Abstract
Vaccines, like drugs and medical procedures, are increasingly amenable to individualization or personalization, often based on novel data resulting from high throughput “omics” technologies. As a result of these technologies, 21st century vaccinology will increasingly see the abandonment of a “one size fits all” approach to vaccine dosing and delivery, as well as the abandonment of the empiric “isolate–inactivate–inject” paradigm for vaccine development. In this review, we discuss the immune response network theory and its application to the new field of vaccinomics and adversomics, and illustrate how vaccinomics can lead to new vaccine candidates, new understandings of how vaccines stimulate immune responses, new biomarkers for vaccine response, and facilitate the understanding of what genetic and other factors might be responsible for rare side effects due to vaccines. Perhaps most exciting will be the ability, at a systems biology level, to integrate increasingly complex high throughput data into descriptive and predictive equations for immune responses to vaccines. Herein, we discuss the above with a view toward the future of vaccinology.
Keywords: Adaptive immunity, Biotechnology, Computational biology, Genomics, Immunogenetics, Individualized medicine, Proteomics, Systems biology, Vaccination, Vaccines, Modeling, Vaccinomics, Adversomics, Predictive equation, Immune response network theory, Individualized vaccinology
1. Introduction
In this paper, we review and expand upon a new direction in vaccinology (defined as the science and study of vaccines), and vaccine development (defined as the use of knowledge that derives from the science of vaccines to construct new vaccine candidates), which we have called “vaccinomics [1].” This new direction represents a novel and holistic scientific paradigm under which vaccine immune responses can be studied, understood, and predicted, and with which new candidate vaccines can be conceived, developed, and tested. This represents a radical departure from the historical methodology by which vaccines were developed—an empiric method we have labeled the “Isolate–Inactivate–Inject” paradigm—and that reigned as the dominant mode of vaccine development until the late 1990s. Issues with such an approach include that, for the most part, it: utilized only whole pathogen (live or inactivated) approaches; ignored population genetics (immunogenomics); is a failed paradigm for vaccines against hyper-variable viruses and more complex pathogens (i.e., parasites, fungi, larger bacteria such as tuberculosis); usually required an intact cold chain for vaccine viability; was a “one dose fits all” population-based approach; required large and expensive clinical efficacy and safety trials of genetically uncharacterized populations; resulted in expensive vaccines and, hence, under-utilization and poor vaccine coverage; did not allow an informed understanding of an individual’s genetically determined risk for an adverse effect due to a vaccine; and other issues.
Now, guided in part by advances in personalized medicine as applied to the use of drugs, the field of vaccinomics provides a conceptual framework for both understanding (and predicting) immune responses to vaccines, and allows the development of new vaccines informed by advances in immunology, immunogenetics (the study of individual host genetic variation associated with individual differences in immune responses to the same antigen(s)) and immunogenomics (the study of population-level genetic variations associated with population-level variations in immune responses), bioinformatics, virology, systems biology, metagenomics (host and non-host), and other fields. We first developed the concept of vaccinomics in 2005, and published an early review in 2007 [2]. Subsequent reviews have enlarged the concept, added more data, and answered questions that have arisen regarding the practical application of vaccinomics, as well as articulating challenges facing the field [3–12]. Others have begun to recognize the importance of vaccinomics in understanding, at a more holistic level, why inter-individual differences in immune responses to vaccines occur, and how these concepts might play a role in new vaccine design [13–15]. Additionally, major initiatives in multiple countries have been proposed using the concept of vaccinomics in studying and evaluating vaccine safety [16]. Additional credibility was lent to the vaccinomics field by a review in Scientific American labeling vaccinomics as “one of the most innovative scientific concepts of the decade [17],” and by The Scientist, which labeled vaccinomics as “one of the hottest omics fields [18],” as well as by other scientists working in the field who have appropriated vaccinomics concepts into subsets of the systems or bioinformatics aspects of vaccine biology [19,20]. Below we elaborate on the concept and implications of vaccinomics and the immune response network theory, its application to understanding vaccine immune responses, initial analytic approaches to the data generated by vaccinomics, application to new vaccine design and development, and progress toward the ultimate goal – that of developing a predictive equation that describes the immune response to a viral vaccine [19,21].
2. Vaccinomics and the immune response network theory: development of a unified theory
Niels Jerne first proposed in the 1970s the “immune network hypothesis,” which theorized how the adaptive immune system worked as an idiotypic network to explain the regulation of clonal immune responses [22]. Later this was expanded into the “symmetrical network theory” in the 1980s through the 1990s by Geoffrey Hoffman in an attempt to solve the “I–J paradox [23].” Incremental additions and changes to these models occurred over the next 20 years, but none were specifically aimed at defining the network as applied to systems-level vaccine-induced immune responses. As such, little consideration of factors outside the immediate immune system itself (strictly defined) were included (e.g., gene polymorphisms, epigenetics, the influence of the host microbiome), and, therefore, such features were not incorporated into the models or theories prevalent at the time. Our proposal of the immune response network theory, while building on the foundations of Jerne’s and Hoffman’s work, as well as other immunologists, is the first proposed in terms of its focus on systems-level vaccine-induced immune responses, its intersection with host genetics and non-host factors (microbiome for example), and its theoretical ability to provide the basis for a mathematical model and predictive equation describing the non-random events that lead to pre-determined immune responses. Since then, others have also proposed systems-level vaccinology approaches [19,20,24–27].
The immune response network theory in its simplest form states that “the response to a vaccine is the cumulative result of interactions driven by a host of genes and their interactions, and is theoretically predictable [2].” We elaborated further on this definition to recognize the impact of epigenetic phenomena, such as gene methylation patterns, the influence of metagenomics (the microbiome), the dominance profile of a given gene/SNP(s), complementarity, epistasis, systems biology and immune profiling, and other factors, including environmental and co-infections [7,9]. This includes a concept we previously introduced termed “polymorphic plasticity,” whereby a specific gene polymorphism could have different measurable genetic (and therefore phenotypic) effects based on concomitant epigenetic effects, effects due to co-infection with other pathogens, or other variables [2,28,29]. As we have previously written:
For an individual to develop a protective immune response to an antigen, a complex series of biologic, molecular, genetic, physiologic, and other processes must be activated (and in some cases suppressed). Antigen recognition, processing, presentation, and activation of innate, adaptive, and cell-mediated immune responses must occur. Protective immunity requires the activation/suppression of specific genes as well as protein transcription, expression, secretion, and function. The immune response network theory seeks to provide a framework for the above described interactive and carefully regulated processes that result in protective responses against pathogen threats [7].
To this traditional view we should add that superantigens can nonspecifically cause massive polyclonal T cell activation without requiring the seemingly orderly steps of the adaptive immune system noted above.
Thus, while the immune response network theory recognizes and incorporates elements of the immune response not yet discovered at the time of Jerne and Hoffman, it includes the immunogenetics of response to antigen, and the growing appreciation of systems biology approaches to understanding more holistically how immune responses are generated and sustained in the host [2,5]. This theory recognizes the roles of individual components (nodes) of the immune system (immune response genes, epigenetic phenomena such as gene methylation patterns and contributory SNPs), as well as networks or pathways composed of groupings of individual genetic components (genes, gene pathways, gene networks, etc.) of the immunogenetic system and other factors determinative of immune response (microbiome, etc.), at both individual and population levels. The immune response network theory and its fundamental application to vaccinomics draws together all of these components and utilizes biostatistics and bioinformatics to deconvolute, visualize, analyze, and understand the individual and group components that together compose immune response phenotypes (neutralizing antibody, cytokine responses, innate immune responses, cell-mediated immune responses, etc.) that are typically measured and labeled “the immune response.”
As of this writing, we conceive the immune response network theory as an encompassing theory that holistically recognizes and explains the temporal, genetic, and immune aspects that together are deterministic and predictive of the immune response(s) to a specific vaccine. Thus, we posit that it can theoretically explain, and eventually predict, the immune response to a vaccine in the form of a mathematical equation that accounts for the complexity of the system. The immune response network theory and the development of a predictive equation implies a deconvoluted yet mechanistic determination of key genetic and other variables that together explain innate, adaptive, and cell-mediated immune phenotype archetypes and individual response patterns [7]. Vaccinomics then uses this and other omics information to engineer novel vaccine candidates that overcome genetic barriers to developing protective immunity. These concepts arose out of our studies demonstrating highly significant associations between specific single nucleotide polymorphisms (SNPs) and gene pathways and networks, human leukocyte antigen (HLA) haplotypes, and immune responses to a variety of viral vaccines, including measles, mumps, rubella (MMR) [30–33], vaccinia (smallpox) [34–37], and influenza [3,4], and one bacterial vaccine (anthrax) [38,39]. In addition, compelling evidence for the predetermined influence of host genetic factors on inter-individual variations in immune response was provided by our work in monozygotic and dizygotic twins demonstrating that the hereditability of the humoral immune response to measles vaccine is almost 90% [40]. Others have reviewed the influence of the microbiome and metagenetics on immune responses to non-self antigens [41,42], and these data add yet another important dimension to the immune response network theory, but will not be further reviewed here.
Like any biologic network, we conceive of the immune response as the outcome of connected components (nodes) that communicate and share information (directed and undirected edges). Data suggest that this network is a neural network in that regulation of activity is possible using feedback loops (transcription and signaling pathways for example, as well as regulation of cytokine responses) [43]. Nonetheless, much work remains to be done to determine attributes of this network, such as scale and scope, topology, methods of communication within the network and between other biologic networks, boundaries of the network, and what governs the behaviors of the network, changes over time (e.g., methylation or microbiome influences), and the details of interactions with other biologic networks. Additional data answering these questions will further define and enlarge the immune response network theory, and some investigators have begun to take aspects of such an approach in understanding immune responses to vaccines [25,26].
Vaccinomics is a holistic field of study whose science base is being built piece-by-piece. As discussed below, it can take advantage of the information derived from the immune response network theory by allowing an understanding of the key drivers of the immune response to a specific or series of antigens, and applies this information to the practical aspects of conceiving, designing, and delivering new vaccine candidates informed by systems-level information about the mechanism(s) by which immune responses are generated, and theoretically by the development of a derived equation describing immune profiles resulting from vaccine antigens [14,15,20,21,44–47]. We and others have described achieving such goals as being the “second golden age” of vaccinology [1,48–50].
Both the immune response network theory and its application to vaccinomics could have multiple practical and beneficial applications. Issues relevant to the feasibility of designing new vaccine candidates include: the need to better understand, at a systems level, how immune responses to specific vaccines and vaccine antigens are generated and sustained across genders, ethnic groups, and with aging of the host; the need for a directed methodology, rather than the historical empiric approach to vaccine design; and methods for vaccine monitoring (by which we imply immune monitoring in clinical trials), which is a hugely expensive aspect of new vaccine development, and, finally, the field we have defined as “adversomics,” which is the application of vaccinomics and system biology approaches to the specific study and discovery of vaccine-induced adverse events [12].
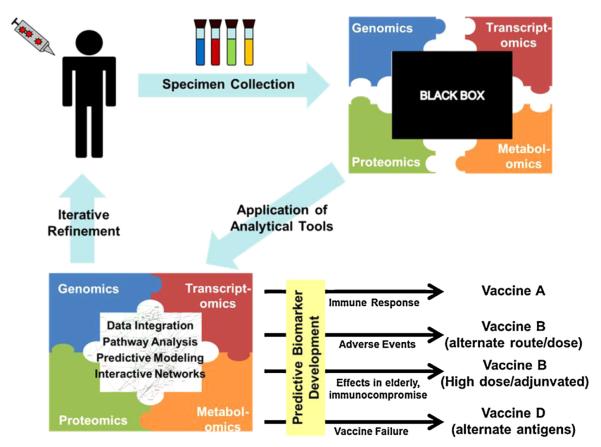
Vaccinomics could inform the rational and directed design of new vaccine candidates by advancing our understanding of the genetic and non-genetic drivers of the immune response to vaccine antigens at the systems or network level. As the importance of personalized medicine, or so-called “precision medicine,” becomes more apparent, it is possible that vaccines will be developed not on a “one size fits all” population-based approach, but rather with a data-driven appreciation for restricting genetic differences between individuals and subgroups within a population [20,21,25,51]. For example, vaccinomics could play an important role in designing new vaccines for an immunosenescent population by defining and understanding, at the systems level, pertubations in the immune, immunogenetic, and metagenomic systems that prevent the development of protective immune responses [46]. Likewise, as discussed in the sections below, understanding the mechanism by which a polymorphism in a critical viral receptor blocks viral binding and uptake, for example, allows the potential ability to engineer around that genetic restriction and allow for improved vaccine immunogenicity. Such an approach would allow a move away from the “isolate–inactivate–inject” paradigm to the “discover–validate–characterize–apply” paradigm of new vaccine discovery and development [7]. A flow chart describing the application of vaccinomics to this new paradigm is presented in Fig. 2. Key elements of this new paradigm are the cutting-edge, “omics” technologies and sophisticated modeling and bioinformatics tools (Fig. 1). As an example, this paradigm starts with the discovery of a key genetic restriction, or series of restrictions, then moves to replication and validation of the effect of that genetic restriction in relevant individuals or sub-groups, progresses to functional studies to uncover the mechanism(s) by which a causal variant results in restriction, then finally “applies” (or engineers) a new vaccine candidate around that restriction such that the new vaccine candidate is immunogenic and efficacious even in the presence of that specific genetic restriction [31].
