Abstract
The estrogen-related receptors (ERRα, ERRβ, and ERRγ) form a family of orphan nuclear receptors that share significant amino acid identity with the estrogen receptors, but for which physiologic roles remain largely unknown. By using a peptide sensor assay, we have identified the stilbenes diethylstilbestrol (DES), tamoxifen (TAM), and 4-hydroxytamoxifen (4-OHT) as high-affinity ligands for ERRγ. In direct binding assays, 4-OHT had a Kd value of 35 nM, and both DES and TAM displaced radiolabeled 4-OHT with Ki values of 870 nM. In cell-based assays, 4-OHT binding caused a dissociation of the complex between ERRγ and the steroid receptor coactivator-1, and led to an inhibition of the constitutive transcriptional activity of ERRγ. ERRα did not bind 4-OHT, but replacing a single amino acid predicted to be in the ERRα ligand-binding pocket with the corresponding ERRγ residue allowed high-affinity 4-OHT binding. These results demonstrate the existence of high-affinity ligands for the ERR family of orphan receptors, and identify 4-OHT as a molecule that can regulate the transcriptional activity of ERRγ.
Nuclear receptors are ligand-activated transcription factors that play critical roles in many aspects of development and adult physiology. Common structural features of nuclear receptors include a central, highly conserved DNA-binding domain (DBD) and a carboxyl-terminal ligand-binding domain (LBD) that contains both a hydrophobic ligand-binding pocket and a transcriptional activation function known as AF-2. Ligand binding induces a conformational change in the LBD, which, in turn, allows the binding of coactivator proteins (1). The binding of coactivators depends on the presence of a short leucine-rich domain with the consensus sequence motif LXXLL (2). Indeed, peptides that contain the LXXLL motif and are as short as eight amino acids can bind to nuclear receptors in a ligand-dependent fashion (3). Many issues concerning the formation and function of nuclear receptor–coactivator complexes remain unresolved, but one emerging theme is that coactivator recruitment brings histone acetyltransferase activity to the transcription complex (4). This activity presumably alters chromatin structure and allows for efficient expression of target genes.
The same general concepts of ligand activation and coactivator interaction apply not only to the classical nuclear receptors but also to a growing list of orphan receptors for which natural or synthetic ligands have recently been identified. This list includes the peroxisome proliferator-activated receptors, the liver X receptors, and the farnesoid X receptor, which activate transcription in response to binding fatty acids (5), oxysterols (6, 7), and bile acids (3, 8), respectively. In addition, two other orphan nuclear receptors, the constitutive androstane receptor and the pregnane X receptor, have been shown to regulate the expression of cytochrome-P450 genes in response to a variety of ligands, including the planar hydrocarbon 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP), the antimycotic clotrimazole, and the steroid 5β-pregnane-3,20-dione (9, 10).
In contrast, the estrogen-related receptors (ERRα, ERRβ, and ERRγ) are orphan nuclear receptors for which natural ligands have yet to be identified. These receptors were initially identified because of their high degree of DNA sequence homology to the estrogen receptor (ER) (11), and each of the three subtypes shows a considerable level of amino acid sequence identity with ER in both the DBD and LBD (11–13). Although the ERRs do not directly respond to β-estradiol (11, 12, 14), they can bind to functional estrogen response elements (EREs) in ER target genes such as lactoferrin (15) and aromatase (16), suggesting a possible overlap between ERR and ER biology. Although natural ligands have not yet been described for the ERRs, two lines of evidence suggest that these receptors may be hormone-regulated. First, Vanacker and colleagues (17) have observed that fetal calf serum contains a factor or factors that can stimulate ERRα basal activity by 12-fold, and second, Yang and Chen (18) have reported that micromolar concentrations of the pesticides toxaphene and chlordane decrease ERRα basal activity.
