Abstract
Burkholderia pseudomallei, the causative agent of melioidosis, is intrinsically resistant to a wide range of antimicrobial agents, including β-lactams, aminoglycosides, macrolides, and polymyxins. An operon, bpeR-bpeA-bpeB-oprB, which encodes a putative repressor, a membrane fusion protein, an inner membrane protein, and an outer membrane protein, respectively, of a multidrug efflux pump of the resistance-nodulation-division family was identified in B. pseudomallei. The divergently transcribed bpeR gene encodes a putative repressor protein of the TetR family which probably regulates the expression of the bpeAB-oprB gene cluster. Comparison of the MICs and minimal bactericidal concentrations of antimicrobials for bpeAB deletion mutant KHWΔbpeAB and its isogenic wild-type parent, KHW, showed that the B. pseudomallei BpeAB-OprB pump is responsible for the efflux of the aminoglycosides gentamicin and streptomycin, the macrolide erythromycin, and the dye acriflavine. Antibiotic efflux by the BpeAB-OprB pump was dependent on a proton gradient and differs from that by the AmrAB-OprA pump in that it did not efflux the aminoglycoside spectinomycin or the macrolide clarithromycin. The broad-spectrum efflux pump inhibitor MC-207,110 did not potentiate the effectiveness of the antimicrobials erythromycin and streptomycin in B. pseudomallei.
Burkholderia pseudomallei, the causative agent of melioidosis, is a gram-negative soil bacillus endemic mainly in Southeast Asia and northern Australia; but cases have also been reported in India, China, Taiwan, and Laos (6). The disease may manifest itself as an acute, subacute, or chronic form; and the acute form of melioidosis is often fatal, despite aggressive antibiotic treatment. Treatment of severe melioidosis includes a combination of cefoperazone-sulbactam plus co-trimoxazole or ceftazidime plus co-trimoxazole (4). A high-dose intravenous ceftazidime regimen was shown to be superior to the conventional four-drug regimen (chloramphenicol, doxycycline, and trimethoprim-sulfamethoxazole) (3). Despite treatment with high-dose ceftazidime, severe melioidosis carries a mortality rate of 40% (2).
Antibiotic-resistant B. pseudomallei strains are known to emerge during the treatment of melioidosis. Such chloramphenicol- and ceftazidime-resistant B. pseudomallei strains were found to be fully virulent and frequently showed cross-resistance to other antimicrobials such as tetracyclines, sulfamethoxazole, trimethoprim, and ciprofloxacin (7).
The low success rate of the treatment of melioidosis is attributed to the fact that B. pseudomallei is intrinsically resistant to a variety of antibiotics, including β-lactams, aminoglycosides, macrolides, and polymyxins (10). Broadly specific efflux systems which are able to accommodate a variety of unrelated antimicrobial agents, including antibiotics, biocides, dyes, detergents, fatty acids, organic solvents, and homoserine lactones, are responsible for much of the intrinsic multidrug resistance in gram-negative bacteria (22). In B. pseudomallei, AmrAB-OprA, an efflux system of the resistance-nodulation-division (RND) family, has been reported to be responsible for the efflux of aminoglycosides and macrolides (18). Other members of the RND family which are responsible for the efflux of antimicrobials in gram-negative bacteria include AcrAB-TolC of Escherichia coli; the AcrAB homologue of Salmonella enterica serovar Typhimurium; MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM of Pseudomonas aeruginosa; and CeoAB-OpcM of Burkholderia cepacia (22).
In this study, we describe the identification in B. pseudomallei of the gene operon bpeR-bpeA-bpeB-oprB, which encodes a multidrug efflux system of the RND family. Gene localization, substrate specificities, and proton gradient dependence distinguish the B. pseudomallei BpeAB-OprB efflux system from that of AmrAB-OprA, although both systems efflux aminoglycosides and macrolides.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise stated, cultures were grown under aerobic conditions at 37°C in Luria-Bertani (LB) agar or LB broth (Becton Dickinson, Cockeysville, Md.). The antibiotic concentrations used for E. coli, when it was used, were as follows: ampicillin, 50 μg/ml; gentamicin, 50 μg/ml; trimethoprim, 25 μg/ml; kanamycin, 10 μg/ml; streptomycin, 50 μg/ml. Those used for B. pseudomallei were as follows: kanamycin, 200 μg/ml; trimethoprim, 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli | ||
| DH5αλpir | DH5α with a λ prophage carrying the gene encoding the p protein; Kans Tmps Gens | 16 |
| LE392 | Permissive host strain for genomic library | Promega |
| B. pseudomallei | ||
| ATCC 23343 | ATCC strain for genomic library generation | ATCC |
| KHW | Wild-type clinical isolate; Kans Tmps Genr | 5 |
| KHWΔbpeAB | KHW with ΔbpeAB; Kanr Tmps Genr | This study |
| KHWΔbpeAB(pUCP28TbpeAB) | KHWΔbpeAB complemented in trans with pUCP28TbpeAB; Kanr Tmpr Genr | This study |
| Plasmids | ||
| pGEM-T | Vector for PCR cloning; Ampr | Promega |
| pJQ200mp18 | Mobilizable allelic exchange vector; traJ sacB Genr | 25 |
| pUCP28T | Broad-host-range vector; IncP OriT; pRO1600; ori Tmpr | 34 |
| pJQ200ΔbpeABKm | pJQ200mp18 derivative carrying a 3.5-kb ApaI-SpeI fragment containing 5′ bpeA-Kmr cassette-3′ bpeB from pGEMTΔbpeABKm inserted into the SmaI site | This study |
| pUCP28TbpeAB | 4.9-kb bpeAB PCR product cloned via blunt-end ligation into pUCP28T; Tmpr | This study |
| pUTKm | Source of kanamycin resistance cassette; oriR6K mobRP4 Kanr Ampr | 8 |
Kan, kanamycin; Tmp, trimethoprim; Gen, gentamicin; Amp, ampicillin; r, resistant; s, sensitive.
