Abstract
NF-E2-related factor 2 (Nrf2) is an important transcription factor involved in antioxidant response. Nrf2 binds antioxidant response elements (ARE) within promoters of genes encoding detoxification enzymes (e.g., NAD (P) H-quinone oxidoreductase 1 (NQO1)) leading to their transcriptional activation. Nrf2 function is regulated post-translationally by its negative regulator Kelch-like ECH-associated protein 1 (Keap1) that binds Nrf2 and induces cytoplasmic Nrf2 degradation. Our present studies provide new evidence that Nrf2 expression can be regulated by a Keap1-independent mechanism. Here, we utilized breast epithelial cells to explore the impact of microRNA (miRNA) on Nrf2 expression. We found that Nrf2 mRNA levels are reversibly correlated with miR-28 expression and that ectopic expression of miR-28 alone reduces Nrf2 mRNA and protein levels. We further investigated the molecular mechanisms by which miR-28 inhibits Nrf2 mRNA expression. Initially, the ability of miR-28 to regulate the 3′ untranslated region (3′UTR) of Nrf2 mRNA was evaluated via luciferase reporter assay. We observed that miR-28 reduces wild-type Nrf2 3′UTR luciferase reporter activity and this repression is eliminated upon mutation of the miR-28 targeting seed sequence within the Nrf2 3′UTR. Moreover, over-expression of miR-28 decreased endogenous Nrf2 mRNA and protein expression. We also explored the impact of miR-28 on Keap1-Nrf2 interactions and found that miR-28 overexpression does not alter Keap1 protein levels and has no effect on the interaction of Keap1 and Nrf2. Our findings, that miR-28 targets the 3′UTR of Nrf2 mRNA and decreases Nrf2 expression, suggest that this miRNA is involved in the regulation of Nrf2 expression in breast epithelial cells.
Keywords: Mammary epithelial cells, miR-28, Nrf2, Chemoprevention
Introduction
Nrf2 is a key transcription factor that regulates the expression of several detoxifying enzymes through binding to AREs within gene promoters [1]. These Nrf2-dependent detoxifying enzymes, including glutathione S-transferases (GSTs), NQO1, γ-glutamylcysteine synthetase (GCL), and glucuronosyl transferases (UDPs), protect cells from carcinogen-induced DNA damage and cytotoxicity [2]. Loss of Nrf2 expression increases the sensitivity of cells to carcinogenesis and promotes tumor formation [3-6]. Several studies suggest that the presence of Nrf2 expression suppresses oxidative stress-induced lung and prostate carcinogenesis [7, 8]. The role of Nrf2 in breast cancer development has been explored in vitro and in vivo. Nrf2 can suppress H2O2-induced oxidative stress by activation of the Nrf2-dependent genes in breast cancer cells [10]. Studies show that the carcinogenic factor, estrogen, inhibits the Nrf2-dependent detoxifying enzymes and promotes estrogen-dependent breast cancer formation in mice [11]. However, Nrf2 levels are increased in some tumor tissues where over-expression may enhance drug resistance through transcription of antioxidant, xenobiotic metabolism, drug efflux pump, and intrinsic chemoresistance genes [12, 13].
Nrf2 expression can be regulated by Keap1-dependent and Keap1-independent mechanisms. It is established that the negative regulatory factor Keap1 binds to and sequesters Nrf2, leading to ubiquitination and proteasomal degradation of Nrf2 [14-16]. Inactive Keap1 releases Nrf2, resulting in Nrf2 nuclear translocation and subsequent activation of Nrf2-dependent gene transcription [16-18]. Nrf2 is also transcriptionally regulated by aryl hydrocarbon receptor (AhR) [19, 20]. In addition, studies have shown that ectodermal-neural cortex 1 (ENC1) represses Nrf2 expression in a Keap1-independent manner by decreasing Nrf2 protein synthesis without affecting Nrf2 transcription or Nrf2 protein ubiquitination [21]. In transgenic adenocarcinoma of the mouse prostate (TRAMP) model, epigenetic mechanisms (DNA methylation and histone deacetylation) contribute to Nrf2 gene silencing [22]. These studies indicate that in addition to post-translational regulation of Nrf2 expression by Keap1, Nrf2 is subject to transcriptional and translational regulation.
MicroRNAs (miRNAs) have drawn a great deal of attention due to increasing understanding of their regulation of genes involved in many important cellular processes. MiRNAs are ~21–23 nucleotides (nt) long, single stranded non-coding RNAs. Transcribed from genomic loci by RNA polymerase II and processed by Drosha, miRNAs are exported from the nucleus as short hairpin precursors upon which miRNAs are cleaved by Dicer to generate their mature form. Once fully processed, miRNAs are loaded onto Argonaute proteins forming the RNA-induced silencing complex (RISC). Through base pairing with the miRNA response element within the 3′UTR of target mRNAs, RISC-complex associated miRNAs inhibit gene expression either through mRNA degradation or inhibition of protein translation [23, 24]. It has been widely documented that aberrant expression of miRNAs is closely associated with various human diseases including cancer [25, 26].
