Abstract
A presumptive chromosomal cephalosporinase (pI, 9.0) from a clinical strain of Acinetobacter genomic species 3 (AG3) is reported. The nucleotide sequence of this β-lactamase shows for the first time the gene encoding an AmpC enzyme in AG3. In addition, the biochemical properties of the novel AG3 AmpC β-lactamase are reported
The bacterial genus Acinetobacter consists of strictly aerobic gram-negative coccobacilli, which are oxidase negative, nonmotile, nitrate negative, and nonfermentative.
Studies based on DNA-DNA hybridization have led to the description of 23 DNA homology groups, or genomospecies, within the genus Acinetobacter (5, 7, 11, 15, 21, 26). Seven species of this genus have been named and are included in the Index of the Bacterial and Yeast Nomenclatural Changes (19): (i) A. calcoaceticus, or genomospecies 1; (ii) A. baumannii, or genomospecies 2; (iii) A. haemolyticus, or genomospecies 4; (iv) A. junii, or genomospecies 5; (v) A. johnsonii, or genomospecies 7; (vi) A. lwoffii, or genomospecies 8; and (vii) A. radioresistens, or genomospecies 12. Groups 1 (A. calcoaceticus), 2 (A. baumannii), and the as-yet-unnamed and closely related genomic DNA groups 3 and 13TU (often referred to as the A. calcoaceticus-A. baumannii complex) are the most frequent species among clinical isolates, particularly A. baumannii and Acinetobacter genomospecies 3 (AG3) (5, 6, 13, 26).
Earlier studies showed statistically significant differences between the distributions in Hong Kong and Europe of genomic DNA groups of isolates obtained from blood cultures and various superficial carriage sites (10). Indeed, in some Hong Kong hospitals, AG3 strains accounted for up to 40% of the blood culture isolates (10). Antimicrobial susceptibilities also differed significantly among members of the A. calcoaceticus-A. baumannii complex, which indicates diversity in the molecular mechanisms involved in the antimicrobial resistance observed (14, 29). In keeping with this trend, A. baumannii was the most resistant genospecies.
The most common mechanism of the resistance of A. baumannii to β-lactam antibiotics is attributed to the presence of β-lactamases encoded either by the chromosome or by plasmids (2). Several class A, B, and D β-lactamases (1, 2, 3, 22, 23, 27), as well as chromosome-mediated cephalosporinases (pI, >8) (2, 3) (which confer different resistance phenotypes to A. baumannii) have been described. However, only one ampC gene (AmpC of RYC52763) encoding an AmpC β-lactamase (4) and two allelic variants (ABAC-1 and ABAC-2) (17) have been reported so far for A. baumannii strains.
Regarding β-lactam resistance in the AG3 group, two metalloenzymes, VIM-2 and IMP-4, have been identified in Hong Kong and Korea (9, 30).
The aim of this study was to elucidate the mechanisms associated with the resistance to β-lactam antibiotics shown by a clinical strain of Acinetobacter genomic DNA group 3 (AJC68081) identified by amplified ribosomal DNA restriction analysis (28) and isolated from a wound exudate of a patient treated at the Juan Canalejo Hospitalary Complex.
The susceptibility testing of the AJC68081 strain was performed by a microdilution method following the recommendations of the National Committee for Clinical Laboratory Standards (20). Antibiotics were kindly provided by their manufacturers as powders of fixed potency. MICs were confirmed by the E-test. The antibiotic susceptibility profiles of all strains included in this study are shown in Table 1.
TABLE 1.