Construction and screening of the B. pseudomallei DNA library.
A genomic library of B. pseudomallei ATCC 23343 (American Type Culture Collection [ATCC], Manassas, Va.) was constructed by using partially digested Sau3AI genomic DNA and XhoI-digested arms of bacteriophage λGem-12 (Promega, Madison, Wis.). DNA manipulation techniques, plaque hybridization, and extraction of bacteriophage DNA were performed as described by Sambrook and Russell (27), while the extraction of bacterial genomic DNA was done by the method described by Pitcher et al. (21). Two genomic clones were identified from a screen by using 32P-radiolabeled B. cepacia ceoA- and ceoB-specific DNA probes (Amersham Biosciences, Little Chalfont, United Kingdom), which were generated with the primers pairs ceoA3F-ceoA4R and ceoB6F-ceoB4R, respectively (Table 2).
TABLE 2.
Primers used for DNA sequencing and PCR
| Primer purpose and primer | Sequence | Purpose |
|---|---|---|
| Sequencing of bpeAB-oprB | ||
| 1. | 5′-CAGAAGCTTCTTGCGCATCGCGGGGCTCGT-3′ | |
| 2. | 5′-GGCAATCAGTTAATATCCGTCTCT-3′ | |
| 3. | 5′-CCGGGCGCGGCGAAGGTC-3′ | |
| 4. | 5′-GCGCATCGAAGAGGGCGTCAAC-3′ | |
| 5. | 5′-TGTGTCTCGCCGCGCTGTATGAAA-3′ | |
| 6. | 5′-CGGAGCACGGCGACGAC-3′ | |
| 7. | 5′-TGCAGGTCACGCAGAACACG-3′ | |
| 8. | 5′-GATCTGTCGGACCTGCGCTACA-3′ | |
| 9. | 5′-CAGAAGCTTCTTGCGCATCGCGGGGCTCGT-3′ | |
| 10. | 5′-GGCCACCGCATCGTCGTA-3′ | |
| 11. | 5′-CAAGCCCTCTTCCGCCATCAC-3′ | |
| 12. | 5′-CGGCAGGCGCACGAACAG-3′ | |
| 13. | 5′-CGCGGCCGGACGCTCGTAG-3′ | |
| 14. | 5′-AGCGTGTTCTGCGTGACCTG-3′ | |
| 15. | 5′-GCGCGATACAGGTCCACGAG-3′ | |
| 16. | 5′-TTCAGCGTGCGTCTCGTGTG-3′ | |
| PCR | ||
| CeoA3F | 5′-GACGGCGCGCTCGTGAAGAAAGG-3′ | 601-bp ceoA probe |
| CeoA4R | 5′-TCGGCCGCCGTGTAGCCGTTGTAG-3′ | |
| CeoB6F | 5′-CAGCTCGAGCGCGGGCATCGTGTT-3′ | 615-bp ceoB probe |
| CeoB4R | 5′-TCGGCCGCCGTGTAGCCGTTGTAG-3′ | |
| 17. AcrAHisF | 5′-CGCGTCGAACGGGTTCC-3′ | Full-length bpeA fragment |
| 18. AcrAHisR | 5′-CCCTGTTATTGCGCGCTCGA-3′ | |
| 19. AcrB3′3716 | 5′-ACTCGGGCCTCGTGTTCGTCA-3′ | 3′ bpeB fragment |
| 20. AcrB3′R1 | 5′-CCGTGCTCCGGCTTGTCGTC-3′ | |
| 21. AcrABpro | 5′-TTCCTCCTTCGTGCGTCTGGC-3′ | Full-length bpeAB for cloning into pUCP28T |
| 20. AcrB3′R1 | 5′-CCGTGCTCCGGCTTGTCGTC-3′ | |
| AcrA3′F3 | 5′-GCGCATCGAAGAGGGCGTCAAC-3′ | Probe for Northern blotting; hybridizes to bpeAB deletion in KHWΔbpeAB |
| AcrB5′R3 | 5′-CAAGCCCTCTTCCGCCCATCAC-3′ | |
| pUCP28TF | 5′-GCCTGCCTTTCAGGCTGCGCAACTG-3′ | Verification of trans complementation of KHWΔbpeAB |
| pUCP28TR | 5′-CGGGCAGTGAGCGCAACGCA-3′ |
The bpeR-bpeAB-oprB contig was assembled from sequences of the ceoB clone obtained by a combination of subcloning into pBluescriptII (Stratagene, La Jolla, Calif.) and primer walking (Fig. 1). Plasmid DNA was purified from overnight cultures with a Qiaprep Spin Miniprep kit (Qiagen, Hilden, Germany). PCR was performed in a PTC-100 Peltier thermal cycler (MJ Research, Waltham, Mass.) in Mg2+-free buffer containing 100 ng of template DNA, 200 μmol (each) deoxynucleoside triphosphate, 50 pmol of each primer, 1.5 mM MgSO4, and 0.5 U of Tth polymerase (Biotools, Madrid, Spain) in a total volume of 50 μl. The cycling parameters included 1 cycle at 94°C for 3 min, followed by 30 cycles at 94°C for 30 s (with 30 s of annealing at the respective temperatures; 1 min/kb of product length at 72°C) and a final extension at 72°C for 10 min. The annealing temperatures and the extension times for the different primer pairs are listed in Table 2.
FIG. 1.