Studies show that miR-144 can inhibit Nrf2 mRNA expression in a myelogenous leukaemia (MLL) cell line [27]. Several other miRNAs including miR-340 and -500 are predicted to target the 3′ UTR of Nrf2 mRNA (mir-db.org/miRDB). However, array-based profiling of miRNAs indicates that those miRNAs may not be expressed in mammary epithelial cells [28-32]. For these reasons, we began to search for specific miRNAs which may regulate Nrf2 in mammary epithelial cells. We utilized the miRDB program to predict miRNAs that might target Nrf2 mRNA and found that miR-28 (mirdb.org/miRDB, target score 70) is a potential regulator for Nrf2. Using mammary epithelial cells, we examined the function of miR-28 in mediating Nrf2 expression. Our study reveals that Nrf2 expression is subject to negative regulation by miR-28.
Methods and materials
Cell culture and reagents
Human breast cancer cells MCF-7 and human embryonic kidney cells 293T (HEK293T) were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS, HyClone; Rockford, IL) and 1% l-glutamine (Invitrogen; Carlsbad, CA). Human normal mammary epithelial cells (HMEC) were grown in Mammary Epithelial Cell Growth Medium (MEGM) (Lonza; Walkersville, MD). MCF-12A cells were maintained in DMEM/F-12 medium (Invitrogen) supplemented with 10 μg/ml insulin (Sigma, St. Louis, MO), 100 ng/ml cholera toxin (Sigma), 0.5 μg/ml hydrocortisone (Sigma), 20 ng/ml epidermal growth factor receptor (Invitrogen), and 5% horse serum (Invitrogen). Cells were incubated in an atmosphere containing 5% CO2 at 37°C. Reagents used in this study include 17-β-Estradiol (E2) (Sigma), Actinomycin D (Act D) (Sigma), cycloheximide (CHX) (Sigma), and miR-28 precursor (pre-miR-28, Mature Accession # MIMAT0000085) (Ambion; Austin, TX).
Plasmids, transfection, and luciferase assay
Expression plasmid for human miR-28 (pri-miR-28, miR-28-pMEGIX-IRES-GFP) was a generous gift from Dr. Stefan N. Constantinescu (Ludwig Institute for Cancer Research Ltd, Brussels, Belgium) [33]. pGL3-Nrf2 3′UTR reporter plasmid contains wild type Nrf2 3′UTR cloned into pGL3 vector. 3′UTR mutant pGL3-Nrf2 reporter plasmid was generated with point mutations within potential miR-28 binding sites. The following primers: 5′tctgagctagtttttttgtactattatactaaaaccacgtactgtgatgtgaaat gc-3′ and 5′gcatttcacatcacagtacgtggttttagtataatagtacaaaa aaactagctcaga-3′, and the Generate Site-Directed Mutagenesis System (Invitrogen) were used to construct Nrf2 3′UTR mutant. The resulting mutant contains three point mutations: CTAAAA (GtoC) C (TtoA) C (CtoG) TACT-GTG and was confirmed by sequencing. HEK293T and MCF-7 cells were transfected with miR-28 (pre-miR-28 or pri-miR-28), pGL3-Nrf2, pGL3-Nrf2-mutant, Keap1-FLAG [34], Nrf2-myc [35], or control vectors using Lipofectamine 2000 (Invitrogen) following manufacturer’s instructions. The luciferase activity was performed using the Dual-Luciferase Reporter Assay System (Promega; Madison, WI) 48 h after the transfection as described previously [11].
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted for mRNA analysis as previously described [11]. In brief, total RNA was extracted with TRIzol reagent (Invitrogen). cDNA was synthesized from 1 μg total RNA using M-MLV Reverse Transcriptase (Invitrogen). qRT-PCR was carried out using a Light Cycler 480II (Roche Diagnostics; Indianapolis, IN) using primers for Nrf2, Forward: 5′-CAAAAGGAGCAAGAGAAAGCC-3′, Reverse: 5′-TCTGATTTGGGAATGTGG GC-3′. Nrf2 mRNA levels were normalized to levels of housekeeping gene GAPDH. In addition, small RNA was converted to complimentary DNA from 1 μg total RNA using the poly A polymerase based First-Strand Synthesis Kit (SABiosciences; Frederick, MD). Follow up miR analysis was performed by qRT-PCR using miR specific (miR-28) primers (SABiosciences) and normalizing to U6 snRNA levels as a control.
Western blotting and co-immunoprecipitation
Western blotting was performed using anti-Nrf2 rabbit polyclonal antibody (Santa Cruz H-300; Santa Cruz, CA.) or anti-Keap1 (Santa Cruz E-20) goat polyclonal antibody. β-actin antibody (Sigma) was used to normalize protein expression. The USCAN-IT program was used to analyze protein expression levels.
