Abstract
The development of resistance to β-lactam antibiotics is a problem in the treatment of chronic Pseudomonas aeruginosa infection in the lungs of patients with cystic fibrosis. The main resistance mechanism is high-level expression of the chromosomally encoded AmpC β-lactamase of P. aeruginosa cells growing in biofilms. Several genes have been shown to influence the level of ampC expression, but little is known about the regulation of ampC expression in P. aeruginosa biofilms. To study the expression of ampC in P. aeruginosa biofilms, we constructed a reporter that consisted of the fusion of the ampC promoter to gfp(ASV) encoding an unstable version of the green fluorescent protein. In vitro biofilms of P. aeruginosa were exposed to the β-lactam antibiotics imipenem and ceftazidime. Sub-MICs of imipenem significantly induced the monitor system of the biofilm bacteria in the peripheries of the microcolonies, but the centers of the microcolonies remained uninduced. However, the centers of the microcolonies were physiologically active, as shown by experiments with another monitor construction consisting of an arabinose-inducible promoter fused to gfp(ASV). The whole biofilm was induced in the presence of increased imipenem concentrations. Ceftazidime induced the monitor system of the biofilm bacteria as well, but only bacteria in the peripheries of the microcolonies were induced in the presence of even very high concentrations. The experiments illustrate for the first time the dynamic and spatial distributions of β-lactamase induction in P. aeruginosa cells growing in biofilms. Thus, our experiments show that P. aeruginosa cells growing in biofilms constitute a heterogeneous population unit which may create different antibiotic-selective environments for the bacteria in the biofilm.
Aggressive antimicrobial treatment of chronic Pseudomonas aeruginosa infections in the lungs of cystic fibrosis (CF) patients improves their lung function and life expectancy (13, 37, 22, 34, 44). However, as a consequence of the intensive treatment, bacteria become increasingly resistant to antipseudomonal drugs (23, 24). The most important mechanism of resistance to β-lactam antibiotics in P. aeruginosa is the production of high levels of chromosomal AmpC β-lactamase (8, 16, 17). Several genes have been shown to influence the level of ampC expression, but little is known about the regulation of ampC expression in P. aeruginosa biofilms (3, 11, 30, 46). Another major reason for the persistence and survival of P. aeruginosa in the lungs of CF patients is the bacterial mode of growth, as P. aeruginosa establishes and grows in biofilm communities (21, 28, 29, 40). P. aeruginosa cells growing in biofilms are embedded in a self-produced polymeric matrix enclosed in microcolonies, and clinical P. aeruginosa isolates often display a mucoid phenotype caused by overproduction of the exopolysaccharide alginate (9, 43).
Mucoid and nonmucoid phenotypes of P. aeruginosa showing differences in their antimicrobial susceptibility patterns are regularly isolated simultaneously from CF patients with chronic lung infections. Ribotyping of phenotypic variants from the same patients (paired isolates) often reveals that these isolates are isogenic, but the majority of the paired isolates show significant differences in their antibiotic resistance patterns (7). Such observations could indicate that the P. aeruginosa cells growing in biofilms encounter different conditions in various compartments of the biofilm, with the selection of subpopulations with different resistance patterns. The spatial distribution of chromosomal β-lactamase gene expression in P. aeruginosa cells growing in biofilms has not been examined before. The aim of the present study was to study in vitro the responses of P. aeruginosa cells growing in biofilms to β-lactam antibiotics and the dynamics of β-lactamase expression.
MATERIALS AND METHODS
Strains and plasmids.
The P. aeruginosa strain PAO1 was used in this study. Plasmid pE59, which contains the translational fusion PampC -gfp(ASV) encoding an unstable variant of the green fluorescent protein (Gfp) (1) fused to the ampC promoter, was transformed into PAO1, resulting in PAO1-J32. All strains, plasmids, and primers used in this study are presented in Table 1.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant genotype or sequence | Reference or source |
|---|---|---|
| P. aeruginosa | ||
| PAO1 | Wild-type P. aeruginosa | 25 |
| PAO1-J32 | PAO1 with plasmid pE59 | This study |
| PAO1-BAD | PAO1 with plasmid pBADGfp | 18 |
| 19676A-3 | Variant of clinical isolate 19676A | 3 |
| E. coli JM105 | thi rpsL (Strr) endA sbcB15 sbcC hsdR4 (rk− mk−) Δ(lac-proAB) [F′ traD36 lacIq Δ(lacZ)M15 proA+B+] | 47 |
| Plasmids | ||
| pUCP22Not | Apr Gmr, Pseudomonas shuttle and cloning vector | 20 |
| pNB100 | Aps, Gmr, Pseudomonas shuttle and cloning vector, derivative of pUCP22Not | 3 |
| pE59 | Derivative of pNB100 including PampC -gfp(ASV) | This study |
| pJBA113 | Apr, pUC18Notl-PA1/04/03-RBSII-gfp(ASV)-T0-T1a | 1 |
| pQ27-1 | Derivative of pJBA113 carrying PampC-PCR fragment | This study |
| pBADGfp | Apr, Gmr, Pseudomonas shuttle vector with araC PBAD-gfp(ASV) | 18 |
| pRK600 | Cmrori ColE1 RK2-Mob+ RK2-Tra+; helper plasmid in triparental matings | 27 |
| Primers | ||
| ampC-primer ext-A2 | 5′-GTCGACCAGTGCCTTCAGGCGATC-3′ | |
| ampC-primer ext-B2 | 5′-GTGGTGGCGAACCGCAGTGTG-3′ | |
| ampC-primer ext-C2 | 5′-CCGCACAGGCAGGGGAATCTG-3′ | |
| ampC1-fwd | 5′-CCGGAATTCCGGGTGGACAGGCGCAGATCGATGAAGG-3′ | |
| ampC1-rev | 5′-ACATGCATGCATGTGATGCCGCACAGGCAGGGGAATCTG-3′ |
T0, transcriptional terminator from phage lambda; T1, transcriptional terminator from the rrnB operon of E. coli.
