Abstract
Background
Genome-wide association studies have identified novel genetic factors that contribute to intracranial aneurysm (IA) susceptibility. We sought to confirm previously reported loci, to identify novel risk factors, and to evaluate the contribution of these factors to familial and sporadic IA.
Method
We utilized 2 complementary samples, one recruited on the basis of a dense family history of IA (discovery sample 1: 388 IA cases and 397 controls) and the other without regard to family history (discovery sample 2: 1095 IA cases and 1286 controls). Imputation was used to generate a common set of single nucleotide polymorphisms (SNP) across samples, and a logistic regression model was used to test for association in each sample. Results from each sample were then combined in a meta-analysis.
Results
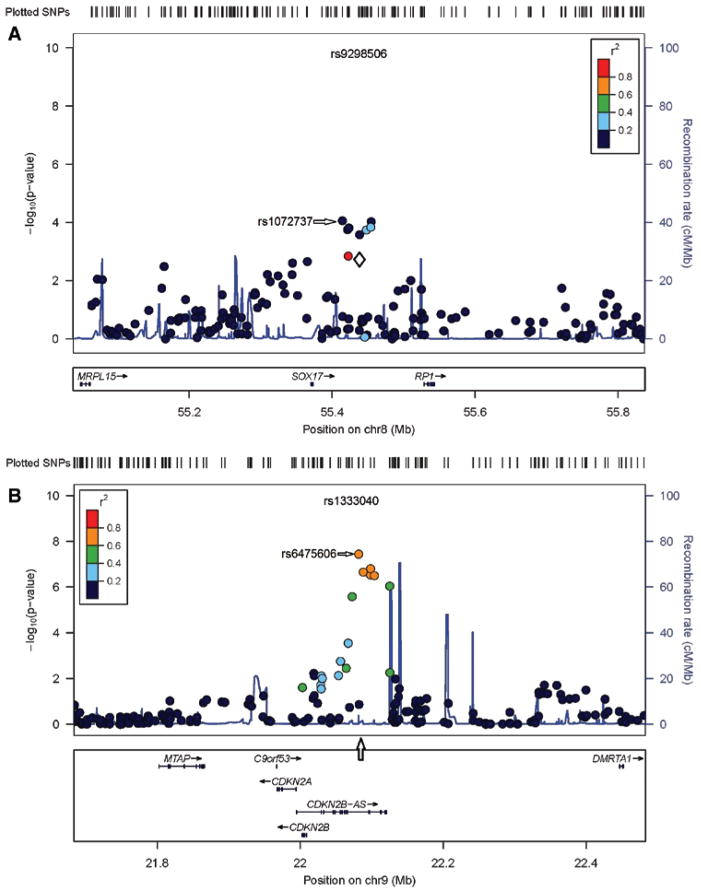
There was only modest overlap in the association results obtained in the 2 samples. In neither sample did results reach genome-wide significance. However, the meta-analysis yielded genome-wide significance for SNP on chromosome 9p (CDKN2BAS; rs6475606; P=3.6×10−8) and provided further evidence to support the previously reported association of IA with SNP in SOX17 on chromosome 8q (rs1072737; P=8.7×10−5). Analyses suggest that the effect of smoking acts multiplicatively with the SNP genotype, and smoking has a greater effect on risk than SNP genotype.
Conclusion
In addition to replicating several previously reported loci, we provide further evidence that the association on chromosome 9p is attributable to variants in CDKN2BAS (also known as ANRIL, an antisense noncoding RNA).
Keywords: genome-wide association study, intracranial aneurysm
The risk of intracranial aneurysm (IA) is increased among individuals with first-degree relatives with history of IA.1 Many approaches have been used to identify genes contributing to the risk of IA.2–4 Genome-wide association studies identified and replicated associations on chromosome 4q31.23 (EDNRA), 8q12.1 (SOX17), 9p213 (CDKN2A/CDKN2B/CDKN2BAS), 10q24.32 (CNNM2), 12q22, 13q13.1 (KL/STARD13), 18q11.2 (RBBP8), and 20p12.1.3–6 Most studies utilized sporadic IA cases; familial cases may potentially involve different genetic risk factors. The present study was undertaken to compare and contrast results from a genome-wide association study of familial and sporadic IA cases.