MICs of β-lactams for clinical strain AJC68081, E. coli TG1, E. coli TG1 (pBE-1), and E. coli TG1 (pGER1)
| Antibiotic | MIC (μg/ml)a
|
|||
|---|---|---|---|---|
| AJC68081 (produces AmpC of AG3) | E. coli TG1 | E. coli TG1 (pBE-1)b | E. coli TG1 (pGER1)c | |
| Amoxicillin | >256 | 3 | 256 | 128 |
| Amoxicillin + clavulanated | 6 | 2 | 24 | 32 |
| Piperacillin | >256 | 0.38 | 64 | 8 |
| Cephalothin | >256 | 3 | >256 | >256 |
| Cefuroxime | >256 | 1.5 | >256 | >256 |
| Cefoxitin | >256 | 2 | 128 | 2 |
| Cefotaxime | 256 | 0.023 | 2 | 4 |
| Cefotaxime + clavulanated | 0.19 | 0.023 | 1 | 1 |
| Ceftazidime | 32 | 0.064 | 0.75 | 16 |
| Ceftazidime + clavulanated | 0.19 | 0.064 | 0.5 | 8 |
| Cefepime | 32 | 0.016 | 0.047 | 0.25 |
| Aztreonam | 12 | 0.032 | 0.25 | 1 |
| Imipenem | 1 | 0.125 | 0.38 | 0.125 |
| Meropenem | 0.75 | 0.008 | 0.023 | 0.012 |
| Clavulanic acid | 8-16 | ND | ND | ND |
| Sulbactam | 16-32 | ND | ND | ND |
ND, not done.
Transformant producing AmpC β-lactamase of AG3.
Transformant producing AmpC β-lactamase of A. baumannii.
Clavulanate was used at 4 μg/ml.
The β-lactamases were analyzed by isoelectric focusing as described by Matthew et al. (18). The sonicated extract of strain AJC68081 contained a single β-lactamase with a pI of ca. 9.0, which may correspond to that of a chromosomal cephalosporinase. Alkaline lysis of the bacteria (24) did not result in plasmid isolation.
The possibility of a certain degree of homology between the present AG3 cephalosporinase and the previously reported ampC gene of A. baumannii allowed us to design oligonucleotides that could specifically amplify our target AG3 cephalosporinase gene by PCR. Chromosomal DNA from strain AJC68081 was purified according to standard protocols (MasterPure DNA purification kit; Epicentre, Madison, Wis.). Five hundred nanograms of the AG3 chromosome was used as a template to be amplified by PCR with the two oligonucleotides ampC1 forward (5′-TAGTATTCGTCGTTAGAAAACAAT) and ampC2 reverse (5′-GCTTAGGATATGTTTGGTTCTT) (Sigma-Genosys Ltd., Cambridge, United Kingdom), which hybridize in the untranslated regions of the A. baumannii (RYC52763) ampC gene (4). The PCR was performed under the following conditions: denaturation, 10 min at 94°C; amplification, 30 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C; and elongation, 16 min at 72°C. An aliquot (20 μl) of the sample was subjected to electrophoresis in a 1.0% agarose gel. The gel showed an amplified product, detected by ethidium bromide staining (50 mg/liter), of 1.3 kb, which was the expected size of the ampC gene in relation to that in A. baumannii. For further analysis, the 1.3-kb amplicon was ligated into pGEM-T easy vector (Promega Corporation, Madison, Wis.), and the recombinant plasmids were introduced into Escherichia coli TG1 by transformation with CaCl2 (24). The selection of transformants on Luria-Bertani agar plates supplemented with ampicillin (50 μg/ml), isopropyl-β-d-thiogalactopyranoside (IPTG, 25 μg/ml; Roche Diagnostics, Mannheim, Germany), and 5′-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal, 100 μg/ml; Roche Diagnostics) resulted in several white colonies, all carrying an identical recombinant plasmid. After alkaline lysis of the transformant (24), enzymatic digestion of the purified pGEM-T plasmid with EcoRI released a 1.3-kb DNA insert, confirming the success of the cloning procedure. The nucleotide sequence of the insert was elucidated by sequencing it with the Taq DyeDeoxiTerminator cycle sequencing kit before analysis with an automatic DNA sequencer (377 Abi-Prims, Perkin-Elmer). The entire sequence of the fragment was 1,321 bp long and contained one open reading frame (Fig. 1) of 1,152 bp (383 amino acids long). GenBank database searches with this open reading frame revealed similarities to several class C chromosome-mediated β-lactamases (Table 2). At a protein level, the highest similarities detected (98.69 to 97.65%) were with the AmpC β-lactamases AmpC ABAC-1, AmpC ABAC-2, and AmpC RYC52763 from A. baumannii, respectively (4, 17). Comparison of the amino acids in the AmpC β-lactamases of AG3 with those in ABAC-1, ABAC-2, and AmpC RYC52763 from A. baumannii yielded five, eight, and nine amino acid differences, respectively (4, 17). The nucleotide sequence of the promoter region of the gene revealed two substitutions (A to G) at positions −21 and −53 with respect to that of ampC in A. baumannii (4). The importance of these changes in the regulation of the expression of the AmpC enzyme remains unknown.