To construct the translational fusion, the ampC promoter (PampC) was amplified by PCR with chromosomal DNA of PAO1 as the template. The position of the transcriptional start site for ampC was verified by primer extension. The PCR fragment, which included the sequence 535 bp upstream of the translational start codon (ATG) of ampC and 33 bp downstream of the start codon, was amplified with primers ampC1-fwd and ampC1-rev, which contained EcoRI and SphI sites, respectively (Table 1). The PCR fragment was inserted into the corresponding sites of pJBA113, resulting in plasmid pQ27-1. Thus, pQ27-1 was a translational fusion of the ampC promoter in which the first 12 codons of ampC were maintained and fused in frame to gfp(ASV). This construct ensured that the native ribosome binding site of ampC and the spacing to the start codon of ampC were preserved and, therefore, that the activity of the reporter gene fusion closely reflected the expression of the ampC gene. The NotI fragment of pQ27-1 (Fig. 1) was inserted into the NotI site of pNB100, resulting in plasmid pE59. Sequencing of the final monitor plasmid construct was performed to verify the sequence. Sequencing was performed on an ABI Prism 377 DNA sequencer (Perkin-Elmer Applied Biosystems) with an ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit and was performed as outlined by the manufacturer.
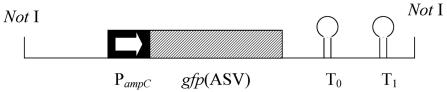
FIG. 1.
Schematic drawing of the translational fusion PampC -gfp(ASV). The NotI fragment indicated was cloned into shuttle vector pNB100, a modified plasmid pUCP22Not. The first 12 codons of ampC were maintained and fused to the gfp(ASV) open reading frame. The 5′ region of the PampC fragment included the whole intergenic region of ampC-ampR. The translational fusion of PampC -gfp(ASV) was followed by translational stop codons in all three reading frames, in addition to the two strong transcriptional terminators T0 (the transcriptional terminator from phage lambda) and T1 (the transcriptional terminator from the rrnB operon of E. coli (1).
Total DNA preparations, restriction enzyme analyses, ligation of DNA, and triparental matings were performed by standard procedures (38). DNA fragments were excised and isolated from agarose gels by using SpinX columns (Costar; Corning Costar Corporation, Corning, N.Y.) and plasmid DNA was isolated by using QiaPrep columns (Qiagen, Hilden, Germany). PCR amplification was performed with the Pwo proofreading DNA polymerase (Roche, Mannheim, Germany). Transformations were performed by electroporation, as described previously (12).
Primer extension.
RNA was isolated from an exponential-phase culture of P. aeruginosa 19676A-3 according to the protocol of the manufacturer of the RNeasy Mini kit (Qiagen). Reverse transcription was performed according to protocol 2 of the manufacturer of the DisplayThermo-RT kit (Display System Biotech, Copenhagen, Denmark). The protocol was slightly modified, as nonlabeled primers were used (Table 1) and 80% of the nonlabeled dATP was replaced by [α-32P]dATP (6,000 Ci/mmol; Amersham Pharmacia Biotech). Reverse transcription was performed at 75°C for 10 min, annealing was performed at 56°C for 40 min after addition of the DisplayThermo-RT Terminator mixture, and finally, the reaction was continued at 65°C for 15 min. Approximately 10 μg of total RNA was used for the reverse transcription reaction.
To determine the transcriptional start site of ampC, the sizes of the reverse transcriptional products were correlated to the DNA sequence upstream of ampC by Sanger sequencing using the same primers. Denaturing gel electrophoresis was done according to the protocol of the manufacturer of the DNA Sequencing kit (U.S. Biochemicals, Cleveland, Ohio).