To facilitate the discovery of ERR ligands, we have adopted a well-established fluorescence resonance energy transfer (FRET) assay (19) to measure changes in the interaction between the ERR LBD and coactivator peptides in response to ligand binding. By using this assay, we determined that the estrogen diethylstilbestrol (DES) and the antiestrogens tamoxifen (TAM) and 4-hydroxytamoxifen (4-OHT) bind to ERRγ with submicromolar affinities. In contrast, DES showed only low-affinity binding to ERRα, whereas TAM and 4-OHT showed no measurable binding. These results were further confirmed in a direct binding assay that uses [3H]4-OHT as the radioligand. Consistent with the biochemical assay results, 4-OHT repressed transactivation mediated by ERRγ, but not ERRα or ERRβ. Changing a single amino acid in the ERRα ligand-binding pocket to the corresponding ERRγ residue conferred binding of ERRγ ligands to the mutant ERRα. These results demonstrate that 4-OHT can bind to and deactivate ERRγ.
Materials and Methods
Constructs.
Full-length ERRα was amplified from a human kidney cDNA library by PCR using primers opc21 (5′-TTGAATTCGCCATGTCCAGCCAGGTGGTGGGC-3′) and ERRαREV (5′- GCCGGATCCTCAGTCCATCATGGCCTCGAG-3′). Full-length ERRβ was amplified from a human kidney cDNA library by using nested PCR and primers opc26 (5′-GAGGGCTGCTGAACAGGATGTC-3′) and opc23 (5′-GGCTCGAGCTAAGCTGCTCTTGGCCAACCTGC-3′) for the first reaction, and primers opc22 (5′-TTGAATTCGCCATGTCCTCGGACGACAGGCACCTG-3′) and opc23 in the second reaction. Full-length ERRγ was amplified from a human heart cDNA library by using primers opc6 (5′-TTGAATTCGCCATGTCAAACAAAGATCGACACATT-3′) and opc8 (5′-GGCTCGAGTTAGCAGACCTTGGCCTCCAACAT-3′). All full-length cDNAs were cloned into pcDNA3 (Invitrogen). For glutathione S-transferase (GST) fusion protein production, each LBD was amplified and cloned into pGEX4T-1 (Amersham Pharmacia). The primers used were: ERRC (5′-TATGAGCCCGGGAAGCAGCAGCCTCGAGGCCATGATGGACTAAA-3′) and ERRAREV for ERRα, opc30 (5′-CCATATGCTGAGCTTACAAATTTCTCCACC-3′) and opc37 (5′-TTCTCGAGCTAGGCCTTGGCCTCCAGCATCTC-3′) for ERRβ, and opc34 (5′-TTCATATGCTGAACCCTCAGCTGGTTCAGCCA-3′) and opc8 for ERRγ. In the case of ERRβ, the F domain was eliminated to achieve better recombinant protein expression. For mammalian two-hybrid experiments, each LBD was amplified and cloned downstream of the herpesvirus VP16 transactivation domain. The primers used were: ERRA241 (5′-GCCGGATCCATGCTCAAGGAGGGAGTGCGC-3′) and ERRAREV for ERRα, opc29 (5′-GGGATCCATGCTGAAGGAAGGTGTGCGCC-3′) and opc23 for ERRβ (includes the F domain), and opc28 (5′-GGGATTCATGCTGAAAGAAGGGGTGCGTC-3′) and opc8 for ERRγ. To generate the 3× ERE-luciferase construct, oligonucleotides opc31 (5′-GATCTAGGTCACAGTGACCTGCG-3′), and opc32 (5′-GATCCGCAGGTCACTGTGACCTA-3′) were annealed, ligated, and digested with BamHI, and then ligated into BglII-digested pGL3TK-Luc. Sequencing confirmed the presence of three copies of the ERE. The ERRαF232A mutant was generated from pGST-ERRα by using the Quickchange mutagenesis kit (Stratagene) and primers opc64 (5′-GGCTACCCTCTGTGACCTCGCTGACCGAGAGATTGTGGTC-3′) and opc65 (5′-GACCACAATCTCTCGGTCAGCGAGGTCACAGAGGGTAGCC-3′). The sequences of all PCR products were verified before use.
FRET Assays.