For co-immunoprecipitation, cell lysates were incubated with anti-myc antibody (Cell Signaling; Boston, MA) or mouse IgG control (Santa Cruz) at 4°C for 2 h followed by overnight incubation of Protein A/G PLUS-Agarose beads (Santa Cruz). The following day, Western blotting was performed to examine Nrf2 and Keap1 interaction by using mouse anti-FLAG antibody (Stratagene; Santa Clara, CA). Keap1 and Nrf2 proteins were immunoblotted using anti-FLAG and anti-myc antibodies, respectively.
Analysis of Nrf2 mRNA stability
Cells were transfected with 50 nM pre-miR-28 or control oligo. Forty-eight hours after transfection, cells were incubated with Act D (5 μg/ml). Cells were harvested at subsequent time intervals (0, 1, 2, 4, and 6 h). Total RNA was extracted and cDNA was synthesized as described above. qRT-PCR analysis was used to monitor Nrf2 mRNA decay. Housekeeping gene GAPDH, which showed little to no decay over 6 h, was used as an internal control. Results from Act D assays were processed using Prism 4.0 software (GraphPad; La Jolla, CA) to calculate the Nrf2 mRNA half-life.
Measurement of Nrf2 protein stability
Cells were transfected with 50 nM pre-miR-28 or control oligo. Forty-eight hours after transfection, cells were treated with 10 μg/ml CHX, harvested at various time points (0, 1, 2, 4, and 6 h), and immunoblotted for Nrf2 with beta-actin as a loading control. Nrf2 protein level was normalized to levels of beta-actin using USCAN-IT program.
Soft agar assay
Soft agar assays were performed, in duplicate, in six-well plates. Each well contained a bottom layer of 0.6% agarose (Bio-Rad; Hercules, CA) and a top layer of 0.3% agarose containing 3.75 × 10 5 cells. Agarose was diluted in DMEM medium. A few drops of DMEM medium were added on the top layer after it solidified. The plates were incubated for 2 weeks. The top layer was replenished every week. After 2 weeks, the cells were stained with 0.05% crystal violet overnight at 37°C. Colonies were visualized and counted with light microscopy. Colonies, larger than 50 μm in diameter, were counted from four random 10× objective fields.
Statistic analysis
Statistical analysis was performed using Student’s t test and P values of <0.01 were considered significant. Data are represented as mean ± SE. GraphPad Prism 4.0 was used for data analysis.
Results
Inverse expression patterns of Nrf2 mRNA and miR-28 in mammary epithelial cells
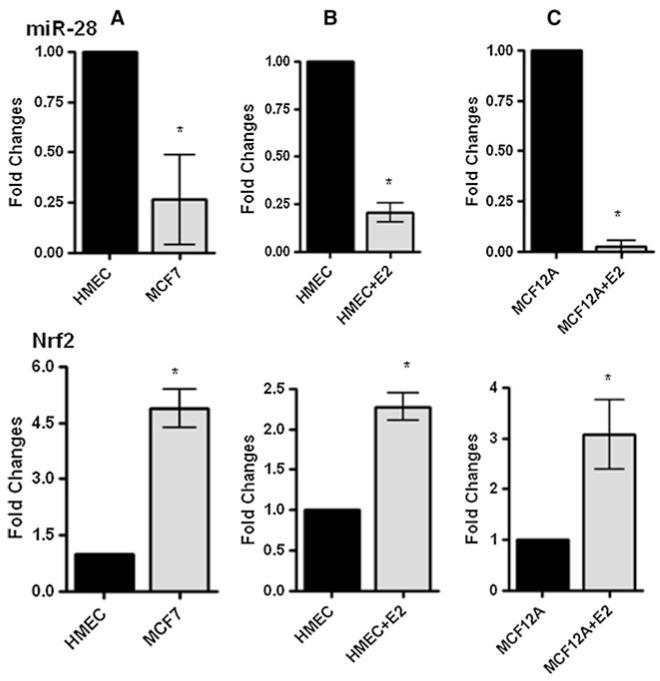
To determine potential miRNAs which regulate Nrf2 expression in breast epithelial cells, we searched miRDB and found miR-28 was among the highest scoring miRNAs predicted to target Nrf2. First we examined miR-28 and Nrf2 expression in normal human mammary epithelial cells (HMEC) and breast cancer cell line MCF-7. We found that MCF-7 cells showed higher levels of Nrf2 mRNA and lower miR-28 levels as compared with HMEC (Fig. 1a). Importantly, both cell lines showed inverse expression pattern of Nrf2 and miR-28. To further confirm the inverse expression pattern between Nrf2 and miR-28, we treated HMEC with estrogen (80nM β-estradiol) for 3 days. In mammary epithelium, stimulation with estrogen and its metabolites can induce oxidative stress ultimately resulting in increasing Nrf2 transcription [9]. As expected, estrogen treated HMEC showed significantly elevated Nrf2 mRNA levels compared with the control cells when measured by qRT-PCR (Fig. 1b). Interestingly, miR-28 levels were decreased in estrogen treated HMEC. Similar results were observed in estrogen treated MCF-12A cells (Fig. 1c). Collectively, these data indicate an inverse correlation between Nrf2 expression and miR-28 levels in mammary epithelial cells.