Flow-cell biofilms.
Biofilms were studied in flow cells at 30°C (5). The procedure for the setup has previously been described by Christensen et al. (5). The flow-cell dimensions were 0.3 by 4.0 by 40 mm, and minimal medium was supplied at a constant flow rate of 3 ml/h with a peristaltic pump (Watson-Marlow, Falmouth, England). The minimal medium consisted of 1 mM MgCl2, 0.1 mM CaCl2, nonchelated trace elements (45), 2 g of (NH4)2SO4 per liter, 6 g of Na2HPO4 · 2H2O per liter, 3 g of KH2PO4 per liter, 3 g of NaCl per liter, and 0.01% glucose (0.55 mM). An exponentially growing culture was diluted to an optical density at 600 nm of 0.1, and inoculation of the flow cell was done with a syringe. The flow-cell channels were exposed to imipenem or ceftazidime at various concentrations. We used SYTO 62 stain (Molecular Probes) to visualize the biofilms. Image acquisition was performed with a scanning confocal laser microscope (model TCS4D; Leica Lasertechnik GmbH, Heidelberg, Germany). Image scanning for detection of Gfp was done at 488 nm (argon laser), while SYTO 62 was detected at 568 nm (He Ne laser). The sensitivities of the photomultipliers and the laser intensity were adjusted and thereafter kept constant throughout the experiment.
Antibiotic stability.
The chemical stability of imipenem was evaluated by measuring the concentration by a microbiological method previously described by Lautrop et al. (31) with Streptococcus sp. strain EB68 grown on Danish blood agar (State Serum Institute, Copenhagen, Denmark).
Susceptibility testing.
MICs were determined with the Etest system (AB Biodisk, Solna, Sweden) according to the instructions of the manufacturer.
RESULTS
Transcriptional start site for ampC.
Primer extension experiments showed that the single transcriptional start site of ampC is located 68 bp upstream of the ATG start codon of ampC. This constituted the basis for construction of the PampC-gfp(ASV) monitor. The exact transcriptional start site has not been determined before. Primer extension experiments were performed with three different primers, and the results obtained with all three primers showed the same transcriptional start site for ampC.
Antibiotic stability.
As we induced the P. aeruginosa isolates growing in biofilms for at least 4 to 5 h, it was of interest to know the degree of degradation of imipenem during the experiment, as the medium containing the antibiotic was not stored on ice. The half-life of imipenem was approximately 30 h at 30°C. Thus, the concentration of imipenem after 5 h was expected to be approximately 0.45 μg/ml and that after 12.3 h was expected to be 0.38 μg/ml, compared to the initial concentration of 0.5 μg/ml. The half-life of ceftazidime was 2.26 days at 30°C. Thus, with an initial concentration of ceftazidime of 100 μg/ml, the concentration was expected to be approximately 95.0 μg/ml after 4 h.
Inducibility of the monitor system.
The translational fusion of PampC to gfp(ASV), established in PAO1, i.e., PAO1-J32, expressed Gfp at an appropriate level for our experiments. Due to the instability of the Gfp encoded by the gfp(ASV) variant, no detectable level of Gfp was expressed in the absence of an inducer, whereas the level of expression in the presence of adequate levels of an inducer was significant. Two different β-lactam antibiotics, ceftazidime and imipenem, were used for induction in our experiments. With ceftazidime as the inducer, the monitor system showed significant expression of an exponentially growing culture of PAO1-J32 in Luria-Bertani (LB) medium when ceftazidime was used at 2.5 to 10 μg/ml. The monitor system did not reveal detectable levels of Gfp expression below the ceftazidime MIC (0.8 μg/ml). In the presence of even sub-MICs of imipenem in LB medium down to 0.05 μg/ml (imipenem MIC, 4.0 μg/ml), the monitor system expressed Gfp at ample levels that were clearly detectable after 1 h (Fig. 2). The bacteria on LB agar plates inoculated with PAO1-J32 and incubated at 37°C for 18 h showed clearly detectable levels of induction in the presence of ≥40 μg of ceftazidime/ml within 2 h after an Etest strip containing a ceftazidime gradient was placed on the agar plate. The lowest concentration of ceftazidime revealing a barely detectable Gfp signal was 2.5 μg/ml. When the bacteria were exposed to an Etest strip with imipenem, the monitor system clearly showed Gfp expression in the presence of concentrations ≥0.10 μg/ml. Imipenem and ceftazidime at concentrations ≥0.10 and 10.0 μg/ml, respectively, allowed detectable Gfp expression by P. aeruginosa PAO1-J32 cells growing in biofilms in the flow cells.
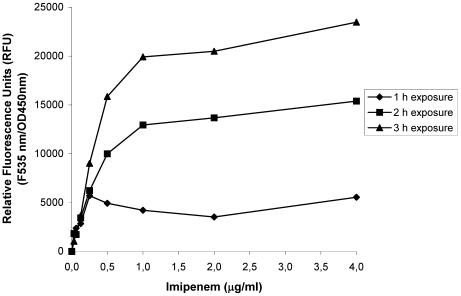
FIG. 2.