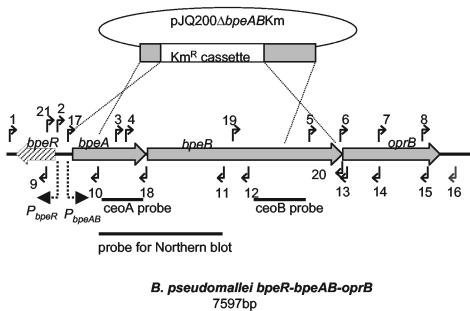
Organization of bpeR-bpeA-bpeB-oprB genes in B. pseudomallei and the locations of primers for (i) sequence determination, (ii) cloning of the full-length bpeAB for trans complementation, and (iii) verification of the allelic exchange. The thin dotted lines illustrate the allelic exchange involved in the construction of KHWΔbpeAB. Solid arrows and their numbers indicate the locations of the primers listed in Table 2, while the predicted promoters for bpeAB-oprB are represented by the thick dotted arrows. The divergently transcribed bpeR is indicated by the stippled arrow. The probes generated with the PCR primers listed in Table 2 are indicated as solid bars at the bottom.
DNA sequencing was performed with a ABI BigDye (version 3) dye terminator kit, and the DNA sequence was analyzed with an automatic sequencer (model 377; Applied Biosystems, Foster City, Calif.) (29). Multiple-sequence alignments of the translated amino acid sequences of B. pseudomallei ATCC 23343 bpeR-bpeAB-oprB with RND efflux pump components from other gram-negative bacteria were performed with the AlignX program in Vector NTI Suite 7 software (Informax Inc., Bethesda, Md.) and the BLASTX program (1). Prediction of the transmembrane domain was performed with TMHMM (version 2.0) software (17).
Construction of KHWΔbpeAB.
The bpeAB deletion was generated in B. pseudomallei KHW, a virulent clinical isolate, by the homologous gene replacement strategy described previously (5). Briefly, bpeA (1.2 kbp) and bpeB (1.3 kbp) PCR products, obtained with primer pairs AcrAHisF-AcrAHisR and AcrB3′3716-AcrB3′R1, respectively, were digested with ClaI, which cleaved the bpeA and bpeB fragments internally. A 271-bp bpeA 5′ fragment and a 989-bp bpeB 3′ fragment were recovered after electrophoresis in 1% agarose with the GeneClean II kit (Bio 101, Inc., Vista, Calif.) and ligated with T-tailed plasmid pGEM-T (Promega) to yield pGEMTΔbpeAB. Next, ClaI-linearized pGEMTΔbpeAB was made blunt ended before ligation with an end-filled 2.3-kbp EcoRI fragment containing the kanamycin resistance gene cassette from pUTKm (GenBank accession number AF102233), yielding pGEMTΔbpeABKm. Conversion of DNA fragments with 3′ recessed or overhanging ends to blunt ends with T4 DNA polymerase was performed as described by the manufacturer (Promega).
A 3.5-kbp ApaI-SpeI fragment containing the 5′ bpeA, 3′ bpeB, and kanamycin resistance cassette from pGEMTΔbpeABKm was made blunt ended and inserted into SmaI-linearized pJQ200mp18, yielding pJQ200ΔbpeABKm. pJQ200ΔbpeABKm was first introduced into E. coli DH5λpir by electroporation with a MicroPulser instrument (Bio-Rad, Hercules, Calif.) and then mobilized into B. pseudomallei KHW by triparental mating with E. coli HB101(pRK600) as the helper strain, as described by de Lorenzo et al. (8). Exconjugants were plated on LB agar containing kanamycin, streptomycin, and 5% (wt/vol) sucrose to select for recombinants which had undergone allelic exchange. The chromosomal deletion of bpeAB was confirmed by PCR and Northern blotting. PCR amplification with primers AcrABpro and AcrB3′R1 (Table 2; Fig. 1) yielded a 4.9-kbp fragment from KHW and a 3.5-kbp fragment from KHWΔbpeAB, consistent with a 3.7-kbp chromosomal deletion of bpeA-bpeB and replacement with a 2.3-kbp kanamycin resistance cassette (data not shown). The insertion of the kanamycin cassette in the bpeA-bpeB deletion was also confirmed by DNA sequencing of the PCR product obtained from KHWΔbpeAB chromosomal DNA with primer AcrABpro, as described above (data not shown). The 2.3-kbp kanamycin resistance cassette from pUTKm and a 1.6-kbp fragment from B. pseudomallei generated by PCR with primers AcrA3′F3 and AcrB5′R3 were used as probes for the Northern blotting (Fig. 1). A 521-bp 16S ribosomal DNA PCR product, generated from KHW with primer pair 16SF2 (5′-GATGACGGTACCGGAAGAATAAGC-3′) and 16SF3 (5′-CCATGTCAAGGGTAGGTAAGGTTT-3′), was used as the probe for the 16S rRNA control.
Complementation of KHWΔbpeAB mutant with wild-type bpeAB.
A 4.9-kbp full-length bpeAB PCR product, amplified from KHW genomic DNA by use of the Expand Long Template PCR system (Roche Diagnostics GmbH, Mannheim, Germany) and primers AcrABpro and AcrB3′R1 (Table 2), was blunt ended with T4 DNA polymerase and was ligated with SmaI-linearized pUCP28T, yielding pUCP28TbpeAB (Table 1). pUCP28TbpeAB was first introduced into DH5αλpir by electroporation and was subsequently mobilized into KHWΔbpeAB by conjugation, as described above. PCR with pUCP28TbpeAB isolated from complemented KHWΔbpeAB and primers pUCP28TF and pUCP28TR (Table 2) produced a 5.3-kbp product which was consistent with the presence of a full-length bpeAB gene product (5 kb), including 300 bp of flanking plasmid DNA (data not shown).
