Abstract
Due to the importance of proteases in regulating cellular processes, the development of protease inhibitors has garnered great attention. Peptide-based aldehydes are a class of compounds that exhibit inhibitory activities against various proteases and proteasomes in the context of anti-proliferative treatments for cancer and other diseases. More than a dozen peptide-based natural products containing aldehydes have been discovered such as chymostatin, leupeptin, and fellutamide; however, the biosynthetic origin of the aldehyde functionality has yet to be elucidated. Herein we describe the discovery of a new group of lipopeptide aldehydes, the flavopeptins, and the corresponding biosynthetic pathway arising from an orphan gene cluster in Streptomyces sp. NRRL-F6652, a close relative of Streptomyces flavogriseus ATCC 33331. This research was initiated using a proteomics approach that screens for expressed enzymes involved in secondary metabolism in microorganisms. Flavopeptins are synthesized through a nonribosomal peptide synthetase containing a terminal NAD(P)H dependent reductase domain likely for the reductive release of the peptide with a C-terminal aldehyde. Solid phase peptide synthesis of several flavopeptin species and derivatives enabled structural verification and subsequent screening of biological activity. Flavopeptins exhibited submicromolar inhibition activities against cysteine proteases such as papain and calpain as well as the human 20S proteasome. They also showed anti-proliferative activities against multiple myeloma and lymphoma cell lines.
INTRODUCTION
Many naturally occurring peptide aldehydes such as chymostatin, leupeptin and fellutamide possess inhibition properties towards various proteases and are widely used in biomedical research to prevent unwanted proteolysis. Notably, some peptide aldehydes that inhibit cathepsins, calpains or proteasomes—associated with human diseases including cancer, osteoporosis, and Alzheimer’s disease— are under extensive study for their use as tool and lead compounds for drug development.1–4 By retaining the aldehyde warhead and varying the peptide backbone residues, chemists have designed and synthesized numerous peptide aldehydes and their boronate analogues that inhibit a spectrum of proteases with varying potencies.5,6 Despite this obvious biological and clinical importance, there has not been a biosynthetic pathway elucidated for a peptide aldehyde.
With the burgeoning of genome sequencing on microbial species, it has become increasingly apparent that only a fraction of their biosynthetic capacities for natural products have been reflected by direct observation of metabolic profiles.7 Many gene clusters for secondary metabolite biosynthesis have no corresponding products identified; these are known as orphan gene clusters. A number of systems biology approaches including genomics, transcriptomics and metabolomics have been employed in attempt to decipher these orphan gene clusters for the corresponding natural products.8–11 A method known as Proteomic Investigation of Secondary Metabolism (PrISM), allows for the screening of the expressed enzymes related to natural product biosynthesis.12 In a PrISM workflow, microbes are cultured under various conditions and their proteomes are analyzed by mass spectrometry. Expressed proteins for secondary metabolite biosynthesis are detected, which enables the biosynthetic gene cluster and associated secondary metabolite to be discovered simultaneously. Two types of secondary metabolites, nonribosomal peptides and polyketides, are of special interest to the PrISM method. The biosynthetic enzymes for nonribosomal peptides and polyketides, nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs), are often enormous in size (> 200 kDa) which permits their targeted selection by size-based separation, such as SDS-PAGE.13 Using the PrISM approach, several new natural products and their corresponding gene clusters have been discovered from bacteria possessing either previously sequenced or non-sequenced genomes.14–16
In this study, we employed the PrISM approach to screen expressed NRPSs and PKSs from Streptomyces species, and identified an orphan NRPS gene cluster from Streptomyces sp. NRRL F-6652. Through bioinformatics analysis of the gene cluster and metabolomics analysis using mass spectrometry, a new class of peptide aldehyde natural products named flavopeptins was discovered and identified as the products of the orphan gene cluster. The biosynthesis of flavopeptins involves an NRPS protein with a relatively rare C-terminal reductase domain catalyzing the formation of the peptide aldehyde. Like other peptide aldehydes, flavopeptins showed inhibitory activities against cysteine proteases including papain and human calpain as well as the human 20S proteasome, with low micromolar to nanomolar inhibition potencies. Flavopeptins also exhibited anti-proliferative activity against multiple myeloma and lymphoma cell lines.
EXPERIMENTAL
Materials
Streptomyces sp. NRRL F-6652 and B- 16185 were acquired from U.S. Department of Agriculture, Agricultural Research Service and Streptomyces flavogriseus ATCC 33331 was obtained from ATCC. Sequencing grade trypsin was from Promega, WI. Stable-isotope labeled amino acids were from Cambridge Isotope Laboratories, MA or CDN isotopes, Canada. Proteases were from Sigma-Aldrich, MO. Human 20S proteasome and its substrates were from Boston Biochem, MA.
Detection of NRPS Expression by Proteomics
Streptomyces sp. NRRL F-6652 was cultured in ATCC 172 medium (10 g/L glucose, 20 g/L soluble starch, 5 g/L yeast extract, 5 g/L N–Z amine type A, 1 g/L CaCO3) at 30 °C for three days before transferring to different screening media. Cells from 24 h, 48 h and 72 h growth were harvested and lysed by sonication. The proteome lysate was separated on a one-dimensional SDS-PAGE, and the proteome region above 150 kDa was excised for in-gel trypsin digestion. The resulting peptides were separated on a self-packed nano-capillary column (5 µm Jupiter C18, 100 mm × 75 µm) using a nanoLC–Ultra system (Eksigent, CA). LC solvents were 5% acetonitrile in water with 0.1% formic acid (mobile phase A) and 95% acetonitrile with 0.1% formic acid (mobile phase B). The LC gradient was set as follows: 0 min, 0% B; 55 min, 45% B; 63 min, 80% B; 67 min, 0% B with re-equilibration of 0% B until 90 min. Peptides were eluted into a nanoelectrospray ionization (nESI) source on a 7 Tesla LTQ–FTICR mass spectrometer (Thermo Scientific). Peptides were first detected in the FT ICR with resolving power setting of 100,000 (at 400 m/z). Intact peptide data were collected in the 400–1800 m/z range, and MS/MS spectra were acquired using data dependent mode where the top three most abundant peaks from the FTMS full scan were selected for collision induced dissociation (CID) fragmentation followed by mass analysis of the fragment ions in the linear ion trap. The LC–MS data were searched against all bacteria in the non-redundant protein database NCBInr using Mascot.