Expression of PampC-gfp(ASV) in planktonic cell cultures of P. aeruginosa exposed to imipenem. The relative fluorescence units in planktonic PAO1 cells growing in response to imipenem were measured after 1, 2, and 3 h. The cells were induced during exponential growth. A relative fluorescence unit of 0 corresponded to completely repressed Gfp expression. Fully induced single cells showed very distinct, visible Gfp expression. OD450nm, optical density at 450 nm.
Dynamics of Gfp expression in PAO1-J32 growing in biofilms.
Pilot experiments were performed with PAO1-J32 to ensure the preservation of plasmid pE59 in the bacteria growing in the biofilms. On day 6 after inoculation, it was found that imipenem (0.5 μg/ml) induced the distinct expression of Gfp by PAO1-J32 cells growing in biofilms.
PAO1-J32 cells growing in biofilms in the flow cells showed significant induction of Gfp expression in the presence of 0.5 μg of imipenem/ml, i.e., one-eighth the MIC (measured with planktonic cells). A decrease in the temperature markedly influenced the level of Gfp expression, and expression was undetectable at, for example, 24°C when the bacteria were exposed to the same concentrations of antibiotics.
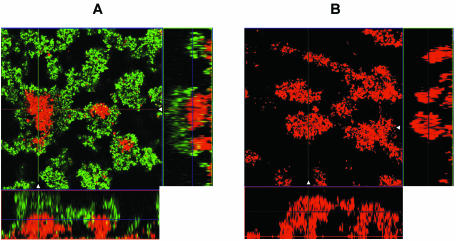
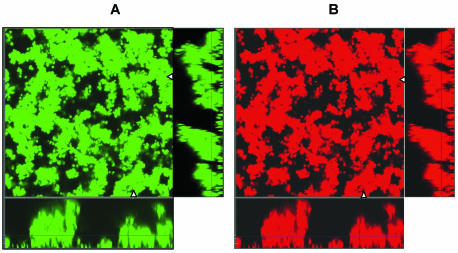
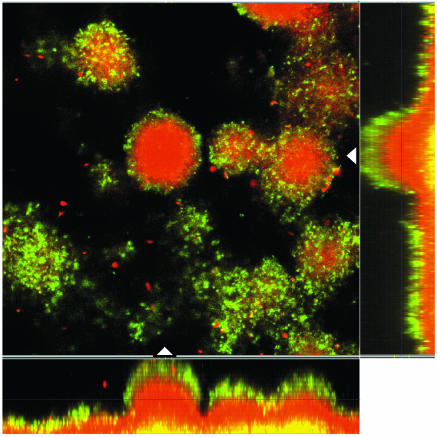
As imipenem is known to be a very strong inducer, it was not surprising that imipenem induced the expression of ampC much more potently, even at sub-MICs, than ceftazidime (26, 39). Figure 3 shows a 6-day-old biofilm of PAO1-J32 after 4 h of induction with imipenem at 0.5 μg/ml (Fig. 3A), as well as a noninduced biofilm showing no Gfp expression (Fig. 3B). It was remarkable that the biofilm was not induced completely, as the basal parts and the centers of the microcolonies remained uninduced (Fig. 3). To examine whether no induction in the center or basal part of the biofilm could be due to a lack of physiological activity, PAO1-BAD was established as a biofilm, and on day 6 the biofilm was induced with 0.5% l-arabinose for 4 h. This experiment verified the physiological activity of the bacteria in the biofilm, as the level of expression of Gfp was pronounced throughout each microcolony (data not shown). As we increased the concentration of imipenem to 10 μg/ml or even higher, PAO1-J32 showed full induction in the entire biofilm, thus verifying that a sufficient level of activity was present in the centers of the microcolonies to respond to the concentration of imipenem used (Fig. 4).
FIG. 3.
Epifluorescence and scanning confocal laser micrographs of PAO1-J32 biofilms on day 6 showing the activity of the ampC promoter in response to imipenem exposure (0.5 μg/ml for 4 h) (A) and the activity of the ampC promoter in a noninduced biofilm (B). Each micrograph consists of a horizontal section and two vertical sections through the biofilms collected at the positions indicated by the white triangles in the horizontal section. The position of the horizontal image is indicated by the crossing lines in the vertical sections. The biofilms were stained red with SYTO 62, demonstrating the presence of biomass in the flow cell. The expression of Gfp indicates the expression of the AmpC β-lactamase in the biofilm.
FIG. 4.