Fig. 2.
Vaccinomics and the development of the Immune Response Network Theory for directed vaccine design. The host response to vaccination leads to the sequential activation of complex and poorly understood biological pathways and networks (black box). The outcome of this response is a consequence of the cascade of biological pathways that are initiated. Because of the inherent complexities of the diseases for which we lack vaccines, it is no longer feasible to take an empirical approach to vaccine development. A logical, directed approach is needed. This begins with the administration of a vaccine to an appropriate animal or human model. Biological specimens consisting of the relevant immune cells or bodily fluids (sera, mucosal secretions) are collected at specified time-points in order to assess immune outcomes over time. High throughput “omics” technologies are utilized in order to comprehensively assess biological processes. This is analogous to opening a puzzle box and spreading out all of the pieces, with each “piece” of the puzzle being a separate assay. This is followed by the use of advanced statistical and bioinformatics tools to make logical sense of the high-throughput data (i.e., putting the pieces together to form a coherent picture of the development of an immune response over time). The resulting systems-level understanding of immune function following vaccination allows us to identify predictive biomarkers, essential immune pathways, select appropriate adjuvants, alter vaccine composition to avoid side effects, include essential epitopes, and circumvent escape variants to deliver a personalized vaccine treatment. The resulting knowledge is also applied to refining the experimental systems used to assess immune function, resulting in an iterative cycle of discover, validate, characterize, and apply this new knowledge.
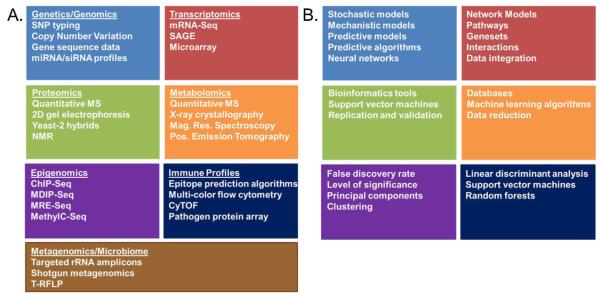
Fig. 1.
High-dimensional “omics” technologies. (A) An overview of high dimensional approaches commonly utilized to comprehensively assess biological systems in response to various stimuli (i.e., vaccination). The resulting datasets provide the “puzzle pieces” necessary to developing a systems level understanding of the biological response being investigated. (B) An overview of the biostatistical and informatics tools required to “assemble the puzzle pieces” in order to make biological sense out of the large datasets generated in panel A.
Our work is different in scope, intent, and process from what has been called “reverse vaccinology [52].” Reverse vaccinology refers to the study and evaluation of the pathogen’s genome in order to discover possible new vaccine candidates. While perhaps complementary, vaccinomics takes the opposite tact – that of study and evaluation of the host’s genome for clues as to how to overcome genetic (or other host) restrictions in devising new vaccine candidates. In fact, the developer of reverse vaccinology has published papers recognizing the limitations of a pathogen genome only approach and has published the need for a theory of “post-genome” vaccine development that incorporates the elements of vaccinomics into a fuller paradigm for understanding and developing vaccines [14,15,53]. Our vaccinomics approach proceeds from an understanding of immune response and its intersection with determinative genetic factors, and then proceeds to development of vaccine candidates. This is a very different approach. Nonetheless, an interesting possibility might be to combine the benefits of both vaccinomics and reverse vaccinology in understanding and identifying new methods of vaccine discovery by understanding and integrating information about both host and pathogen genetics, respectively [13,15]. Vaccinomics and reverse vaccinology are only two recent theories and approaches relevant to vaccinology (see Table 1).
Table 1.
Recent theories/approaches in vaccinology.
| Theory/model | Description | Tools used |
|---|---|---|
| Reverse vaccinology [52,156] | The use of genomic data and in silico analyses to rapidly identify antigens for vaccine use. |
Transcriptomics, proteomics, epitope prediction algorithms, immune monitoring |
| Immune response network theory [2] | Describes immunity as the predictable result of sequential activation/interaction of genes and gene pathways. |
Transcriptomics, proteomics, pathway analysis |
| Vaccinomics [2,9] | A comprehensive study of immune responses to vaccination such that vaccine-induced immunity can be understood and predicted and then applied to the rationale and directed development of vaccines. |
Transcriptomics, proteomics, epigenomics, Immunogenetics/immunogenomics, computational modeling, immune monitoring |
| Systems vaccinology [19,157] | The application of systems biology methods to understanding and predicting vaccine-induced immune responses. |
Transcriptomics, proteomics, epigenomics, computational modeling |
| Structural vaccinology [27,53] | The use of structural biology studies to facilitate the selection of vaccine epitopes. |
Proteomics, NMR, X-ray crystallography, immune monitoring |
| Vaccine informatics [24,158] | The use of bioinformatics approaches to facilitate vaccine development, production, testing, and licensure. |
Computational modeling, epitope prediction algorithms, HLA-binding algorithms, data mining and integration, mathematical simulations of immune response |
This table outlines several recent approaches being applied to the understanding of vaccine-induced immune responses and to the development of improved vaccines. As science advances and new tools are developed, these approaches undergo continual refinement. For example: (1) The initial reverse vaccinology approach has evolved into several closely-related approaches (i.e., classical, comparative, subtractive genome); (2) Vaccinomics was first conceived as immunogenetics/genomics applied to understanding and improving vaccine responses and now incorporates additional ‘omic’ technologies in order to move beyond host genetic variations and include the functional effects (epigenomic/transcriptomic/proteomic) of those variations; (3) Structural vaccinology is now beginning to use of synthetic protein mimetics to mimic three-dimensional peptide/protein epitopes for use in vaccine formulations.
In the design of new vaccines, we have developed a proof of principle approach for vaccinomics and the resulting “discover–validate–characterize–apply” paradigm. By utilizing the immune response network theory and vaccinomics approaches, we developed a mass spectrometry approach to isolating and identifying naturally presented and processed pathogen-derived peptides from the HLA peptide binding groove of individuals of interest (for example hyper- or hypo-responders to a specific vaccine) [54]. Because some peptides from the grooves of HLA molecule supertypes are promiscuous and can bind to a wide spectrum of HLA alleles, we reasoned that such peptides could form the basis for a peptide-based vaccine approach among individuals with that HLA supertype. Indeed, both measles and vaccinia peptides derived using this methodology have raised recall immune responses among HLA discordant individuals [54,55], and are now being used in preclinical vaccine development. Thus, this approach can be used to identify immunogenic peptides, compare and contrast peptide sets among hyper- and non-responders to viral vaccines, and allow the development of HLA supertype vaccines. By extension, adding peptide sets from additional HLA supertypes could allow coverage of >90% of a given population [56–58].
Another potential application of vaccinomics could be in relation to clinical trials. Under the current usual method of developing and testing new vaccine candidates and therapies, they are tested wholesale, again using a “one size fits all” population-based approach, among the general (and genetically uncharacterized) population. We posit that by using an uncharacterized population, subgroups that may respond vigorously—or, alternately, poorly—are unlikely to be identified within the context of the entire experimental group [59–63]. This has been seen with clinical trials of drugs, and we speculate also occurs in clinical trials of vaccines.
In this review, we discuss these and other practical uses and examples of the application of vaccinomics.
3. Practical uses of vaccinomics
3.1. A tool for evaluation of immune responses
Immunologists typically analyze discrete areas of immune function using reductionist approaches. This type of approach has been tremendously successful in furthering our understanding of discrete components of the immune system such as: individual signaling pathways; B-cell receptor (BCR) rearrangements; CD4T cell development; and Th1/Th2 differentiation. As increasingly sophisticated tools become available, our ability to examine complex networks and the important interactions between these discrete components has also expanded. From this point, we can then move beyond quantifying individual immune outcomes to understanding the specific pathways necessary for the development and regulatory control of those specific immune outcomes. These powerful “omics” technologies also provide a systems-level view of immune function, allowing us to identify important relationships between outcomes, pathways, cells, and even whole tissues.
These vaccinomics tools are being applied toward a deeper understanding of immune function in multiple ways. For example, the use of polychromatic flow cytometry allows for the simultaneous detection of multiple cell surface and/or functional markers on immune cells at precise time points during the generation of an immune response. One can analyze specific cell types (CD14+ monocytes, CD4+ or CD8+ T cells, CD56+ natural killer (NK) cells), distinguish between naïve (CCR7+CD45RA+), central/effector memory (CCR7+CD45RA−/CCR7−CD45RA−), and terminal effector (CCR7−CD45RA+) subsets, plasmacytoid or myeloid dendritic cells, or between IgA and IgG-producing CD19+CD38+CD27+ plasmablasts. The same experiment can identify cells secreting cytokines (TNFa, IL-2, IFNg), chemokines (MIP-1a), degranulation markers (CD107a, Granzyme B), as well as specific activation markers (CD38, CD69). Due to overlapping excitation/emission spectra, flow cytometry is currently limited to ~20 different markers [64]. Recent advances in single-cell, mass spectrometric cytometry (CyTOF) have the potential to significantly increase the number of parameters that can be measured [65]. Data from these types of experiments indicate that T cell responses display a high degree of diversity, which, in turn, provides the host’s immune response with tremendous functional flexibility. Other labs are merging flow cytometry with histology techniques to allow multispectral imaging of cells capable of evaluating both visual/fluorescence traits and spatial separation [66]. As another example, high dimensional transcriptomic analysis of alterations in gene expression before and after vaccination has identified specific gene signatures predictive of functional immune outcomes for yellow fever vaccine [67].
Adaptive immunity is not the only area where vaccinomics approaches have yielded important insights. Over the last several years, innate immunity has begun to take center stage not only for its ability to provide non-specific defenses against pathogens, but also for its ability to recognize and differentiate between diverse bacterial, viral, fungal, and parasitic pathogens [68–70], as well as to prime appropriate adaptive responses tailored toward specific pathogens [71]. Vaccinomics approaches are perfectly suited toward unraveling the intricacies of innate pattern recognition receptors, which control a complex system of signaling pathways that exhibit additive, synergistic, cross-regulatory, and even inhibitory effects. The outcome of pathogen detection by innate immune components is controlled by the combinatorial and integrated outcome of the cell type recognizing the pathogen, the anatomic location and microenvironment present during pathogen recognition, and the pathogen recognition receptors activated. In this regard, genome/proteome-level analyses have been used to identify key innate pathways activated by various pathogens and can be applied toward the directed selection of adjuvants stimulating critical innate pathways [71]. An example of this type of analysis is a recent study examining global gene expression patterns in the genital tract upon administration of experimental adjuvants (CpG and aGalCer) [72]. The authors performed both pathway and co-expression analyses to identify adjuvant-induced functional and coordinated gene expression activities. Their findings indicated that both adjuvants led to enhanced neutrophil chemotaxis into the vaginal tissues with corresponding inflammatory responses, although CpG administration induced a greater degree of inflammation and has been linked to epithelial damage [73]. aGalCer, on the other hand, also upregulated genes involved in PD-1 signaling. The authors hypothesized that the concomitant upregulation of PD-1 limited the excessive neutrophil-induced inflammation [74]. This information could be used to identify mucosal adjuvants that selectively activate pathways necessary for robust innate responses while limiting immunopathology.