Natural Product Detection and Structure Elucidation by Mass Spectrometry
NRRL F-6652 was grown in ATCC 172 medium for three days at 30 °C before transferring to mineral based media (MSB)17 supplemented with 10 mM sodium succinate and 0.05% casamino acids. Stable isotope-labeled amino acids were added to the medium to a final concentration of 1 mM when necessary. Culture supernatants from 24 h, 48 h, 72 h and 96 h of growth were analyzed by LC–MS. The samples were separated by a Phenomenex Luna C18 column (250 × 2 mm, 5 µm) eluting with acetonitrile/water with 0.1% formic acid from 0 to 100%. An Exactive Orbitrap (Thermo Scientific) was used for mass detection from m/z 200 to 1500. The LC–MS data were processed using Thermo SIEVE software for chromatographic alignment, feature identification and quantitation.
Isolation of Flavopeptins
Streptomyces sp. NRRL F-6652 was grown in 30 × 500 mL ATCC 172 medium in 2 L baffled flasks at 30 °C for seven days. The culture supernatant was separated from cells by centrifugation and extracted with one volume of ethyl acetate twice. The organic extract (~2.2 g) was dried down, re-dissolved in 50% methanol in H2O, and underwent solvent partitioning between 50% methanol/hexane (1:9) followed by 50% methanol/dichloromethane (3:7). The dichloromethane portion (~1 g) was separated by solid phase extraction (SPE) using a Strata C18-E column (Phenomenex, CA). SPE fractions eluted with 70–100% acetonitrile (~60 mg) were further separated on a Phenomenex Luna C18 column (150 × 10 mm, 5 µm) using 1:1 acetonitrile/ isopropanol–H2O with 0.1% formic acid to yield ~1.5 mg of flavopeptins. Purified flavopeptins were analyzed by LC−MS/MS on a 7 Tesla LTQ–FTICR MS instrument.
Total Synthesis of Flavopeptins and Analogues
Flavopeptins were synthesized with H-Phe-H NovaSyn® TG resin (EMD Millipore, MA) using standard Fmoc coupling procedures. The resin (0.1 mmol) was allowed to swell in DMF (2 mL) for 30 minutes before use. Fmoc protecting groups were removed after treatment with 5 mL of DBU/piperidine/DMF (2:2:96) for 15 min. The amino acid chain was extended using a 3-fold excess of Fmoc-amino acids that had been previously activated with PyBOP® (0.3 mmol), HOBt (0.05 mmol), and DIPEA (0.6 mmol) in DMF. After assembly of the target sequence, the resin was washed with DMF, i-PrOH, THF, MeOH, and then dried overnight. Side-chain protecting groups were removed by three treatments with anhydrous TFA (0.5 mL) in dry DCM (2.5 mL) for 10 min. The product was cleaved from the resin by three treatments with a mixture of AcOH/H2O/DCM/MeOH (10:5:64:21) for 30 min to afford the crude product. The crude residue was purified on a Luna C18 column (150 × 21.2 mm, 5 µm) using acetonitrile– H2O with 0.1% formic acid. Compounds 1 (6.8 mg, 8%), 2 (10.4 mg, 12%), and 3 (6.3 mg, 9%) were obtained through this method.
The carboxylic acid analogue 4 was synthesized with a Wang Resin (EMD Millipore, MA). Fmoc-Phe-OH (0.1 mmol) was loaded on to the resin with HOBt (0.1 mmol), DCC (0.1 mmol), and DMAP (0.01 mmol). Standard Fmoc coupling procedures were used as described above. A mixture of TFA (1.0 mL) and DCM (1.0 mL) with TIPS (0.10 mL) was added to the resin for cleavage. The resin was allowed to shake for 3 h after which the crude product was collected from the resin and dried, washed with cold ether, and dried again. HPLC purification was performed using the same condition as above (19.4 mg, 22%).1H and13C NMR spectra of the synthetic peptides 1 through 4 are shown in Figures S12–S13 and S16–S21. 2D NMR (TOCSY, HSQC and HMBC) spectra of 2 are shown in Figures S14–S15.
Labeling the Aldehyde Group with PFBHA
Flavopeptins isolated from NRRL F-6652 (~1.5 µg) were treated with 5 µL of pentafluorobenzylhydroxylamine hydrochloride (PFBHA; 10 mg/mL) in methanol at RT for 3 h, and analyzed by LC–MS.
Determination of Amino Acid Stereochemistry
Flavopeptins (~100 µg) were dissolved in constant-boiling 6 N HCl (Thermo Scientific) and heated at 110 °C for 24 h for hydrolysis. After lyopholization, the hydrolyzed product was dissolved in 50 µL water, 50 µL of 1 M NaHCO3 and 100 µL of 1% Marfey’s reagent (1-fluoro-2–4- dinitrophenyl-5-l-alanine amide, FDAA, Thermo Scientific) in acetone and heated at 37 °C for 1 h. The reaction was quenched with 50 µL of 1 M HCl and diluted in 1:1 H2O/acetonitrile for LC–MS analysis. The derivatized amino acid mixture was separated on a Luna C18 column (150 × 2 mm, 5 µm) using 25–60% acetonitrile/H2O with 0.1% formic acid over 40 min, and detected by UV absorbance at 340 nm and mass spectrometry using an Ex-active Orbitrap.