Epifluorescence and scanning confocal laser photomicrographs of PAO1-J32 biofilms on day 6 showing the activity of the ampC promoter in response to imipenem exposure (10 μg/ml for 4 h) (A) and the same biofilm stained red with SYTO 62 to demonstrate the total biomass of the biofilm (B). Each micrograph consists of a horizontal section and two vertical sections through the biofilms collected at the positions indicated by the white triangles in the horizontal section. The position of the horizontal image is indicated by the crossing lines in the vertical sections. The expression of Gfp indicates the expressions of the AmpC β-lactamase in the biofilm.
The level of induction of PampC-gfp(ASV) caused by the administration of ceftazidime to PAO1-J32 cells growing in biofilms was significantly less efficient than that caused by imipenem when we compared the concentrations efficient for induction and the MICs of imipenem and ceftazidime. For ceftazidime to induce a significant Gfp signal, the biofilm had to be exposed to drug concentrations of at least 40 to 100 μg/ml, which corresponds to 50 to 125 times the MIC, respectively. The administration of 100 μg of ceftazidime/ml (which corresponds to >100 times the MIC) for 4 h caused the induction of only the superficial layers of a 6-day-old biofilm (Fig. 5).
FIG. 5.
Epifluorescence and scanning confocal laser photomicrographs of PAO1-J32 biofilms on day 6 showing the activity of the ampC promoter in response to ceftazidime exposure (100 μg/ml for 4 h). The micrograph consists of a horizontal section and two vertical sections through the biofilms collected at the positions indicated by the white triangles in the horizontal section. The position of the horizontal image is indicated by the crossing lines in the vertical sections. The biofilms were stained red with SYTO 62, demonstrating the presence of biomass in the flow cell. The expression of Gfp indicates the expression of the AmpC β-lactamase in the biofilm.
Dynamics of ampC expression.
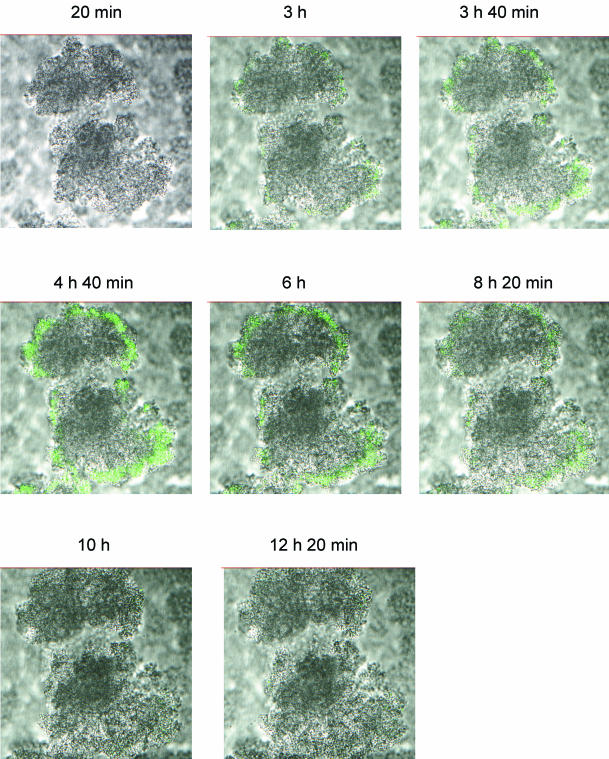
To study the activity of ampC expression over time, PAO1-J32 was established in the flow cells and induced from days 6 to 7; i.e., the biofilm was continuously exposed to imipenem overnight. As shown in Fig. 6, the induction of the ampC promoter was optimal after 4 to 5 h, whereupon the induction faded out and was nonexistent after approximately 10 h. The initial concentration of imipenem was 0.5 μg/ml, but the concentration gradually decreased to 0.38 μg/ml at the end of the experiment due to the instability of imipenem.
FIG. 6.
Epifluorescence and scanning confocal laser photomicrographs showing a time series of images of a horizontal section of a 6-day-old PAO1-J32 biofilm during induction with imipenem. Imipenem was administered from time zero (0 h), and the image of one microcolony was scanned every 10 min for 14 h. The result was increasing levels of AmpC β-lactamase expression in the first 5 h, as demonstrated by Gfp expression, followed by decreased levels of expression that faded out completely after 10 h.
DISCUSSION
We have shown for the first time the heterogeneity of β-lactamase induction in P. aeruginosa cells growing in biofilms. The mechanism of induction of β-lactamase production in communities of P. aeruginosa cells growing in biofilms has previously been measured in in vitro biofilm experiments (with a modified Robbins device) after exposure to imipenem or piperacillin (15). It is widely recognized that, due to the biofilm mode of growth, the bacteria established in biofilms are much more resistant to antimicrobial chemotherapy than their isogenic counterparts cultured as planktonic cells (2, 9, 14, 43). Accordingly, the concentrations of imipenem and ceftazidime that induced the PampC-gfp(ASV) reporter in our experiments were highest in the biofilms. Three hypotheses for the high-level resistance of biofilms to antibiotics have been proposed: (i) the biofilm may act as a barrier to antibiotics, slowing the diffusion of antibiotics through the biofilm matrix; (ii) the biofilm may cause an altered microenvironment in the individual cells, such as nutrient and oxygen depletion and waste product accumulation; and (iii) the bacteria may differentiate into a protected phenotype, e.g., the mucoid phenotype (43).