MIC and MBC determinations.
MIC determinations were carried out in 96-well microtiter plates by a standard broth microdilution method (19). Muller-Hinton broth (MHB; 5 ml; Becton Dickinson) was inoculated with 50 μl of an overnight culture, and the mixture was incubated for 4 h at 37°C with shaking. After adjustment of the culture with MHB to a 2 McFarland nephelometer standard (∼5 × 108 cells/ml), the culture was further diluted 100-fold before inoculation of 10 μl into 100 μl of MHB, yielding a final inoculum density of ∼5 × 105 cells/ml. Bacterial growth was determined 24 h after incubation at 35°C. The minimal bactericidal concentration (MBC) was determined by plating serially diluted cultures from the MIC test medium. The MBC was defined as the lowest concentration of antibiotic required to kill 99.9% of the inoculum. All antibiotics were purchased from Sigma Chemical Co. (St. Louis, Mo.).
Erythromycin accumulation assay.
The efflux of erythromycin was studied by monitoring the intracellular levels of [14C]erythromycin (NEN, Boston, Mass.) in intact cells. Overnight cultures (10 ml each) of KHW, KHWΔbpeAB, and KHWΔbpeAB(pUCP28TbpeAB) were washed three times in LB medium and were resuspended in equal volumes of fresh antibiotic-free LB medium before inoculation of 50 μl into 10 ml of antibiotic-free LB medium (optical density at 600 nm [OD600], ∼0.05). [14C]erythromycin (final concentration, 0.1 μg/ml) was added to the culture at early log phase (OD600, ∼0.5), and 1-ml aliquots were removed at 30-min intervals. The cells were washed three times in 1 ml of cold 0.9% (wt/vol) NaCl containing 1 μg of erythromycin per ml, air dried, and solubilized in 2 ml of scintillation cocktail (Amersham Biosciences) for liquid scintillation counting with an LS6500 multipurpose scintillation counter (Beckman Instruments Inc., Fullerton, Calif.). The effect of carbonyl cyanide m-chlorophenylhydrazone (CCCP; Sigma) on macrolide efflux was tested by adding CCCP at 20 μM (final concentration) to the culture for 10 min at 37°C prior to the addition of [14C]erythromycin.
Checkerboard titration assay of pump inhibitor MC-207,110.
Interactions between streptomycin, gentamicin, or erythromycin and MC-207,110 (Phe-Arg-β-naphthylamide dihydrochloride; Sigma) were assessed by a checkerboard titration assay in a 96-well microtiter plate, as described by Lomovskaya et al. (13). The antibiotics were tested at 12 twofold serial dilutions (2,048 to 0 μg/ml,), while MC-207,110 was tested at 7 twofold serial dilutions (40 to 0.625 μg/ml, including 0 μg/ml). A total of 0.2 ml of LB medium containing 5 × 105 cells/ml was added to each well, and the plate was incubated for 24 h at 37°C.
Nucleotide sequence accession number.
The GenBank nucleotide sequence accession number for bpeR-bpeA-bpeB-oprB of B. pseudomallei ATCC 23343 is AY325270.
RESULTS
B. pseudomallei bpeR-bpeAB-oprB encodes an efflux pump belonging to the RND family.
A contig of 7,597 bp containing four open reading frames (ORFs) was obtained. The ORFs were mapped to nucleotide positions 941806 to 949450 on chromosome 1 of the recently completed B. pseudomallei strain K96243 genome sequence (www.sanger.ac.uk). A difference between the ATCC 23343 sequence and that of the K96243 sequence was a 49-nucleotide GC-rich sequence located at positions 944037 to 944084 on the K96243 genome that is absent from strain ATCC 23343. The four ORFs are bpeR (636 bp), which encodes a 211-amino-acid (aa) repressor protein of the TetR family; bpeA (1,209 bp), which encodes a 402-aa periplasmic linker protein; bpeB (3,201 bp), which encodes a 1,066-aa inner membrane protein; and oprB (1,599 bp), which encodes a 532-aa outer membrane protein.
Several homologues of the RND family of efflux pumps, identified from a search of the National Center for Biotechnology Information protein database with the BLASTX program by using the BpeA, BpeB, OprB, and BpeR protein sequences as queries, were analyzed by multiple-amino-acid-sequence alignment. BpeA shared 54, 52, 42, and 22% amino acid similarities with AcrA (E. coli), MexA (P. aeruginosa), AmrA (B. pseudomallei), and CeoA (B. cepacia), respectively, while BpeB shared 65, 62, 49, and 40% amino acid similarities with AcrB (E. coli), MexB (P. aeruginosa), AmrB (B. pseudomallei), and CeoB (B. cepacia), respectively. The OprB protein shared 18, 52, and 28% amino acid similarities with TolC (E. coli), OprM (P. aeruginosa), and OpcM (B. cepacia), respectively; and BpeR shared 60, 57, and 41% amino acid similarities with AcrR (E. coli), AmrR (B. pseudomallei), and MexR (P. aeruginosa), respectively. These homologues belonged to the AcrAB-TolC, MexAB-OprM, AmrAB-OprA, and CeoAB-OpcM RND efflux pumps of E. coli, P. aeruginosa, B. pseudomallei, and B. cepacia, respectively. BpeB was also predicted to contain the conserved motifs and the characteristic structure of the inner membrane component of the RND family by having a 12-transmembrane helical domain structure with two large periplasmic loops between transmembrane segments 1-2 and 7-8 (17, 24). Putative promoter regions are located 15 and 10 bp from the start codons of bpeA-bpeB-oprM and bpeR, respectively (Fig. 1) (26).
