Abstract
Recent studies using an internal transcribed spacer (ITS)-based real-time polymerase chain reaction (PCR) for the detection of Schistosoma DNA in urine samples has shown high sensitivity and specificity when performed on controls and known microscopy-positive samples. In this study, using 730 urine samples collected from children in five primary schools from different communities in the Greater Accra region of Ghana, specific detection of Schistosoma DNA showed excellent sensitivity of 100% and 85.2% in urines with > 50 eggs/10 mL urine and ≤ 50 eggs/10 mL of urine, respectively. Additionally, Schistosoma-specific DNA was amplified in 102 of 673 samples in which Schistosoma eggs could not be detected with microscopy. Taking microscopy and/or PCR-positive samples as true positives, the negative predictive value calculated was 94.6–100% for each school sampled as compared with 54.3–95.7% using microscopy. This ITS-based real-time PCR proves to be a powerful tool in epidemiological surveys of schistosomiasis providing more precise and sensitive results than microscopy.
Introduction
Schistosomiasis is one of the most important human helminth infections in terms of morbidity and mortality.1 The disease is endemic in many developing countries affecting largely children, farmers, and women who depend primarily on water bodies, which may harbor intermediate host snails. Millions of people are infected worldwide, with schistosomes of various species leading to the loss of 1.53 million disability-adjusted life years, with the majority of morbidity (85%) and mortality occurring in sub-Saharan Africa.2
Diagnosis plays a vital role in the identification of schistosomiasis infections generating results that influence decisions on individual and community treatment, estimations of prognosis and assessment of morbidity, evaluation of chemotherapy, and other control measures.3 However, there are limitations with some of the diagnostic techniques. Although tedious and time-consuming, most epidemiological assessments of the burden of schistosomiasis have relied on microscopy, providing a relatively easy and cheap tool for detecting and estimating the intensity of schistosome eggs in fecal and urine samples in many schistosomiasis-endemic countries.4 However, inadequate sensitivity as a result of great fluctuations of egg output in urine or stool are well known, and the difficulties in meeting the multiple sampling requirements for classical parasitological diagnosis often lead to suboptimal results.5
Antibody-based assays are sensitive but cannot distinguish history of exposure from active infection; their cross-reaction with other helminths has been shown, and these assays are not easily applicable under field conditions.2 Antigen-based assays, such as circulating cathodic antigen detection in urine, have proven to be a valuable, field applicable method for detecting Schistosoma mansoni infections, unfortunately, the test has shown to be less sensitive for infections with Schistosoma haematobium.6,7
Polymerase chain reaction (PCR)-based methods have shown high sensitivity and specificity for the detection of parasitic DNA and have been used for the detection of a broad range of parasites.5,8,9 Until now their use in human epidemiological surveys has been limited with more studies exploring the potential of schistosome-specific PCR focusing more on the snail vector rather than its definitive host.10,11 Recent developments in the simplification of DNA isolation procedures and PCR technology, especially real-time PCR, have made DNA amplification a worthwhile alternative for microscopy-based diagnostic methods.8,12–14
In this study, the prevalence and intensity of urinary schistosomiasis in Ghanian school children was assessed using real-time PCR.
Methods
Study area and sample collection.
The study was conducted in five primary schools (children 5–19 years of age, median age 11 years) from different communities in Greater Accra region of Ghana. The schools selected included Anyamam Presby Primary (AN), Goi Presby Primary (GP), Pantang D/A Primary (PA), Nii Okai Basic (NB), and DeYoungsters (DY) representing schools with a schistosomiasis prevalence based on microscopy ranging from ≈0% to 18%. Three of these schools (i.e., AN, GP, and PA) are located in rural areas where as the other two can be found in urban areas of Accra. There had been no mass treatment of schistosomiasis in these areas in the 3 years before sample collection. Urine samples (N = 730) were collected in 50 mL containers between 10.00 and 14.00 when schistosome eggs excretion is known to be highest15 and left at ambient temperature between the collection sites and the Noguchi Memorial Institute for Medical Research (NMIMR) laboratory. A 2 mL aliquot of each sample was kept frozen at −80°C for PCR. The study was performed within a larger epidemiological study on allergy and parasitic infections (“GLOFAL GHANA”) approved by the Institutional Review Board of the NMIMR, Accra, Ghana. A written informed consent was received from parents or guardian of each child. Mass drug administration took place in the same year after the samples were collected.