Biological Assays
To test the protease inhibitory action of the flavopeptins, the EnzChek protease assay kit (Invitrogen, NY) was used. Proteases were dissolved in their corresponding digestion buffer. For trypsin, chymotrypsin, thermolysin and subtilisin, 10 mM Tris-HCl, pH 7.8; for pepsin, 10 mM HCl, pH 2.0; for papain, 10 mM MES with 5 mM L-cysteine, pH 6.2; for elastase, 10 mM Tris-HCl, pH 8.8; for calpain-I, 10 mM HEPES, 10 mM DTT, 1 mM EDTA, 2 mM CaCl2, pH 7.2. The proteases (1 µg/mL) were incubated with flavopeptins or DMSO for 30 min at RT before an equal amount of substrate BODIPYFL casein (10 µg/mL) was added. After 1 h incubation at RT, the fluorescence at 485/528 nm was read on a Synergy H4 fluorospectrometer (BioTek, VT). To measure the IC50 value against papain, the fluorescence over reaction time was recorded and the initial rates were plotted against log [inhibitor] and fitted to the variable slope model using GraphPad Prism software. For calpain IC50 calculation, flavopeptins were incubated with human calpain-I (20 nM) for 30 min before the substrate Suc-LLVY-AMC (40 µM) was added and the fluorescence at 380/460 nm was continuously recorded for 30 min.
For the proteasome inhibition assay, human 20S proteasome (1 nM) in assay buffer (10 mM Tris-HCl, 0.035% SDS, pH 7.4) was incubated with flavopeptins for 15 min at RT before substrates (Suc-LLVY-AMC for chymotrypsin- like and Z-LLE-AMC for caspase-like activities, 5 µM) were added and the fluorescence was recorded using a 380/460 nm filter. The initial rates were used for IC50 calculation. Cell viability assays were performed using the CellTiter-Glo luminescent cell viability assay kit (Promega, WI). Cancer cell lines (100 µL) were plated in 96-well plates to a density of 5,000 cells per well. Flavopeptins dissolved in DMSO were added after 24 h followed by incubation at 37 °C for another 72 h. CellTiter-Glo reagent (100 µL) was added to each well and the luminescence was recorded.
RESULTS AND DISCUSSION
Detection of NRPS Expression by Proteomics
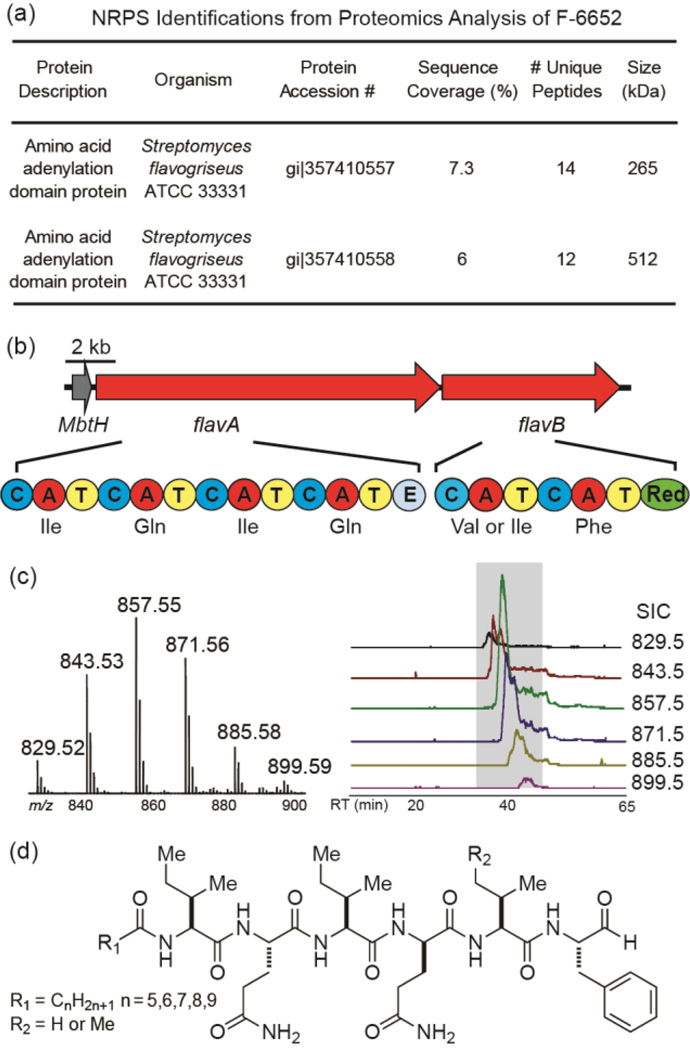
In previous reports, we described a proteomics based approach called PrISM to screen for expressed NRPS or PKS proteins and applied this approach to discover new natural products from Bacillus and Actinobacterium species.12,14–16 In one such screen, the proteomics data for Streptomyces sp. NRRL F-6652 grown in MSB medium for 48 h identified peptides originating from two NRPS proteins from Streptomyces flavogriseus ATCC 33331 (Figure 1a).