Fig. 1.
The inverse expression pattern of Nrf2 mRNA and miR-28 in mammary epithelial cells. a Fold changes of Nrf2 mRNA and miR-28 in HMEC and MCF-7 cells were measured by qRT-PCR. b and c HMEC and MCF-12A cells were treated with 80nM β-estradiol (E2) for 3 days, and fold changes of Nrf2 and miR-28 were measured by qRT-PCR analysis. N = 2 ± SE. * represents statistical significance (P < 0.01)
MiR-28 targets the 3′UTR of Nrf2 mRNA
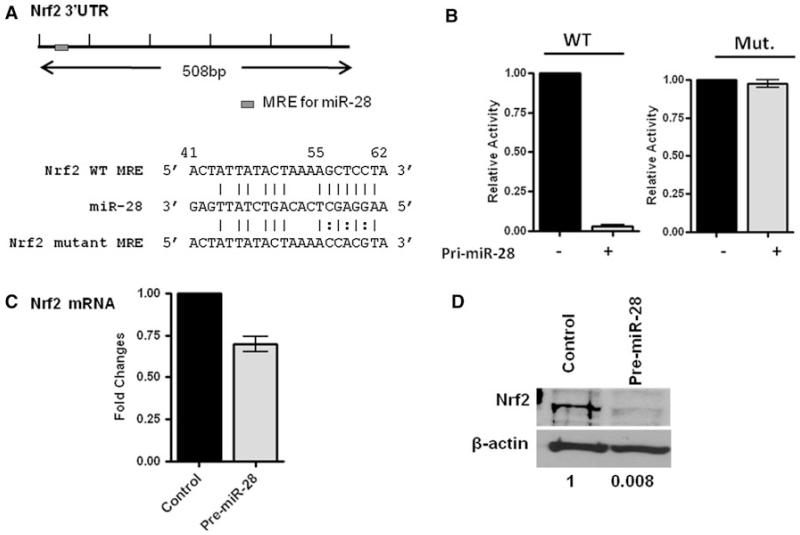
We hypothesized that miR-28 interacts with the 3′UTR of the Nrf2 mRNA to suppress Nrf2 expression. To test this hypothesis, the ability of miR-28 to regulate the 3′UTR of Nrf2 mRNA was evaluated via luciferase reporter assays. Within the Nrf2 3′UTR, a single miR response element (MER) containing an 8-mer miR-28 seeding region was predicted by miRDB (Fig. 2a). Wild type Nrf2 3′UTR and mutant Nrf2 3′UTR (MRE: CTAAAA (GtoC) C (TtoA) C (CtoG) TACTGTG) was cloned into the pGL3 luciferase reporter plasmid. HEK293T cells, which do not express detectable levels of miR-28 (data not shown), were transfected with wild type or mutant Nrf2 3′UTR pGL3 luciferase vectors along with the pfRG-B renilla luciferase vector. Pri-miR-28 was also co-transfected into HEK293T cells and was found to decrease wild type Nrf2 3′UTR reporter activity by more than 90% (P < 0.001) compared to control transfections (Fig. 2b). Pri-miR-28 transfection did not alter mutant Nrf2 3′UTR reporter activity (P >0.1) (Fig. 2b). We examined Nrf2 expression with qRT-PCR and Western blotting in MCF-7 cells and found pre-mir-28 transfection resulted in a 35% decrease in Nrf2 mRNA levels (Fig. 2c) and a more than 90% decrease in Nrf2 protein levels (P < 0.001) (Fig. 2d). These results confirm that miR-28 negatively regulates Nrf2 expression by targeting the 3′ UTR of Nrf2 mRNA.
Fig. 2.