Construction of bpeAB deletion mutant KHWΔbpeAB and trans complementation with pUCP28bpeAB.
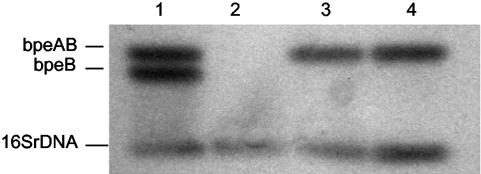
Northern blotting was performed to verify that the bpeAB deletion in KHWΔbpeAB resulted in a null mutation (Fig. 2). The mutation in KHWΔbpeAB could be complemented in trans by using plasmid pUCP28TbpeAB, which carried the full-length bpeA-bpeB sequence, with restoration of bpeAB mRNA expression (Fig. 2). No significant increase in the level of mRNA expression was detected in the complemented mutant compared to that in the wild type, even though pUCP28T was a multicopy plasmid (Fig. 2).
FIG. 2.
Northern blot analysis of bpeB mRNA expression in B. pseudomallei KHW, KHWΔbpeAB, and KHWΔbpeAB(pUCP28TbpeAB). Total RNA was extracted with the TRIzol reagent (Invitrogen, Carlsbad, Calif.). Total RNA (10 μg) was resolved on a 1% formaldehyde-agarose gel, transferred onto a nitrocellulose membrane, and probed with a 32P-radiolabeled partial bpeAB PCR product generated with AcrA3′F3-AcrB5′R3 according to the instructions of the manufacturer (Amersham) (Fig. 1; Table 2). Lane 1, a mixture of 0.5 μg each of full-length bpeAB, bpeB, and 16S RNA PCR products included as a positive control; lanes 2 to 4, 10 μg each of total RNA from KHWΔbpeAB, KHW, and KHWΔbpeAB (pUCP28TbpeAB), respectively.
Substrate specificities of the B. pseudomallei BpeAB-OprB efflux pump.
The susceptibilities of KHW and KHWΔbpeAB to a variety of antimicrobial agents were compared in an attempt to identify the substrates of the B. pseudomallei BpeAB-OprB pump. Table 3 summarizes the MICs and MBCs of the different antimicrobial agents for KHW, KHWΔbpeAB, and trans-complemented mutant KHWΔbpeAB(pUCP28TbpeAB). The parental strain, KHW, was resistant to a variety of antibiotics and agents, including aminoglycosides, macrolides, polymyxins, β-lactams, and sodium dodecyl sulfate. Intermediate resistance to DNA intercalators like ethidium bromide, crystal violet, and acriflavine was observed. Deletion of bpeAB resulted in about 1,000-fold increased susceptibilities to the aminoglycosides gentamicin and streptomycin and the macrolide erythromycin. KHWΔbpeAB remained resistant to the aminoglycoside spectinomycin and the macrolides clarithromycin and oleandomycin, showing that there was substrate selectivity even among the aminoglycosides and macrolides. KHWΔbpeAB was also more susceptible to acriflavine.
TABLE 3.
Susceptibilities of B. pseudomallei KHW, KHWΔbpeAB, and KHWΔbpeAB(pUCP28TbpeAB) to antimicrobial and other agents
| Antimicrobial or other agent | KHW
|
KHWΔbpeAB
|
KHWΔbpeAB(pUCP28TbpeAB)
|
|||
|---|---|---|---|---|---|---|
| MIC (μg/ml) | MBC (μg/ml) | MIC (μg/ml) | MBC (μg/ml) | MIC (μg/ml) | MBC (μg/ml) | |
| Aminoglycosides | ||||||
| Gentamicin | 128 | 512 | 0.125 | 0.5 | 256 | 512 |
| Streptomycin | 1,024 | 1,024 | 1 | 1 | 1,024 | 1,024 |
| Spectinomycin | 512 | >2,000 | 512 | >2,000 | 512 | >2,000 |
| Macrolides | ||||||
| Erythromycin | 128 | 1,024 | 0.125 | 0.5 | >2,000 | >2,000 |
| Clarithromycin | 1,024 | 1,024 | 1,024 | 1,024 | 1,024 | 1,024 |
| Oleandomycin | 512 | >2,000 | 512 | >2,000 | 512 | >2,000 |
| β-Lactams | ||||||
| Cloxacillin | 64 | 64 | 64 | 64 | NDa | ND |
| Amoxicillin | >2,000 | >2,000 | >2,000 | >2,000 | ND | ND |
| Piperacillin | 0.5 | 2 | 0.5 | 2 | ND | ND |
| Ceftazidime | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| Cefotaxime | 0.5 | 4 | 0.5 | 4 | ND | ND |
| Polymyxin B | 512 | 1,024 | 512 | 1,024 | ND | ND |
| Tetracyclines | ||||||
| Doxycycline | 0.5 | 256 | 0.5 | 256 | 0.5 | 256 |
| Tetracycline | 1 | 4 | 1 | 4 | 1 | 4 |
| Fluoroquinolones | ||||||
| Ofloxacin | 8 | 32 | 8 | 32 | ND | ND |
| Enoxacin | 1 | 2 | ND | ND | ND | ND |
| Chloramphenicol | 4 | 128 | 4 | 128 | 4 | 128 |
| Rifampin | 1 | 4 | 1 | 4 | 1 | 4 |
| Novobiocin | 0.5 | 1 | 0.5 | 1 | ND | ND |
| Sulfodiazine | 0.5 | >2,000 | 0.5 | >2,000 | ND | ND |
| Sulfamethoxazole | 128 | >2,000 | 128 | >2,000 | 128 | >2,000 |
| Others | ||||||
| Ethidium bromide | 128 | 128 | 128 | 128 | ND | ND |
| Crystal violet | 16 | 128 | 16 | 128 | ND | ND |
| Acriflavine | 16 | 16 | 4 | 4 | 16 | 16 |
| SDSb | 256 | 512 | 256 | 512 | ND | ND |
ND, not determined.
