Abstract
The ability to achieve immunologic tolerance after transplantation is a therapeutic goal. Here, we report interim results from an ongoing trial of tolerance in HLA-identical sibling renal transplantation. The immunosuppressive regimen included alemtuzumab induction, donor hematopoietic stem cells, tacrolimus/mycophenolate immunosuppression converted to sirolimus, and complete drug withdrawal by 24 months post-transplantation. Recipients were considered tolerant if they had normal biopsies and renal function after an additional 12 months without immunosuppression. Of the 20 recipients enrolled, 10 had at least 36 months of follow-up after transplantation. Five of these 10 recipients had immunosuppression successfully withdrawn for 16–36 months (tolerant), 2 had disease recurrence, and 3 had subclinical rejection in protocol biopsies (nontolerant). Microchimerism disappeared after 1 year, and CD4+CD25highCD127−FOXP3+ regulatory T cells and CD19+IgD/M+CD27− B cells were increased through 5 years post-transplantation in both tolerant and nontolerant recipients. Immune/inflammatory gene expression pathways in the peripheral blood and urine, however, were differentially downregulated between tolerant and nontolerant recipients. In summary, interim results from this trial of tolerance in HLA-identical renal transplantation suggest that predictive genomic biomarkers, but not immunoregulatory phenotyping, may be able to discriminate tolerant from nontolerant patients.
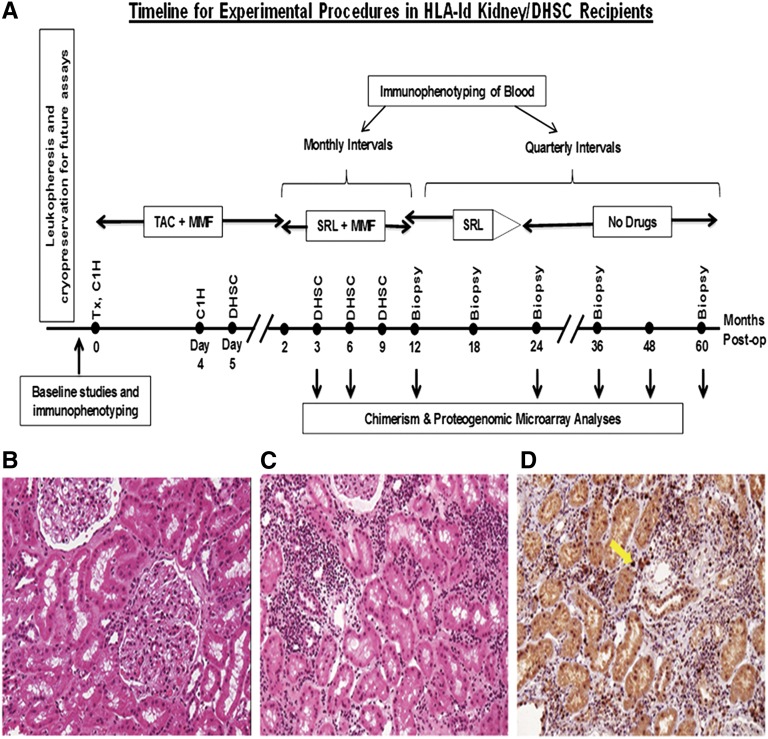
We describe an HLA-identical tolerance trial the without any myeloablation,1–4 but with four immunoselected CD34+ donor hematopoietic stem cell (DHSC) infusions in the first 9 months after renal transplantation.5,6 Temporary immunosuppression consisted of alemtuzumab induction, tacrolimus/mycophenolate converted to sirolimus, followed by complete withdrawal after a normal 24-month biopsy (Figure 1). We herein evaluate the first 10 of 20 patients enrolled, 5 of which passed the 36-month post-transplant milestone, with normal biopsies one year after complete immunosuppression withdrawal, designated as tolerant (Figure 1B and Tables 1 and 2). Two of the remaining five patients, designated as nontolerant (see clinical summaries in the Supplemental Material), had renal disease recurrence, and continued immunosuppression. The remaining three patients had (subclinical) biopsy rejection after complete withdrawal (Figure 1, C and D, and Tables 1 and 2), and immunosuppression was reinstated, solely based on the biopsies, without increase in panel reactive antibodies (PRAs) or positive donor-specific crossmatches, and with unchanged serum creatinine concentrations. The other 10 enrollees have not yet reached 36 months.
Figure 1.