Microscopy.
For each subject, the urine sample was examined for ova of S. haematobium by the urine-filtration method, which allowed the intensity of infection to be expressed as the numbers of S. haematobium eggs/10 mL urine with counts of 1–49 and > 50 eggs/10 mL indicative of low and high intensity of infections, respectively.
DNA extraction.
The DNA was isolated from all urine samples using QIAamp DNA mini kit (QIAGEN, Hilden, Germany) protocol after a 200 μL subsample of each urine sample was heated for 10 min at 100°C before treatment with sodium dodecyl sulphate and proteinase K for 2 h at 55°C. As an internal control, phocin herpes virus 1 (PhHV-1) was added to the lysis buffer (at 1,000 plaque-forming units/mL).
Real-time PCR.
Schistosoma genus-specific primers amplifying a 77-bp fragment of the internal transcribed spacer-2 (ITS2) sub unit described by Obeng and others,15 consisting of Ssp48F (5′-GGT CTA GAT GAC TTG ATY GAG ATG CT-3′) and Ssp124R (5′-TCC CGA GCG YGT ATA ATG TCA TTA-3′) detected by the probe, Ssp78T [FAM-5′-TGG GTT GTG CTC GAG TCG TGGC-3′-Black Hole Quencher (Biolegio, Nijmegen, The Netherlands)], were selected for amplification and detection of Schistosoma DNA.
Amplification of each DNA sample was performed in a 25 μL reaction mixture containing PCR buffer (HotstarTaq mastermix; QIAGEN), 5 mM MgCl2, 12.5 pmol of each Schistosoma genus-specific primer, 15 pmol of each PhHV-1-specific primer, 2.5 pmol each of the Schistosoma genus-specific and PhHV-1-specific double-labeled probes, and 5 μL of the DNA sample. The thermocycler used was set to give 15 minutes at 95°C, followed by 50 cycles, each of 15 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C. Amplification, amplicon detection and the related data analysis were performed with the CFX96 real-time PCR detection system (Bio-Rad, Veenendaal, The Netherlands) using the CFX Manager version 1.6.514 (Bio-Rad). The PCR output from this system consists of a cycle-threshold (Ct) value, representing the amplification cycle in which the level of fluorescent signal exceeded the background fluorescence, indicating the parasite-specific DNA load in the urine sample tested.
The DNA isolation and setup of the PCR reactions were performed using a custom-made Hamilton robot platform.
Data analysis.
Statistical analysis was done using the version 16.0 of the SPSS software package (SPSS, Chicago, IL). Real-time PCR results were stratified into high (Ct < 30), moderate (30 ≤ Ct ≥ 35), low (Ct > 35) DNA load and negative (no amplification detected in 50 cycles). The positive and negative predictive values of microscopy and real-time PCR were calculated with true positives as microscopy and/or PCR positive and microscopy and PCR negatives assumed to be true negatives.
Results
Microscopy.
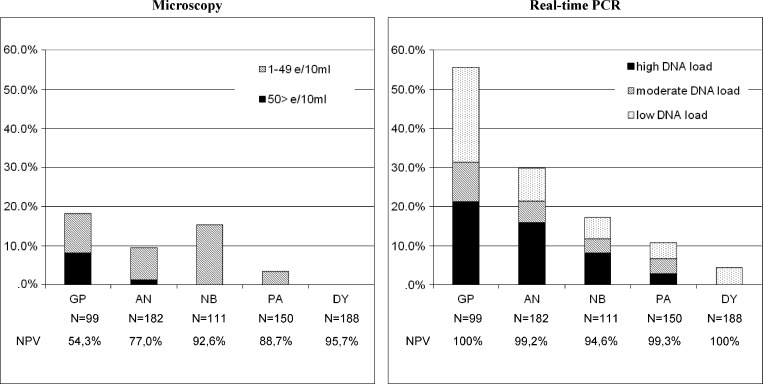
Microscopy revealed an overall prevalence of 7.8% (57 of 730) for S. haematobium eggs in the total samples used in this study. Prevalence and intensity within schools is represented graphically in Figure 1 showing GP with 18.2% infection followed by NB with 15.3% infection.