Reactions contained europium-labeled anti-GST antibody and streptavidin-conjugated allophycocyanin (both from Perkin-Elmer), ERR GST-LBD fusion proteins (2 nM), and biotinylated SRC1.2 sensor peptide. The sequence of SRC-1.2 is SLTERHKILHRLLQEGSPSDI. Reactions were incubated at room temperature for 1 h in FRET buffer (10 mM Hepes, pH 7.9/150 mM NaCl/2 mM MgCl2/1 mM EDTA/0.1 mg/ml BSA). FRET was measured on a Victor 1420 multilabel counter (Wallac, Gaithersburg, MD).
Radioligand Binding Assays.
Binding assays were performed in the presence of 0.1 mg per well glutathione-coated scintillation proximity assay (SPA) beads (Amersham Pharmacia) and 1 μg per well GST-ERRγ or GST-ERRαF232A LBD in SPA buffer [10 mM Hepes (pH 7.5)/50 mM NaCl/2 mM MgCl2/1 mM EDTA/2 mM CHAPS/0.1 mg/ml BSA/1 mM DTT]. For competition binding assays, [3H]4-OHT (70 Ci/mmol, Amersham Pharmacia; 1 Ci = 37 GBq) was diluted in SPA buffer and added to the wells at a final concentration of 1 nM. Reactions were incubated for 3 h at room temperature and measured on a Packard Topcount. Saturation binding experiments were performed in the presence or absence of 30 μM unlabeled 4-OHT.
Reporter Gene Assays.
All transfection experiments were performed in CV-1 cells in 24 well plates, by using Lipofectamine (Life Technologies, Rockville, MD) according the manufacturer's protocol. In each case, 500 ng of DNA and 5 μl of Lipofectamine were used per well, and pCMVβGal (CLONTECH) was included as a control for transfection efficiency. The total amount of DNA added was kept constant by adding the appropriate amount of pBluescript (Stratagene) to the transfection reaction. For mammalian two-hybrid experiments, 100 ng ERR LBD-VP16 construct, 10 ng of GAL4 DBD-SRC fusion construct, 100 ng of pG5-Luc (3), and 50 of ng pCMVβgal were added per well. For transfections with full-length receptors, 50 ng of receptor, 100 ng of 3× ERE, and 50 ng of pCMVβgal were added to each well. Cells were transfected for 5 to 16 h. Compound treatment lasted for 20 h.
Results and Discussion
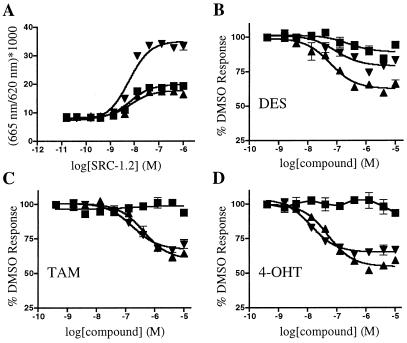
In an effort to identify ERR ligands, we adopted a biochemical FRET assay used by others to characterize nuclear receptor-ligand interactions (3, 8, 19). GST ERR-LBD fusion proteins were labeled with anti-GST antibody coupled to europium chelate and incubated with allophycocyanin coupled to streptavidin and biotinylated peptides of 23 amino acids containing an LXXLL motif. An optimal signal was obtained with a peptide that matched the sequence of the second nuclear receptor-box of the steroid receptor coactivator-1 (SRC-1), and this peptide, SRC-1.2, was used in all subsequent FRET experiments. Consistent with previous findings (13, 20), all three ERR LBDs interacted constitutively with the SRC-1.2 peptide, although ERRγ showed a higher level of FRET than ERRα or ERRβ (Fig. 1A). In comparison, significant interaction between the ER LBD and the SRC-1.2 peptide was seen only in the presence of β-estradiol (data not shown). Dose–response analysis showed that the SRC-1.2 peptide bound to all three ERR LBDs with similar EC50 values of ≈10 nM. None of the receptors interacted with SRC-1.2m, a mutant peptide where the sequence LHRLLhad been changed to LHRAA (data not shown).