The biofilm as a diffusion barrier for antibiotics (the first hypothesis) has been subject to extensive research (10, 14, 19, 41, 42). For positively charged antibiotics, such as aminoglycosides (e.g., tobramycin), the biofilm affects the ability of the antibiotic to reach the cells deep within the biofilm matrix, as the exopolysaccharides of the biofilm are negatively charged and are known to function as an ion-exchange resin capable of binding to a large number of antibiotics attempting to reach the cells embedded in the biofilm (2, 14, 43). For relatively uncharged agents, such as the β-lactam antibiotics, such binding to the exopolysaccharide matrix is unlikely to occur (14, 33). Nichols et al. (33) predicted from mathematical models that the biofilm would not afford protection against diffusion of β-lactam antibiotics into the bacteria embedded in the biofilm as long as the level of chromosomal expression of β-lactamase is low. However, P. aeruginosa cells growing in biofilms can secrete β-lactamase to the surrounding environment, with β-lactamase thereby accumulating within the extracellular polymer matrix, or β-lactamase may be released to the polymer matrix due to lysis of the cells in the biofilm (6, 14, 17). Accordingly, Nichols et al. (33) reported that P. aeruginosa cells growing in biofilms and expressing high levels of chromosomal β-lactamase would be exposed to reduced concentrations of β-lactam antibiotics in the biofilm.
To correlate the previous results to the present observations, there may be multiple explanations for the differential levels of ampC expression in PAO1-J32 cells growing in biofilms exposed to 0.5 μg of imipenem/ml. That concentration of imipenem was found to be close to the lower limit of the concentration that causes detectable levels of Gfp expression by our monitor system. Therefore, even relatively small reductions in the inducing concentrations of imipenem present in the biofilm may cause the monitor system to cease to detect detectable levels of Gfp. The detection level of our monitor system was affected by the short half-life of gfp(ASV). In Pseudomonas putida it was measured to be approximately 3 h, and in Escherichia coli it was measured to be approximately 2 h (1). Although PAO1 does not produce high levels of chromosomal β-lactamase, the accumulation of β-lactamase in the biofilm due to secretion or lysis of cells may trap and hydrolyze the β-lactam antibiotics and retard their penetration into the biofilm, thereby causing the concentration in the centers of the microcolonies to be lower and at levels below the detection limit of our monitor system.
Alginate has not been shown to be a key constituent in the resistance of P. aeruginosa cells growing in biofilms to β-lactam antibiotics (14, 33). However, the accumulation of β-lactamase may be a major mechanism of resistance to β-lactam antibiotics in P. aeruginosa cells growing in biofilms (43). Therefore, as we used nonmucoid strain PAO1-J32 for our experiments, we have highlighted the importance of β-lactamase in biofilm resistance to β-lactam antibiotics.
The resistance of biofilms may be affected by altered microenvironments in the different compartments of the biofilm, as mentioned in the second hypothesis above. We registered physiological activity in the whole biofilm by means of the arabinose-induced Gfp monitoring system. However, there may still be a reduced level of activity in the center and basal compartments of the biofilm, as such differences may not be registered by the arabinose-induced Gfp monitoring system. Reduced oxygen tension, reduced accessibility to nutrients, and the accumulation of waste products may reduce the growth rate of the bacteria within the biofilm (32, 43). A retarded growth rate affects the bactericidal actions of the β-lactam antibiotics because transpeptidase inhibition, which induces cellular injury, is directly related to the growth rate (14, 43). We could therefore expect both β-lactamase accumulation and the differences in the growth rates of P. aeruginosa cells growing in biofilms to affect the differential gene expression illustrated in this study. The experiments illustrating the dynamic expression of β-lactamase over time could be due to the reduced levels of imipenem in the biofilm caused by the accumulation of the AmpC β-lactamase, which hydrolyzes imipenem at the surface of the biofilm, although the reduced level of imipenem over time due to chemical instability may also have caused a reduced level of induction of the AmpC β-lactamase.
The antibiotic pressure exerted in the microbial environment selects for antibiotic resistance (35, 36). The different antibiotic concentrations in the different compartments of the biofilm may result in different selective pressures on the bacteria (4). Differences in the growth rates of the bacteria in the different compartments of the biofilm, differences in stress levels (e.g., due to nutrient or oxygen depletion), and differences in the selective effects of antibiotics may promote bacterial diversity. In the lungs of CF patients, the selection process is also affected by host factors, such as the effects of the immune system. A complex and heterogeneous environmental landscape provides the basis for different selective pressures that allow organisms with differences in fitness due to newly acquired characteristics to survive and proliferate (4). Thus, the results presented here emphasize the differential effects of two widely used β-lactam antibiotics on communities of P. aeruginosa cells growing in biofilms.