Subjects and Methods
Discovery Sample 1
Discovery sample (DS) 1 consisted of individuals recruited through the Familial Intracranial Aneurysm (FIA) I study,7 which recruited familial cases appropriate for linkage analysis8 through an international consortium and applied rigorous inclusion and exclusion criteria. A set of independent unrelated cases was obtained by selecting 1 self-reported white individual with definite IA from each family (n=389). White control subjects were obtained from 2 population-based studies, Genetic and Environmental Risk Factors for Hemorrhage Stroke and the Cincinnati Control Cohort. All studies were approved by the appropriate Institutional Review Boards. Additional information is provided in the Online Supplement.
DS2
During FIA II study recruitment, the requirement for family history of IA was removed and both familial and sporadic IA cases were enrolled. The same exclusion criteria were in place, and all case subjects underwent the same rigorous review; 829 white IA case subjects from FIA II and 61 white sporadic aneurysmal subarachnoid hemorrhage case subjects were selected.
The sample was augmented by white case subjects and control subjects identified from other studies. Subjects with incident cases of subarachnoid hemorrhage secondary to documented or presumed ruptured IA (n=160) who were frequency-matched to control subjects (n=168) were obtained from the Australasian Cooperative Research on Subarachnoid Hemorrhage Study (ACROSS).9 An additional 184 white IA case subjects were selected from a prospective San Francisco cohort study of adult patients with spontaneous subarachnoid hemorrhage attributable to IA confirmed by noncontrast computed tomography and cerebral angiogram.10 All studies were approved by the appropriate Institutional Review Boards.
Genotypic data from 1148 white control subjects were obtained through a collaborative agreement with the Atherosclerosis Risk in Communities (ARIC) study. In the ARIC sample, a subset of subjects who never had a stroke or transient ischemic attack was matched to the DS2 case subjects by sex and, when possible, by age (±5 years).
Genotyping, Quality Review, and Imputation
Genotyping was performed using the Axiom array for all samples except the ARIC control subjects. All released genotypes underwent a common quality review pipeline. A principal component analysis was performed to identify and remove samples from subjects with African, Asian, or Hispanic admixture (n=47). Genotypic data for the ARIC samples were obtained from the Affy 6.0 array11 and underwent a similar quality review.
Imputation for the Axiom samples was performed for all autosomes using IMPUTE2.12 All samples genotyped on the Axiom array (n=2115) were imputed together using the 1000 Genomes haplotypes (n=1094) as the phased reference panel. The ARIC samples were included as another unphased reference panel to maximize available information at each imputed single nucleotide polymorphism (SNP). We did not impute genome-wide SNP genotypes for the ARIC sample. Extensive and detailed quality review13 was performed to ensure that spurious association was not detected based on platform effects. Using an aggressive filtering approach, we retained 453 699 SNP for analysis of DS2.
Statistical Analysis
The genotyped and imputed SNP were used to test for association with IA susceptibility using a logistic regression model. No additional covariates were necessary. Data from DS1 were analyzed initially using only the genotyped SNP. Data from DS2 were analyzed using the genotyped data from ARIC, along with both genotyped and imputed data from the samples genotyped on the Axiom array using the SNPTEST version 2 software. Imputed genotypes were encoded in the logistic model as the expected allele count.14
Meta-analysis was performed by combining results from DS1 and DS2 for the common set of 451 088 SNP genotyped or imputed in both samples. METAL15 was used using an inverse-variance weighting scheme. Genomic control was used before and after meta-analysis to do weight results in each sample when the genomic inflationwn-coefficient exceeded 1.00. We evaluated results to identify findings meeting genome-wide significance criteria (P<5×10−8), and we also sought to replicate previous associations using a more modest significance level (P<10−4).