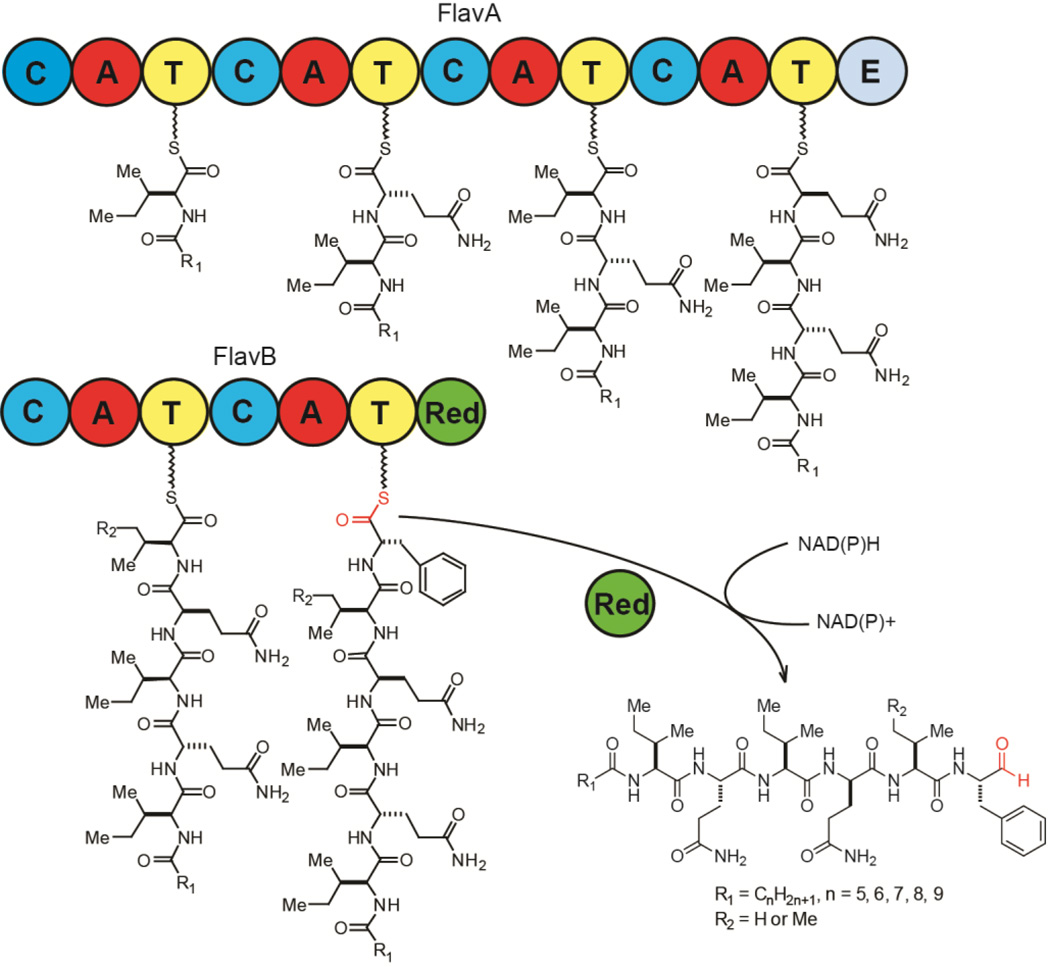
Figure 1.
The sequential proteomic, genomic and metabolomic analysis that led to the discovery of flavopeptins. (a) Two NRPS proteins were identified through LC–MS/MS analysis of the proteome of F-6652. (b) The genomic context of the two identified NRPS genes (flavA and flavB), their domain organizations and the predicted amino acid substrates. A, adenylation; T, thiolation; C, condensation; E, epimerization; Red, reductase. (c) Detection of flavopeptins from culture supernatant of F-6652: left, mass spectrum created by compiling spectra of all flavopeptin species detected; right, Selected ion chromatograms (SICs) for six flavopeptin species (m/z 829.5–899.5). (d) The chemical structure of flavopeptins.
The two NRPS proteins, weighing 265 kDa and 512 kDa respectively, are located next to each other in the genome and very likely within a gene cluster. However, despite an extensive literature search, we were not able to find any natural products associated with this orphan gene cluster.
Oligonucleotide primers were designed using the reverse translation of the detected peptide sequences and used to verify the genomic DNA sequence from Streptomyces sp. NRRL F-6652 is identical or similar to ATCC 33331 by PCR. All the PCR amplified DNA sequences showed 100% sequence identity to the corresponding genomic regions of S. flavogriseus ATCC 33331. The Mascot search results of the F-6652 proteome sample showed that 81% of the identified protein families were coming from the ATCC strain, indicating a high sequence similarity between the strain NRRL F-6652 and ATCC 33331 (Table S1). Therefore, all the bioinformatics analysis was performed based on the genomic sequence from S. flavogriseus ATCC 33331.
Product Prediction Based on the NRPS Gene Cluster
The 22.6 kb orphan NRPS gene cluster from S. flavogriseus ATCC 33331 encodes two modular NRPS proteins FlavA and FlavB, which can be further divided into six NRPS biosynthetic modules (Figure 1b). Bioinformatics analysis of this NRPS gene cluster predicted the six adenylation domains incorporate Ile, Gln, Ile, Gln, Val/Ile and Phe.18,19 The most salient features of the cluster are an apparent ‘starter’ condensation domain often involved in lipopeptide biosynthesis (Figure S1),20 and a thioester reductase at the final domain of FlavB. This reductase domain aligns with other known NRPS reductases, including the biosynthetic enzymes for myxochelin, saframycin, nostocyclopeptide, linear gramicidin, koranimine, peptaibol and aureusimine (Figure S2). From FlavB, conserved NAD(P)H binding domains were identified and the conserved catalytic triad of Thr-Tyr-Lys common to short chain reductase (SDR) is present. The NRPS reductase domain has been associated with the reductive release of peptide thioesters from their biosynthetic enzymes, forming aldehydes, alcohols, or cyclicimines.21 Based on the domain organization, we predicted the product from this cluster was a hexapeptide likely containing isoleucine, glutamine, valine and phenylalanine, an N-terminal aliphatic chain, and a C-terminal aldehyde, alcohol or imine.