miR-28 targets the 3′UTR of Nrf2 mRNA. a Schematics of Nrf2 mRNA 3′UTR and its potential miR-28 binding site. Nrf2 3′UTR mutant was generated with point mutations in the miR-28 binding site referred to as the MRE. b HEK293T cells transfected with wild type (WT) or mutant Nrf2 mRNA 3′UTR reporter plasmids with vehicle control or pre-miR-28. The luciferase activities were measured 48 h after transfection. The luciferase activities were normalized to those of renilla luciferase activity of a co-transfected reporter. The relative luciferase activities were calculated by normalizing to that of vehicle controls. c qRT-PCR showing the fold changes in mRNA levels after pre-miR-28 transfection of MCF-7 cells compared with control transfected cells. N = 2 ± SE, * P < 0.001. The expression level of normalized control cells is arbitrarily set as 1. d Western blotting showing Nrf2 protein expression in the pre-miR-28 transfected and vehicle-control transfected MCF-7 cells. β-actin was used as a loading control. A representative experiment was shown
MiR-28 decreases Nrf2 mRNA and protein stability
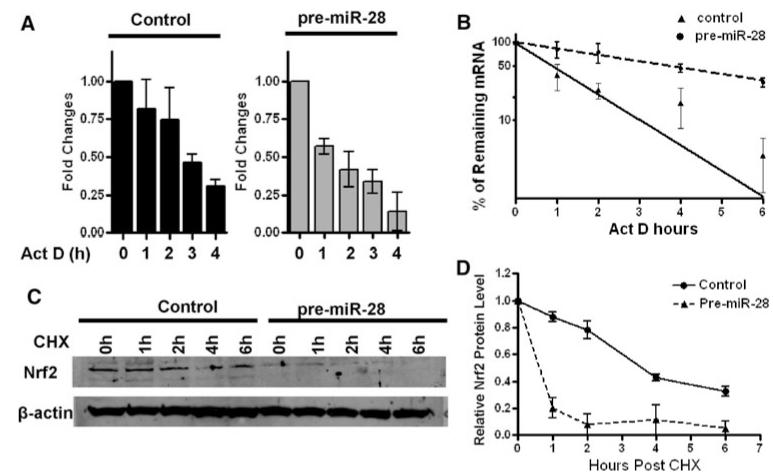
MiRNAs can inhibit gene expression by causing mRNA degradation or by inhibiting protein translation [23]. In order to dissect the potential mechanisms through which miR-28 inhibits Nrf2, pre-miR-28, or control transfected MCF-7 cells were treated with the transcriptional inhibitor Act D [36]. Nrf2 mRNA level was significantly decreased after 6 h of Act D treatment (Fig. 3a). We calculated half-life of Nrf2 mRNA in pre-miR-28 transfected MCF-7 cells to be 0.86 h (Fig. 3b); however, in vehicle-control transfected cells Nrf2 mRNA half-life was found to be 2.92 h. This indicates that miR-28 inhibits Nrf2 expression through Nrf2 mRNA degradation.
Fig. 3.
miR-28 decreases Nrf2 mRNA and protein stability. a Nrf2 mRNA stability assay was performed in MCF-7 cells transfected with pre-miR-28 or vehicle-control. The cells were treated with 5 μg/ml Actinomycin D (Act D) 48 h after transfection, and total RNA was extracted 0, 1, 2, 4, and 6 h after Act D treatment. qRT-PCR was used to examine Nrf2 mRNA levels. GAPDH was used as an internal control. Mean values from two independent experiment were shown. b Quantitation of Nrf2 mRNA decay in both MCF-7 control cells and MCF-7/pre-miR-28 cells. c Nrf2 protein stability assay was performed in pre-miR-28 transfected and vehicle-control transfected MCF-7 cells. The cells were treated with 10 μg/ml CHX 48 h after transfection and protein extract was prepared 0, 1, 2, 4, and 6 h after CHX treatment. Nrf2 protein expression was detected by Western blotting and β-actin was used as a loading control. A representative experiment was shown. d Normalized Nrf2 protein expression was analyzed using UN-SCAN-IT program. The relative protein expression was calculated by comparing normalized Nrf2 expression in treated cells to 0 h control
We sought to also examine if decreased Nrf2 protein level might be due to a change in Nrf2 protein stability, in addition to miR-28 mediated Nrf2 mRNA decay. Pre-miR-28 or control transfected cells were treated with the translation inhibitor CHX [37]. The half-life of Nrf2 protein in pre-miR-28 transfected MCF-7 cells was ~0.7 h, but in the vehicle-control transfected cells, the half-life of Nrf2 protein was ~5 h (Fig. 3c, d). Remarkably, this indicated that miR-28 also reduces Nrf2 protein stability. Collectively, these data demonstrate that Nrf2 is subject to regulation by miR-28 through alterations of mRNA and protein stability.
MiR-28 inhibits Nrf2 expression through a Keap1-independent manner
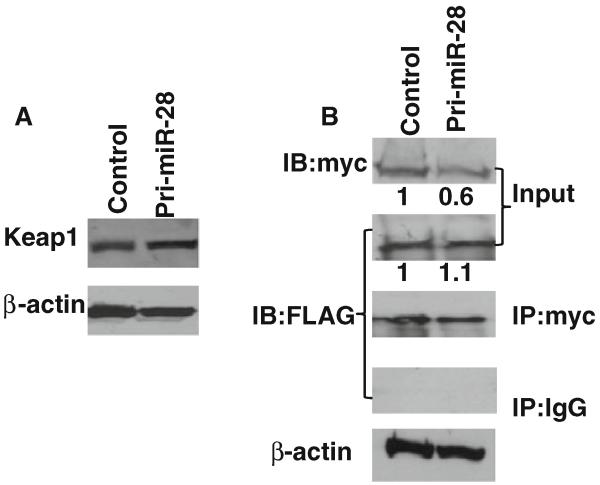
As previously discussed, Nrf2 can be degraded in the cytoplasm through ubiquitination resulting from Keap1/Nrf2 interaction. In order to examine the possibility that changes in Nrf2 protein stability after miR-28 transfection might be due to differences in Keap1 expression or Keap1/Nrf2 interaction, Keap1-FLAG along with pri-miR-28 or vehicle controls were co-transfected into MCF-7 cells. Western blotting revealed that the pri-miR-28 transfection does not alter Keap1 protein levels in MCF-7 cells (Fig. 4a). To examine any possible impact of miR-28 on Keap1/Nrf2 interaction, HEK293T cells were co-transfected with Keap1-FLAG/Nrf2-myc/pri-miR28 or Keap1-FLAG/Nrf2-myc/vehicle-control plasmids. Again, miR-28 decreased Nrf2 levels but did not affect Keap1 levels (input in Fig. 4b). Co-immunoprecipitation (CoIP) was performed to examine interactions of Keap1 and Nrf2. CoIP experiments revealed no significant changes in Keap1/Nrf2 interaction regardless of miR-28 level (Fig. 4b). These results demonstrate that miR-28 mediated changes in Nrf2 protein stability were not due to the regulation of Keap1 expression level or altered Keap1/Nrf2 interaction.