To test whether there might be >1 risk variant in a particular gene or gene region contributing to the association, we performed conditional analyses. For the statistically significant region on chromosome 9, we identified the SNP with the most extreme probability value in the meta-analysis. We then modified the logistic regression model to include the genotype at the most significant SNP in the region in each of the samples and performed meta-analysis to combine the results from each sample. Finally, we reviewed the probability value for the other SNP in the region to determine whether any other SNP remained statistically significant.
Because cigarette smoking is a strong risk factor in IA,16,17 we examined the possible interaction of the most highly associated SNP on chromosomes 8 and 9 with cigarette smoking. A logistic regression model, with age and sex as covariates, was used to test for departures from a multiplicative relationship between the 3 genotypes for that SNP and cumulative exposure to smoking, as measured by pack-years. Because the distribution of pack-years was highly skewed, we used the log of pack-years in the logistic model, with 0.05 assigned to never-smokers. Results from DS1 and DS2 were combined in a meta-analysis weighted by the inverse variance of the sample estimates.
Results
DS1
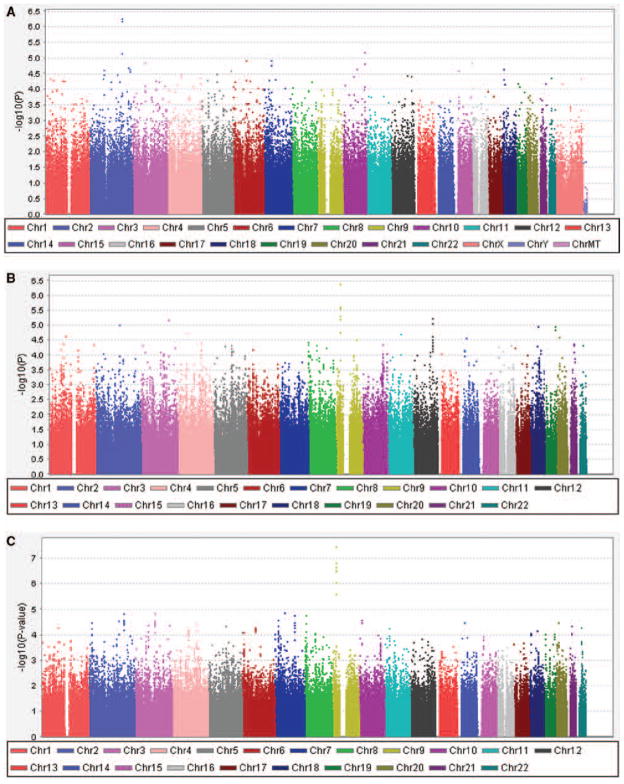
The final sample included 388 familial IA case subjects and 397 control subjects (Table 1) with genotypes for 492 284 SNP. Results are summarized in Figure 1A and Table 2. Although no results achieved traditional criteria for genome-wide significance, several SNP yielded interesting results within phosphodiesterase 1A (PDE1A; rs3769801; P=5.5×10−7), with little evidence of increased false-positive results (λ =1.047).
Table 1.
Sample Demographics
| Discovery Sample 1
|
Discovery Sample 2
|
|||
|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |
| Sample size | 388 | 397 | 1095 | 1286 |
| % Male | 31.4 | 45.8 | 26.1 | 29.2 |
| Mean (SD) age at onset (cases)* or at recruitment (controls) | 50.7 (11.9) | 63.4 (14.8) | 53.9 (12.2) | 53.9 (9.0) |
| % Family history of IA† | 100 | 5.5 | 27.0 | … |
| % Cigarette smoker (current or former)‡ | 82.0 | 52.9 | 67.7 | 54.8 |
| % Cigarette smoker (current only) | 45.6 | 17.4 | 39.5 | 27.0 |
| Mean (SD) pack years of cigarette smoking§ | 25.7 (26.0) | 14.9 (22.8) | 21.1 (25.0) | 14.1 (19.8) |
DS indicates discovery sample; IA, intracranial aneurysm; SD, standard deviation; UCSF, University of California San Francisco.