SDS, sodium dodecyl sulfate.
Wild-type bpeAB expression, with concomitant resistance to the antimicrobials gentamicin, streptomycin, erythromycin, and acriflavine, was restored in the complemented KHWΔbpeAB mutant, although the level of resistance to erythromycin was significantly higher in the trans-complemented strain than in the wild type (Table 3). The complemented strain was also slightly more resistant to gentamicin; but its susceptibilities to spectinomycin, streptomycin, clarithromycin, oleandomycin, ceftazidime, doxycycline, tetracycline, chloramphenicol, rifampin, and acriflavine were comparable to those of the wild type. This suggests that overexpression of BpeAB-OprB is unlikely to affect the susceptibility of B. pseudomallei to antimicrobials of therapeutic importance for melioidosis. The successful complementation of the mutant with pUCP28TbpeAB also demonstrated either that the mutation in KHWΔbpeAB did not have a polar effect on oprB or that another outer membrane efflux pump component could compensate for the OprB function.
Efflux of [14C]erythromycin.
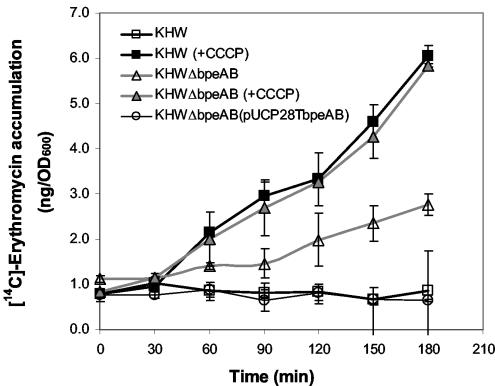
A threefold higher intracellular level of [14C]erythromycin (∼3.0 ng/OD600 unit) was observed in KHWΔbpeAB compared to the levels observed in KHW and the complemented KHWΔbpeAB mutant (0.8 ng/OD600 unit) after 3 h (Fig. 3), suggesting the involvement of the BpeAB-OprB pump in the efflux of erythromycin. The ability to restore intracellular levels of erythromycin to the wild-type levels through trans complementation of KHWΔbpeAB with full-length bpeAB DNA supports the role of bpeAB in the efflux of erythromycin. The addition of 20 μM CCCP, a proton conductor, resulted in rapid intracellular accumulation of [14C]erythromycin to about 6 ng/OD600 unit after 3 h in both the wild type and the KHWΔbpeAB mutant.
FIG. 3.
Intracellular accumulation of [14C]erythromycin by B. pseudomallei KHW, KHWΔbpeAB, and KHWΔbpeAB(pUCP28TbpeAB) in the presence and absence of 20 μM CCCP. Closed and open symbols, intracellular levels of [14C]erythromycin in CCCP-treated and untreated samples, respectively. The average standard deviation of the data was 16.5%.
Effect of broad-spectrum efflux pump inhibitor MC-207,110 on BpeAB-OprB.
The potentiating effect of the broad-spectrum efflux pump inhibitor MC-207,110 on the antimicrobial substrates of the B. pseudomallei BpeAB-OprB pump was assessed by checkerboard assays, in which the MICs of erythromycin and streptomycin were determined in the presence of different concentrations of MC-207,110 (13). In contrast to its potentiating effect on fluoroquinolones in P. aeruginosa, the addition of MC-207,110 at concentrations up to 40 μg/ml did not potentiate the antimicrobial activities of erythromycin and streptomycin in B. pseudomallei. MC-207,110 also did not have any antimicrobial activity.
DISCUSSION
The B. pseudomallei bpeR-bpeAB-oprB operon encodes a repressor of the TetR family (BpeR) and a three-component antimicrobial efflux system comprising a periplasmic linker protein (BpeA), an inner membrane protein (BpeB), and an outer membrane protein (OprB). Multiple-amino-acid-sequence alignments with homologues and secondary structure prediction showed that BpeAB-OprB pump belongs to the RND superfamily of transporters, which are known to have extremely broad substrate specificities. The large periplasmic loops of RND transporters, located between transmembrane segments 1,2 and 7,8, are believed to be involved in multidrug recognition and efflux (9, 15, 30).
In B. pseudomallei, at least one other efflux system, AmrAB-OprA, contributes to resistance to aminoglycosides and macrolides (18). Both the BpeAB-OprM pump and the AmrAB-OprA pump share some similarity with respect to substrate specificity but are distinct in their chromosomal locations and amino acid sequences. Both pumps efflux the aminoglycosides gentamicin and streptomycin and the macrolide erythromycin. We could not compare the efflux of kanamycin, which is also a substrate of the AmrAB-OprA pump, because KHWΔbpeAB was kanamycin resistant. Although both pumps have some substrates in common, their substrates differed in that B. pseudomallei strains which had either amrA or amrB deletions displayed increased susceptibilities to a wider range of aminoglycosides and macrolides, including gentamicin, kanamycin, streptomycin, spectinomycin, tobramycin, neomycin, erythromycin, and clarithromycin (18). The B. pseudomallei bpeAB deletion, in contrast, resulted in increased susceptibility to erythromycin, streptomycin, and gentamicin but not to spectinomycin and clarithromycin. Although this was likely to be attributed to differences in the substrate specificities of these two pumps, it was also possible that the inactivation of the BpeAB-OprB pump in KHWΔbpeAB might have consequentially upregulated the AmrAB-OprA pump, resulting in the higher levels of efflux of clarithromycin and spectinomycin from this strain. However, limited data on the inducibility of the AmrAB-OprA pump showed that it is not inducible by its substrate, and it is difficult to explain how the efflux of clarithromycin and spectinomycin could be selectively affected by this upregulation (18). A study on the effect of the bpeAB deletion in a strain that already lacks amrAB in order to elucidate whether these pumps have an additive effect on aminoglycoside and macrolide resistance in B. pseudomallei would be useful, since additive or multiplicative effects on drug resistance have been reported for P. aeruginosa, which has multiple efflux pumps with overlapping substrate specificities (12).