Figure 1.
Development of a FRET assay for ERR family members. (A) Coactivator peptide interacts with ERR LBDs in the FRET assay. SRC-1.2 peptide shows a dose-dependent, constitutive interaction with ERRα (■), ERRβ (▴), and ERRγ (▾) LBDs. (B–D) DES, TAM, and 4-OHT disrupt the interaction between SRC-1.2 and ERR LBDs in the FRET assay. Various concentrations of DES (B), TAM (C), or 4-OHT (D) were added to 2 nM of ERRα (■), ERRβ (▴), and ERRγ (▾) and 300 nM SRC-1.2. EC50 values for each compound are listed in the text. For each panel, the data are from a single experiment preformed in duplicate. At least two additional experiments gave similar results.
By using the ERRγ LBD fusion protein and 300 nM SRC-1.2 peptide (an amount that produced maximal interaction with the unliganded receptor; see Fig. 1A), we screened a large panel of known nuclear receptor ligands and structurally related compounds at a concentration of 10 μM. We found that DES, TAM, and 4-OHT disrupted the ERRγ–SRC-1.2 interaction, whereas none of the other compounds, including glucocorticoids, retinoids, androstanes, and bile acids, showed any significant effect (data not shown). In dose–response experiments (Fig. 1), the EC50 values for DES, TAM, and 4-OHT were 700 nM, 400 nM, and 50 nM, respectively. When tested on ERRβ, the EC50 values for DES, TAM, and 4-OHT were 700 nM, 950 nM, and 150 nM, respectively. In contrast, on ERRα the EC50 value for DES was 10 μM, and TAM and 4-OHT showed no activity. Similar results were also obtained with a biotinylated SRC-1 peptide fragment (amino acids 215–442) that contains all three nuclear receptor boxes (data not shown). In comparison, DES increased, and TAM and 4-OHT decreased, the interaction between ERβ and the SRC-1.2 peptide, each with an EC50 value of ≈1 nM, the limit of sensitivity of this assay (data not shown). These values are in good agreement with previously determined binding constants for these compounds (21, 22). Together, these data demonstrate that the stilbenes DES, TAM, and 4-OHT function as ERRβ and ERRγ inverse agonists, and indicate a close relationship between the ligand binding properties of ERs and ERRs.
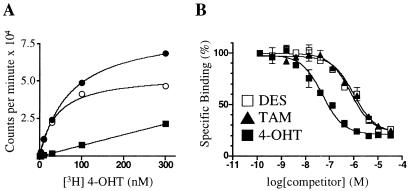
To confirm the results of the FRET experiments and to measure ligand binding directly, we established a scintillation proximity assay (23) for ERRγ that uses [3H]4-OHT as the radioligand. As shown in Fig. 2, the binding affinity, or Kd value, of [3H]4-OHT for ERRγ was 35 nM. In competition binding experiments, both DES and TAM displaced the radioligand with Ki values of 870 nM, whereas 4-OHT had a Ki of 75 nM. These binding data are in good agreement with the results of the FRET experiments. We then tested the ability of several endogenous and synthetic estrogens, including β-estradiol, 2- and 4-hydroxyestradiol, estriol, genistein, resveratrol, and raloxifene to bind ERRγ, but none showed any significant radioligand displacement at concentrations up to 30 μM, the highest tested (data not shown). Consistent with the results of the FRET experiments, [3H]4-OHT showed no specific binding to ERRα at concentrations up to 1 μM, the highest concentration tested (data not shown).
Figure 2.
A radioligand binding assay for ERRγ. (A) Saturation binding curve of [3H]4-OHT and GST-ERRγ. The graph shows total (●), specific (○), and nonspecific (■) binding. Excess unlabeled 4-OHT (30 μM) was used to determine nonspecific binding. The Kd of 4-OHT was 35 nM. (B) Nonradioactive DES, TAM, and 4-OHT compete with [3H]4-OHT for binding to ERRγ. The Ki value for DES and TAM was 870 nM; for 4-OHT the Ki was 75 nM. These data are from a single experiment preformed in duplicate. Two additional experiments gave similar results.