Age at onset defined as age when aneurysm was identified via imaging or age at rupture if there is no previous information on which to base diagnosis.
Positive family history for cases in DS1 was validated. For most DS2 case subjects and for control subjects in DS1, positive family history was based on self-report by the subject.
Percentage of DS2 case subjects is an underestimate because UCSF case subjects (N=128) recorded data for current but not former smoking.
Percentage of DS2 cases is based on 963 cases. UCSF case subjects (N=128) did not record pack-years, and 4 other case subjects had unknown pack-year data.
Figure 1.
Results of the genome-wide association analysis for intracranial aneurysm (IA) susceptibility. A, Discovery sample 1. B, Discovery sample 2. C, Meta-analysis.
Table 2.
Summary of Top Association Results
| Chr | Position (bp) | SNP | Gene (Nearest Gene) | EA | AF | Axiom (G/I) | Discovery Sample 1
|
Discovery Sample 2
|
Meta-analysis
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P Value | OR | P Value | OR | P Value | OR | |||||||
| 2 | 128796641 | rs11693075 | SAP130 (11kb) | A | 0.75 | G | 0.8008 | 1.03 | 9.85×10−6 | 1.37 | 0.000267 | 1.26 |
| 2 | 183026612 | rs1897472 | PDE1A | A | 0.69 | G | 6.66×10−7 | 1.78 | 0.24* | N/A | ||
| 2 | 183043380 | rs3769801 | PDE1A | G | 0.69 | G | 5.53×10−7 | 1.79 | 0.13* | N/A | ||
| 9 | 22072719 | rs10757270 | CDKN2BAS | G | 0.42 | G | 0.03686 | 1.24 | 2.81×10−6 | 1.32 | 2.62×10−6 | 1.30 |
| 9 | 22081850 | rs6475606 | CDKN2BAS | T | 0.48 | G | 0.001332 | 1.39 | 4.29×10−7 | 1.34 | 3.59×10−8 | 1.35 |
| 9 | 22088260 | rs10757272 | CDKN2BAS | T | 0.48 | G | 0.001899 | 1.37 | 2.77×10−6 | 1.31 | 2.20×10−7 | 1.33 |
| 9 | 22098574 | rs4977574 | CDKN2BAS | G | 0.49 | G | 0.001319 | 1.39 | 5.12×10−6 | 1.31 | 2.95×10−7 | 1.32 |
| 9 | 22098619 | rs2891168 | CDKN2BAS | G | 0.48 | G | 0.001294 | 1.39 | 2.54×10−6 | 1.32 | 1.56×10−7 | 1.33 |
| 9 | 22125347 | rs1333048 | CDKN2BAS (4kb) | C | 0.49 | G | 0.005015 | 1.33 | 6.27×10−6 | 1.30 | 9.06×10−7 | 1.32 |
| 10 | 124045887 | rs911774 | BTBD16 | A | 0.20 | G | 6.69×10−6 | 1.73 | 0.63125 | 1.03 | 0.0118 | 1.18 |
| 12 | 105933442 | rs11112585 | C12orf75 (168kb) | C | 0.84 | I | 0.081172 | 0.78 | 8.61×10−6 | 1.45 | 0.01071 | 1.22 |
| 12 | 106007300 | rs2374513 | C12orf75 (242kb) | C | 0.86 | G | 0.2955 | 0.86 | 5.88×10−6 | 1.49 | 0.00226 | 1.28 |
AF indicates allele frequency; Chr, chromosome; DS, discovery sample; EA, effective allele; G/I, genotyped or imputed; OR, odds ratio; N/A, not applicable; SNP, single nucleotide polymorphism.
N/A: this SNP either was not genotyped in ARIC or the genotypic data was removed as part of the SNP cleaning process; therefore, the SNP was not tested in DS2 or the meta analysis.
Position (bp): from buildGRCh37.p1; P<10−5 in either DS1 or DS2.