Acknowledgments
We thank Jette Teglhus Møller for excellent technical assistance.
This work was supported by grants from H:S (Hovedstadens Sygehusfaellesskab, Copenhagen, Denmark), The Danish Medical Research Council, The Danish Technical Research Council, and The Villum Kann Rasmussen Foundation (Denmark).
REFERENCES
- 1.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjørn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anwar, H., J. L. Strap, and J. W. Costerton. 1992. Establishment of aging biofilms: possible mechanism of bacterial resistance to antimicrobial therapy. Antimicrob. Agents Chemother. 36:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagge, N., O. Ciofu, M. Hentzer, J. I. A. Campbell, M. Givskov, and N. Høiby. 2002. Constitutive high expression of chromosomal beta-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob. Agents Chemother. 46:3406-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baquero, F., M.-C. Negri, M.-I. Morosini, and J. Blázquez. 1998. Antibiotic-selective environments. Clin. Infect. Dis. 27:S5-S11. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools in physiological studies of microbial biofilms. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 6.Ciofu, O., T. J. Beveridge, J. Kadurugamuwa, J. Walther-Rasmussen, and N. Høiby. 2000. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 45:9-13. [DOI] [PubMed] [Google Scholar]
- 7.Ciofu, O., V. Fussing, N. Bagge, C. Koch, and N. Høiby. 2001. Characterization of paired mucoid/non-mucoid Pseudomonas aeruginosa isolates from Danish cystic fibrosis patients: antibiotic resistance, beta-lactamase activity and RiboPrinting. J. Antimicrob. Chemother. 48:391-396. [DOI] [PubMed] [Google Scholar]
- 8.Ciofu, O., B. Giwercman, S. S. Pedersen, and N. Høiby. 1994. Development of antibiotic resistance in Pseudomonas aeruginosa during two decades of antipseudomonal treatment at the Danish CF Center. APMIS 102:674-680. [PubMed] [Google Scholar]
- 9.Costerton, J. W., Z. Lewandowski, D. DeBeer, D. Caldwell, D. Korber, and G. James. 1994. Biofilms, the customized microniche. J. Bacteriol. 176:2137-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dibdin, G. H., S. J. Assinder, W. W. Nichols, and P. A. Lambert. 1996. Mathematical model of beta-lactam penetration into a biofilm of Pseudomonas aeruginosa while undergoing simultaneous inactivation by released beta-lactamase. J. Antimicrob. Chemother. 38:757-769. [DOI] [PubMed] [Google Scholar]
- 11.Dietz, H., D. Pfeifle, and B. Wiedemann. 1997. The signal molecule for beta-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob. Agents Chemother. 41:2113-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diver, J. M., L. E. Bryan, and P. A. Sokol. 1990. Transformation of Pseudomonas aeruginosa by electroporation. Anal. Biochem. 189:75-79. [DOI] [PubMed] [Google Scholar]
- 13.Frederiksen, B., S. Lanng, C. Koch, and N. Høiby. 1996. Improved survival in the Danish center-treated cystic fibrosis patients: results of aggressive treatment. Pediatr. Pulmonol. 21:153-158. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert, P., and M. R. W. Brown. 1998. Biofilms and beta-lactam activity. J. Antimicrob. Chemother. 41:571-578. [DOI] [PubMed] [Google Scholar]
- 15.Giwercman, B., E. T. Jensen, N. Høiby, A. Kharazmi, and J. W. Costerton. 1991. Induction of beta-lactamase production in Pseudomonas aeruginosa biofilm. Antimicrob. Agents Chemother. 35:1008-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giwercman, B., P. A. Lambert, V. T. Rosdahl, G. H. Shand, and N. Høiby. 1990. Rapid emergence of resistance in Pseudomonas aeruginosa in cystic fibrosis due to in vivo selection of stable derepressed beta-lactamase producing strains. J. Antimicrob. Chemother. 26:247-259. [DOI] [PubMed] [Google Scholar]
- 17.Giwercman, B., C. Meyer, P. A. Lambert, C. Reinert, and N. Høiby. 1992. High-level beta-lactamase activity in sputum samples from cystic fibrosis patients during antipseudomonal treatment. Antimicrob. Agents Chemother. 36:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hentzer, M., K. Riedel, T. B. Rasmussen, A. Heydorn, J. B. Andersen, M. R. Parsek, S. A. Rice, L. Eberl, S. Molin, N. Høiby, S. Kjelleberg, and M. Givskov. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87-102. [DOI] [PubMed] [Google Scholar]