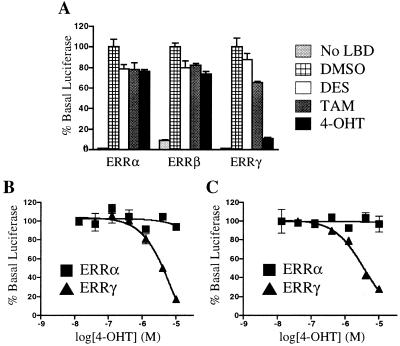
Next we wanted to determine whether DES, TAM, and 4-OHT also disrupt ERRγ–coactivator interaction in the context of a whole cell. For this experiment, we used the mammalian two-hybrid system, in which the coactivator SRC-1 is fused to the DNA-binding domain of the yeast transcription factor GAL4, and the herpes simplex virus VP16 transactivation domain is fused to the ERRγ LBD. As expected, the ERRγ LBD showed strong constitutive interaction with the GAL4-SRC fusion protein (Fig. 3A). Although DES and TAM showed a modest activity (35% inhibition at 10 μM), 4-OHT caused a significant (75%) disruption of the receptor-coactivator complex, with half-maximal inhibition achieved at a concentration of 2 μM (Fig. 4). These compounds did not reproducibly disrupt the interaction of coactivator with ERRα or ERRβ at concentrations up to 10 μM (Fig. 3 and data not shown). Thus, although all three stilbenes bind to ERRγ, only the most potent ligand, 4-OHT, showed significant disruption of the constitutive interaction between ERRγ and SRC-1.
Figure 3.
Cell-based activity of DES, TAM, and 4-OHT on ERR family members. CV1 cells were transfected as described in Materials and Methods. (A) Mammalian two-hybrid assay. Compounds were added at 10 μM. (B) Dose-response curves of 4-OHT on ERRα and ERRγ in the mammalian two-hybrid assay. (C) Dose-response curve of 4-OHT on full-length ERRα and ERRγ when a 3× ERE promoter-driven reporter gene was used. In all cases, luciferase activity was normalized to β-galactosidase activity. These data are from a single experiment preformed in triplicate. Two additional experiments gave similar results.
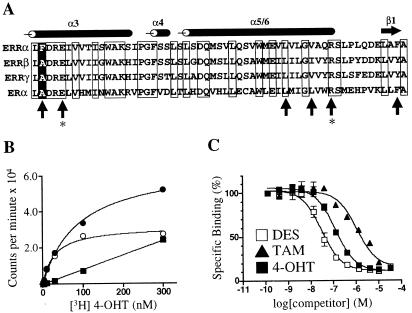
Figure 4.
ERRαF232A has binding characteristics similar to those of ERRγ. (A) Alignment of the ERR and ER LBDs. The sequence begins with amino acid 231 for ERRα, 246 for ERRβ, 248 for ERRγ, and 349 for ERα. The positions of the α-helices and β-sheet are noted above the alignment. Conserved residues are boxed. Upright arrows indicate ER residues involved in recognition of DES and 4-OHT (26). The positions of glutamic acid-353 and arginine-394 in ERα, responsible for hydrogen bonding to the phenolic hydroxyl of the A ring of 4-OHT, are indicated by asterisks. The position of phenylalanine-232 in ERRα is indicated by a white letter on a black background. (B) Saturation binding curve of [3H]4-OHT and GST-ERRαF232A. The graph shows total (●), specific (○), and nonspecific (■) binding. Excess unlabeled 4-OHT (30 μM) was used to determine nonspecific binding. The Kd of 4-OHT was 40 nM. (C) Nonradioactive DES, TAM, and 4-OHT compete with [3H]4-OHT for binding to ERRαF232A. Ki values were 30 nM, 750 nM, and 110 nM, respectively. These data are from a single experiment preformed in duplicate. Two additional experiments gave similar results.