We believe that next generation sequencing technologies will prove to be powerful tools for understanding both innate and adaptive immune function [4,8,9,75]. These technologies provide comprehensive and holistic data on simultaneous gene expression, miRNA levels, epigenetic events, and/or DNA/RNA sequence information. Although these technologies can be used in a targeted manner (specific genetic regions, isolated pathways, selected miRNA species), the global nature of the resulting data can be a considerable advantage as it allows the investigator to observe multifaceted interactions between critical immune elements/pathways. It also provides the opportunity to discover novel features of immune function that cannot be identified using reductionist approaches. These novel features can then be incorporated into the design of new vaccine candidates. One example would be the appropriate use of adjuvants stimulating necessary pathways involved in generating protective immune responses (i.e., aGalCer for mucosal immunity). In addition, rather than measuring vaccine efficacy solely on neutralizing antibody titer or the number of pathogen-specific T cells, one could develop an integrated model to identify better correlates of immunity (such as the aforementioned multifunctional T cells), or even predictive models of immune protection (such as the TNFRSF17 gene signature proposed as being predictive of antibody responses to the yellow fever vaccine) [67]. Another example is the use of transcriptional profiling to identify vaccine vectors capable of appropriate activation of key antigen presenting cells as was done with the poxvirus vector NYVAC containing HIV antigens (NYVAC-C) [76]. The authors found that NYVAC-C-KC (with restored expression of the host range genes K1L and C7L) activated antigen processing pathways without concomitant expression of inflammatory and IFN-induced genes, whereas NYVAC-C-ΔB19R (missing the viral type I IFN receptor homolog) induced inflammatory type I IFN and IL-1R gene pathways. By combining the two mutations, the authors were able to construct a vaccinia vector (NYVAC-C-KC-ΔB19R) capable of activating strong inflammatory and IFN responses, while also fully activating antigen processing and presentation pathways.
Thus, high dimensional “omics” tools provide not only the capability to study biologic responses in a comprehensive fashion, but also the opportunity to uncover novel interrelationships and to deconvolute the intricacies of the immune response to specific antigens.
3.2. A tool in developing new vaccine candidates
Lower costs, and an overall consensus that novel technologies must be applied in the process of vaccine discovery, have led more researchers to adopt vaccinomics-like approaches. The logical first step is to deploy vaccinomics technologies to profile immune responses to model vaccine systems [25,26]. For example, we are currently applying this approach to investigate immune responses to seasonal influenza vaccine in older adults with the intent of: (a) developing a predictive model of immunity to seasonal influenza vaccine; (b) elucidating potential mechanisms behind the influence of immunosensescence on vaccine response; and (c) discovering biomarkers associated with vaccine efficacy. By utilizing multiple “omics” platforms (genomics, proteomics, transcriptomics, and epigenomics), a definitive model of host response to seasonal influenza vaccine can be developed.
Recently, the National Institutes of Health funded the “Human Immunopehnotyping Consortium” to accelerate these efforts (http://www.immuneprofiling.org). The elegance of this approach is that it can be adapted for use with other model and novel vaccine systems. Additionally, if specific host factors are involved in immunity to more than one vaccine candidate, then perhaps these factors could be incorporated into constructs that recapitulate or enhance the immune response in low responder subjects to the levels observed in high responders. For example, we have observed polymorphisms/haplotypes in the vitamin D receptor (VDR) that influence the immune response across two different live viral vaccines—measles and rubella [77,78]. Genetic variations in the VDR influence the subsequent immune response. The active form of vitamin D (1,25(OH)2D3) binds to VDR and mediates the expression of vitamin D responsive genes [79]. VDR is expressed on multiple immune cells [80] and has been shown to influence immunity by regulating the expression of antimicrobial peptides, MHC expression, and polarize toward a Th2 immune response [81]. A very important observation is that baseline levels of 1,25(OH)2D3 in subjects with prostate cancer correlate with the serological response to seasonal influenza vaccine [82]. An indepth analysis of genes and gene pathways that links the expression of vitamin D receptor and its influence on the immune response to live viral vaccines will allow us to elucidate host-derived factors that could, for example, be used as adjuvants with a vaccine candidate.
Our own work with elucidating naturally processed, HLA-derived peptides from measles and vaccinia viruses [54,83] can be taken a step further with the advent of next generation sequencing for rapid HLA-typing [84] and TCR repertoire profiling [85]. Ideally, this would lead to a highly antigenic peptide cocktail that would cover the majority of HLA haplotypes present in the population. Another vaccinomics approach to elucidating naturally processed or mathematically predicted peptides is to use HLA-specific MHC tetramers, coupled with high-throughput flow cytometric analyses [86,87]. An advantage to this approach is the ability to quantify how many effector cells are specific for each peptide and the phenotype of these cells. Using MHC tetramers, Wagar et al. was able to identify a late effector subset of CD8T cells (KLRG1(+)CD57(+) influenza M1-specific) that were negatively associated with humoral immunity to influenza vaccine in older adults [87].
There has been some success in pathogen genomic analysis to determine potentially immunogenic antigens. One example is work with Group B meningococci and the synthesis of a vaccine containing five identified antigens [88]. Using these novel assays in concordance with high-throughput sequencing and proteomics will allow researchers to determine if a potential candidate will be effective across diverse genetic backgrounds and immunological phenotypes, and prior to committing large sums of money to empiric clinical trials.
A very promising use of “omic” technologies is to elucidate mechanisms behind vaccine escape variants. Golubchik et al. deployed genome sequence tracking of 62 pneumococcal vaccine escape isolates to determine what recombination events lead to vaccine escape [89]. The real-time surveillance of epidemiologically relevant strains could also be used with other highly variable pathogens to determine if putative vaccines or novel candidates would be effective. Researchers are utilizing immunoinformatics tools to develop vaccines that target conserved regions of circulating strains. One pitfall in the development of an effective HIV-1 vaccine is the high levels of genetic diversity between clades. One approach was to construct a cocktail of bioinformatically predicted T-cell epitopes from 43,282 HIV-1 sequences. Twenty-seven of these sequences were tested for binding and immunogenicity on PBMCs isolated from HIV-infected individuals [90]. Isolating immunogenic epitopes using a reductionist-based approach would prove to be near impossible with such a highly variable virus.
Not only can vaccinomics be applied to understanding immune responses and to developing novel vaccine candidates, but this approach can also be applied to investigate why historically successful candidates are effective [91–93]. Vaccinomics as a practice is not a complete abandonment of reductionist-based science, but a supplement and enhancement of the foundations built by earlier works. In retrospect, we can elucidate the mechanisms behind successful vaccine candidates against infectious diseases and apply them to the more difficult, elusive targets.
3.3. A tool in developing individualized vaccinology approaches
Vaccinomics could play a significant role in the development of personalized vaccines and in individualized therapeutic strategies. Understanding the genetics of host–pathogen interactions, particularly how immunogenetics and metagenomics impact the heterogeneity of vaccine-induced immunity, could allow the production of new vaccine candidates and perhaps, one day, individualized vaccines (or vaccines for specific sub-groups of interest – such as individuals who are immunosenescent or who carry common genetic variants that currently preclude development of protective immune responses). Further, integrating immunogenetics with functional SNP polymorphisms (including high-dimensional studies to quantitate mRNA gene transcripts of polymorphic genes) will add significant information to the rapidly developing concept of personalized vaccinology. Indeed, these novel concepts (the immune response network theory, vaccinomics, and adversomics), combined with the use of novel high-throughput technology (next generation sequencing and transcriptomics, proteomics, and epigenetics), will help elucidate, measure, and predict vaccine-induced immune responses, and bring personalized vaccinology closer to reality. In addition, advances in new high-throughput technology will allow identification of novel targets to be applied toward individualized vaccinology, such as cytokines (or cytokine antagonists) and cytokine adjuvants, hypomethylating agents (aberrant methylation), immune-relevant known and unknown genes and immune response-associated polymorphisms and networks, and/or pathways. Gathering information from metagenomic studies of microbial communities and their contribution to the health of the host will be an important step in comprehending the roles of microorganisms in immune response to vaccines [94]. Such advances will allow development of specific and general principles that advance personalized vaccinology and provide support for a new paradigm of vaccine development at individual (and population) levels.
An individualized vaccine approach results from application of the vaccinomics concept and an understanding of the HLA and other immune response genes’ contributions to immunity [95]. For example, identification of the antigenic peptides identified in the context of specific HLA proteins and associated with certain HLA alleles has allowed utilization of an HLA-based approach as a vaccinomics tool to identify novel vaccine candidates [45]. Our population-based study of smallpox vaccine-induced immune responses suggested that the HLA-B*1501 allele (p = 0.016) was highly correlated with vaccinia virus-specific cellular IFN-γ Elispot T cell responses after smallpox vaccination [34]. By applying an HLA-based approach, we have identified multiple class I A*0201 (A2 supertype), B*1501 (B15 supertype) and C*03-restricted T-cell epitopes in vaccinia virus proteins [54]. These naturally processed peptides are currently being assessed as new smallpox vaccines—either as synthetic epitope vaccines or as long synthetic epitope-adjuvanted vaccines—while having the benefits of immunogenicity and effectiveness, reduced frequency of side effects, and the ability to produce them quickly and at low cost [6,96]. An alternative strategy to overcome genetic restrictions is to create a vaccine that also includes a “universal” immunodominant T-cell epitope to induce strong immune responses in all individuals, regardless of genetic background, such as what is being done with influenza vaccines [97]. When considering peptide-based vaccines for potential use in the majority of the population, a relationship between HLA “supertype” alleles associated with overlapping peptide-binding specificities should be considered [98,99]. As an example, a relationship between the HLA-B7 supertype and higher measles vaccine-induced antibody levels, and a relationship between the HLA-B27 supertype and lower rubella antibody levels were successfully replicated between two independent vaccine immunogenetic studies [30,32]. Personalized selection of viral peptides presented by these B7 and B27 supertype alleles could be a promising strategy for novel MMR vaccines.
Several population-based vaccine studies have addressed the association between genetic polymorphisms and MMR, influenza, vaccinia, hepatitis B and HIV vaccines [34,100–107,59]. A recent Anthrax Vaccine Adsorbed (AVA) study analyzing vaccine-induced antibody responses to Bacillus anthracis protective antigen (PA) has suggested a genetic basis for interindividual variations in immune responses following vaccination [38,39]. Pajewski et al. [38] reported a significant association between DRB1–DQA1–DQB1 haplotypes (*1501–*0102–*0602, *0101–*0101–*0501, and *0102–*0101–*0501) and lower production of PA-specific antibodies in AVA-immunized subjects. Furthermore, significant associations with lower PA-specific T lymphocyte proliferation levels and HLA homozygosity (i.e., at the A, DQA1, and DQB1 loci) were also discovered [159]. Studies suggest that individuals who carry homozygous HLA alleles are more likely to fail to develop antibody and cytotoxic T cells responses to HIV vaccines, and are more likely to be susceptible to HIV-1 infection [59,108]. The effects of gene polymorphisms in MMR vaccine-induced immunity have been recognized and replicated for several gene systems: antigen-presenting class I (B) and class II (DR, DQ and DP) genes [30,32]; cytokine (IL2, IL10) and cytokine receptor (IL12B, IL12RB1) genes [33,109]; measles viral receptor CD46 genes [31]; and innate TLR4 genes [78,110,111]. Other examples include the HLA class IB loci and CCR5-delta receptor, which is correlated with susceptibility to HIV-1 infection and many previously unrecognized loci, such as killer Ig-like receptors (KIR), nucleotide-binding oligomerization domain-containing cytosolic proteins (NOD), tripartite motif protein TRIM5alpha, and mannose-binding lectin (MBL) [59,112–117]. Thus, understanding complex relationships between host genetic diversity and vaccine-induced immune response can assist in designing future personalized vaccines. In this respect, combined analyses of SNPs (n = 726) that tag multiple (n = 84) candidate genes also demonstrated that a multigenic assessment (p = 0.017) may explain variations in rubella vaccine antibody levels [118]. In our recent rubella vaccine population study, a search for the SNPs that influenced the antibody levels identified a set of 29 SNPs in 23 candidate immune response genes (such as IL6, IL4, IL4R, TNF, IL10RA, LTA, LST1, OAS2, ADAR, EIF2AK2, RARB, TRIM5, TRIM22, and others) that were jointly associated with significant variation in rubella antibody titers, while only four SNPs in three candidate genes (LTA, IL6, and IL4R) were implicated when SNPs were analyzed in isolation [118]. These data show that multigenic models may allow enhanced identification of additional significant SNP associations otherwise unobservable by simple univariate models.
As another example, we have recently identified and replicated a CD46 gene rs2724384 polymorphism that was associated with a two-fold decrease in measles vaccine-induced (Edmonston strain) antibody levels [31,119]. While functional studies are not yet complete, we speculate that this SNP impacts the capacity of measles virus to attach to its receptor and, therefore, prevents immune response. Understanding these genetic relationships allows the rational and directed design of a vaccine that evades immunogenetic limitations such as these. It terms of applicability to cellular immune responses, a group of SNPs in the cytokine and cytokine receptor genes, such as IL1RN, IL2RB, IL4R, IL6, IL10RB, IL12A, and IL12RB2, demonstrated consistent associations with vaccinia virus-specific cytokine production after smallpox vaccination among Caucasians and African-Americans [120]. In different ethnic groups, allele frequencies have been shown to vary significantly; thus, discovering common SNPs associated with vaccine-induced immunity among different racial or ethnic groups may allow for rational vaccine design across sub-groups of individuals.