In addition to the two NRPS genes, there is a small protein with 84 amino acids that belongs to the MbtHlike protein superfamily in the gene cluster (54% identical and 67% similar to MbtH protein from Mycobacterium tuberculosis). MbtH-like proteins have been found in many other NRPS gene clusters, and previous in vitro and in vivo studies have shown that they are required for the function of adenylation domains and correlated with high production of secondary metabolites.22,23
Targeted Identification and Structure Elucidation of Flavopeptins
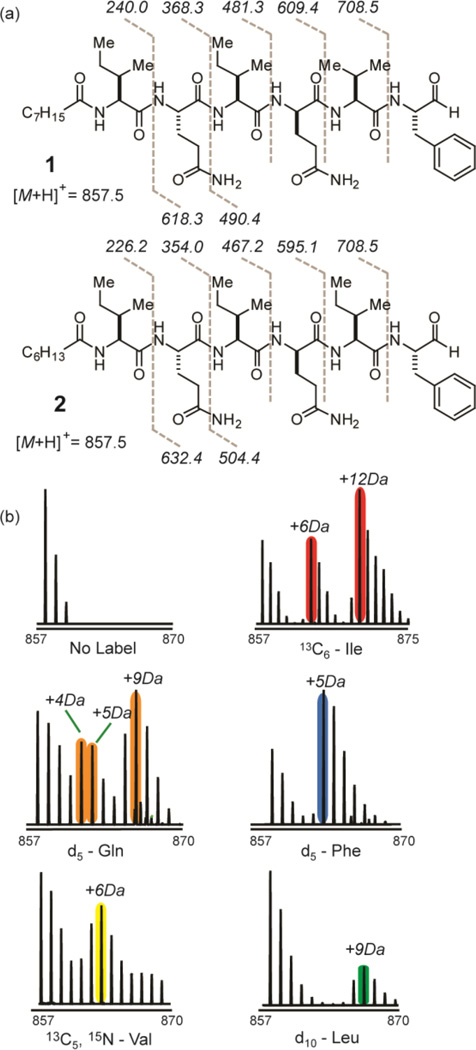
In order to identify the natural products arising from the NRPS gene cluster, we performed a targeted metabolomics analysis of F-6652 grown in MSB medium, the same medium in which the NRPS proteins were detected. In addition to monitoring metabolites that show increased intensities over time course, a predicted amino acid precursor with stable isotope labeling, d5- phenylalanine, was added to the medium to monitor metabolites with heavy isotope incorporation. Six ion species (m/z 829.5, 843.5, 857.5, 871.5, 885.5 and 899.5, Figure 1c) showed clear evidence of phenylalanine incorporation as 5 Da mass shifts (Figure 2b). These six ion species also generated peptide-like fragmentation patterns, all yielding a common amino acid sequence tag of Gln-Ile/Leu- Gln-Val (or Ile/Leu)-Phe(CHO) (Figures 2a and S3). This sequence tag provided a reasonable match to the predicted amino acid substrates for the detected NRPS gene cluster. MS4 fragmentation on the target mass was able to reveal the N-terminal structure to be an acylated isoleucine/ leucine residue (Figure S4). The six masses are spaced by 14.0162 Da, which is unambiguously a CH2 group, and their retention times are slightly increasing with the mass (Figure 1c), which leads to the prediction that these six species are differentiated by the length of the N-terminal aliphatic chain. All six masses pointed to related lipopeptide aldehyde structures that we named flavopeptins (Figure 1d; cal. 856.5422 Da, obs. 856.5427 Da, C44H72N8O9 for R1 = C7H15 and R2 = H).
Figure 2.
Partial structure elucidation of flavopeptins by MS and tandem MS. (a) Two isobaric structures of flavopeptin with mass 856.5 Da, with the observed MS2 fragments shown. (b) Metabolic labeling of flavopeptins with amino acid precursors containing stable isotopes. Shown are the mass spectra for flavopeptins from F-6652 grown in MSB medium with no labeling and with13C6-isoleucine, 2,3,3,4,4-d5-glutamine, ring-d5 phenylalanine,13C5,15N-valine and d10-leucine.
To corroborate structural assignments, several metabolic feeding experiments were performed using stable isotope-labeled amino acids. In addition to phenylalanine, LC–MS analysis of the labeled samples also showed the incorporation of two isoleucines, two glutamines and one valine into flavopeptins (Figure 2b). When supplemented with13C6-isoleucine, +6 Da and +12 Da mass shifts were observed, indicating at least two isoleucine residues were incorporated into the final product. MS2 fragmentation of these +6 Da and +12 Da mass species revealed that isoleucine can be incorporated at the first, the third and the fifth amino acid positions (Figure S5). Similarly, the incorporation of the phenylalanine and valine into the predicted positions was confirmed. Labeling with 2,3,3,4,4- d5-glutamine showed species with +4 Da, +5 Da and +9 Da mass shifts (instead of straight +5 Da and +10 Da), indicating one deuterium from one of the labeled glutamine exchanged with environmental H2O when incorporated into the final product. It is known that the first step of epimerization involves the removal the α-H from the loaded amino acid to generate a carbanion. Thus, one glutamine residue of the final product is likely epimerized to a d-glutamine based on the loss of one deuterium, which agrees with the presence of the epimerization domain in the fourth NRPS module that incorporates glutamine. Only a minor amount of d10-leucine was incorporated into the product, likely due to the relaxed substrate specificity of one of the adenylation domains activating valine or isoleucine.
Flavopeptins were isolated in 0.1 mg/L scale from the producing strain NRRL F-6652. All flavopeptin species with different aliphatic chain lengths were isolated as one fraction due to the difficulty of separation. To determine the absolute configuration of the amino acid building blocks, the isolated flavopeptins were hydrolyzed in strong acid and the resulting amino acids were derivatized with Marfey’s reagent (FDAA). Reverse-phase LC–MS analysis indicated the presence of l-Ile, l-Val and l-Phe, an equal amount of l-Gln and d-Gln, as well as a minor amount of l-Leu, which agrees with the previous assignments (Figure S6).