SNP was imputed in the ARIC sample. The logistic regression analysis only included SNP genotype as the main effect. No additional covariates were included.
DS2
The final sample included 1095 IA case subjects and 1286 control subjects (Table 1) with genotypes for 453 699 SNP (Figure 1B and Table 2). Although no results attained genome-wide significance, 2 regions had SNP with P<10−5. These included a region on chromosome 9p nominated in previous genome-wide association studies (CDKN2BAS; rs6475606; P=4.3×10−7) as well as the region near c12orf75 on chromosome 12q (rs2374513; P=5.9×10−6). The Q-Q plot suggested that the rate of false-positive results was acceptable (λ =1.072).
Meta-Analysis
Combining the results from DS1 and DS2 in a meta-analysis yielded genome-wide significance for the region on chromosome 9p (CDKN2BAS; rs6475606; P=3.6×10−8; Figure 1C). No other region attained this level of significance. We systematically compared our results with those in previous genome-wide association studies5,6 and replicated not only the association on chromosome 9p but also the association with SOX17 on chromosome 8q (rs1072737; P=8.7×10−5; Figure 2). We did not find substantial support for the previously reported associations in other chromosomal regions (Supplemental Figure I). However, at least on chromosome 20, our study did not include any SNP with at least moderate linkage disequilibrium with the most significant SNP previously reported. Therefore, we cannot definitely evaluate whether our sample provides support for the previously reported association.3,4
Figure 2.
Comparison with selected previously reported results from genome-wide association studies.5,6 A, Chromosome 8q12.1. B, Chromosome 9p213. When the most significant single nucleotide polymorphism (SNP) from the initial report was available in the meta-analysis, a white diamond indicates the SNP, and the SNP is listed at the top of the Figure. If the SNP was not available in the meta-analysis sample, then an arrow is used to denote the position of that SNP. Each circle indicates the probability value for a SNP at that position in the meta-analysis. The color of the square symbol denotes the extent of linkage disequilibrium (as computed by r2) with the SNP reported in the initial report.
Conditional Analysis
Given previous reports of association of several genes in the chromosome 9 region, we performed conditional analysis to test for evidence of >1 risk factor in the region. We performed analyses in DS1 and DS2 separately, including the most significant SNP (rs6475606) in the logistic regression model. After meta-analysis of the results in the 2 samples in this region, no SNP remained significant when rs6475606 was included in the model (all P>0.54), suggesting there is only 1 risk factor for IA in this chromosomal region.
Gene and Smoking Relationship
Statistical testing of the SNP on chromosomes 8 (rs1072737) and 9 (rs6475606) did not find evidence for an interaction of either SNP with log (pack-years). Rather, the data were consistent with a multiplicative relationship between the SNP genotype and smoking (Table 3). Importantly, when both genotype and smoking are modeled together, the effect of smoking on disease risk remains substantially greater than that of the SNP genotypes (smoking odds ratio, 2.5; SNP odds ratio, 1.25–1.36).
Table 3.
Logistic Regression Model to Evaluate the Relationship Between Single Nucleotide Polymorphism Genotype and Smoking on the Relative Risk of Intracranial Aneurysm
| SNP rs1072737 (Chromosome 8)
|
Smoking*
|
|||||
|---|---|---|---|---|---|---|
| Sample | OR | 95% CI | P Value | OR | 95% CI | P Value |
| Sample 1 | 1.39 | 1.06 to 1.81 | 0.017 | 5.34 | 3.57 to 7.95 | <0.001 |
| Sample 2 | 1.22 | 1.07 to 1.39 | 0.002 | 2.12 | 1.76 to 2.56 | <0.001 |
| Meta-analysis | 1.25 | 1.11 to 1.40 | <0.001 | 2.51 | 2.14 to 2.96 | <0.001 |
| SNP rs6475606 (Chromosome 9)
|
Smoking*
|
|||||
| Sample | OR | 95% CI | P Value | OR | 95% CI | P Value |
|
| ||||||
| Sample 1 | 1.52 | 1.18 to 1.95 | 0.001 | 5.77 | 3.96 to 8.39 | <0.001 |
| Sample 2 | 1.33 | 1.18 to 1.50 | <0.001 | 2.11 | 1.77 to 2.52 | <0.001 |
| Meta-analysis | 1.36 | 1.22 to 1.52 | <0.001 | 2.53 | 2.17 to 2.94 | <0.001 |
CI indicates confidence interval; OR, odds ratio; SNP, single nucleotide polymorphism.