Figure 1.
Prevalence and intensity of infection as measured with detection of Schistosoma eggs using microscopy and Schistosoma DNA using real-time PCR in urine samples from five schools in Ghana (N = 730) with the negative predicted values as calculated with a positive result of microscopy and/or real-time PCR as true positive.
Real-time PCR.
Amplification of the internal control was detected at the expected Ct value of ≈30 in all samples and Schistosoma-specific DNA was detected in 152 of 730 samples. Prevalence and intensity of infection as reflected by low, moderate, and high DNA load in the schools are shown in Figure 1. The GP showed the highest prevalence of 55.6%, with the majority of subjects showing a high to moderate DNA load with Ct-values < 35. The AN followed next with 29.7% positives with 21.3% containing high to moderate DNA load. In DY, which was revealed as negative by microscopy, Schistosoma-specific DNA amplification was detected in 8 of 188 samples (4.3%) all showing a low DNA load.
Comparing microscopy detection and PCR amplification.
The results of microscopy and PCR are summarized in Table 1. Cycle threshold values were significantly higher for urine samples in which S. haematobium eggs were not found by microscopy (N = 102, median 35.3, range 20.5–39.4) than for those from urines in which S. haematobium eggs were seen by microscopy (N = 50, median Ct 24.7, range 16.1–39.1) (Mann–Whitney test; P < 0.0001).
Table 1.
Results of microscopy with the numbers of S. haematobium eggs/10 mL urine divided into egg counts of 1–49 and > 50 eggs/10 mL indicative of low and high intensity of infections respectively and real-time PCR results divided into high (Ct < 30), moderate (30 ≤ Ct ≥ 35), and low (Ct > 35) DNA loads and negative (no amplification detected in 50 cycles)*
| PCR Ct Values | ||||||
|---|---|---|---|---|---|---|
| Negative | Ct > 35 | 30 > Ct < 35 | Ct < 30 | Total | Median Ct-value in PCR (range) | |
| Microscopy egg class | ||||||
| Negative | 571 (84.6%) | 56 (8.3%) | 21 (3.1%) | 25 (3.7%) | 673 (100%) | 35.3 (20.5–39.4) |
| 1–49 eggs/10 mL | 7 (14.8%) | 3 (6.3%) | 9 (19.1%) | 28 (59.6%) | 47 (100%) | 25.8 (17.8–39.1) |
| ≥ 50 eggs/10 mL | 0 (0%) | 0 (0%) | 0 (0%) | 10 (100%) | 10 (100%) | 23.1 (16.1–28.1) |
| Total | 578 (79.2%) | 59 (8.1%) | 30 (4.1%) | 63 (8.6%) | 730 (100%) | 31.9 (16.1–39.4) |
PCR = polymerase chain reaction; Ct = cycle-threshold.
Discussion
In a previous study, using well-defined DNA and urine samples as controls, and 74 urine samples known to contain S. haematobium eggs, the Schistosoma real-time PCR achieved 100% specificity and sensitivities of 100% and 87% in urines with > 50 eggs/10 mL urine and ≤ 50 eggs/10 mL of urine, respectively.15 In this study using 730 urine samples collected from children in five primary schools from different communities in Greater Accra region of Ghana, the real-time PCR for the specific detection of Schistosoma DNA again showed excellent sensitivities of 100% and 85.2%, in urines with > 50 eggs/10 mL urine and ≤ 50 eggs/10 mL of urine, respectively (Table 1). Additionally, Schistosoma DNA was detected with PCR using 200 μL of urine in samples in which eggs were not detected by microscopy using 10 mL of urine. Although it is not clear why Schistosoma DNA could not be amplified in seven of the microscopy positive samples (1–12 eggs/10 mL) apparently Schistosoma DNA, sometimes with high DNA loads, can be present even in the absence of eggs in the urine sample. A higher sensitivity of the PCR in samples with low egg counts might be achievable by using a larger volume of the urine sample by applying filter-based DNA isolation methods16,17 In the schools included in this study, the higher sensitivity of the real-time PCR results in much higher negative predictive values (> 94.6%) as compared with microscopy (54.3–95.7%).