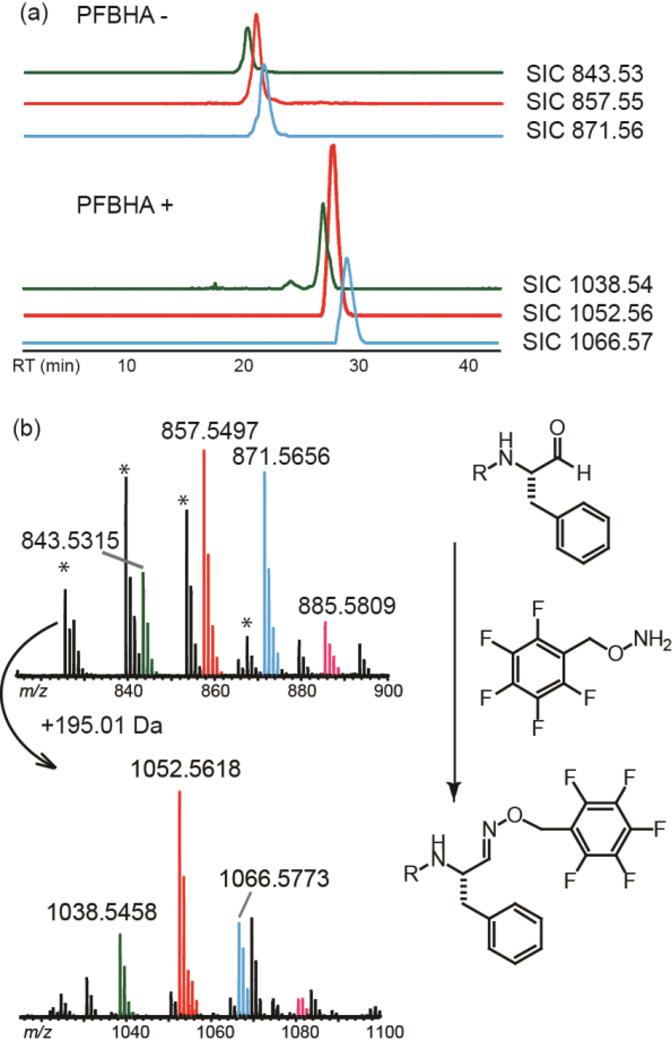
To confirm the presence of the aldehyde group, a selective aldehyde derivatization agent O-(2,3,4,5,6- pentafluorobenzyl) hydroxylamine hydrochloride (PFBHA) was allowed to react with the isolated flavopeptins. LC–MS analysis of the reaction mixture showed a series of new peaks with 195.01 Da increases for the pentafluorobenzyloximine derivatives (e.g. m/z 1052.5618 for the PFBHA derivative of 1, calculated 1052.5603, Δ = 1.4 ppm) (Figure 3).
Figure 3.
Detection of the aldehyde group of flavopeptins by derivatization with PFBHA. (a) LC–MS traces of flavopeptins before (top) and after (bottom) reacting with PFBHA. A series of masses with 195.01 Da increases (represented by SICs of m/z 1038.54, 1052.56 and 1066.57) appeared after PFBHA derivatization. (b) The high-resolution mass spectra of flavopeptins before and after derivatization, together with the labeling reaction mechanism. Peaks with ‘*’ indicate dehydrated products from the original sample.
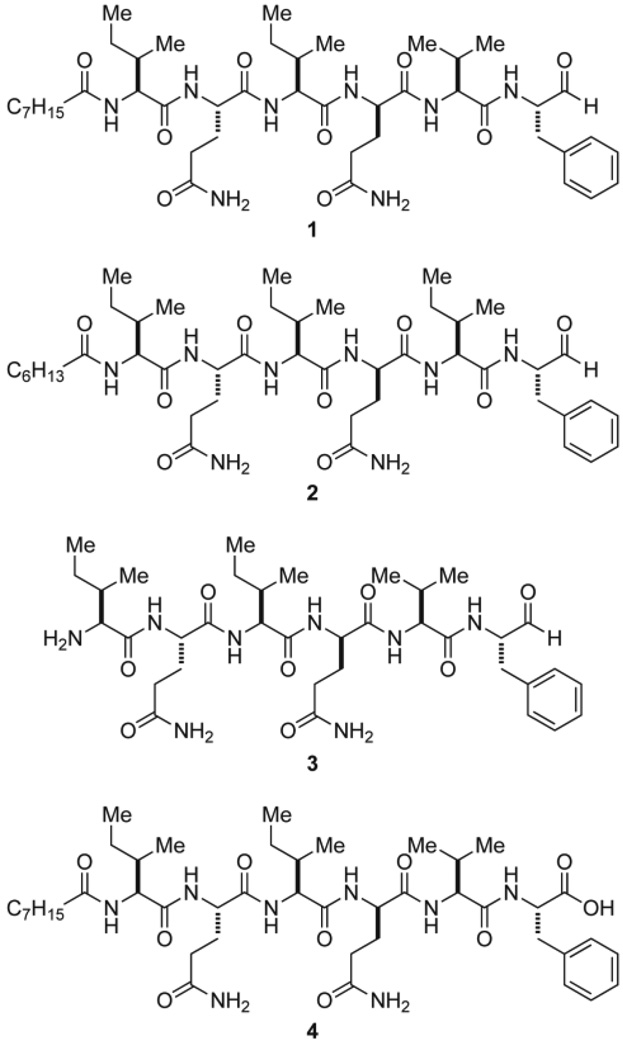
Attempts to obtain sufficient quantities of pure flavopeptins from the natural producer proved difficult due to low yield, co-elution with other compounds, and the presence of multiple flavopeptin species that resist separation. Rather than solve these major problems, solid phase peptide synthesis was used to synthesize two flavopeptin species with the same mass 856.6 Da (1 and 2) as well as two additional derivatives; one with an N-terminal free amine (3) and the other with C-terminal carboxylic acid (4) (Figure 4). Compound 1 showed the same LC retention time, the same intact mass and MS2 fragmentation masses as the corresponding flavopeptin species isolated from the natural source (Figure S7).
Figure 4.
Flavopeptins (1 and 2), the N-terminal amine derivative (3), and the C-terminal carboxylic acid derivative (4) synthesized by solid phase peptide synthesis.
Biosynthesis of Flavopeptins
The predicted biosynthetic mechanism for the flavopeptins is shown in Figure 5. The most unusual aspect is the presence of a relatively rare reductase domain at the C-terminus of FlavB instead of the more common thioesterase. A number of previous reports have shown that NAD(P)H-dependent reductase domains catalyze the reductive off-loading of peptide thioesters to produce linear aldehydes.21 In all the previous cases, however, the linear aldehyde only exists as an intermediate that undergoes subsequent reactions, such as further reduction to an alcohol (lyngbyatoxin), transamination to form an amine (myxochelin B and lysine biosynthesis in yeast), intramolecular cyclization to form a cyclic imine (koranimine and nostocyclopeptide), and intramolecular Pictet- Spengler reaction (saframycin).14,24–28 Flavopeptins are the first examples in which a stable peptide aldehyde is released as the end product from an NRPS assembly line. There were no masses observed in the culture broth corresponding to an alcohol, amine or imine product. Only after organic extraction followed by drying did we observe masses with an 18 Da loss, which corresponded to a dehydrated form (Figure S8). Tandem MS analysis indicated the dehydration occurred between Gln2 and the terminal aldehyde, possibly forming an imino group between the side chain amide of the glutamine and the aldehyde. Drying the synthetic flavopeptins in organic solvent also showed the dehydration product with the same MS2 fragmentation patterns, indicating the dehydrated compound was an artifact produced during purification.