Fig. 4.
miR-28 regulation of Nrf2 expression is Keap1 independent. a MCF-7 cells were co-transfected with Keap1-FLAG and pri-miR-28 or vehicle-control. Western blotting was used to detect Keap1 expression with anti-FLAG antibody. β-actin was used as a loading control. b Keap1-FLAG and Nrf2-myc were co-transfected into HEK293T cells with pri-miR-28 or vehicle control. Co-immunoprecipitation was performed to examine Nrf2-Keap1 interaction status. The anti-myc antibody was used to pull down antigens and the mouse IgG was used as negative control; anti-FLAG or anti-myc antibody was used for Western blotting. A representative experiment was shown. Normalized protein expression in input was analyzed using UN-SCAN-IT program
Loss of Nrf2 increases anchorage-independent growth in MCF-7 cells
Elevated levels of Nrf2 were found to enhance protection against H2O2 induced oxidative stress in breast cancer cells [10]. To determine the function of miR-28 and its target Nrf2 in breast cancer progression, we utilized soft agar anchorage-independent cell growth assays to examine the impact of Nrf2 on colony formation. MCF-7 cells were transfected with pre-miR-28 or infected with a short-hairpin lentiviral vector targeting Nrf2 expression or vehicle controls. Infected cells were selected by puromycin. The ability of cells to grow in an anchorage-independent environment was examined in MCF-7/pre-miR-28 and MCF-7/shNrf2 cells or control cells using soft-agar assays. After 2 weeks of cell proliferation, colony formation was detected. Colonies with a diameter greater than 50 μm were counted. A significant increases in colonies were found in MCF-7/pre-miR-28 and MCF-7/shNrf2 cells compared with control cells (Fig. 5a-c P < 0.01). Figure 5d shows the effectiveness of Nrf2 knockdown (more than 80%) in MCF-7/shNrf2 cells compared to control cells and the down-regulation of Nrf2 expression after transfection of MCF-7 cells with pre-miR-28 was previously shown (Fig. 2d). shNrf2 reduction of Nrf2 was confirmed in Fig. 5c. These results suggest that loss of Nrf2 expression increases the ability of colony formation in MCF-7 cells.
Fig. 5.
Loss of Nrf2 increases anchorage-independent growth in MCF-7 cells. a MCF-7/shNrf2 cells, MCF-7/pre-miR-28 cells, and control cells were examined by soft agar assay. After 2 weeks, colonies larger than 50 μm in diameter were counted. b Quantification of colony formation in MCF-7/shNrf2 cells, MCF-7/pre-miR-28, and control cells. N = 2 ± SE. * represents statistical significance (P < 0.01). c Western blotting was used to confirm down-regulation of Nrf2 in MCF-7/shNrf2 cells compared to control cells. β-actin was used as the loading control. The expression level of normalized control cells is arbitrarily set as 1
Discussion
The dynamic functions of Nrf2 in tumor initiation and progression have been previously reported [9, 38]. However, the mechanisms responsible for regulating Nrf2 expression are in need of further elucidation. The well-known Keap1-Nrf2 pathway only regulates Nrf2 protein by ubiquitination [18]. Possible epigenetic modulation of Nrf2 expression has only recently been studied in a mouse model [22, 27]. Previously, miR-144 was shown to target the 3′UTR of Nrf2 mRNA and modulate Nrf2 expression in blood cells, which is associated with sickle cell disease [27]. The differential expression of miR-28 was documented in several cancers, including lymphoma [39], glioma [32] and squamous carcinoma [40]. However, in breast cancer cells the role of miRNAs in the regulation of Nrf2 expression remains largely unclear. Our study is the first to study the impact of miR-28 on normal human mammary epithelial cells as well as breast cancer cells. We found that miR-28 regulates Nrf2 expression at the posttranscriptional level by binding to the 3′UTR of Nrf2 mRNA and resulting in Nrf2 mRNA degradation. In addition, miR-28 promoted Nrf2 protein degradation. This miR-28-induced protein degradation was not due to changes in Keap1 protein expression or Keap1/Nrf2 interaction. As discussed above, miRNAs can suppress target gene expression through cleavage of mRNA or inhibition of translation. Concerning the later mechanism of action, miRNA-dependent suppression of target gene translation may involve multiple steps of protein translation including: (1) the repression of 7-methyl-guanosine (m7G) cap-dependent mRNA translation at the initiation step, (2) the repression of mRNA translation by preventing joining of the 60S subunit, (3) repression by termination of translation, or (4) a possible involvement in the proteolysis of nascent polypeptide chains (reviewed in [23]). It is speculated that specific proteases, although unknown, might be associated with the nascent polypeptides degradation through 3′ UTR-tethered miRNA-RISC complex [41, 42]. This possible mechanism may explain our observations that miR-28 reduces Nrf2 protein stability leading to inhibition of protein production. Our further studies will determine the exact mechanism by which miR-28 mediates Nrf2 protein degradation.