The apparent poor sensitivity of the microscopy in this study might have resulted in an overestimate of the PCR sensitivity. Analysis using a (Bayesian) latent class model for analysis without a “true gold standard,” performing PCR and microscopy on multiple samples from the same individuals, and standardization of converting DNA-loads into egg counts is subject for further study. Further evaluation is needed in particular in low-prevalence communities. It would also be interesting to assess the application of the Schistosoma genus PCR testing urine samples in areas where only S. mansoni is endemic.
Facilities for real-time PCR are available in an increasing number of research centers in low- to middle-income countries. Sample collection and transportation to a central laboratory for PCR results into simplification of the complex organization of labor-intensive field studies performing microscopy on the spot, especially when thinking about integrated control in which urine samples can be collected together with stool samples for molecular screening for intestinal schistosomiasis and soil-transmitted helminths.
In conclusion, the results of this study indicate that real-time PCR can serve as a powerful alternative in determining the prevalence and intensity of Schistosoma infections using urine samples as template. This can lead to a more accurate assessment of the prevalence and intensity of the disease and evaluating the effectiveness of treatment programs in endemic areas.
ACKNOWLEDGMENTS
We wish to acknowledge the community leaders, head teachers, and the parents and children involved in this study as well as the employees and students from the Noguchi Memorial Institute for Medical Research and the Leiden University Medical Centre who were involved in the field collection of samples.
Footnotes
Financial support: European Research Council FP6 (TRANCHI, GLOFAL, and COINFECT), FP7 (SCHISTOVAC), and the Prof. Dr. P.C. Flu Foundation.
Authors' addresses: Yvonne A. Aryeetey, Irene A. Larbi, Kwaku Ahmed, Abena S. Amoah, Benedicta B. Obeng, and Daniel A. Boakye, Parasitology Department, Noguchi Memorial Institute for Medical Research, University of Ghana, Accra, Ghana, E-mails: yaryeetey@noguchi.mimcom.org, i.larbi@lancaster.ac.uk, kahmed@noguchi.mimcom.org, a.s.Amoah@lumc.nl, baffoah@yahoo.com, and DBoakye@noguchi.mimcom.org. Samuel Essien-Baidoo, Department of Laboratory Sciences, University of Cape Coast, Ghana, E-mail: sbessien@yahoo.com. Lisette van Lieshout and Maria Yazdanbakhsh, Department of Parasitology, Leiden University Medical Center, Leiden, The Netherlands, E-mails: lvanlieshout@lumc.nl and m.yazdanbakhsh@lumc.nl. Jaco J. Verweij, Laboratory for Medical Microbiology and Immunology, St. Elisabeth Hospital, Tilburg, The Netherlands, E-mail: j.verweij@elisabeth.nl.