Figure 5.
Predicted biosynthetic mechanism for the flavopeptins. The aldehyde group is predicted to be formed through reductive release of the peptide thioester using the NRPS reductase domain.
Unlike nostocyclopeptides and koranimine, where the aldehyde intermediates spontaneously undergo macrocyclization to form an imine, flavopeptins maintain the linear form in nature. We speculated that the N-terminal fatty acid “cap” serves to protect the primary amine from condensation with the aldehyde, thus blocking imine formation. However, the synthetic analogue of flavopeptin with an N-terminal free amine (3) also retains its linear form, with only a very minor proportion of dehydrated product observed (less than 10% by MS analysis, Figure S7). This result demonstrated that the peptide sequence of flavopeptins itself is not prone to inducing the macrocyclic conformation. Our observations were in accordance with a previous study on nostocyclopeptide, where certain amino acids in the peptide sequence were found essential for the spontaneous macrocyclization of the peptidyl aldehyde intermediate into a cyclic imine.24
A BLAST search of FlavA and FlavB against the database revealed that two other strains, Kitasatospora setae KM-6054 (= NRRL B-16185) and Streptomyces sp. Sirex AA-E, contain similar NRPS gene clusters with 69% and 80% sequence identity to F-6652 at the protein level (Figure S9). These two strains also share the same NRPS domain organization and same predicted amino acid substrates, leading us to hypothesize that K. setae and S. sp. Sirex AA-E would have the genetic potential to produce flavopeptins. Indeed, the same masses corresponding to flavopeptins were detected from NRRL B-16185. Other neighboring genes are not highly conserved among these three strains, except for the MbtH protein (74% and 79% identical to F-6652), indicating the MbtH, FlavA and FlavB likely constitute the core genes responsible for flavopeptin biosynthesis.
Biological Activities of Flavopeptins
Many naturally occurring peptide aldehydes are known to exhibit inhibitory activities against proteases. The aldehyde warhead undergoes nucleophilic attack by the active site residue of the protease (serine, threonine or cysteine) to form a reversible covalent hemiacetal/ hemithioacetal adduct with the enzyme, while the peptide portion specifically interacts with the substrate binding site. By comparing the peptide sequence of flavopeptins with known peptide aldehyde protease inhibitors, we predicted the two hydrophobic amino acids valine and phenylalanine at the C-terminus of the peptide could make flavopeptins potential inhibitors against cysteine proteases and the mammalian proteasome (Figure S10). When tested, flavopeptins strongly inhibited cysteine proteases like papain and calpain with ≥ 90% inhibition at 15 µg/mL. Proteases of other types, including serine, aspartate and metallo-proteases, were inhibited by < 50% (Figure S11). The IC50 values of flavopeptins were measured using synthetic compounds (Table 1). Compound 1 showed low-micromolar to nanomolar inhibition against cysteine proteases (IC50 = 0.16 ± 0.04 µM for papain and 2.9 ± 0.3 µM for calpain).
Table 1.
Protease, Proteasome and Cell Inhibition Activities of Flavopeptins
| IC50 (µM) | 1 | 2 | 3 | |
|---|---|---|---|---|
| cysteine proteases | papain | 0.16 ± 0.04 | 0.35 ± 0.03 | 0.07 ± 0.01 |
| calpain | 2.9 ± 0.3 | 2.7 ± 0.4 | 1.8 ± 0.3 | |
| proteasome1 | chymotrypsin-like | 4.9 ± 0.3 | 3.3 ± 0.3 | 0.43 ± 0.08 |
| caspase-like | 2.3 ± 0.2 | 2.3 ± 0.7 | 0.33 ± 0.07 | |
| cell line | MM.M1S | 33 ± 2 | Not tested | No inhibition |
| FR4 | 35 ± 6 | 30 ± 10 | No inhibition | |
| U-937 | 13 ± 1 | Not tested | No inhibition | |
For comparison with another natural peptide aldehyde, fellutamide B inhibits the proteasome with IC50 values of ~9 nM and ~2 µM for chymotrypsin-like and caspase-like activities, respectively.4
Given the structural similarity between flavopeptins and known peptide aldehyde proteasome inhibitors, we investigated whether they could inhibit the human 20S proteasome. As shown in Table 1, compound 1 inhibited the chymotrypsin-like activity of the human 20S proteasome with an IC50 of 4.9 ± 0.3 µM, and the caspase-like activity with an IC50 of 2.3 ± 0.2 µM. As a negative control, the flavopeptin analogue with a C-terminal carboxylic acid group rather than an aldehyde (4) was tested, but showed no inhibition up to 100 µM (Figure S11). Compound 2 (containing Ile5 instead of Val5) exhibited similar inhibition potencies as 1, while compound 3 (with an N-terminal free amine) showed 6 and 10-fold higher potencies. The stronger activity of 3 is possibly due to a greater affinity for the proteasome substrate-binding site.
In view of the anti-proliferative activities of some known proteasome inhibitors such as bortezomib,29 we tested flavopeptins for activity against human multiple myeloma cell lines MM.M1S and FR4, and a histiocytic lymphoma cell line U-937. Using a cell viability assay that quantifies cellular ATP, compound 1 displayed IC50 values of ~30 µM and 13 µM against multiple myeloma and lymphoma cell lines, respectively. The N-terminal free amine analogue (i.e., 3) which exhibited higher in vitro activities showed no in vivo inhibition up to 80 µM, most likely due to its low membrane permeability and low cellular stability compared to the N-acylated compounds. A noteworthy feature within the flavopeptins is the presence of a d-glutamine in the peptide sequence. Although d-amino acids are commonly seen in nonribosomally synthesized peptides, they are rarely present in protease/proteasome inhibitors.