- 19.Hentzer, M., G. M. Teitzel, G. J. Baltzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Høiby, N. 1977. Pseudomonas aeruginosa infection in cystic fibrosis. Diagnostic and prognostic significance of Pseudomonas aeruginosa precipitins determined by means of crossed immunoelectrophoresis. A survey. Acta Pathol. Microbiol. Scand. Suppl. 262:1-96. [PubMed] [Google Scholar]
- 22.Høiby, N. 2000. Prospects for the prevention and control of pseudomonal infection in children with cystic fibrosis. Paediatr. Drugs 2:451-463. [DOI] [PubMed] [Google Scholar]
- 23.Høiby, N. 2002. New antimicrobials in the management of cystic fibrosis. J. Antimicrob. Chemother. 49:235-238. [DOI] [PubMed] [Google Scholar]
- 24.Høiby, N., H. K. Johansen, C. Moser, Z. Song, O. Ciofu, and A. Kharazmi. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microb. Infect. 3:23-35. [DOI] [PubMed] [Google Scholar]
- 25.Holloway, B. W. 1955. Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13:572-581. [DOI] [PubMed] [Google Scholar]
- 26.Jones, R. N. 1998. Important and emerging beta-lactamase-mediated resistances in hospital-based pathogens: the AmpC enzymes. Diagn. Microbiol. Infect. Dis. 31:461-466. [DOI] [PubMed] [Google Scholar]
- 27.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter in the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293-301. [DOI] [PubMed] [Google Scholar]
- 28.Koch, C., and N. Høiby. 1993. Pathogenesis of cystic fibrosis. Lancet 341:1065-1069. [DOI] [PubMed] [Google Scholar]
- 29.Lam, J., R. Chan, K. Lam, and J. W. Costerton. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28:546-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langaee, T. Y., L. Gagnon, and A. Huletsky. 2000. Inactivation of the ampD gene in Pseudomonas aeruginosa leads to moderate-basal-level and hyperinducible AmpC beta-lactamase expression. Antimicrob. Agents Chemother. 44:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lautrop, H., N. Høiby, A. Bremmelgaard, and B. Korsager. 1979. Bakteriologiske undersøgelsesmetoder. FADL, Copenhagen, Denmark.
- 32.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichols, W. W., M. J. Evans, M. P. E. Slack, and H. L. Walmsley. 1989. The penetration of antibiotics into aggregates of mucoid and non-mucoid Pseudomonas aeruginosa. J. Gen. Microbiol. 135:1291-1303. [DOI] [PubMed] [Google Scholar]
- 34.Pedersen, S. S., T. Jensen, N. Høiby, C. Koch, and E. W. Flensborg. 1987. Management of Pseudomonas aeruginosa lung infection in Danish cystic fibrosis patients. Acta Paediatr. Scand. 76:955-961. [DOI] [PubMed] [Google Scholar]
- 35.Pedersen, S. S., C. Koch, N. Høiby, and K. Rosendal. 1986. An epidemic spread of multiresistant Pseudomonas aeruginosa in a cystic fibrosis centre. J. Antimicrob. Chemother. 17:505-516. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen, S. S., T. Pressler, T. Jensen, V. T. Rosdahl, M. W. Bentzon, N. Høiby, and C. Koch. 1987. Combined imipenem/cilastatin and tobramycin therapy of multiresistant Pseudomonas aeruginosa in cystic fibrosis. J. Antimicrob. Chemother. 19:101-107. [DOI] [PubMed] [Google Scholar]
- 37.Regelmann, W. E., G. R. Elliott, W. J. Warwick, and C. C. Clawson. 1990. Reduction of sputum Pseudomonas aeruginosa density by antibiotics improves lung function in cystic fibrosis more than do bronchodilators and chest physiotherapy alone. Am. Rev. Respir. Dis. 141:914-921. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Sanders, C. C. 1987. Chromosomal cephalosporinases responsible for multiple resistance to newer beta-lactam antibiotics. Annu. Rev. Microbiol. 41:573-593. [DOI] [PubMed] [Google Scholar]
- 40.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 41.Stewart, P. S. 1994. Biofilm accumulation model that predicts antibiotic resistance of Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 38:1052-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart, P. S. 1996. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob. Agents Chemother. 40:2517-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 44.Szaff, M., N. Høiby, and E. W. Flensborg. 1983. Frequent antibiotic therapy improves survival of cystic fibrosis patients with chronic Pseudomonas aeruginosa infection. Acta Paediatr. Scand. 72:651-657. [DOI] [PubMed] [Google Scholar]
- 45.Widdel, F., and F. Bak. 1992. The procaryotes. Springer-Verlag, New York, N.Y.
- 46.Wiedemann, B., H. Dietz, and D. Pfeifle. 1998. Induction of beta-lactamase in Enterobacter cloacae. Clin. Infect. Dis. 27:S42-S47. [DOI] [PubMed] [Google Scholar]
- 47.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]