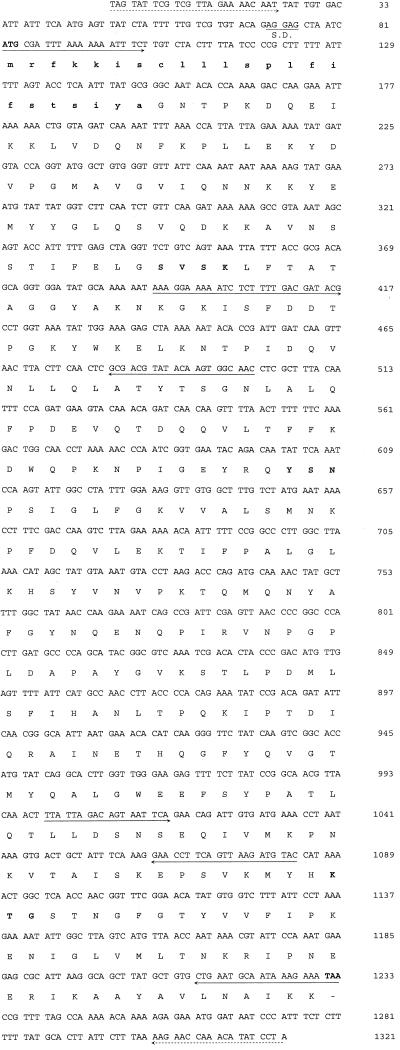
FIG. 1.
Nucleotide sequence of the 1.3-kb fragment. The amino acid sequence deduced for AmpC G3 β-lactamase is shown on the linebelow the nucleotide triplets. The ATG and TAA shown in boldface type represent the initiation and termination codons, respectively. A putative Shine-Dalgarno (S.D.) ribosomal recognition site is indicated. The positions of the primers used to sequence the gene and to detect the ampC gene in several AG3 strains are indicated by arrows. The positions of the primers used for amplification and further cloning of the gene are indicated by discontinuous arrows. The putative sequence of the signal peptide is indicated by boldface type and lowercase letters. The β-lactamase active site SVSK, the conserved triad KTG, and the typical class C motif YXN are shown in boldface type (12).
TABLE 2.
Percent identity between amino acid sequences of AG3 AmpC and other class C β-lactamases
| β-Lactamasea | % Identity with:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| AG3 AmpC | A. baumannii AmpC | Aeromonas hydrophila AmpC | CMY-1 | Serratia marcescens AmpC | Aeromonas sobria AmpC | FOX-5 | Pseudomonas aeruginosa AmpC | |
| AG3 AmpC | 100 | 97 | 46 | 45 | 45 | 44 | 44 | 42 |
| A. baumannii AmpC | 100 | 46 | 45 | 43 | 44 | 43 | 40 | |
| A. hydrophila AmpC | 100 | 82 | 48 | 79 | 75 | 57 | ||
| CMY-1 | 100 | 48 | 77 | 72 | 56 | |||
| S. marcescens AmpC | 100 | 48 | 49 | 50 | ||||
| A. sobria AmpC | 100 | 77 | 56 | |||||
| FOX-5 | 100 | 55 | ||||||
| P. aeruginosa AmpC | 100 | |||||||
To determine the MICs and the biochemical properties of the β-lactamase encoded by the 1.3-kb DNA insert, the insert was cloned in the pBGS18− plasmid (25) (which carries a kanamycin resistance gene), resulting in the pBE-1 plasmid. The β-lactam patterns of resistance to amoxicillin, cephalothin, cefuroxime, and cefoxitin and, to a lesser extent, amoxicillin-clavulanic acid and piperacillin of several transformants containing the pBE-1 plasmid were identical, whereas the MICs of ceftazidime, cefotaxime, cefepime, aztreonam, and carbapenems were slightly higher than those for the host E. coli TG1 strain (Table 1).