CONCLUSIONS
In this work, we described the discovery of the flavopeptins, a class of novel peptide aldehydes, from an orphan gene cluster in a Streptomyces species using the proteomics-based method, PrISM. Although there are previous examples of NRPS reductase domains catalyzing the formation of aldehydes as intermediates, flavopeptins are the first examples of stable linear peptide aldehydes arising definitively from an NRPS biosynthetic pathway. Similar to other peptide aldehydes, flavopeptins inhibit cysteine proteases and the human 20S proteasome by forming a covalent adduct between their aldehyde group and the active site cysteine or threonine residue of the target enzyme. There will be continued interest in elucidating whether other naturally occurring peptide aldehydes are also synthesized through a similar mechanism involving NRPS reductase domains. This can be realized by sequencing the genome of strains known to produce peptide aldehydes and locating the corresponding gene cluster.
Supplementary Material
ACKNOWLEDGMENT
We thank the Agricultural Research Service, United States Department of Agriculture for providing us the bacterial strains. We thank the ChemCore facility at Northwestern University for providing HPLC purification services, and the High-Throughput Analysis Lab for providing fluorospectroscopy services. Research reported in this publication was supported by the National Institute of Health under award number GM067725 to N.L.K. and by the National Science Foundation under award number CHE0845063 to R.J.T.
Footnotes
ASSOCIATED CONTENTS
Supporting Information
Mascot search result, bioinformatics analysis of protein sequences, extended mass spectrometry data, bioassay results and NMR spectra for synthetic peptides. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Storr SJ, Carragher NO, Frame MC, Parr T, Martin SG. Nat. Rev. Cancer. 2011;11:364–374. doi: 10.1038/nrc3050. [DOI] [PubMed] [Google Scholar]
- 2.Lecaille F, Kaleta J, Bromme D. Chem. Rev. 2002;102:4459–4488. doi: 10.1021/cr0101656. [DOI] [PubMed] [Google Scholar]
- 3.Kisselev AF, van der Linden WA, Overkleeft HS. Chem Biol. 2012;19:99–115. doi: 10.1016/j.chembiol.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hines J, Groll M, Fahnestock M, Crews CM. Chem Biol. 2008;15:501–512. doi: 10.1016/j.chembiol.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim KB, Crews CM. J Med Chem. 2008;51:2600–2605. doi: 10.1021/jm070421s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rentsch A, Landsberg D, Brodmann T, Bulow L, Girbig AK, Kalesse M. Angew. Chem. Int. Ed. Engl. 2013;52:5450–5488. doi: 10.1002/anie.201207900. [DOI] [PubMed] [Google Scholar]
- 7.Nett M, Ikeda H, Moore BS. Nat. Prod. Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lautru S, Deeth RJ, Bailey LM, Challis GL. Nat. Chem. Biol. 2005;1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- 9.Forseth RR, Amaike S, Schwenk D, Affeldt KJ, Hoffmeister D, Schroeder FC, Keller NP. Angew. Chem. Int. Ed. Engl. 2013;52:1590–1594. doi: 10.1002/anie.201207456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kersten RD, Yang YL, Xu Y, Cimermancic P, Nam SJ, Fenical W, Fischbach MA, Moore BS, Dorrestein PC. Nat. Chem. Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang FY, Brady SF. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2478–2483. doi: 10.1073/pnas.1218073110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bumpus SB, Evans BS, Thomas PM, Ntai I, Kelleher NL. Nat. Biotechnol. 2009;27:951–956. doi: 10.1038/nbt.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischbach MA, Walsh CT. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 14.Evans BS, Ntai I, Chen Y, Robinson SJ, Kelleher NL. J. Am. Chem. Soc. 2011;133:7316–7319. doi: 10.1021/ja2015795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Ntai I, Ju KS, Unger M, Zamdborg L, Robinson SJ, Doroghazi JR, Labeda DP, Metcalf WW, Kelleher NL. J. Proteome Res. 2012;11:85–94. doi: 10.1021/pr2009115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Unger M, Ntai I, McClure RA, Albright JC, Thomson RJ, Kelleher NL. Med. Chem. Commun. 2013;4:233–238. doi: 10.1039/C2MD20232H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stanier RY, Palleroni NJ, Doudoroff M. J. Gen. Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 18.Bachmann BO, Ravel J. Methods Enzymol. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- 19.Rottig M, Medema MH, Blin K, Weber T, Rausch C, Kohlbacher O. Nucleic Acids Res. 2011;39:W362–W367. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rausch C, Hoof I, Weber T, Wohlleben W, Huson DH. BMC Evol. Biol. 2007;7:78. doi: 10.1186/1471-2148-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du L, Lou L. Nat Prod Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- 22.Lautru S, Oves-Costales D, Pernodet JL, Challis GL. Microbiology. 2007;153:1405–1412. doi: 10.1099/mic.0.2006/003145-0. [DOI] [PubMed] [Google Scholar]
- 23.Felnagle EA, Barkei JJ, Park H, Podevels AM, McMahon MD, Drott DW, Thomas MG. Biochemistry. 2010;49:8815–8817. doi: 10.1021/bi1012854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopp F, Mahlert C, Grunewald J, Marahiel MA. J. Am. Chem. Soc. 2006;128:16478–16479. doi: 10.1021/ja0667458. [DOI] [PubMed] [Google Scholar]
- 25.Ehmann DE, Gehring AM, Walsh CT. Biochemistry. 1999;38:6171–6177. doi: 10.1021/bi9829940. [DOI] [PubMed] [Google Scholar]
- 26.Koketsu K, Watanabe K, Suda H, Oguri H, Oikawa H. Nat. Chem. Biol. 2010;6:408–410. doi: 10.1038/nchembio.365. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Weissman KJ, Muller R. J. Am. Chem. Soc. 2008;130:7554–7555. doi: 10.1021/ja8025278. [DOI] [PubMed] [Google Scholar]
- 28.Read JA, Walsh CT. J. Am. Chem. Soc. 2007;129:15762–15763. doi: 10.1021/ja077374d. [DOI] [PubMed] [Google Scholar]
- 29.Teicher BA, Ara G, Herbst R, Palombella VJ, Adams J. Clin. Cancer Res. 1999;5:2638–2645. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.