell T, Lin X, Griesemer S, Kumar S, Bose M, Viboud C, Holmes E, Henrickson K. The early diversification of influenza A/H1N1pdm. PLoS Curr. 2009;1:RRN1126. doi: 10.1371/currents.RRN1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nzussouo NT, Michalove J, Diop OM, Njouom R, Monteiro M de L, Adje HK, Manoncourt S, Amankwa J, Koivogui L, Sow S, Elkory MB, Collard JM, Dalhatu I, Niang MN, Lafond K, Moniz F, Coulibaly D, Kronman KC, Oyofo BA, Ampofo W, Tamboura B, Bara AO, Jusot JF, Ekanem E, Sarr FD, Hwang I, Cornelius C, Coker B, Lindstrom S, Davis R, Dueger E, Moen A, Widdowson MA. Delayed 2009 pandemic influenza A virus subtype H1N1 circulation in West Africa, May 2009–April 2010. J Infect Dis. 2012;206((Suppl 1)):S101–S107. doi: 10.1093/infdis/jis572. [DOI] [PubMed] [Google Scholar]
- 15.Lowen AC, Steel J, Mubareka S, Palese P. High temperature (30 degrees C) blocks aerosol but not contact transmission of influenza virus. J Virol. 2008;82:5650–5652. doi: 10.1128/JVI.00325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steel J, Palese P, Lowen AC. Transmission of a 2009 pandemic influenza virus shows similar sensitivity to temperature and humidity as an H3N2 seasonal strain. J Virol. 2010;85:1400–1402. doi: 10.1128/JVI.02186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jusot JF, Adamou L, Collard JM. Influenza transmission during a one-year period (2009–2010) in a Sahelian city: low temperature plays a major role. Influenza Other Respi Viruses. 2012;6:87–89. doi: 10.1111/j.1750-2659.2011.00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karageorgopoulos DE, Vouloumanou EK, Korbila IP, Kapaskelis A, Falagas ME. Age distribution of cases of 2009 (H1N1) pandemic influenza in comparison with seasonal influenza. PLoS One. 2011;6:e21690. doi: 10.1371/journal.pone.0021690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falagas ME, Koletsi PK, Baskouta E, Rafailidis PI, Dimopoulos G, Karageorgopoulos DE. Pandemic A(H1N1) 2009 influenza: review of the Southern Hemisphere experience. Epidemiol Infect. 2011;139:27–40. doi: 10.1017/S0950268810002037. [DOI] [PubMed] [Google Scholar]
- 20.Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, Brammer TL, Cox NJ, Tumpey TM, Katz JM. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361:1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 21.Niang MN, Dosseh A, Ndiaye K, Sagna M, Gregory V, Goudiaby D, Hay A, Diop OM. Sentinel surveillance for influenza in Senegal, 1996–2009. J Infect Dis. 2012;206((Suppl 1)):S129–S135. doi: 10.1093/infdis/jis576. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira JL, Borborema SE, Brígido LF, Oliveira MI, Paiva TM, Santos CL. Sequence analysis of the 2009 pandemic influenza A H1N1 virus haemagglutinin gene from 2009–2010 Brazilian clinical samples. Mem Inst Oswaldo Cruz. 2011;106:613–616. doi: 10.1590/s0074-02762011000500014. [DOI] [PubMed] [Google Scholar]
- 23.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 24.World Health Organization . Pandemic (H1N1): Update 76. Geneva: World Health Organization; 2009. [Google Scholar]
- 25.Reid AH, Fanning TG, Hultin JV, Taubenberger JK. Origin and evolution of the 1918 ‘Spanish’ influenza virus hemagglutinin gene. Proc Natl Acad Sci USA. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glinsky GV. Genomic analysis of pandemic (H1N1) 2009 reveals association of increasing disease severity with emergencof novel hemagglutinin mutations. Cell Cycle. 2010;9:958–970. doi: 10.4161/cc.9.5.10913. [DOI] [PubMed] [Google Scholar]
- 27.Meijer A, Lackenby A, Hungnes O, Lina B, van-der-Werf S, Schweiger B, Opp M, Paget J, van-de-Kassteele J, Hay A, Zambon M. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007–08 season. Emerg Infect Dis. 2009;15:552–560. doi: 10.3201/eid1504.081280. [DOI] [PMC free article] [PubMed] [Google Scholar]