Chromosomal AmpC β-lactamases may be inducible in some gram-negative rods, and genes such as the repressor ampR and ampD may be involved in this pathway (16). The sequences of the flanking regions of the gene encoding the AmpC β-lactamase of A. baumannii did not show any homology with the ampR gene. Similarly, induction experiments with cefoxitin (at one-half the MIC) performed with the original AJC68081 strain did not show an increase in the synthesis of the AmpC β-lactamase, measured as specific enzymatic activity (in micromoles of nitrocefin hydrolyzed per minute per microgram of protein) when the inducer was added (159 ± 38 μmol/min/μg of protein without the inducer, 145 ± 15 μmol/min/μg of protein in the presence of inducer). These experiments indicated that the AG3 AmpC β-lactamase in this strain is noninducible.
To purify the AmpC enzyme, the blaampC gene was cloned in the pGEX-6P-1 vector, which allows a fusion protein between glutathione S-transferase and the AmpC enzyme. The β-lactamase was purified at homogeneity with the GST gene fusion system (Amersham Pharmacia Biotech, Europe GmbH) according to the manufacturer's directions. With sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the purified protein appeared as a band of ca. 40 kDa (≥95% purity) (data not shown). Kinetic experiments were performed as previously described (4). The specific activity of semipurified AmpC β-lactamase was 49,000 μmol of nitrocefin/min/μg of protein. The relative enzymatic efficiency (Vmax/Km) values indicated that cephalothin was hydrolyzed with higher hydrolytic efficiency than ampicillin, as expected for a class C β-lactamase (Table 3). This enzyme also showed moderate hydrolytic activity against cefuroxime and cefoxitin and very little hydrolytic activity against cefotaxime and imipenem; therefore, we could not obtain reliable Km and Vmax values for these antibiotics. Hydrolytic activity against ceftazidime was not detected. Fifty percent inhibitory concentrations were calculated as previously reported (4) (Table 3).
TABLE 3.
Kinetic parameters of the Acinetobacter genomic species 3 AmpC β-lactamase
| Antibiotic | Km (μM) | Relative Vmax (%)a | Vmax/Km | Relative Vmax/Km (%)a | Hydrolysis rate (μmol/min/μl)b | Relative hydrolysis rate (%)a | IC50 (μM)c |
|---|---|---|---|---|---|---|---|
| Ampicillin | 53 | 100 | 31 | 100 | 420 | 100 | |
| Cephalothin | 100 | 20,000 | 3330 | 10,600 | 28,400 | 6,770 | |
| Cefoxitin | 1 | 6 | 152 | 482 | 111 | 26 | |
| Cefuroxime | 52 | 46 | 15 | 47 | 63 | 15 | |
| Cefotaxime | NDd | ND | ND | ND | 19 | 4 | |
| Imipenem | ND | ND | ND | ND | 9 | 2 | |
| Ceftazidime | ND | ND | ND | ND | <0.1 | NHe | |
| Clavulanic acid | >250 | ||||||
| Sulbactam | 13 |
Normalized with respect to value for ampicillin (taken as 100%).
Hydrolysis rates were determined by using 100 μM concentrations of the indicated substrates.
IC50, 50% inhibitory concentration.
ND, not done.
NH, no hydrolysis detected.