Gender-based differences may also influence the development of a personalized approach to vaccine practice. Several studies have demonstrated a connection between gender and immune responses to vaccine antigens [121–123]. For example, our population-based vaccine studies demonstrate significantly higher humoral (IgG and neutralizing antibody) immune responses to rubella, mumps and vaccinia vaccines in females versus males [101,124,125]. A meta-analysis of several vaccine studies (n = 1800 influenza vaccinees) also demonstrated gender disparities in both local and systemic adverse events following inactivated influenza vaccination [126]. Though vaccine studies have focused mainly on gender-based differences in the humoral aspects of immunity, there is growing interest regarding gender disparities in cellular immunity, an important factor for vaccine development and individualized vaccine development [127]. Of interest, age- and gender-associated disparities in circulating NK T cell levels, as well as CD4+ NK T cell subsets, were recently found in healthy Korean individuals [128]. A better understanding of genetic background and gender-based differences in the development of vaccine-specific immunity may, for example, lead to the development of prophylactic vaccines customized to gender.
3.4. A tool in adversomics
Routine vaccination has been identified as a one of the greatest public health achievements of the 20th century [129] and is recognized in the scientific and medical community for its very low risk-to-benefit ratio [130]. Nonetheless, adverse events associated with vaccination represent important problems, in part because of the impact of disproportionate publicity and its impact on uptake [131], but, more importantly, because of vaccination’s use in otherwise healthy individuals as a preventive—rather than a therapeutic—intervention [132]. However, the determination of causality with rarely occurring adverse events is fraught with difficulties for both pre- and post-licensure study [133,134]. Vaccines are biologics, and the human response to biologics varies widely across individuals and not just in the desired, salutary immune response, but also in untoward reactions. This variation depends upon a complex interaction of past exposures and infections, current physical and emotional health, and the individual’s genome and microbiome [135], among other factors. With respect to the variation that results in the rare, but serious, adverse events associated with vaccines, Relman wrote the following in 2008:
If these aberrant responses could be predicted and understood mechanistically (the first element does not necessarily require the second), we would be able to reduce the number and/or severity of vaccine adverse events (AEs), improve vaccine design, and create personalized strategies for eliciting immune protection. [135]
We are moving toward an era where we may measure and predict the complex interactions that result in adverse events associated with vaccination or “adversomics [12,136].” This is not science fiction. Several recent studies, as reviewed below, demonstrate the feasibility and utility of such investigations.
The first example is an investigation of sodium channel gene SCN1A mutations in alleged vaccine encephalopathy [137]. Berkovic et al. studied 14 patients with alleged vaccine encephalopathy that was made manifest by seizures occurring within 72 h of vaccination after one of the first three doses of diphtheria–pertussis–tetanus (DPT) or diphtheria–pertussis–tetanus-inactivated polio-Haemophilus influenzae type b (DPT-IPV-Hib) vaccine given in infancy. Of the 14 patients, eight had Dravet’s syndrome or severe myoclonic epilepsy in infancy (SMEI), four had borderline severe myoclonic epilepsy in infancy (SMEB), and two had Lennox–Gastaut syndrome. Investigators conducted PCR amplification on all 26 exons of the sodium channel gene SCN1A along with flanking intronic primers and ascertained the presence of heterozygous mutations of SCN1A in 11 of 14 cases. None of the mutations found occurred in the 130–149 control samples obtained from blood donors. In nine of 11 patients, studies of the parents demonstrated that the mutations occurred de novo. Parental DNA was not available in the other two, so de novo mutations could not be confirmed in those. The 11 cases included eight of the eight original SMEI cases, three of the four original SMEB cases, and none of the two Lennox–Gastaut cases. As the authors wrote, while epidemiologic studies have not supported associations of childhood vaccination with encephalopathic disorders, individual cases with their coincidental associations pose real difficulties for the parents, the physicians, and the public. Investigations into causation such as these not only provide explanation, but also solace as well – and potentially inform the design of newer, safer vaccines.
The second example concerned an investigation by Ball et al. with regard to the association of arthritis with Lyme disease vaccination [138]. The investigators compared the results of HLA-DRB1 allele typing, Western blots for Lyme disease, and T cell reactivity in 27 cases of individuals who developed arthritis following their receipt of Lyme disease vaccine and 162 matched controls. The investigators found no differences across HLA-DRB1 alleles or T cell response to outer surface protein A (Osp-A), but did find lower levels of IgG antibodies to Osp-A. The results point away from the hypothesis that the Lyme disease vaccine was a major factor in the development of these cases of arthritis [138]. This association of arthritis and vaccination was rejected by the analyses conducted by U.S. Food and Drug Administration and other investigators [139].
The preceding examples, while not genomic studies, demonstrate the potential to use broad genomic evaluations in addressing important questions of vaccine-associated adverse events—the first to uncover an alternate explanation and the second to test a proposed theory. A third example illustrated the actual use of modern methods of adversomics. Reif et al. conducted genome-wide SNP genotyping of subjects participating in two clinical trials of smallpox vaccine [140]. In both trials, the subjects were healthy adults receiving the smallpox vaccine for the first time, and were followed closely to document any adverse events. In the first trial of 96 subjects, 16 had systematic adverse events. A total of 36 SNPs representing 26 genes were identified in the first trial to have significant associations with systematic adverse events. The investigators then used the same SNP panel on those in the second trial including 46 subjects where 24 had systematic adverse events. Of the original 36 SNPs, three SNPs were confirmed to have statistically significant associations with systematic adverse events. These SNPs were associated with genes [5,10-methylenetetrahydrofolate reductase gene (MTHFR) and interferon regulatory factor-1 (IRF1)] that are quite possibly involved in pathways in line with hypothesized mechanisms for adverse events resulting from imbalances in inflammatory stimulation and tissue damage repair. The study not only demonstrated the power of the SNP arrays, but also the need for confirmatory replication [135]. Further work with systemic adverse events associated with smallpox vaccine used advanced computational techniques and specifically an analytic approach labeled Random Forest™, which utilizes both genetic and proteomic data to create a biologic model that included the intercellular adhesion molecule-1, interleukin-10, colony stimulating factor-, and interleukin-4, and fit well with the previous findings and the proposed underlying mechanism [141].
A fourth example concerned the combination of microarray with immunofluorescence to monitor the effects of vaccine adjuvants in mouse muscle [142]. The adjuvants studied included MF59, CpG, and alum. The authors examined the expression of genes over time to demonstrate that MF59 affected the up or downregulation of 891 genes, whereas CpG and alum up or down regulated 387 and 312 genes, respectively. All regulated a common set of 168 genes, but MF59 had the strongest effect in inducing cytokines, cytokine receptors, adhesion molecules, and antigen presentation cells. The authors concluded from their investigation that MF59 represented the best of the three adjuvants compared because MF59 not only had the strongest, but had the earliest effects.
In a fifth example, Mizukami et al. evaluated the utility of cloned complementary DNA microarrays in the quality control of influenza vaccine manufacture, with particular reference to novel strains [143]. Specifically, they demonstrated—with DNA microarrays—that they were able to identify 76 genes that distinguish whole-virus H5N1 vaccines from split-virion H5N1 vaccines, as well as saline controls. From their analyses of gene expression, they concluded that this rapid and highly sensitive method produced results consistent with the standard toxicity tests currently employed in quality control. This novel approach would reduce the time necessary for safety tests, as well as the number of animals sacrificed.
These examples illustrate a diverse set of applications of adversomics. Clearly, the combination of high-throughput genomics, transcriptomics, proteomics, and others can—with the new advances in computational capacity and techniques—open new frontiers of investigation. We have learned that gene expression studies of vaccines and their adjuvants as tested in animals may fail to translate to humans [132]. Furthermore, despite the rarity of the most concerning adverse events, replication of findings in subsequent samples is critical; investigations of genomic associations in single samples are only hypothesis-generating [135,140]. Investigators will need access to multiple samples whose clinical status (phenotype) is well characterized and from whom great care is taken in collecting samples in terms of timing, as well as in collecting data with respect to exposures and outcomes [132,135].
Not only will such investigations require financial resources; consideration must be given to creating shared repositories of data (freed from proprietary interests) [132], as well as to centralized authorities to monitor and account for vaccine adverse events [136]. Such large-scale phenotype–genotype databases are critical to developing multigenetic models of adverse effects from vaccines and to achieving the promise of adversomics in both developing safer vaccines and in predicting/preventing serious adverse reactions in vaccine recipients.
3.5. Development of systems level models to support vaccinomics
The development and application of bioinformatics and systems biology tools will be essential to meeting the goals of vaccinomics in the 21st century. In this section, we discuss analytical tools and strategies for their application in two areas of vaccinomics. First, the prediction of immune response outcomes (immune response markers such as antibody response and adverse events) based on a complex signature of biomarkers. The goal of immune response prediction is to infer a statistical or machine learning classifier that incorporates one or a combination of genetic and other biomarkers to answer the question “will a particular vaccine ‘work’ for a given individual?” Personalized vaccination will use these classifiers to predict an individual’s quantitative response to different vaccination types, doses, or routes, and to suggest the safest and most effective route, given the individual’s unique combination of genotype, gene expression levels, epigenetic and other markers. Second, controllability of the immune response through the use of statistical and mechanistic network models that can be manipulated in silico to guide modifications to achieve safer, more effective vaccines. The goal of controllability is very similar to the goal of vaccination itself—to guide the immune response trajectory to the optimum state. The difference lies in the controllability theory’s use of mechanistic models, based on known interactions and signaling of cells and molecules in immune system pathways, to identify drivers within the model that may be used to achieve the precise biological outcome. That is, understanding how the immune system works, and what key elements are required to achieve a response. A combination of mechanistic and statistical/machine learning approaches will likely emerge to achieve robust system identification and target a desired immunological state.
Machine-learning algorithms have become popular in bioinformatics for class prediction and regression in high-dimensional data [144]. For class prediction, a rule is learned from training data to predict class labels, such as case-control status, in unseen test data. Similarly, machine-learning regression establishes a rule for fitting a quantitative outcome variable, such as immune response. Among the most popular class prediction and regression algorithms are linear discriminant analysis, neural networks, support vector machines (SVM), classification tress (CART), random forests, penalized regression, and nearest neighbor methods. An integrative, high-dimensional framework is needed to meet the challenge of immune system prediction following vaccination because no single data domain (genome-wide association studies or GWAS, gene expression, epigenetic, environmental covariate) or single marker will be sufficient to predict a complex outcome such as vaccine immune response. Indeed, for GWAS it has been shown that when replicated disease-associated SNPs are used as input, the resulting classifiers provide poor accuracy [145]. In large part, poor classification is due to the low relative risks of validated disease loci, typically in the range of 1.1–1.2, whereas very large relative risks are required for accurate prediction [146]. Thus, in order to infer classifiers that can predict personalized outcomes in the absence of characteristics with large relative risks, classifiers likely will need to include a large number of biomarkers and include interactions among them. However, increasing the number of markers included generally reduces the generalizability of a predictive model. Alternatively, integration of signatures from other molecular biology levels, such as the protein or transcript level, together with DNA-level information may be required for accurate prediction [141]. Integration may take place in a hierarchy or on multiple levels (e.g., SNP to gene expression with feedback loops, etc.) or without regard to any structure that assumes the biological relationships between data types. The advantage of a hierarchical network structure is the potential to identify causal relationships. In addition, the no-free-lunch theorem of data mining tells us that multiple statistical methods will need to be combined to find consensus biomarkers and signatures for immune response prediction [147,148].
A disadvantage of many of the modern methods mentioned for class prediction is the difficulty of interpreting the classification model. The SVM provides some degree of geometric interpretability as a separating hyperplane; however, the ability to understand or visualize such a plane is lost in higher dimensional feature space. Thus, alternative and complementary approaches are needed to rank important markers and interpret biological relationships. Network and pathway approaches lend themselves to interpretability because network relationships can be visualized in two dimensions and pathway annotation can be mapped onto nodes and edges (Fig. 2).
A network is simply a set of nodes connected by edges. For a biological network, the nodes are typically genes or gene products. However, there are numerous ways to define the edges in a biological network, including correlation, regulatory, biochemical reaction, statistical interaction, protein–protein interaction, and others. All of these ways of defining edge information content will help vaccinomics; however, given the currently discussed goals of personalized immune response prediction and control, we focus on edges that carry information about immune response on the population level. These epistasis networks or genetic association interaction networks (GAIN) have been applied to SNP data for vaccine immune response [149] and demonstrated pathway replication in GWAS of mental disorders [150].