To further investigate the effect of 4-OHT on ERRγ activity, we transiently transfected CV-1 cells with an expression plasmid encoding full-length ERRγ together with a reporter plasmid containing three copies of the vitellogenin A2 ERE (24). Consistent with the results obtained in the mammalian two-hybrid experiment, 4-OHT decreased the transcriptional activity of ERRγ by more than 75%, with an EC50 value of 2 μM (Fig. 3C). Similar decreases in ERRγ basal activity in response to 4-OHT were also seen when a different reporter plasmid containing three copies of the SF-1 response element was used (25) (data not shown). No significant repression of reporter gene activity was seen in cells transfected either with full-length ERRα (Fig. 3C) or with a cytomegalovirus-luciferase reporter plasmid (data not shown), indicating that the repression was not the result of a nonspecific decrease in luciferase activity.
The LBDs of ERRα and ERRγ share about 60% amino acid identity, yet ERRα did not bind to 4-OHT. Sequence alignment of the ERR and ERα LBDs (Fig. 4A) pointed to one residue as potentially playing a significant role in this difference in binding affinity. This residue, located at the bottom of helix 3, is an alanine in ERα, ERRβ, and ERRγ, each of which bound 4-OHT, but is a phenylalanine in ERRα, which did not bind 4-OHT. A homology model of the ERRα LBD derived from the crystal structure of ERα bound to 4-OHT (26) provided a possible explanation for the lack of binding of 4-OHT to ERRα. Replacing the alanine at ER position 350 with phenylalanine would be predicted to effectively block the phenolic hydroxyl of the A ring of 4-OHT from hydrogen bonding to the arginine at position 394 and glutamic acid at position 353. Conversely, replacing the phenylalanine at ERRα position 232 with alanine would presumably open that part of the pocket and allow 4-OHT to bind to ERRα. To test this model, we established a [3H]4-OHT radioligand binding assay with recombinant ERRαF232A protein, under the same conditions used previously for ERRγ (see Fig. 2). As shown in Fig. 4, ERRαF232A bound to [3H]4-OHT with a Kd of 40 nM, similar to the affinity shown for ERRγ. The Ki value for TAM was 750 nM, again similar to ERRγ. For DES, however, the Ki value was 30 nM, ≈25× lower than observed with ERRγ, indicating that the ligand binding properties of these two receptors are similar but not identical. These results are consistent with the model presented above, and provide further evidence that 4-OHT is binding in the ligand-binding pocket.
It is not clear what physiologic effects would be expected of a compound that regulates ERRγ activity, as there have been no reports of specific ERRγ target genes. Several target genes have been proposed for ERRα, however, including lactoferrin (15), aromatase (16), osteopontin (27, 28), medium chain acyl-CoA dehydrogenase (29, 30), and thyroid hormone receptor α (31). Because of the ≈90% amino acid identity in the ERRα, ERRβ, and ERRγ DBDs, these genes are also potential ERRγ and/or ERRβ target genes. The tissue expression patterns of ERRγ and ERRα are widespread and, at least in part, overlapping, with high levels of expression seen in heart, brain, kidney, and placenta (11–13, 29). ERRβ is predominately expressed during embryonic development in the extra-embryonic ectoderm and chorion, and in the adult in the heart, brain, and kidney, but at levels 10–100× lower than ERRα (11, 32). The overlap seen in the tissue expression patterns and in the DNA and ligand-binding characteristics of the ERRs highlights the need for potent and selective compounds to determine the roles of the individual receptors in ERR and, possibly, ER biology. The identification of 4-OHT as a potent ERRγ ligand with cell-based activity represents an important first step toward meeting this need.
Surprisingly, 4-OHT was the only compound tested that showed significant activity in cell-based assays, and this activity was seen only with ERRγ. It is not clear why this should be the case, but a simple explanation is that 4-OHT was the most potent compound tested, and its affinity for ERRγ (35 nM) represents the minimum needed for activity in these assays. It is also possible that the cell-based assays we have used, although well established for studying the ligand regulation of other nuclear receptors, are not optimized for the ERRs. This possibility may explain the ≈50-fold difference we see in the Kd of 4-OHT in the radioligand binding assay and the EC50 in the cell-based assays. Development of new assays, perhaps by using reporter genes driven by physiologically relevant promoter elements, reporter genes stably integrated into the host chromosome, or measuring the transcriptional regulation of endogenous genes, may result in more potent cell-based activities. Developments of this sort may also result in the identification of additional compounds that regulate ERR activity in tissue culture cells or in animals.