The data on the susceptibility of the complemented KHWΔbpeAB strain to antimicrobials showed that, apart from erythromycin, multiple copies of bpeAB-oprB did not increase the level of resistance of B. pseudomallei to any of the antimicrobials tested. Coupled with the absence of any significant increase in the level of bpeAB mRNA expression in the complemented mutant, this would suggest that the expression of bpeAB-oprB is tightly regulated, perhaps by an abundance of the BpeR repressor. The successful complementation of the bpeAB mutation in trans with a plasmid carrying full-length bpeAB genes showed that the bpeAB deletion in KHWΔbpeAB did not have a polar effect on oprB expression, or alternatively, another outer membrane efflux pump component could compensate for the absence of OprB in KHWΔbpeAB. For instance, the OprM outer membrane component is shared by MexAB and MexXY in P. aeruginosa, and TolC is shared by AcrAB and AcrEF in E. coli (22).
Although B. pseudomallei is intrinsically resistant to a number of antibiotics, it is also highly susceptible to many others, including piperacillin, ceftazidime, tetracycline, doxycycline, and chloramphenicol; but the choice of antimicrobials for effective treatment of melioidosis remains limited. Combinations of chloramphenicol, doxycycline, and trimethoprim-sulfamethoxazole, which have been used previously to treat confirmed cases of acute severe melioidosis (3), were ineffective because of their bacteriostatic rather than bactericidal properties and their potential toxicities. At present, ceftazidime-containing regimens, imipenem, and amoxicillin-clavulanate are the preferred therapies for acute melioidosis; but the emergence of chloramphenicol- and ceftazidime-resistant strains which are fully virulent is a cause for concern (7). The use of a combination of quinolones, such as ciprofloxacin, and macrolides has also been suggested for melioidosis therapy because ciprofloxacin could penetrate phagocytic cells and the macrolide could reduce or inhibit biofilm formation, both mechanisms of which are relevant for the treatment of melioidosis relapses (31, 32, 33). The contribution of BpeAB-OprB, as well as AmrAB-OprA, to the intrinsic resistance of B. pseudomallei to the antimicrobials gentamicin, streptomycin, and erythromycin would explain why aminoglycoside-β-lactam combinations, which are commonly used to treat suspected cases of community-acquired sepsis in many parts of the world, would be ineffective for the treatment of melioidosis (12, 14, 18, 20, 23).
It is noteworthy that in the complemented KHWΔbpeAB mutant, which carried multiple copies of bpeAB, the overexpression of BpeAB-OprB did not affect the organism's susceptibilities to the antibiotics of therapeutic importance, such as chloramphenicol, doxycycline, tetracycline, and ceftazidime, although the level of resistance to erythromycin was increased in the complemented mutant (Table 3). We also attempted to determine the frequency of occurrence of spontaneous mutants overexpressing BpeAB-OprB using the single-exposure method described by Gilbert et al. (11) but were unsuccessful in obtaining any mutants on selection media containing erythromycin at greater than twice the MIC for (data not shown). Although mutants which were twice as resistant to erythromycin, gentamicin, and streptomycin occurred at a frequency of 5 × 10−8, none of them overexpressed BpeAB-OprB in Western blots with polyclonal anti-BpeA antibodies or in promoter assays with a plasmid carrying a PbpeAB promoter-lacZ gene fusion (data not shown).
Since multidrug efflux by gram-negative bacteria is an energy-dependent process that is driven by the proton motive force (PMF), an increase in the intracellular level of accumulation of erythromycin by B. pseudomallei KHW was expected when CCCP, a proton conductor which dissipates the PMF, was added (Fig. 3) (24). However, it was unexpected that KHWΔbpeAB, which had a higher intracellular level of erythromycin than KHW, would also respond likewise when it was treated with CCCP. An explanation might be that erythromycin, which is a weak base (pKa = 8.8), was excluded from the cytoplasm due to the pH gradient across the energized cytoplasmic membrane of KHWΔbpeAB; and spontaneous influx occurred when the PMF was dissipated by CCCP, which would account for the observed increase (28). Another interpretation might be the disruption of another PMF-dependent system, such as AmrAB-OprA, which shared the same substrates with BpeAB-OprB, with the two systems perhaps functioning additively, although preliminary evidence showed that AmrAB-OprA is not sensitive to CCCP (18). It was not clear if AmrAB-OprA efficiently effluxed CCCP at the concentration used in that study. Whether AmrAB-OprA and BpeAB-OprB function additively in the efflux of aminoglycosides and macrolides in B. pseudomallei could be addressed by using an isogenic derivative with deletions in both amrAB and bpeAB.