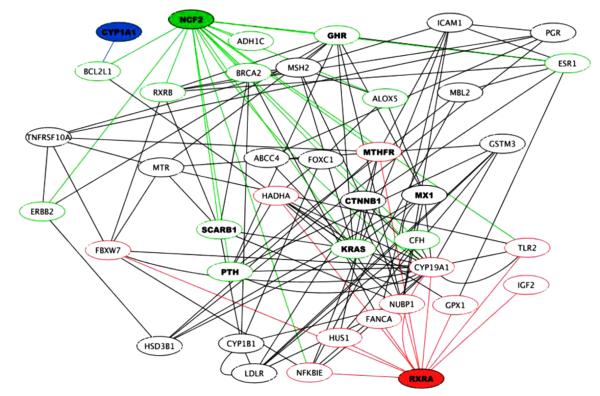
Fig. 3 shows an epistasis network from Davis et al. [149] for a genetic association study of human antibody response to smallpox vaccine. Empirical node and edge weights for the GAIN are determined by main effects and gene–gene interactions in the context of post-vaccination antibody response. The most important SNP node, based on network centrality (importance based on main effect associations with response, together with interactions with other SNPs conditional on response), is in the retinoid X receptor alpha (RXRA) gene, highlighted in red along with its connections in Fig. 3. The RXRA gene mediates vitamin D signaling, and recent studies show that RXRA is involved in innate immune response [77–82]. Other important genes based on their network centrality are highlighted, such as Cytochrome P450, family 1, subfamily a, polypeptide 1 (CYP1A1) (highlighted in blue), and neutrophil cytosolic factor 2 (NCF2) in green. The individual effect of RXRA in this data would have been below the threshold of significance in a standard statistical analysis, but our SNPrank algorithm identified RXRA as the most important association with smallpox vaccine antibody response due to its many gene–gene interactions in the network. Driven by RXRA, GAIN and SNPrank provide evidence that other vitamin regulation pathway genes play an important role in immune response to smallpox vaccination.
Fig. 3.
Genetic association epistasis network for human antibody response to smallpox vaccine. Data-driven, vaccine-specific interaction networks will be an important component of personalized, predictive vaccinomics. The properties of such networks can provide target information for vaccine or therapeutic design, and the connectivity of such networks can guide causal and mechanistic models of vaccine immune response. This network was inferred from a genetic association study of individuals who received the smallpox vaccine. The nodes and edges carry genetic variation that explains an individual’s response to smallpox vaccine. The most influential hub gene in the network is retinoid-X receptor-alpha (RXRA), labeled in red at the bottom. This gene is involved in mediating vitamin D signaling and innate immune response. However, the importance of RXRA would not be recognized in this data by a univariate analysis. The identification of this gene through a genome-wide network analysis indicates the importance of statistical modeling that reflects the underlying complexity of the biological system. Adapted from [149].
The epistasis network approach may lead to greater sensitivity because it ranks genes based on their connections in the system-wide network as well as their individual importance to the immune response. The connections in these population-level statistical epistasis networks will likely have some degree of isomorphism with the connections in cellular, signaling and regulatory networks. The isomorphism will be imperfect because the connectivity in epistasis networks is specifically conditioned on risk of low immune response or risk of adverse event. However, identifying isomorphisms will help us understand both statistical networks and mechanistic networks. In addition to refining our understanding of biological relationships involved in immune response, knowledge of the complexity and topology of the epistasis network and centrality of nodes will improve our ability to control the individual immune response.
Controllability of a network refers to the number of nodes required to reach a desired state of the system. The history of vaccination has demonstrated the controllability of the immune system. However, there is a need for a mathematical theory of control that quantifies how controllable the immune system network is and predicts the drivers of control. Drivers are nodes required to guide a system to a desired state. In the study of dynamic networks, Liu et al. [151] identified sets of driver nodes for complex networks. The main determinant of the number of driver nodes was the network’s degree distribution, and results suggested that dense and homogeneous networks may be controlled using a few driver nodes. Surprisingly, networks became more difficult to control as the degree of heterogeneity increased. These dynamic networks are fundamentally different from some other types of biological networks, such as epistasis networks. Thus, an important goal of vaccinomics is to determine whether similar controllability properties exist for epistasis networks and other biological networks related to the immune system response. However, these results for dynamic networks suggest that one may be able to identify new drivers for improved vaccine design.
A challenge to development of these class prediction models is that many individuals’ immune responses may be truly unique, and a personalized classifier may require a unique panel of rare variants and other markers to predict who will be a poor vaccine responder. Unique signatures pose a challenge to machine learning and statistical methods in general because the goal is to learn patterns and trends from training data that generalize to test data containing characteristics on independent people. As discussed in the prior section on adversomics, large sample sizes will be required in order to incorporate both common and rare markers that span multiple molecular levels in the body into class prediction models, ensuring accuracy and generalizability of the classifiers. Establishment of biobanks containing clinical specimens together with clinical characteristics, together with public deposit of data into public data bases such as GEO (Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/) or Imm-Port (http://immport.niaid.nih.gov) will help to move this field forward.
3.6. The ultimate goal: development of descriptive and predictive immune response equations
Given the aforementioned theoretical and current models, we have established the goal of developing descriptive and predictive equations, whereby it is theoretically possible to devise a meta-equation that describes the role and degree of importance of key drivers/variables that in toto define the immune response to a given antigen. Such an equation will need to characterize key genetic restrictions at the SNP/gene/haplotype level, epigenetic phenomena, complementarity, epistasis, polymorphic plasticity, the role of adjuvants, and, more recently, the current microbiome state [118,152]—all at the systems-integrated level.
The usefulness of such equations includes a method to organize thinking about the components and interactions of these components on immune response, and to provide initial hypotheses as to how the immune system works. As Box so elegantly stated, “all models are wrong, but some are useful [153].” We suspect that while the initial equations we propose are overly simplified, additional data will make clear that such a system (and its descriptive equation) will rapidly become quite complex. With more data and increasingly refined iterations of mathematical models, we anticipate insights that will allow increasing simplicity of such an equation – leveraged on increased understandings of what variables are of sufficient magnitude to be descriptive and predictive of immune response, and what the interactions among such variables might be when considered within the confines of the human system. A general form of this model of immune response can be written as y = fv(HF, VF, GF, …) where y indicates a measure of a person’s immune response (such as humoral immunity or circulating antibody level), fv indicates a function (linear or nonlinear) of the indicated variables for the vth vaccine, HF indicates human demographic factors, VF indicates vaccine variables, GF indicates genomics factors, and so on. Based on past findings, we are confident that demographic variables such as age and gender will be a part of this model. If we assume this model can be linearized for a given vaccine, it could be written in the form , where y is a measure of a person’s immune response, β0 is the intercept, βi is the coefficient or slope for the ith variable xi and indicates the amount of change in y for a 1 unit change in xi, and ε represents random deviations from this model. The xi variables are a mathematical representation of the human demographic and other factors described above. In reality the model is likely more complex than can be represented by a linear equation and will be a nonlinear function of these human characteristics defined by a complex system of differential equations, and may differ according to properties of the vaccine.
Of course, each of the variables (host factors, genetic factors, etc.) are themselves sub-models that can be mathematically characterized. Some may be linear, though likely most biologic systems will not be. By virtue of building such equations, we are forced to confront such issues, and incrementally design experiments and provide data to inform belter model building.
While building such equations and models is conceptually possible, significant work remains to be accomplished in being able to characterize each such component and its role in immune phenotypes and profiles of interest. Much more research into immunogenetics, immunogenomics, metagenomics, and bioinformatics models will be needed, and readers should understand this quest as a “forward-looking” statement. Nonetheless, given adequate characterization of the role and strength of influence of each component influencing or driving the immune system, it should be possible to derive a descriptive, and later predictive, equation that, through successive refinements informed by ongoing research findings, allows definition and prediction of a specific immune phenotype of interest at both the individual, and perhaps, the population or “sub-population” level. Key to these efforts will be the integration of new bioinformatic approaches to such studies, and the development of newer systems level analytical approaches as noted above [11].
Development of such an equation(s) represents the “holy grail” for vaccinologists and could enable more rapid, directed, and rational design of new vaccines, more efficacious vaccines, safer vaccines, and could potentially remove or minimize much of the time and financial risk inherent in new vaccine development. Such an approach is urgently needed for developing vaccine candidates against pathogenic hyper-variable viruses, parasites, fungi, and bacteria that plague humans. Too, such an equation(s) would allow early determination of vaccine immunogenicity, dosing, and safety in a given individual or subgroup of highly characterized individuals. In addition to developing vaccines against pathogens that cause significant morbidity and mortality, such “individualized vaccinology” is a major and compelling goal for the 21st century practice of vaccinology.
4. Summary
Vaccinomics and the immune response theory are poised to be the major drivers of how vaccines are developed, administered, and monitored in the 21st century. In addition, the application of these concepts to vaccine safety (adversomics) will enable the concept of personalized vaccinology, and allow administration of vaccines informed by more refined estimates of potential adverse side effects to a given vaccine. The application of these concepts may result in significantly diminishing the current anti-vaccine and general fear of vaccines that seem so prevalent today, and that measurably—and adversely—affect population coverage with vaccines [154,155].
In addition, we have described within this article other practical applications of vaccinomics in terms of creating a tool that will assist us in determining how vaccines are evaluated and developed; this will provide further impetus for the development of genetic and systems levels models and analysis tools.
These concepts will emerge as critical, we believe, as we move into the future where we: must better understand how to manipulate the immune systems for the benefit of individuals and populations; are increasingly compelled to develop vaccines against hyper-variable viruses and newly emerging infectious and non-infectious threats; are engaging in efforts to develop new vaccine adjuvants to increase immunogenicity and vaccine efficacy in the increasingly older and immunosenescent population, as well as the increasing numbers of immunosuppressed individuals who remain at high-risk for morbidity and mortality due to common pathogens. Finally, as issues of vaccine-induced selective pressures over long periods of time begin to be recognized, new vaccines and new approaches will be required to replace currently successful vaccines. Despite these challenges, the future looks bright, the technology and science base ever more useful and enlightening, and the willingness to apply new scientific concepts invigorating.
Acknowledgements
Research reported in this publication was supported by the National Institute of Allergy And Infectious Diseases of the National Institutes of Health under award/contract numbers U01AI089859, R37AI048793, (which recently received a MERIT Award) R01AI033144 and HHSN266200400065C (AI40065). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosures Dr. Poland is the chair of a Safety Evaluation Committee for novel investigational vaccine trials being conducted by Merck Research Laboratories. Dr. Poland offers consultative advice on vaccine development to Merck & Co. Inc., CSL Biotherapies, Avianax, Sanofi Pasteur, Dynavax, Novartis Vaccines and Therapeutics, and PAXVAX Inc and Emergent Biosolutions. These activities have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies. This research has been reviewed by the Mayo Clinic Conflict of Interest Review Board and was conducted in compliance with Mayo Clinic Conflict of Interest policies.
Dr. Jacobson is a member of the Safety Review Committee for Kaiser-Permanente, supporting two of its studies regarding the safety of Gardasil, a Merck product. He also serves as principal investigator for three studies including one by Novartis for its Menveo meningococcal conjugate vaccine and two by Pfizer for its Prevnar 13 pneumococcal conjugate vaccine.