Smoking refers to 20 pack-years of smoking.
rs1072737 test of interaction, z=0.40, P=0.689; rs6475606 test of interaction: z=0.42, P=0.674.
A logistic regression model was used to test for departures from a multiplicative relationship between the risk allele scores (no risk allele=0; 1 risk allele=1; and 2 risk alleles=2) and cumulative exposure to smoking as measured by log (pack-years). Age and sex were included as covariates in the model. More details on the interpretation of the model are available in Supplementary Materials.
Discussion
We found the strongest evidence of association with a SNP in CDKN2BAS, also known as ANRIL. Previous studies reported an association of IA,2–6,18,19 as well as myocardial infarction,18 large-vessel ischemic stroke subtype, 20 and aortic aneurysm,18,21 with SNP in this region. Others6 examined the association in multiplex families as well as sporadic IA and found consistent evidence of association with rs1333040 (allele T), located in intron 12 of CDKN2BAS and having limited linkage disequilibrium spanning introns 7 through 15 of CDKN2BAS. The association of this SNP with both sporadic and familial IA parallels our findings. In DS1, we found evidence for association with the high-risk T allele (P=0.02; odds ratio, 1.26). We imputed this SNP in the ARIC sample and saw even greater evidence of this association in DS2 (P=1.4×10−5; odds ratio, 1.29). When the 2 samples were combined, the association strengthened (P=7.6×10−6; odds ratio, 1.29).
The linkage disequilibrium structure of the chromosome 9p21 region has at least 2 major linkage disequilibrium blocks. The first is associated with vascular diseases and the other is associated with diabetes mellitus.18 SNP in the region are associated with several types of cancer as well as open-angle glaucoma.22 Using conditional analyses in our sample, we did not find evidence of >1 risk factor for IA within this chromosomal region.
ANRIL is a long intergenic noncoding RNA. A mutant mouse with a deletion corresponding to this region has significantly greater suppression of the RNA encoded by Cdkn2a and Cdkn2b.23 Cultured aortic smooth muscle cells from these mutant mice had increased proliferative activity. These data suggest that the association of ANRIL with both atherosclerosis and IA may be attributable to similar pathophysiologic mechanisms, because both involve thinning of the tunica media layer in affected vessels.24
Previous reports found association of IA with SNP in both the 5′ and 3′ regions of SOX17.2,4 The SNP in the 2 regions were not in linkage disequilibrium and appear to have independent effects. The association with both regions of SOX17 was found in samples of European descent; however, a Japanese cohort demonstrated association only with the 5′ SNP.2 A subsequent study found that the association with the 5′ SNP varied among different populations, whereas the 3′ association remained robust in all but the Japanese cohorts.4 We were able to detect association only with SNP in the 3′ end of SOX17.
Smoking effects on IA may be moderated in part via genes that contain high-risk genetic variants. We found no evidence of an interaction between the most significant SNP in CDKN2BAS and SOX 17 and smoking. The data were consistent with the multiplicative effect of 2 variables in a logistic regression model in which the predicted risk is obtained by multiplying the 2 odds ratios. Thus, subjects who have the SNP risk allele and who also smoke have a greatly increased risk of IA as compared with subjects who have only the SNP risk allele. However, smoking remains the dominant risk factor and, because genotype cannot be altered, cessation of smoking would dramatically decrease the risk of IA, even among those having inherited a high-risk variant.