The broad-spectrum efflux pump inhibitor MC-207,110, which was active against the MexAB-OprM, MexCD-OprJ, and MexEF-OprN pumps of P. aeruginosa, as well as the AcrAB-TolC pump of E. coli, did not have any effect on erythromycin or streptomycin efflux by BpeAB-OprM in B. pseudomallei (13). It potentiated the antimicrobial effect of the fluoroquinolone levofloxacin in P. aeruginosa but not that of fluoroquinolones in B. cepacia, a close relative of B. pseudomallei (O. Lomovskaya, personal communication). The possibility of identifying compounds which might inhibit BpeAB-OprB and potentiate the effects of antimicrobials, such as aminoglycosides, macrolides, and β-lactams, for the treatment of melioidosis is being investigated.
Acknowledgments
This work was funded by the National Medical Research Council of Singapore (grant NMRC/0426/2000). Y. Y. Chan was the recipient of a National University of Singapore Graduate Scholarship.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angus, B. J., M. D. Smith, Y. Suputtamongkol, H. Mattie, A. L. Walsh, V. Wuthiekanun, W. Chaowagul, and N. J. White. 2000. Pharmacokinetic-pharmacodynamic evaluation of ceftazidime continuous infusion vs intermittent bolus injection in septicaemic melioidosis. Br. J. Clin. Pharmacol. 49:445-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaowagul, W. 2000. Recent advances in the treatment of severe melioidosis. Acta Trop. 74:133-137. [DOI] [PubMed] [Google Scholar]
- 4.Chetchotisakd, P., S. Porramatikul, P. Mootsikapun, S. Anunnatsiri, and B. Thinkhamrop. 2001. Randomized, double-blind, controlled study of cefoperazone-sulbactam plus cotrimoxazole versus ceftazidime plus cotrimoxazole for the treatment of severe melioidosis. Clin. Infect. Dis. 33:29-34. [DOI] [PubMed] [Google Scholar]
- 5.Chua, K. L., Y. Y. Chan, and Y. H. Gan. 2003. Flagella are virulence determinants of Burkholderia pseudomallei. Infect. Immun. 71:1622-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dance, D. A. 2002. Melioidosis. Curr. Opin. Infect. Dis. 15:127-132. [DOI] [PubMed] [Google Scholar]
- 7.Dance, D. A., V. Wuthiekanun, W. Chaowagul, and N. J. White. 1989. The antimicrobial susceptibility of Pseudomonas pseudomallei. Emergence of resistance in vitro and during treatment. J. Antimicrob. Chemother. 24:295-309. [DOI] [PubMed] [Google Scholar]
- 8.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eda, S., H. Maseda, and T. Nakae. 2002. An elegant means of self-protection in gram-negative bacteria by recognizing and extruding xenobiotics from the periplasmic space. J. Biol. Chem. 278:2085-2088. [DOI] [PubMed] [Google Scholar]
- 10.Eickhoff, T. C., J. V. Bennett, P. S. Hayes, and J. Feeley. 1970. Pseudomonas pseudomallei: susceptibility to chemotherapeutic agents. J. Infect. Dis. 121:95-102. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert, D. N., S. J. Kohlhepp, K. A. Slama, G. Grunkemeier, G. Lewis, R. J. Dworkin, S. E. Slaughter, and J. E. Leggett. 2001. Phenotypic resistance of Staphylococcus aureus, selected Enterobacteriaceae, and Pseudomonas aeruginosa after single and multiple in vitro exposures to ciprofloxacin, levofloxacin, and trovafloxacin. Antimicrob. Agents Chemother. 45:883-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, A., W. Mao, M. S. Warren, A. Mistry, K. Hoshino, R. Okumura, H. Ishida, and O. Lomovskaya. 2000. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J. Bacteriol. 182:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKeegan, K. S., M. I. Borges-Walmsley, and A. R. Walmsley. 2003. The structure and function of drug pumps: an update. Trends Microbiol. 11:21-29. [DOI] [PubMed] [Google Scholar]
- 16.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moller, S., M. D. Croning, and R. Apweiler. 2001. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646-653. [DOI] [PubMed] [Google Scholar]
- 18.Moore, R. A., D. DeShazer, S. Reckseidler, A. Weissman, and D. E. Woods. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43:465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7-A6 and MIC testing supplemental tables M100-S13, 6th ed., vol. 23, no. 2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Nikaido, H. 1998. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin. Infect. Dis. 27(Suppl. 1):S32-S41. [DOI] [PubMed] [Google Scholar]
- 21.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 22.Poole, K. 2001. Multidrug resistance in gram-negative bacteria. Curr. Opin. Microbiol. 4:500-508. [DOI] [PubMed] [Google Scholar]
- 23.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Putman, M., H. W. Van Veen, J. E. Degener, and W. N. Konings. 2000. Antibiotic resistance: era of the multidrug pump. Mol. Microbiol. 36:772-773. [DOI] [PubMed] [Google Scholar]
- 25.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 26.Reese, M. G. 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 26:51-56. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sanchez, L., W. Pan, M. Vinas, and H. Nikaido. 1997. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J. Bacteriol. 179:6855-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tikhonova, E. B., Q. Wang, and H. I. Zgurskaya. 2002. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 184:6499-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vorachit, M., P. Chongtrakool, S. Arkomsean, and S. Boonsong. 2000. Antimicrobial resistance in Burkholderia pseudomallei. Acta Trop. 74:139-144. [DOI] [PubMed] [Google Scholar]
- 32.Vorachit, M., K. Lam, P. Jayanetra, and J. W. Costerton. 1995. Electron microscopy study of the mode of growth of Pseudomonas pseudomallei in vitro and in vivo. J. Trop. Med. Hyg. 98:379-391. [PubMed] [Google Scholar]
- 33.Vorachit, M., K. Lam, P. Jayanetra, and J. W. Costerton. 1993. Resistance of Pseudomonas pseudomallei growing as a biofilm on silastic discs to ceftazidime and co-trimoxazole. Antimicrob. Agents Chemother. 37:2000-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]