References
- [1].Tuells J. Vaccinology: the name, the concept, the adjectives. Vaccine. 2012;30:5491–5. doi: 10.1016/j.vaccine.2012.06.059. [DOI] [PubMed] [Google Scholar]
- [2].Poland GA, Ovsyannikova IG, Jacobson RM, Smith DI. Heterogeneity in vaccine immune response: the role of immunogenetics and the emerging field of vaccinomics. Clinical Pharmacology & Therapeutics. 2007;82:653–64. doi: 10.1038/sj.clpt.6100415. [DOI] [PubMed] [Google Scholar]
- [3].Poland GA, Ovsyannikova IG, Jacobson RM. Application of pharmacogenomics to vaccines. Pharmacogenomics. 2009 May;10:837–52. doi: 10.2217/PGS.09.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ovsyannikova IG, Poland GA. Vaccinomics: current findings, challenges and novel approaches for vaccine development. American Association of Pharmaceutical Scientists. 2011;13:438–44. doi: 10.1208/s12248-011-9281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Poland GA, Kennedy RB, Ovsyannikova IG. Vaccinomics and personalized vaccinology: is science leading us toward a new path of directed vaccine development and discovery? PLoS Pathogens. 2011;7:e1002344. doi: 10.1371/journal.ppat.1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Poland GA, Ovsyannikova IG, Jacobson RM. Personalized vaccines: the emerging field of vaccinomics. Expert Opinion on Biological Therapy. 2008;8:1659–67. doi: 10.1517/14712598.8.11.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Poland GA, Ovsyannikova IG, Kennedy RB, Haralambieva IH, Jacobson RM. Vaccinomics, a new paradigm for the development of preventive vaccines against viral infections. Omics. 2011;15:625–36. doi: 10.1089/omi.2011.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kennedy RB, Poland GA. The top five game changers in vaccinology: toward rational and directed vaccine development. Omics. 2011;15:533–7. doi: 10.1089/omi.2011.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Oberg AL, Kennedy RB, Li P, Ovsyannikova IG, Poland GA. Systems biology approaches to new vaccine development. Current Opinion in Immunology. 2011;23:436–43. doi: 10.1016/j.coi.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Poland GA, Ovsyannikova IG, Jacobson RM. Vaccinomics and personalized vaccinology. U.S. Department of Health and Human Services, The Jordan Report. 2012:11–8. [Google Scholar]
- [11].Poland GA, Oberg AL. Vaccinomics and bioinformatics: accelerants for the next golden age of vaccinology. Vaccine. 2010;28:3509–10. doi: 10.1016/j.vaccine.2010.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Poland GA, Ovsyannikova IG, Jacobson RM. Adversomics: the emerging field of vaccine adverse event immunogenetics. Pediatric Infectious Disease Journal. 2009;28:431–2. doi: 10.1097/INF.0b013e3181a6a511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bernstein A, Pulendran B, Rappuoli R. Systems vaccinomics: the road ahead for vaccinology. Omics. 2011;15:529–31. doi: 10.1089/omi.2011.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rinaudo CD, Telford JL, Rappuoli R, Seib KL. Vaccinology in the genome era. Journal of Clinical Investigation. 2009;119:2515–25. doi: 10.1172/JCI38330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Seib KL, Dougan G, Rappuoli R. The key role of genomics in modern vaccine and drug design for emerging infectious diseases. PLoS Genetics. 2009;5:e1000612. doi: 10.1371/journal.pgen.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Joly Y, Koutrikas G, Ramos-Paque E, Zawati M, Gardy J, Hayden MR, et al. Diagnostic testing for vaccinomics: is the regulatory approval framework adequate? A comparison of Canada, the United States, and Europe. OMICS. 2011;15:597–605. doi: 10.1089/omi.2010.0135. [DOI] [PubMed] [Google Scholar]
- [17].Moyer MW. [Last updated June 24, 2010. [Accessed 25.09.12]];Vaccinomics scientists are devising your personal vaccine. Scientific American. http://www.scientificamerican.com/article.cfm?id=vaccinomics-personal-vaccine.
- [18].Scudellari M. [Last updated October 2011. [Accessed 25.09.12]];Data deluge the scientist. http://the-scientist.com/2011/10/01/data-deluge/
- [19].Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33(4):516–29. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nakaya HI, Li S, Pulendran B. Systems vaccinology: learning to compute the behavior of vaccine induced immunity. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2012;4:193–205. doi: 10.1002/wsbm.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Buonaguro L, Pulendran B. Immunogenomics and systems biology of vaccines. Immunological Reviews. 2011;239:197–208. doi: 10.1111/j.1600-065X.2010.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jerne NK. Towards a network theory of the immune system. Annals d Immunologie (Paris) 1974;125C:373–89. [PubMed] [Google Scholar]
- [23].Hoffman GW. [Accessed 20.03.13];Immune network theory. The University of British Columbia. 2008 http://www.phas.ubc.ca/~hoffmann/book/first%20section.pdf.
- [24].He Y, Rappuoli R, De Groot AS, Chen RT. Emerging vaccine informatics. Journal of Biomedicine and Biotechnology. 2010;2010:218590. doi: 10.1155/2010/218590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nature Reviews Immunology. 2009;9:741–7. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- [26].Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, et al. Systems biology of seasonal influenza vaccination in humans. Nature Immunology. 2011;12:786–95. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dormitzer PR, Grandi G, Rappuoli R. Structural vaccinology starts to deliver. Nature Reviews Microbiology. 2012;10:807–13. doi: 10.1038/nrmicro2893. [DOI] [PubMed] [Google Scholar]
- [28].Yang J, Loos RJ, Powell JE, Medland SE, Speliotes EK, Chasman DI, et al. FTO genotype is associated with phenotypic variability of body mass index. Nature. 2012;490:267–72. doi: 10.1038/nature11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Eder W, Klimecki W, Yu L, von Mutius E, Riedler J, Braun-Fahrländer C, et al. Opposite effects of CD 14/-260 on serum IgE levels in children raised in different environments. Journal of Allergy and Clinical Immunology. 2005;116:601–7. doi: 10.1016/j.jaci.2005.05.003. [DOI] [PubMed] [Google Scholar]
- [30].Ovsyannikova IG, Jacobson RM, Vierkant RA, O’Byrne MM, Poland GA. Replication of rubella vaccine population genetic studies: validation of HLA genotype and humoral response associations. Vaccine. 2009;27:6926–31. doi: 10.1016/j.vaccine.2009.08.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ovsyannikova IG, Haralambieva IH, Vierkant RA, O’Byrne MM, Jacobson RM, Poland GA. The association of CD46, SLAM, and CD209 cellular receptor gene SNPs with variations in measles vaccine-induced immune responses—a replication study and examination of novel polymorphisms. Human Heredity. 2011;72:206–23. doi: 10.1159/000331585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA. Consistency of HLA associations between two independent measles vaccine cohorts: a replication study. Vaccine. 2012;30:2146–52. doi: 10.1016/j.vaccine.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Haralambieva IH, Ovsyannikova IG, Kennedy RB, Vierkant RA, Pankratz VS, Jacobson RM, et al. Associations between single nucleotide polymorphisms and haplotypes in cytokine and cytokine receptor genes and immunity to measles vaccination. Vaccine. 2011;29:7883–95. doi: 10.1016/j.vaccine.2011.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ovsyannikova IG, Vierkant RA, Pankratz VS, Jacobson RM, Poland GA. Human leukocyte antigen genotypes in the genetic control of adaptive immune responses to smallpox vaccine. Journal of Infectious Diseases. 2011;203:1546–55. doi: 10.1093/infdis/jir167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ovsyannikova IG, Kennedy RB, O’Byrne M, Jacobson RM, Pankratz VS, Poland GA. Genome-wide association study of antibody response to smallpox vaccine. Vaccine. 2012;30:4182–9. doi: 10.1016/j.vaccine.2012.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kennedy RB, Ovsyannikova IG, Shane P, Haralambieva VIH, Vierkant RA, Poland GA. Genome-wide analysis of polymorphisms associated with cytokine responses in smallpox vaccine recipients. Human Genetics. 2012;131:1403–21. doi: 10.1007/s00439-012-1174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kennedy RB, Ovsyannikova IG, Pankratz VS, Haralambieva IH, Vierkant RA, Jacobson RM, et al. Genome-wide genetic associations with IFNgamma response to smallpox vaccine. Human Genetics. 2012;131:1433–51. doi: 10.1007/s00439-012-1179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pajewski NM, Parker SD, Poland GA, Ovsyannikova IG, Song W, Zhang K, et al. The role of HLA-DR-DQ haplotypes in variable antibody responses to anthrax vaccine adsorbed. Genes and Immunity. 2011;12:457–65. doi: 10.1038/gene.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pajewski NM, Shrestha S, Quinn CP, Parker SD, Wiener H, Aissani B, et al. A genome-wide association study of host genetic determinants of the antibody response to Anthrax Vaccine Adsorbed. Vaccine. 2012;30:4778–84. doi: 10.1016/j.vaccine.2012.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tan PL, Jacobson RM, Poland GA, Jacobsen SJ, Pankratz SV. Twin studies of immunogenicity – determining the genetic contribution to vaccine failure. Vaccine. 2001;19:2434–9. doi: 10.1016/s0264-410x(00)00468-0. [DOI] [PubMed] [Google Scholar]
- [41].Virgin HW, Todd JA. Metagenomics and personalized medicine. Cell. 2011;147:44–56. doi: 10.1016/j.cell.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Varela FJ, Coutinho A. Second generation immune networks. Immunology Today. 1991;12:159–66. doi: 10.1016/S0167-5699(05)80046-5. [DOI] [PubMed] [Google Scholar]
- [44].Korth MJ, Tchitchek N, Benecke AG, Katze MG. Systems approaches to influenza–virus host interactions and the pathogenesis of highly virulent and pandemic viruses. Seminars in Immunology. 2012 doi: 10.1016/j.smim.2012.11.001. [Epub ahead] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sette A, Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 2010;33:530–41. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Duraisingham SS, Rouphael N, Cavanagh MM, Nakaya HI, Goronzy JJ, Pulendran B. Systems biology of vaccination in the elderly. Current Topics in Microbiology and Immunology. 2013;363:117–42. doi: 10.1007/82_2012_250. [DOI] [PubMed] [Google Scholar]
- [47].Barocchi MA, Censini S, Rappuoli R. Vaccines in the era of genomics: the pneumococcal challenge. Vaccine. 2007;25:2963–73. doi: 10.1016/j.vaccine.2007.01.065. [DOI] [PubMed] [Google Scholar]
- [48].Poland GA. Pharmacology, vaccinomics, and the second golden age of vaccinology. Clinical Pharmacology & Therapeutics. 2007;82:623–6. doi: 10.1038/sj.clpt.6100379. [DOI] [PubMed] [Google Scholar]
- [49].Plotkin SA, Orenstein WA, Offit PA, editors. Vaccines. 4th ed Elsevier; Netherlands: 2004. A short history of vaccination; pp. 1–17. [Google Scholar]
- [50].Galambos L. What are the prospects for a new golden era in vaccines. Euro-health. 2007;14:12–4. [Google Scholar]
- [51].Garcia-Cordero JL, Nembrini C, Stano A, Hubbell JA, Maerkl SJ. A high-throughput nanoimmunoassay chip applied to large-scale vaccine adjuvant screening. Integrative Biology (Camb) 2013;5:650–8. doi: 10.1039/c3ib20263a. [DOI] [PubMed] [Google Scholar]
- [52].Rappuoli R. Reverse vaccinology. Current Opinions in Microbiology. 2000;3:445–50. doi: 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- [53].Serruto D, Rappuoli R. Post-genomic vaccine development. FEBS Letters. 2006;580:2985–92. doi: 10.1016/j.febslet.2006.04.084. [DOI] [PubMed] [Google Scholar]
- [54].Johnson KL, Ovsyannikova IG, Mason CJ, Bergen HR, III, Poland GA. Discovery of naturally processed and HLA-presented class I peptides from vaccinia virus infection using mass spectrometry for vaccine development. Vaccine. 2009;28:38–47. doi: 10.1016/j.vaccine.2009.09.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Johnson KL, Ovsyannikova IG, Madden BJ, Poland GA, Muddiman DC. Accurate mass precursor ion data and tandem mass spectrometry identify a class I Human Leukocyte Antigen A*0201-presented peptide originating from vaccinia virus. Journal of American Society for Mass Spectrometry. 2005;16:1812–7. doi: 10.1016/j.jasms.2005.07.015. [DOI] [PubMed] [Google Scholar]
- [56].Reche PA, Reinherz EL. Definition of MHC supertypes through clustering of MHC peptide-binding repertoires. Methods in Molecular Biology. 2007;409:163–73. doi: 10.1007/978-1-60327-118-9_11. [DOI] [PubMed] [Google Scholar]
- [57].Ou D, Mitchell LA, Tingle AJ. A new categorization of HLA DR alleles on a functional basis. Human Immunology. 1998;59:665–76. doi: 10.1016/s0198-8859(98)00067-6. [DOI] [PubMed] [Google Scholar]
- [58].Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. Journal of Immunology. 1998;160:3363–73. [PubMed] [Google Scholar]
- [59].Kaslow RA, Rivers C, Tang J, Bender TJ, Goepfert PA, El Habib R, et al. Polymorphisms in HLA class I genes associated with both favorable prognosis of human immunodeficiency virus (HIV) type 1 infection and positive cytotoxic T-lymphocyte responses to ALVAC-HIV recombinant canarypox vaccines. Journal of Virology. 2001;75:8681–9. doi: 10.1128/JVI.75.18.8681-8689.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].McDermott AB, Cohen SBA, Zuckerman JN, Madrigal JA. Human leukocyte antigens influence the immune response to a pre-S/S hepatitis B vaccine. Vaccine. 1999;17:330–9. doi: 10.1016/s0264-410x(98)00203-5. [DOI] [PubMed] [Google Scholar]
- [61].Kruskall MS, Alper CA, Awdeh Z, Yunis EJ, Marcus-Bagley D. The immune response to hepatitis B vaccine in humans: inheritance patterns in families. Journal of Experimental Medicine. 1992;175:495–502. doi: 10.1084/jem.175.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tebas P, Stein D. [Updated March 20, 2013. [Accessed 21.03.13]];Autologous T-cells genetically modified at the CCR5 gene by zinc finger nucleases SB-728 for HIV (Zinc Finger) http://clinicaltrials.gov/ct2/show/NCT00842634?term=zinc+finger+HIV&rank=1.