References
- 1.McManus DP, Loukas A. Current status of vaccine for schistosomiasis. Clin Microbiol Rev. 2008;21:225–242. doi: 10.1128/CMR.00046-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gryseels B, Polman K, Clerinx J, Kestens L. Human schistosomiasis. Lancet. 2006;368:1106–1118. doi: 10.1016/S0140-6736(06)69440-3. [DOI] [PubMed] [Google Scholar]
- 3.Feldmeier H, Poggensee G. Diagnostic techniques in schistosomiasis control: a review. Acta Tropica. 1993;52:205–220. doi: 10.1016/0001-706x(93)90009-z. [DOI] [PubMed] [Google Scholar]
- 4.Lengeler C, Utzinger J, Tanner M. Questionnaires for rapid screening of schistosorniasis in sub-Saharan Africa. Bull World Health Organ. 2002;80:235–242. [PMC free article] [PubMed] [Google Scholar]
- 5.ten Hove RJ, Verweij JJ, Vereecken K, Polman K, Dieye L, van Lieshout L. Multiplex real-time PCR for the detection and quantification of Schistosoma mansoni and S. haematobium infection in stool samples collected in northern Senegal. Trans R Soc Trop Med Hyg. 2008;102:179–185. doi: 10.1016/j.trstmh.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 6.van Dam GJ, Wichers JH, Ferreira TM, Ghati D, van Amerongen A, Deelder AM. Diagnosis of schistosomiasis by reagent strip test for detection of circulating cathodic antigen. J Clin Microbiol. 2004;42:5458–5461. doi: 10.1128/JCM.42.12.5458-5461.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stothard JR, Kabatereine N, Tukahebwa EM, Kazibwe F, Rollinson D, Mathieson W, Webster JP, Fenwick A. Use of circulating cathodic antigen (CCA) dipsticks for detection of intestinal and urinary schistosomiasis. Acta Trop. 2006;97:219–228. doi: 10.1016/j.actatropica.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Verweij JJ, Brienen EA, Ziem J, Yelifari L, Polderman AM, van Lieshout L. Simultaneous detection and quantification of Ancylostoma duodenale, Necator americanus, and Oesophagostomum bifurcum in fecal samples using multiplex real-time PCR. Am J Trop Med Hyg. 2007a;77:685–690. [PubMed] [Google Scholar]
- 9.Verweij JJ, Ten Hove R, Brienen EA, van Lieshout L. Multiplex detection of Enterocytozoon bieneusi and Encephalitozoon spp. in fecal samples using real-time PCR. Diagn Microbiol Infect Dis. 2007b;57:163–167. doi: 10.1016/j.diagmicrobio.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Barber KE, Mkoji GM, Loker ES. PCR-RFLP analysis of The ITS2 region to identify Schistosoma haematobium and S. bovis from Kenya. Am J Trop Med Hyg. 2000;62:434–440. doi: 10.4269/ajtmh.2000.62.434. [DOI] [PubMed] [Google Scholar]
- 11.Abbasi I, King CH, Sturrock RF, Kariuki C, Muchiri E, Hamburger J. Differentiation of Schistosoma haematobium from related schistosomes by PCR amplifying an inter-repeat sequence. Am J Trop Med Hyg. 2007;76:950–955. [PMC free article] [PubMed] [Google Scholar]
- 12.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JDC, Wengenack NL, Rosenblatt JE, Cockerill FR, Smith TF. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clinical Microbiology. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein D. Quantification using real-time PCR technology: applications and limitations. Trends Mol Med. 2002;8:257–260. doi: 10.1016/s1471-4914(02)02355-9. [DOI] [PubMed] [Google Scholar]
- 14.Verweij JJ, Blange RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, van Lieshout L, Polderman AM. Simultaneous detection of Entamoeba histolytica, Giardia lamblia and Cryptosporidium parvum in fecal samples using multiplex real-time PCR. J Clin Microbiol. 2004;3:1220–1223. doi: 10.1128/JCM.42.3.1220-1223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obeng BB, Aryeetey YA, de Dood CJ, Amoah AS, Larbi IA, Deelder AM, Yazdanbakhsh M, Hartgers FC, Boakye DA, Verweij JJ, van Dam GJ, van Lieshout L. Application of a circulating-cathodic-antigen (CCA) strip test and real-time PCR, in comparison with microscopy, for the detection of Schistosoma haematobium in urine samples from Ghana. Ann Trop Med Parasitol. 2008;102:625–633. doi: 10.1179/136485908X337490. [DOI] [PubMed] [Google Scholar]
- 16.Ibironke OA, Phillips AE, Garba A, Lamine SM, Shiff C. Diagnosis of Schistosoma haematobium by detection of specific DNA fragments from filtered urine samples. Am J Trop Med Hyg. 2011;84:998–1001. doi: 10.4269/ajtmh.2011.10-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibironke O, Koukounari A, Asaolu S, Moustaki I, Shiff C. Validation of a new test for Schistosoma haematobium based on detection of Dra1 DNA fragments in urine: evaluation through latent class analysis. PLoS Negl Trop Dis. 2012;6:1464. doi: 10.1371/journal.pntd.0001464. [DOI] [PMC free article] [PubMed] [Google Scholar]