In the clinic, TAM is indicated for the treatment of breast cancer, and patients taking the recommended dose of 20 mg per day achieve steady-state plasma levels that average 320 nM (33), within 3-fold of the 870 nM Ki for ERRγ. For DES, indicated for the treatment of certain breast and prostate cancers, the recommended dose ranges from 1 to 15 mg per day. At these levels of exposure, TAM and DES may be physiologically relevant ERR ligands, and some changes in gene expression in response to TAM or DES treatment could be mediated by ERRγ and/or ERRβ, adding an additional level of complexity to the pharmacology of ER ligands.
Acknowledgments
We thank Miki Rich for DNA sequencing, and Drs. Tim Hoey, Andy Shiau, and Hui Tian for helpful discussions and critical review of the manuscript. The member who communicated this paper is Chief Executive Officer and major stockholder of Tularik, Inc.
Abbreviations
- ERR
estrogen-related receptor
- DES
diethylstilbestrol
- TAM
tamoxifen
- 4-OHT
4-hydroxytamoxifen
- SRC-1
steroid receptor coactivator-1
- LBD
ligand-binding domain
- ER
estrogen receptor
- FRET
fluorescence resonance energy transfer
- ERE
estrogen response element
- GST
glutathione S-transferase
Note
While this paper was in review, Tremblay et al. (34) reported that DES functions as an inverse agoinst on all three ERRs, and that pregnant wild-type mice treated with DES, but not estrogen, exhibit placental abnormalities similar to those seen in ERRβ-null mice. These results provide additional evidence for the ability of stilbene compounds to regulate the activity of the ERRs.
References
- 1.Freedman L P. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 2.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 3.Makishima M, Okamoto A Y, Repa J J, Tu H, Learned R M, Luk A, Hull M V, Lustig K D, Mangelsdorf D J, Shan B. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 4.Xu L, Glass C K, Rosenfeld M G. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 5.Willson T M, Brown P J, Sternbach D D, Henke B R. J Med Chem. 2000;43:527–550. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann J M, Kliewer S A, Moore L B, Smith-Oliver T A, Oliver B B, Su J L, Sundseth S S, Winegar D A, Blanchard D E, Spencer T A, Willson T M. J Biol Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- 7.Janowski B A, Willy P J, Devi T R, Falck J R, Mangelsdorf D J. Nature (London) 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 8.Parks D J, Blanchard S G, Bledsoe R K, Chandra G, Consler T G, Kliewer S A, Stimmel J B, Willson T M, Zavacki A M, Moore D D, Lehmann J M. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 9.Moore L B, Parks D J, Jones S A, Bledsoe R K, Consler T G, Stimmel J B, Goodwin B, Liddle C, Blanchard S G, Willson T M, et al. J Biol Chem. 2000;275:15122–15127. doi: 10.1074/jbc.M001215200. [DOI] [PubMed] [Google Scholar]
- 10.Waxman D J. Arch Biochem Biophys. 1999;369:11–23. doi: 10.1006/abbi.1999.1351. [DOI] [PubMed] [Google Scholar]
- 11.Giguere V, Yang N, Segui P, Evans R M. Nature (London) 1988;331:91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- 12.Heard D J, Norby P L, Holloway J, Vissing H. Mol Endocrinol. 2000;14:382–392. doi: 10.1210/mend.14.3.0431. [DOI] [PubMed] [Google Scholar]