- [63].Maier DA, Brennan AL, Jiang S, Binder-Scholl GK, Lee G, Plesa G, et al. Efficient clinical scale gene modification via Zinc finger nuclease-targeted disruption of the HIV Co-receptor CCR5. Human Gene Therapy. 2013;24:245–58. doi: 10.1089/hum.2012.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends in Immunology. 2012;33:323–32. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–52. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].McGrath KE, Bushnell TP, Palis J. Multispectral imaging of hematopoietic cells: where flow meets morphology. Journal of Immunological Methods. 2008;336:91–7. doi: 10.1016/j.jim.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nature Immunology. 2009;10:116–25. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. Journal of Leukocyte Biology. 2007;81:1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- [69].Osterloh A, Breloer M. Heat shock proteins: linking danger and pathogen recognition. Medical Microbiology and Immunology. 2008;197:1–8. doi: 10.1007/s00430-007-0055-0. [DOI] [PubMed] [Google Scholar]
- [70].Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. International Reviews of Immunology. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- [71].Steinman RM, Hemmi H. Dendritic cells: translating innate to adaptive immunity. Current Topics in Microbiology and Immunology. 2006;311:17–58. doi: 10.1007/3-540-32636-7_2. [DOI] [PubMed] [Google Scholar]
- [72].Lindqvist M, Nookaew I, Brinkenberg I, Samuelson E, Thörn K, Nielsen J, et al. Unraveling molecular signatures of immunostimulatory adjuvants in the female genital tract through systems biology. PLoS ONE. 2011;6:e20448. doi: 10.1371/journal.pone.0020448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lindqvist M, Navabi N, Jansson M, Samuelson E, Sjöling A, Orndal C, et al. Local cytokine and inflammatory responses to candidate vaginal adjuvants in mice. Vaccine. 2009;28:270–8. doi: 10.1016/j.vaccine.2009.09.083. [DOI] [PubMed] [Google Scholar]
- [74].Brown KE, Freeman GJ, Wherry EJ, Sharpe AH. Role of PD-1 in regulating acute infections. Current Opinions in Immunology. 2010;22:397–401. doi: 10.1016/j.coi.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Dhiman N, Smith DI, Poland GA. Next-generation sequencing: a transformative tool for vaccinology. Expert Reviews of Vaccines. 2009;8:963–7. doi: 10.1586/erv.09.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Quakkelaar ED, Redeker A, Haddad EK, Harari A, McCaughey SM, Duhen T, et al. Improved innate and adaptive immunostimulation by genetically modified HIV-1 protein expressing NYVAC vectors. PLoS ONE. 2011;6:e16819. doi: 10.1371/journal.pone.0016819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ovsyannikova IG, Haralambieva IH, Vierkant RA, O’Byrne MM, Jacobson RM, Poland GA. Effects of Vitamin A and D receptor gene polymophisms/haplotypes on immune responses to measles vaccine. Pharmacogenetics and Genomics. 2012;22:20–31. doi: 10.1097/FPC.0b013e32834df186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ovsyannikova IG, Dhiman N, Haralambieva IH, Vierkant RA, O’Byrne MM, Jacobson RM, et al. Rubella vaccine-induced cellular immunity: evidence of associations with polymorphisms in the Toll-like, vitamin A and D receptors, and innate immune response genes. Human Genetics. 2010;127:207–21. doi: 10.1007/s00439-009-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Dusso AS, Brown AJ, Slatopolsky E, Vitamin D. American Journal of Physiology: Renal Physiology. 2005;289:F8–28. doi: 10.1152/ajprenal.00336.2004. [DOI] [PubMed] [Google Scholar]
- [80].Bikle D. Nonclassic actions of vitamin D. Journal of Clinical Endocrinology & Metabolism. 2009;94:26–34. doi: 10.1210/jc.2008-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Baeke F, Takiishi T, Korf H, Gysemans C, Mathieu C. Vitamin D: modulator of the immune system. Current Opinion in Pharmacology. 2010;10(4):482–96. doi: 10.1016/j.coph.2010.04.001. [DOI] [PubMed] [Google Scholar]
- [82].Chadha MK, Fakih M, Muindi J, Tian L, Mashtare T, Johnson CS, et al. Effect of 25-hydroxyvitamin D status on serological response to influenza vaccine in prostate cancer patients. Prostate. 2011;71:368–72. doi: 10.1002/pros.21250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Johnson KL, Ovsyannikova IG, Poland G, Muddiman DC. Identification of class II HLA-DRB1*03-bound measles virus peptides by 2D-liquid chromatography tandem mass spectrometry. Journal of Proteome Research. 2005;4:2243–9. doi: 10.1021/pr0501416. [DOI] [PubMed] [Google Scholar]
- [84].Wang C, Krishnakumar S, Wilhelmy J, Babrzadeh F, Stepanyan L, Su LF, et al. High-throughput, high-fidelity HLA genotyping with deep sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8676–81. doi: 10.1073/pnas.1206614109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Bolotin DA, Mamedov IZ, Britanova OV, Zvyagin IV, Shagin D, Ustyugova SV, et al. Next generation sequencing for TCR repertoire profiling: platform-specific features and correction algorithms. European Journal of Immunology. 2012;42:3073–83. doi: 10.1002/eji.201242517. [DOI] [PubMed] [Google Scholar]
- [86].Hombrink P, Hadrup SR, Bakker A, Kester MG, Falkenburg JH, von dem Borne PA, et al. High-throughput identification of potential minor histocompatibility antigens by MHC tetramer-based screening: feasibility and limitations. PLoS ONE. 2011;6:e22523. doi: 10.1371/journal.pone.0022523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wagar LE, Gentleman B, Pircher H, McElhaney JE, Watts TH. Influenza-specific T cells from older people are enriched in the late effector subset and their presence inversely correlates with vaccine response. PLoS ONE. 2011;6:e23698. doi: 10.1371/journal.pone.0023698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Gossger N, Snape MD, Yu LM, Finn A, Bona G, Esposito S, et al. Immunogenicity and tolerability of recombinant serogroup B meningococcal vaccine administered with or without routine infant vaccinations according to different immunization schedules: a randomized controlled trial. JAMA. 2012;307:573–82. doi: 10.1001/jama.2012.85. [DOI] [PubMed] [Google Scholar]
- [89].Golubchik T, Brueggemann AB, Street T, Gertz RE, Jr, Spencer CC, Ho T, et al. Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nature Genetics. 2012;44:352–5. doi: 10.1038/ng.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].De Groot AS, Levitz L, Ardito MT, Skowron G, Mayer KH, Buus S, et al. Further progress on defining highly conserved immunogenic epitopes for a global HIV vaccine: HLA-A3-restricted GAIA vaccine epitopes. Human Vaccine Immunotherapies. 2012;8:987–1000. doi: 10.4161/hv.20528. [DOI] [PubMed] [Google Scholar]
- [91].Haralambieva IH, Oberg AL, Dhiman N, Ovsyannikova IG, Kennedy RB, Grill DE, et al. High-dimensional gene expression profiling studies in high and low responders to primary smallpox vaccination. The Journal of Infectious Diseases. 2012;206:1512–20. doi: 10.1093/infdis/jis546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kennedy RB, Oberg AL, Ovsyannikova IG, Haralambieva IH, Grill DE, Poland GA. Transcriptomic profiles of high and low antibody responders to smallpox vaccine. Genes and Immunity. 2013 doi: 10.1038/gene.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Davies DH, Molina DM, Wrammert J, Miller J, Hirst S, Mu Y, et al. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics. 2007;7:1678–86. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- [94].National Research Council Committee on Metagenomics . The new science of metagenomics: revealing the secrets of our microbial planet. National Academies Press; Washington, DC: 2007. [PubMed] [Google Scholar]
- [95].Davenport MP, Hill AVS. Reverse immunogenetics: from HLA-disease associations to vaccine candidates. Molecular Medicine Today. 1996;2:38–45. doi: 10.1016/1357-4310(96)88757-0. [DOI] [PubMed] [Google Scholar]
- [96].Ovsyannikova IG, Johnson KL, Bergen HR, III, Poland GA. Mass spectrometry and peptide-based vaccine development. Clinical Pharmacology & Therapeutics. 2007;82:644–52. doi: 10.1038/sj.clpt.6100389. [DOI] [PubMed] [Google Scholar]
- [97].Alexander J, Bilsel P, del Guercio MF, Marinkovic-Petrovic A, Southwood S, Stewart S, et al. Identification of broad binding class I HLA supertype epitopes to provide universal coverage of influenza A virus. Human Immunology. 2010;71:468–74. doi: 10.1016/j.humimm.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Sette A, Sidney J. HLA supertypes and supermotifs: a functional perspective on HLA polymorphism. Current Opinions in Immunology. 1998;10:478–82. doi: 10.1016/s0952-7915(98)80124-6. [DOI] [PubMed] [Google Scholar]
- [99].Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201–12. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- [100].Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA. Human leukocyte antigen haplotypes in the genetic control of immune response to measles-mumps-rubella vaccine. The Journal of Infectious Diseases. 2006;193:655–63. doi: 10.1086/500144. [DOI] [PubMed] [Google Scholar]
- [101].Ovsyannikova IG, Jacobson RM, Dhiman N, Vierkant RA, Pankratz VS, Poland GA. Human leukocyte antigen and cytokine receptor gene polymorphisms associated with heterogeneous immune responses to mumps viral vaccine. Pediatrics. 2008;121:e1091–9. doi: 10.1542/peds.2007-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Poland GA, Ovsyannikova IG, Jacobson RM. Immunogenetics of seasonal influenza vaccine response. Vaccine. 2008;26S:D35–40. doi: 10.1016/j.vaccine.2008.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Haralambieva IH, Ovsyannikova IG, Dhiman N, Kennedy RB, O’Byrne M, Pankratz VS, et al. Common SNPs/Haplotypes in IL18R1 and IL18 genes are associated with variations in humoral immunity to smallpox vaccination in Caucasians and African-Americans. Journal of Infectious Diseases. 2011;204:433–41. doi: 10.1093/infdis/jir268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Milich DR, Leroux-Roels GG. Immunogenetics of the response to HBsAg vaccination. Autoimmunity Reviews. 2003;2:248–57. doi: 10.1016/s1568-9972(03)00031-4. [DOI] [PubMed] [Google Scholar]
- [105].Wang C, Tang J, Song W, Lobashevsky E, Wilson CM, Kaslow RA. HLA and cytokine gene polymorphisms are independently associated with responses to hepatitis B vaccination. Hepatology. 2004 Apr;39(4):978–88. doi: 10.1002/hep.20142. [DOI] [PubMed] [Google Scholar]
- [106].Yucesoy B, Sleijffers A, Kashon M, Garssen J, de Gruijl FR, Boland GJ, et al. IL-1beta gene polymorphisms influence hepatitis B vaccination. Vaccine. 2002;20:3193–6. doi: 10.1016/s0264-410x(02)00267-0. [DOI] [PubMed] [Google Scholar]
- [107].Chen J, Liang Z, Lu F, Fang X, Liu S, Zeng Y, et al. Toll-like receptors and cytokines/cytokine receptors polymorphisms associate with non-response to hepatitis B vaccine. Vaccine. 2011;29:706–11. doi: 10.1016/j.vaccine.2010.11.023. [DOI] [PubMed] [Google Scholar]
- [108].Tang J, Costello C, Keet IP, Rivers C, Leblanc S, Karita E, et al. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type 1 infection. AIDS Research and Human Retroviruses. 1999;15:317–24. doi: 10.1089/088922299311277. [DOI] [PubMed] [Google Scholar]