The function of Nrf2 in preventing oxidative stress-induced breast cancer was well-documented [43, 44]. In hepatocellular carcinoma and adenocarcinoma cell lines, Nrf2 knock-down was found to increase transforming growth factor β/Smad signaling and promote cell migration and plasticity [45]. Activation of the Nrf2 pathways by antioxidants treatment inhibits cell growth by induction of cyclin A degradation and cell cycle arrest in late-G1-phase in glioma cells [46]. In our study, we investigated the impact of Nrf2 on breast cancer motility and growth through examination of anchorage-independent cell growth in MCF-7 cells. We found that Nrf2 has ability to decrease colony formation in MCF-7 cells by soft agar assay. Our findings further support that Nrf2 may inhibit tumor cell growth [10].
In conclusion, our study has demonstrated a novel mechanism for regulation of Nrf2 expression in breast cancer, through miR-28 targeting of the 3′UTR of Nrf2 mRNA. Our results provide new insights into the functions of miR-28 in breast cancer gene regulation. Moreover, our findings suggest that miR-28 may control breast cancer progression by targeting the Nrf2 pathway. Revealing a new mechanism regulating Nrf2 expression may provide implications for understanding breast cancer progression and may aid in development of new chemoprevention strategies.
Acknowledgments
This study was supported by grants from Flight Attendants Medical Research Institute (FAMRI YCSA072084 to QZ), and Maryland Stem Cell Research Fund (2010-MSCRFE-0179-00 to QZ).
Footnotes
Conflict of interest None.
References
- 1.Yu S, Kong A. Targeting carcinogen metabolism by dietary cancer preventive compounds. Curr Cancer Drug Targets. 2007;7:416–424. doi: 10.2174/156800907781386669. [DOI] [PubMed] [Google Scholar]
- 2.Fields WR, Morrow CS, Doehmer J, et al. Expression of stably transfected murine glutathione S-transferase A3–3 protects against nucleic acid alkylation and cytotoxicity by aflatoxin B1 in hamster V79 cells expressing rat cytochrome P450–2B1. Carcinogenesis. 1999;20:1121–1125. doi: 10.1093/carcin/20.6.1121. [DOI] [PubMed] [Google Scholar]
- 3.Henderson CJ, Smith AG, Ure J, et al. Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases. Proc Natl Acad Sci USA. 1998;95:5275–5280. doi: 10.1073/pnas.95.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramos-Gomez M, Kwak MK, Dolan PM, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci USA. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramos-Gomez M, Dolan P, Itoh K, et al. Interactive effects of nrf2 genotype and oltipraz on benzo[a]pyrene-DNA adducts and tumor yield in mice. Carcinogenesis. 2003;24:461–467. doi: 10.1093/carcin/24.3.461. [DOI] [PubMed] [Google Scholar]
- 6.McMahon M, Itoh K, Yamamoto M, et al. The Cap’n’Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- 7.Harvey CJ, Thimmulappa RK, Singh A, et al. Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic Biol Med. 2009;46:443–453. doi: 10.1016/j.freeradbiomed.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barve A, Khor T, Nair S, et al. Gamma-tocopherol-enriched mixed tocopherol diet inhibits prostate carcinogenesis in TRAMP mice. Int J Cancer. 2009;124:1693–1699. doi: 10.1002/ijc.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acharya A, Das I, D Chandhok, et al. Redox regulation in cancer: a double-edged sword with therapeutic potential. Oxid Med Cell Longev. 2010;3(1):23–34. doi: 10.4161/oxim.3.1.10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh T, Elangovan S, Wu J. Differential suppression of proliferation in MCF-7 and MDA-MB-231 breast cancer cells exposed to alpha-, gamma- and delta-tocotrienols is accompanied by altered expression of oxidative stress modulatory enzymes. Anticancer Res. 2010;30:4169–4176. [PubMed] [Google Scholar]
- 11.Yao Y, Brodie AMH, Davidson N, et al. Inhibition of estrogen signaling activates the NRF2 pathway in breast cancer. Breast Cancer Res Treat. 2010;124:585–591. doi: 10.1007/s10549-010-1023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh A, Misra V, Thimmulappa R, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Medicine. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta T, Iijima K, Miyamoto M, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 14.Dinkova-Kostova A, Holtzclaw WD, Cole R, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakabayashi N, Dinkova-Kostova A, Holtzclaw WD, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Niture SK, Kaspar JW, Shen J, Jaiswal AK. Nrf2 signaling and cell survival. Toxicol Appl Pharmacol. 2010;244(1):37–42. doi: 10.1016/j.taap.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaspar J, Niture S, Jaiswal A. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao W, Hu L, Scrivens PJ, et al. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem. 2005;280:20340–20348. doi: 10.1074/jbc.M412081200. [DOI] [PubMed] [Google Scholar]