We did not replicate the other SNP found associated with IA in previous genome-wide association studies.2–4 Insufficient power may be an explanation for the failure to detect such an association, compared with the replication of associations on chromosome 8 and 9. Another possible reason may be that the associated risk is heterogeneous, and these risk variants were less important in our samples.
We used 2 discovery samples to allow us to test the hypothesis that unique risk variants segregate in familial IA. The most promising association from the familial IA analysis was with PDE1A. The same SNP were not genotyped in ARIC so we imputed them. However, there was no evidence of association with either of these SNP (rs1897472: P=0.24; rs3769801: P=0.13). This lack of replication suggests that this finding may have been a false-positive association.
This study had several important strengths. All case subjects underwent a uniform and rigorous evaluation that included review of medical records. The case subjects from DS1 came from densely affected, multiplex families who underwent a thorough evaluation of all putative cases, even when biological samples were not available. The control subjects, although recruited through multiple studies, also underwent a careful review to identify any evidence of a previous stroke or IA. Detailed data regarding smoking and hypertension, key risk factors for IA, were obtained from case subjects and control subjects, allowing us to evaluate whether our associations were independent of these critical covariates. This study also had some weaknesses. DS1 is of modest size and best-suited to detect risk factors of moderate effect. DS2 was larger, but still only modestly powered. Thus, both samples were best-suited to replicate previous findings rather than primary gene discovery. Furthermore, we used case subjects and control subjects from a variety of studies to achieve the critical sample size necessary to obtain genome-wide significance, which may have introduced genetic heterogeneity.
In summary, we provide further evidence that the association on chromosome 9p is attributable to variants in CDKN2BAS rather than CDKN2A or CDKN2B as initially reported. Furthermore, smoking increases the effect of the risk allele on chromosomes 8 and 9, and cessation of smoking may dramatically decrease the risk of IA, even among those who inherited a gene variant associated with small to moderate risk.
Supplementary Material
Acknowledgments
The authors thank the subjects and their families for participating in this research study.
Sources of Funding
This project was supported by R01NS39512, R01NS36695, K02NS060892, HHMI P0012375, HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, HHSN268201100012C, R01HL087641, R01HL59367 R01HL086694, U01HG004402, HHSN268200625226C N01HC55015, N01HC55016, N01HC55018, N01HC55019, N01HC55020, N01HC55021, N01HC55022, R01HL087641, UL1RR025005, U01 HL096917, and R01HL093029, and the Intramural Research Program of NGHRI (NIH). Addition funding was provided by the National Health and Medical Research Council of Australia and the Health Research Council of New Zealand.
Footnotes
Disclosures
None.
References
- 1.Ronkainen A, Miettinen H, Karkola K, Papinaho S, Vanninen R, Puranen M, et al. Risk of harboring an unruptured intracranial aneurysm. Stroke. 1998;29:359–362. doi: 10.1161/01.str.29.2.359. [DOI] [PubMed] [Google Scholar]
- 2.Bilguvar K, Yasuno K, Niemela M, Ruigrok YM, von Und Zu Fraunberg M, van Duijn CM, et al. Susceptibility loci for intracranial aneurysm in European and Japanese populations. Nat Genet. 2008;40:1472–1477. doi: 10.1038/ng.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yasuno K, Bakircioglu M, Low SK, Bilguvar K, Gaal E, Ruigrok YM, et al. Common variant near the endothelin receptor type A (EDNRA) gene is associated with intracranial aneurysm risk. Proc Natl Acad Sci U S A. 2011;108:19707–19712. doi: 10.1073/pnas.1117137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yasuno K, Bilguvar K, Bijlenga P, Low SK, Krischek B, Auburger, et al. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat Genet. 2010;42:420–425. doi: 10.1038/ng.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deka R, Koller DL, Lai D, Indugula SR, Sun G, Woo D, et al. The relationship between smoking and replicated sequence variants on chromosomes 8 and 9 with familial intracranial aneurysm. Stroke. 2010;41:1132–1137. doi: 10.1161/STROKEAHA.109.574640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashikata H, Liu W, Inoue K, Mineharu Y, Yamada S, Nanayakkara S, et al. Confirmation of an association of single-nucleotide polymorphism rs1333040 on 9p21 with familial and sporadic intracranial aneurysms in Japanese patients. Stroke. 2010;41:1138–1144. doi: 10.1161/STROKEAHA.109.576694. [DOI] [PubMed] [Google Scholar]
- 7.Broderick JP, Sauerbeck LR, Foroud T, Huston J, III, Pankratz N, Meissner I, et al. The familial intracranial aneurysm (FIA) study protocol. BMC Med Genet. 2005;6:17. doi: 10.1186/1471-2350-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foroud T, Sauerbeck L, Brown R, Anderson C, Woo D, Kleindorfer D, et al. Genome screen to detect linkage to intracranial aneurysm susceptibility genes: The Familial Intracranial Aneurysm (FIA) study. Stroke. 2008;39:1434–1440. doi: 10.1161/STROKEAHA.107.502930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mhurchu CN, Anderson C, Jamrozik K, Hankey G, Dunbabin D Australasian Cooperative Research on Subarachnoid Hemorrhage Study (ACROSS) Group. Hormonal factors and risk of aneurysmal subarachnoid hemorrhage: An international population-based, case-control study. Stroke. 2001;32:606–612. doi: 10.1161/01.str.32.3.606. [DOI] [PubMed] [Google Scholar]
- 10.Ko NU, Rajendran P, Kim H, Rutkowski M, Pawlikowska L, Kwok PY, et al. Endothelial nitric oxide synthase polymorphism (−786t->C) and increased risk of angiographic vasospasm after aneurysmal subarachnoid hemorrhage. Stroke. 2008;39:1103–1108. doi: 10.1161/STROKEAHA.107.496596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, et al. Genomewide association studies of stroke. N Engl J Med. 2009;360:1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinnott JA, Kraft P. Artifact due to differential error when cases and controls are imputed from different platforms. Hum Genet. 2012;131:111–119. doi: 10.1007/s00439-011-1054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longstreth WT, Jr, Nelson LM, Koepsell TD, van Belle G. Cigarette smoking, alcohol use, and subarachnoid hemorrhage. Stroke. 1992;23:1242–1249. doi: 10.1161/01.str.23.9.1242. [DOI] [PubMed] [Google Scholar]
- 17.Woo D, Khoury J, Haverbusch MM, Sekar P, Flaherty ML, Kleindorfer DO, et al. Smoking and family history and risk of aneurysmal subarachnoid hemorrhage. Neurology. 2009;72:69–72. doi: 10.1212/01.wnl.0000338567.90260.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 19.Olsson S, Csajbok LZ, Jood K, Nylen K, Nellgard B, Jern C. et Association between genetic variation on chromosome 9p21 and aneurysmal subarachnoid haemorrhage. J Neurol Neurosurg Psychiatry. 2011;82:384–388. doi: 10.1136/jnnp.2009.187427. [DOI] [PubMed] [Google Scholar]
- 20.Gschwendtner A, Bevan S, Cole JW, Plourde A, Matarin M, Ross-Adams H, et al. Sequence variants on chromosome 9p21.3 confer risk for atherosclerotic stroke. Ann Neurol. 2009;65:531–539. doi: 10.1002/ana.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biros E, Cooper M, Palmer LJ, Walker PJ, Norman PE, Golledge J. Association of an allele on chromosome 9 and abdominal aortic aneurysm. Atherosclerosis. 2010;212:539–542. doi: 10.1016/j.atherosclerosis.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holdt LM, Teupser D. Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler Thromb Vasc Biol. 2012;32:196–206. doi: 10.1161/ATVBAHA.111.232678. [DOI] [PubMed] [Google Scholar]
- 23.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stehbens WE. Histopathology of cerebral aneurysms. Arch Neurol. 1963;8:272–285. doi: 10.1001/archneur.1963.00460030056005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.