A PCR assay was performed with seven genotypically different AG3 strains (repetitive extragenic palindromic-PCR tested) to study the presence of the ampC gene. The reactions were carried out with a 50-μl volume of a reaction mixture containing 20 mM Tris-HCl (pH 8.8), 100 mM potassium chloride, 2.0 mM magnesium chloride, 200 μM deoxynucleotide triphosphate, 50 pmol of each oligonucleotide, 0.5 μg of the chromosomal DNA, and 2.5 U of Ecotaq polymerase (Group 3, Vigo, Spain). The primers for the ampC-coding region, P1 forward (5′-ACTTACTTCAACTCGCGACG) and P2 reverse (5′-TAAACACCACATATGTTCCG), were used in the amplification reaction. Amplification conditions were as follows: 10 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C, followed by a final step of 10 min at 72°C. The amplified 663-bp product was resolved by electrophoresis in a 1.5% (wt/vol) agarose gel containing ethidium bromide (50 μg/ml). The ampC gene was disseminated among all of the AG3 isolates (data not shown). This result strongly suggests that AmpC β-lactamase may play a role in the β-lactam resistance of AG3. Moreover, sequence identities among the ampC genes of the seven AG3 isolates studied were between 85 to 95% with respect to the ampC gene of A. baumannii.
The susceptibility of the clinical strain of AJC68081 to the combination of cefotaxime and ceftazidime with clavulanic acid is also remarkable (with a clavulanic acid MIC of 8 to 16 mg/liter [Table 1]). This unusual phenomenon has been detected in several strains of A. baumannii and AG3 and is independent of the presence of any extended-spectrum β-lactamases, which are well inhibited by clavulanic acid (8). It is thought that this finding may be related to penicillin-binding protein alterations which increase the susceptibility to clavulanic acid (unpublished data).
Another important consideration is the role of the β-lactamase under study in resistance to β-lactams in AG3. The MICs for clinical strain AJC68081 were higher than those for the E. coli transformant expressing the G3 AmpC enzyme (Table 1). An explanation for this result could be that other antibiotic resistance mechanisms, such as a loss or a reduction of porin expression, a constitutively basal expression of some efflux pump, or penicillin-binding protein modifications, operate at the same time in A. baumannii and/or AG3, as no other β-lactamases have been detected in clinical strain AJC68081.
The results obtained in the present study show that this AmpC β-lactamase (i) may play an important role in β-lactam resistance in AG3, (ii) was not inducible when cefoxitin was added (at one-half the MIC) and thus can be considered noninducible, and (iii) shows a typical cephalosporinase substrate profile, corresponding to that of a class C β-lactamase.
In summary, we report for the first time the cloning, sequencing, and analysis of the ampC gene and the biochemical characterization of the AmpC β-lactamase from a clinical strain of AG3.
Nucleotide sequence accession number.
The GenBank accession number for the AmpC β-lactamase of AG3 is AJ575184.
Acknowledgments
We thank Instituto de Salud Carlos III for amplified ribosomal DNA restriction analysis of the AG3 clinical and nonclinical strains.
This work was supported by Secretaria Xeral de Investigación e Desenvolvemento, Xunta de Galicia (grant PGIDT01PXR90101PR), and Fondo de Investigaciones Sanitarias (grant PI021415).
REFERENCES
- 1.Afzal-Shah, M., N. Woodford, and D. M. Livermore. 2001. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D β-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 45:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergogne-Bérézin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blechschmidt, B., P. Borneleit, and H. P. Kleber. 1992. Purification and characterization of an extracellular β-lactamase produced by Acinetobacter calcoaceticus. J. Gen. Microbiol. 138:1197-1202. [DOI] [PubMed] [Google Scholar]
- 4.Bou, G., and J. Martinez-Beltran. 2000. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 44:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvet, P. J. M., and P. A. D. Grimont. 1986. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. Nov., Acinetobacter haemolyticus sp. Nov. and amended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 36:228-240. [Google Scholar]
- 6.Bouvet, P. J. M., and P. A. D. Grimont. 1987. Identification and biotyping of clinical isolates of Acinetobacter. Ann. Inst. Pasteur Microbiol. 138:569-578. [DOI] [PubMed] [Google Scholar]