- 20.Kwak M, Itoh K, Yamamoto M, Kensler T. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol Cell Biol. 2002;22:2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Zhang D. Ectodermal-neural cortex 1 downregulates Nrf2 at the translational level. PLoS ONE. 2009;4:e5492. doi: 10.1371/journal.pone.0005492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu S, Khor T, Cheung K, et al. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PLoS ONE. 2010;5:e8579. doi: 10.1371/journal.pone.0008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filipowicz W, Bhattacharyya S, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 24.Winter J, Jung S, Keller S, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 25.Zimmerman A, Wu S. MicroRNAs, cancer and cancer stem cells. Cancer Lett. 2011;300:10–19. doi: 10.1016/j.canlet.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voorhoeve PM, Agami R. Classifying microRNAs in cancer: the good, the bad and the ugly. Biochim Biophys Acta. 2007;1775:274–282. doi: 10.1016/j.bbcan.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Sangokoya C, Telen M, Chi J. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood. 2010;116:4338–4348. doi: 10.1182/blood-2009-04-214817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 29.Corcoran C, Friel A, Duffy M, et al. Intracellular and extracellular microRNAs in breast cancer. Clin Chem. 2011;57:18–32. doi: 10.1373/clinchem.2010.150730. [DOI] [PubMed] [Google Scholar]
- 30.Farazi T, Spitzer J, Morozov P, et al. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottardo F, Liu C, Ferracin M, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 32.Malzkorn B, Wolter M, Liesenberg F, et al. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20:539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girardot M, Pecquet C, Boukour S, et al. miR-28 is a thrombopoietin receptor targeting microRNA detected in a fraction of myeloproliferative neoplasm patient platelets. Blood. 2010;116:437–445. doi: 10.1182/blood-2008-06-165985. [DOI] [PubMed] [Google Scholar]
- 34.Fan W, Tang Z, Chen D, et al. Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy. 2010;6:614–621. doi: 10.4161/auto.6.5.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furukawa M, Xiong Y. BTB protein Keap1 targets anti-oxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou H, Mazan-Mamczarz K, Martindale J, et al. Post-transcriptional regulation of androgen receptor mRNA by an ErbB3 binding protein 1 in prostate cancer. Nucleic Acids Res. 2010;38:3619–3631. doi: 10.1093/nar/gkq084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agoston A, Argani P, Yegnasubramanian S, et al. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem. 2005;280:18302–18310. doi: 10.1074/jbc.M501675200. [DOI] [PubMed] [Google Scholar]
- 38.Kensler T, Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31:90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu W, Li J, Fan L, et al. Expressions of bcl-6, lpp and miR-28 genes in diffuse large B cell lymphoma cell lines. Zhong Guo Shi Yan Xue Ye Xue Za Zhi. 2009;17:83–87. [PubMed] [Google Scholar]
- 40.Li L, Zhang Z, Liu Y, et al. DNA microarrays-based microRNA expression profiles derived from formalin-fixed paraffin-embedded tissue blocks of squammous cell carcinoma of larynx. Zhong Hua Bing Li Xue Za Zhi. 2010;39:391–395. [PubMed] [Google Scholar]
- 41.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 42.Nottrott S, Simard M, Richter J. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 43.Klaunig J, Kamendulis L, Hocevar B. Oxidative stress and oxidative damage in carcinogenesis. Toxicol Pathol. 2010;38:96–109. doi: 10.1177/0192623309356453. [DOI] [PubMed] [Google Scholar]
- 44.Kwak M, Kensler T. Targeting NRF2 signaling for cancer chemoprevention. Toxicol Appl Pharmacol. 2010;244:66–76. doi: 10.1016/j.taap.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rachakonda G, Sekhar KR, Jowhar D, et al. Increased cell migration and plasticity in Nrf2-deficient cancer cell lines. Oncogene. 2010;29:3703–3714. doi: 10.1038/onc.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Havens C, Ho A, Yoshioka N, et al. Regulation of late G1/S phase transition and APC Cdh1 by reactive oxygen species. Mol Cell Biol. 2006;26:4701–4711. doi: 10.1128/MCB.00303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]