- [109].White SJ, Haralambieva IH, Ovsyannikova IG, Vierkant RA, O’Byrne MM, Poland GA. Replication of associations between cytokine and cytokine receptor single nucleotide polymorphisms and measles-specific adaptive immunophenotypic extremes. Human Immunology. 2012;73:636–40. doi: 10.1016/j.humimm.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Ovsyannikova IG, Haralambieva IH, Dhiman N, O’Byrne MM, Pankratz VS, Jacobson RM, et al. Polymorphisms in the vitamin A receptor and innate immunity genes influence the antibody response to rubella vaccination. Journal of Infectious Diseases. 2010;201:207–13. doi: 10.1086/649588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Ovsyannikova IG, Haralambieva IH, Vierkant RA, Pankratz VS, Poland GA. The role of polymorphisms in toll-like receptors and their associated intracellular signaling genes in measles vaccine immunity. Human Genetics. 2011;130:547–61. doi: 10.1007/s00439-011-0977-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–52. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- [113].Smith MW, Dean M, Carrington M, Winkler C, Huttley GA, Lomb DA, et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277:959–65. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- [114].Singh P, Kaur G, Sharma G, Mehra NK. Immunogenetic basis of HIV-1 infection, transmission and disease progression. Vaccine. 2008;26:2966–80. doi: 10.1016/j.vaccine.2008.01.012. [DOI] [PubMed] [Google Scholar]
- [115].Boniotto M, Crovella S, Pirulli D, Scarlatti G, Spanò A, Vatta L, et al. Polymorphisms in the MBL2 promoter correlated with risk of HIV-1 vertical transmission and AIDS progression. Genes and Immunity. 2000;1:346–8. doi: 10.1038/sj.gene.6363685. [DOI] [PubMed] [Google Scholar]
- [116].Speelmon EC, Livingston-Rosanoff D, Li SS, Vu Q, Bui J, Geraghty DE, et al. Genetic association of the antiviral restriction factor TRIM5alpha with human immunodeficiency virus type 1 infection. Journal of Virology. 2006;80:2463–71. doi: 10.1128/JVI.80.5.2463-2471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].van Manen D, Rits MA, Beugeling C, van Dort K, Schuitemaker H, Kootstra NA. The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathogens. 2008;4:e18. doi: 10.1371/journal.ppat.0040018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Pankratz VS, Vierkant RA, O’Byrne MM, Ovsyannikova IG, Poland GA. Associations between SNPs in candidate immune-relevant genes and rubella antibody levels: a multigenic assessment. BMC Immunology. 2010;11:48. doi: 10.1186/1471-2172-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Dhiman N, Cunningham JM, Jacobson RM, Jacobson RM, Ovsyannikova IG, Vierkant RA, et al. Variations in measles vaccine-specific humoral immunity by polymorphisms in SLAM and CD46 measles virus receptors. Journal of Allergy and Clinical Immunology. 2007;120:666–72. doi: 10.1016/j.jaci.2007.04.036. [DOI] [PubMed] [Google Scholar]
- [120].Ovsyannikova IG, Haralambieva IH, Kennedy RB, Pankratz VS, Vierkant RA, Jacobson RM, et al. Impact of cytokine and cytokine receptor gene polymorphisms on cellular immunity after smallpox vaccination. Gene. 2012;510:59–65. doi: 10.1016/j.gene.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Green MS, Shohat T, Lerman Y, Cohen D, Slepon R, Duvdevani P, et al. Sex differences in the humoral antibody response to live measles vaccine in young adults. International Journal of Epidemiology. 1994;23:1078–81. doi: 10.1093/ije/23.5.1078. [DOI] [PubMed] [Google Scholar]
- [122].Mitchell LA. Sex differences in antibody- and cell-mediated immune response to rubella re-immunisation. Journal of Medical Microbiology. 1999;48:1075–80. doi: 10.1099/00222615-48-12-1075. [DOI] [PubMed] [Google Scholar]
- [123].Klein SL, Jedlicka A, Pekosz A. The Xs and Y of immune responses to viral vaccines. Lancet Infectious Diseases. 2010;10:338–49. doi: 10.1016/S1473-3099(10)70049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Ovsyannikova IG, Jacobson RM, Vierkant RA, Jacobsen SJ, Pankratz VS, Poland GA. The contribution of HLA class I antigens in immune status following two doses of rubella vaccination. Human Immunology. 2004;65:1506–15. doi: 10.1016/j.humimm.2004.07.001. [DOI] [PubMed] [Google Scholar]
- [125].Kennedy RB, Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Ryan MA, et al. Gender effects on humoral immune responses to smallpox vaccine. Vaccine. 2009;27:3319–23. doi: 10.1016/j.vaccine.2009.01.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Beyer WE, Palache AM, Kerstens R, Masurel N. Gender differences in local and systemic reactions to inactivated influenza vaccine, established by a meta-analysis of fourteen independent studies. European Journal of Clinical Microbiology & Infectious Diseases. 1996;15:65–70. doi: 10.1007/BF01586187. [DOI] [PubMed] [Google Scholar]
- [127].Klein NP, Holmes TH, Sharp MA, Heineman TC, Schleiss MR, Bernstein DI, et al. Variability and gender differences in memory T cell immunity to varicellazoster virus in healthy adults. Vaccine. 2006;24:5913–8. doi: 10.1016/j.vaccine.2006.04.060. [DOI] [PubMed] [Google Scholar]
- [128].Kee SJ, Park YW, Cho YN, Jin HM, Kim MJ, Lee SJ, et al. Age- and gender-related differences in circulating natural killer T cells and their subset levels in healthy Korean adults. Human Immunology. 2012;73:1011–6. doi: 10.1016/j.humimm.2012.07.335. [DOI] [PubMed] [Google Scholar]
- [129].Centers for Disease Control and Prevention Ten Great Public Health Achievements-United States, 1900–1999. MMWR. 1999;48(12):241–64. [PubMed] [Google Scholar]
- [130].Institute of Medicine . Adverse effects of vaccines: evidence and causality. The National Academics Press; Washington, DC: 2011. [PubMed] [Google Scholar]
- [131].Salmon DA, Sotir MJ, Pan WK, Berg JL, Omer SB, Stokley S, et al. Parental vaccine refusal in Wisconsin: a case-control study. Wisconsin Medical Journal. 2009;108:17–23. [PMC free article] [PubMed] [Google Scholar]
- [132].Ahmed SS, Plotkin SA, Black S, Coffman RL. Assessing the safety of adjuvanted vaccines. Science Translational Medicine. 2011;3:93rv2. doi: 10.1126/scitranslmed.3002302. [DOI] [PubMed] [Google Scholar]
- [133].Jacobson RM, Adegbenro A, Pankratz VS, Poland GA. Adverse events and vaccination – the lack of power and predictability of infrequent events in pre-licensure study. Vaccine. 2001;19:2428–33. doi: 10.1016/s0264-410x(00)00467-9. [DOI] [PubMed] [Google Scholar]
- [134].Vaccines Kwok R. The real issues in vaccine safety. Nature. 2011;473:436–8. doi: 10.1038/473436a. [DOI] [PubMed] [Google Scholar]
- [135].Relman DA. Learning to appreciate our differences. Journal of Infectious Diseases. 2008;198:4–5. doi: 10.1086/588672. [DOI] [PubMed] [Google Scholar]
- [136].Poland GA. Vaccidents and adversomics. Vaccine. 2010;28:6549–50. doi: 10.1016/j.vaccine.2010.08.032. [DOI] [PubMed] [Google Scholar]
- [137].Berkovic SF, Harkin L, McMahon JM, Pelekanos JT, Zuberi SM, Wirrell EC, et al. De-novo mutations of the sodium channel gene SCN1A in alleged vaccine encephalopathy: a retrospective study. Lancet Neurology. 2006;5:488–92. doi: 10.1016/S1474-4422(06)70446-X. [DOI] [PubMed] [Google Scholar]
- [138].Ball R, Shadomy SV, Meyer A, Huber BT, Leffell MS, Zachary A, et al. HLA type and immune response to Borrelia burgdorferi outer surface protein a in people in whom arthritis developed after Lyme disease vaccination. Arthritis & Rheumatism. 2009;60:1179–86. doi: 10.1002/art.24418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Poland GA. Vaccines against Lyme disease: what happened and what lessons can we learn? Clinical Infectious Diseases. 2011;52(3):s253–8. doi: 10.1093/cid/ciq116. [DOI] [PubMed] [Google Scholar]
- [140].Reif DM, McKinney BA, Motsinger AA, Chanock SJ, Edwards KM, Rock MT, et al. Genetic basis for adverse events after smallpox vaccination. Journal of Infectious Diseases. 2008;198:16–22. doi: 10.1086/588670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Reif DM, Motsinger-Reif AA, McKinney BA, Rock MT, Crowe JE, Jr, Moore JH. Integrated analysis of genetic and proteomic data identifies biomarkers associated with adverse events following smallpox vaccination. Genes and Immunity. 2009;10:112–9. doi: 10.1038/gene.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Mosca F, Tritto E, Muzzi A, Monaci E, Bagnoli F, Iavarone C, et al. Molecular and cellular signatures of human vaccine adjuvants. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10501–6. doi: 10.1073/pnas.0804699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Mizukami T, Imai J, Hamaguchi I, Kawamura M, Momose H, Naito S, et al. Application of DNA microarray technology to influenza A/Vietnam/1194/2004 (H5N1) vaccine safety evaluation. Vaccine. 2008;26:2270–83. doi: 10.1016/j.vaccine.2008.02.031. [DOI] [PubMed] [Google Scholar]
- [144].McKinney BA, Reif DM, Ritchie MD, Moore JH. Machine learning for detecting gene–gene interactions: a review. Applied Bioinformatics. 2006;5:77–88. doi: 10.2165/00822942-200605020-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, Weeks DE. Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genetics. 2009;5:e1000337. doi: 10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. American Journal of Epidemiology. 2004;159:882–90. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- [147].McKinney BA, Reif DM, Rock MT, Edwards KM, Kingsmore SF, Moore JH, et al. Cytokine expression patterns associated with systemic adverse events following smallpox immunization. The Journal of Infectious Diseases. 2006;194:444–53. doi: 10.1086/505503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].McKinney BA, Crowe JE, Guo J, Tian D. Capturing the spectrum of interaction effects in genetic association studies by simulated evaporative cooling network analysis. PLoS Genetics. 2009;5:e1000432. doi: 10.1371/journal.pgen.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Davis NA, Crowe JE, Jr, Pajewski NM, McKinney BA. Surfing a genetic association interaction network to identify modulators of antibody response to smallpox vaccine. Genes and Immunity. 2010;11:630–6. doi: 10.1038/gene.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Pandey A, Davis NA, White BC, Pajewski NM, Savitz J, Drevets WC, et al. Epistasis network centrality analysis yields pathway replication across two GWAS cohorts for bipolar disorder. Translational Psychiatry. 2012;2:e154. doi: 10.1038/tp.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Liu YY, Slotine JJ, Barabasi AL. Controllability of complex networks. Nature. 2011;473:167–73. doi: 10.1038/nature10011. [DOI] [PubMed] [Google Scholar]
- [152].Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature Reviews Genetics. 2012;13:260–70. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Box GEPB, Draper NR. Robustness in the strategy of scientific model building. In: Launer R, Wilkinson G, editors. Robustness in statistics. Wisconsin-Univ-Madison Mathematics Research Center; Madison, WI: 1979. [Google Scholar]
- [154].Poland GA, Jacobson RM. The age-old struggle against the antivaccinationists. The New England Journal of Medicine. 2011;364(2):97–9. doi: 10.1056/NEJMp1010594. [DOI] [PubMed] [Google Scholar]
- [155].Poland GA, Jacobson RM. The clinician’s guide to the anti-vaccinationists’ galaxy. Human Immunology. 2012;73:859–66. doi: 10.1016/j.humimm.2012.03.014. [DOI] [PubMed] [Google Scholar]
- [156].Seib KL, Zhao X, Rappuoli R. Developing vaccines in the era of genomics: a decade of reverse vaccinology. Clinical Microbiology and Infection. 2012;18(Suppl 5):109–16. doi: 10.1111/j.1469-0691.2012.03939.x. [DOI] [PubMed] [Google Scholar]
- [157].Zak DE, Aderem A. Overcoming limitations in the systems vaccinology approach: a pathway for accelerated HIV vaccine development. Current Opinion in HIV and AIDS. 2012;7:58–63. doi: 10.1097/COH.0b013e32834ddd31. [DOI] [PubMed] [Google Scholar]
- [158].McGarvey PB, Huang H, Mazumder R, Zhang J, Chen Y, Zhang C, et al. Systems integration of biodefense omics data for analysis of pathogen-host interactions and identification of potential targets. PLoS ONE. 2009;4:e7162. doi: 10.1371/journal.pone.0007162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Ovsyannikova IG, Pankratz VS, Vierkant RA, Pajewski NM, Quinn CP, Kaslow RA, et al. Human leukocyte antigens and cellular immune responses to anthrax vaccine adsorbed. Infect Immun. 2013 doi: 10.1128/IAI.00269-13. [DOI] [PMC free article] [PubMed] [Google Scholar]