- 7.Bouvet, P. J. M., and S. Jeanjean. 1989. Delineation of new proteolytic genomic species in the genus Acinetobacter. Res. Microbiol. 140:291-299. [DOI] [PubMed] [Google Scholar]
- 8.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu, Y. W., M. Afzal-Shah, E. T. Houang, M. I. Palepou, D. J. Lyon, N. Woodford, and D. M. Livermore. 2001. IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45:710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu, Y. W., C. M. Leung, E. T. S. Houang, K. C. Ng, C. B. Leung, H. Y. Leung, and A. F. B. Cheng. 1999. Skin carriage of acinetobacters in Hong Kong. J. Clin. Microbiol. 37:2962-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerner-Smidt, P., and I. Tjernberg. 1993. Acinetobacter in Denmark: II. Molecular studies of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex. APMIS 101:826-832. [PubMed] [Google Scholar]
- 12.Ghuysen, J. M. 1991. Serine β-lactamases and penicillin-binding proteins. Annu. Rev. Microbiol. 45:37-67. [DOI] [PubMed] [Google Scholar]
- 13.Horrevorts, A., K. Bergman, L. Kollée, I. Breuker, I. Tjernberg, and L. Dijkshoorn. 1995. Clinical and epidemiological investigations of Acinetobacter genomospecies 3 in a neonatal intensive care unit. J. Clin. Microbiol. 33:1567-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houang, E. T. S., Y. W. Chu, K. Y. Chu, K. C. Ng, C. M. Leung, and A. F. Cheng. 2003. Significance of genomic DNA group delineation in comparative studies of antimicrobial susceptibility of Acinetobacter spp. Antimicrob. Agents Chemother. 47:1472-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibrahim, A., P. Gerner-Smidt, and W. Liesack. 1997. Phylogenetic relationship of the twenty-one DNA groups of the genus Acinetobacter as revealed by 16S ribosomal DNA sequence analysis. Int. J. Syst. Bacteriol. 47:837-841. [DOI] [PubMed] [Google Scholar]
- 16.Lodge, J. M., S. D. Minchin, L. Piddock, and J. W. Busby. 1990. Cloning, sequencing and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal AmpC β-lactamase. Biochem. J. 272:627-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mammeri, H., L. Poirel, N. Mangeney, and P. Nordmann. 2003. Chromosomal integration of a cephalosporinase gene from Acinetobacter baumannii into Oligella urethralis as a source of acquired resistance to β-lactams. Antimicrob. Agents Chemother. 47:1536-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthew, M., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 19.Moore, W. E. C., and L. V. H. Moore. 1989. Index of the bacterial and yeast nomenclatural changes, p. 2-3. American Society for Microbiology, Washington, D.C.
- 20.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 21.Nishimura. Y., T. Ino, and H. Hzuka. 1988. Acinetobacter radioresistens sp. nov. isolated from cotton and soil. Int. J. Syst. Bacteriol. 38:209-211. [Google Scholar]
- 22.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 23.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Spratt, B., P. J. Hedge, T. S. Heesen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene 41:337-342. [DOI] [PubMed] [Google Scholar]
- 26.Tjernberg, I., and J. Ursing. 1989. Clinical strains of Acinetobacter classified by DNA-DNA hybridization. APMIS 79:595-605. [DOI] [PubMed] [Google Scholar]
- 27.Vahaboglu, H., R. Ozturk, G. Aygun, F. Coskunkan, A. Yaman, A. Kaygusuz, H. Leblebicioglu, I. Balik, K. Aydin, and M. Otkun. 1997. Widespread detection of PER-1-type extended-spectrum beta-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob. Agents Chemother. 41:2265-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaneechoutte, M., L. Dijkshoorn, I. Tjernberg, A. Elaichouni, P. de Vos, G. Claeys, and G. Verschraegen. 1995. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 33:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visalli, M. A., M. R. Jacobs, T. D. Moore, F. A. Renzi, and P. C. Appelbaum. 1997. Activities of β-lactams against Acinetobacter genospecies as determined by agar dilution and E-test MIC methods. Antimicrob. Agents Chemother. 41:767-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yum, J. H., K. Yi, H. Lee, D. Yong, K. Lee, J. M. Kim, G. M. Rossolini, and Y. Chong. 2002. Molecular characterization of metallo-beta-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospecies 3 from Korea: identification of two new integrons carrying the bla (VIM-2) gene cassettes. J. Antimicrob. Chemother. 49:837-840. [DOI] [PubMed] [Google Scholar]