- 13.Hong H, Yang L, Stallcup M R. J Biol Chem. 1999;274:22618–22626. doi: 10.1074/jbc.274.32.22618. [DOI] [PubMed] [Google Scholar]
- 14.Vanacker J M, Pettersson K, Gustafsson J A, Laudet V. EMBO J. 1999;18:4270–4279. doi: 10.1093/emboj/18.15.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang N, Shigeta H, Shi H, Teng C T. J Biol Chem. 1996;271:5795–5804. doi: 10.1074/jbc.271.10.5795. [DOI] [PubMed] [Google Scholar]
- 16.Yang C, Zhou D, Chen S. Cancer Res. 1998;58:5695–5700. [PubMed] [Google Scholar]
- 17.Vanacker J M, Bonnelye E, Chopin-Delannoy S, Delmarre C, Cavailles V, Laudet V. Mol Endocrinol. 1999;13:764–773. doi: 10.1210/mend.13.5.0281. [DOI] [PubMed] [Google Scholar]
- 18.Yang C, Chen S. Cancer Res. 1999;59:4519–4524. [PubMed] [Google Scholar]
- 19.Zhou G, Cummings R, Li Y, Mitra S, Wilkinson H A, Elbrecht A, Hermes J D, Schaeffer J M, Smith R G, Moller D E. Mol Endocrinol. 1998;12:1594–1604. doi: 10.1210/mend.12.10.0176. [DOI] [PubMed] [Google Scholar]
- 20.Xie W, Hong H, Yang N N, Lin R J, Simon C M, Stallcup M R, Evans R M. Mol Endocrinol. 1999;13:2151–2162. doi: 10.1210/mend.13.12.0381. [DOI] [PubMed] [Google Scholar]
- 21.Kuiper G G, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, Gustafsson J A. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Teng C T. J Biol Chem. 2000;275:20837–20846. doi: 10.1074/jbc.M001880200. [DOI] [PubMed] [Google Scholar]
- 23.Nichols J S, Parks D J, Consler T G, Blanchard S G. Anal Biochem. 1998;257:112–119. doi: 10.1006/abio.1997.2557. [DOI] [PubMed] [Google Scholar]
- 24.Klein-Hitpass L, Schorpp M, Wagner U, Ryffel G U. Cell. 1986;46:1053–1061. doi: 10.1016/0092-8674(86)90705-1. [DOI] [PubMed] [Google Scholar]
- 25.Wilson T E, Fahrner T J, Milbrandt J. Mol Cell Biol. 1993;13:5794–5804. doi: 10.1128/mcb.13.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 27.Bonnelye E, Vanacker J M, Dittmar T, Begue A, Desbiens X, Denhardt D T, Aubin J E, Laudet V, Fournier B. Mol Endocrinol. 1997;11:905–916. doi: 10.1210/mend.11.7.9948. [DOI] [PubMed] [Google Scholar]
- 28.Vanacker J M, Delmarre C, Guo X, Laudet V. Cell Growth Differ. 1998;9:1007–1014. [PubMed] [Google Scholar]
- 29.Sladek R, Bader J A, Giguere V. Mol Cell Biol. 1997;17:5400–5409. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vega R B, Kelly D P. J Biol Chem. 1997;272:31693–31699. doi: 10.1074/jbc.272.50.31693. [DOI] [PubMed] [Google Scholar]
- 31.Vanacker J M, Bonnelye E, Delmarre C, Laudet V. Oncogene. 1998;17:2429–2435. doi: 10.1038/sj.onc.1202167. [DOI] [PubMed] [Google Scholar]
- 32.Luo J, Sladek R, Bader J A, Matthyssen A, Rossant J, Giguere V. Nature (London) 1997;388:778–782. doi: 10.1038/42022. [DOI] [PubMed] [Google Scholar]
- 33.Anonymous. Physician's Desk Reference. 52nd Ed. Inc., Montvale, NJ: Medical Economics Company; 1998. pp. 3175–3177. [Google Scholar]
- 34.Tremblay G B, Kunath T, Bergeron D, Lapointe L, Champigny C, Bader J A, Rossant J, Giguere V. Genes Dev. 2001;15:833–838. doi: 10.1101/gad.873401. [DOI] [PMC free article] [PubMed] [Google Scholar]