Experimental tolerance inducing protocol and representative milestone tolerant and nontolerant biopsies. (A) Planned tolerance trial protocol timeline for treatment procedures and monitoring of HLA-identical recipients up to 5 years postoperatively. (B) H&E stain of the 36-month transplant biopsy of patient 2 (Table 1) reads as normal 12 months after immunosuppression is withdrawn. This is also typical for patients 5, 6, 8, and 9 (i.e., the tolerant group) (Tables 1 and 2). (C) H&E stain of the transplant biopsy of patient 1 (Table 1) reads as Banff 1A acute rejection (with normal renal function) obtained after 12 months off immunosuppression. (D) FOXP3 stain of this biopsy. Note that the stain appears positive (arrow) in approximately 10% of the infiltrating cells (approximately 50% CD4+ cells, not shown), possibly indicative of a less severe inflammatory component. Similar findings occur in the other Banff 1A biopsies (patients 3 and 4; Table 1). However, there are very few cells to stain for FOXP3 in the tolerant recipients. H&E, hematoxylin and eosin.
Table 1.
Patient demographics and clinical courses in the first 10 renal transplant recipients in the HLA-identical DHSC tolerance protocol
| Patient | Recipient | Donor | Preoperative Diagnosis of Native Disease | Peak PRA Pre-Tx (%) | Time Post-Tx (mo) | Cr (mg/dl)b | Latest Urine Protein per 24 h/ cells | Latest Tx Biopsy Results (mo) | Current Daily IS Dose | Time off of IS (mo) | Maximum Microchimerism in Year 1 Postoperativelyc | Current Microchimerism in Blood (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (yr)a | Sex | Race | Age (yr)a | Sex | Race | % in Blood | % in Bone Marrow | ||||||||||
| 1 | 51 | M | Caucasian | 49 | F | Caucasian | Polycystic | 5 | 60 | 1.2 | 0/0 | Banff 1A: 54 | Sirolimus 2 mgd | 12d | 0.016 | 0.02 | 0 |
| 2 | 54 | M | Caucasian | 56 | M | Caucasian | Multicystic | 9 | 57 | 1.1 | 0/0 | Neg: 36 (Tol)e,f | 0 (Tol)f | 32f | 0.1 | NT | 0 |
| 3 | 46 | F | Caucasian | 45 | M | Caucasian | IgAN | 29 | 50 | 0.8 | 0/0 | Banff 1A: 25 | MMF 1 g 2×d | 1d | 0.1 | NT | 0 |
| 4 | 41 | M | Hispanic | 39 | F | Hispanic | Type 2 diabetes mellitus | 0 | 48 | 1.6 | 0/0 | Banff 1A: 36 | Tacro 2 mg 2×, MMF 1 g 2×d | 12d | 0.01 | 0.03 | 0 |
| 5 | 39 | M | Caucasian | 33 | F | Caucasian | IgAN | 0 | 45 | 1.4 | 0/0 | Neg: 36 (Tol)e,f | 0 (Tol)f | 20f | 2.3 | NT | 0 |
| 6 | 38 | M | Caucasian | 35 | M | Caucasian | IgAN | 0 | 45 | 1.2 | 0/0 | Neg: 36 (Tol)e,f | 0 (Tol)f | 20f | 1.7 | 3 | 0 |
| 7 | 20 | M | Caucasian | 22 | F | Caucasian | Unknowng | 0 | 42 | 1.1 | <1 g/0 | Disease recurrence: 18 | MMF 1 g 2×d | 0d | 0.06 | NT | 0 |
| 8 | 24 | M | Hispanic | 21 | M | Hispanic | Obstructive | 11 | 41 | 0.9 | 0/0 | Neg: 36 (Tol)e,f | 0 (Tol)f | 17f | 0.47 | 8.5 | 0 |
| 9 | 39 | M | Caucasian | 30 | M | Caucasian | IgAN | 0 | 39 | 1.4 | 0/0 | Neg: 36 (Tol)e | 0 (Tol)f | 15f | 0.01 | NT | 0 |
| 10 | 45 | M | African American | 38 | F | African American | Nephrosclerosis | 15 | 36 | 2.1 | 4 g/0h,i | FSGS: 24i | MMF 1 g 2×d | 0d | 0.21 | 0.43 | 0 |
Tx, transplantation; Cr, creatinine; IS, immunosuppression; M, male; F, female; IgAN, IgA nephropathy; Tol, tolerant; NT, not tested; MMF, mycophenolate mofetil; Tacro, Tacrolimus.
At transplantation.
Most recent serum creatinine; all values still reflect postoperative nadirs.
PBMC and bone marrow microchimerism (up to 8.5% [patient 8] by short tandem repeat analysis) lasted up to 1 year only.
IS reinstated, or never withdrawn (see text), but with no change in renal function; also see text (Supplemental Material) describing adverse event postponing 24-month biopsy of patient 1–30 months.
Neg includes no acute or chronic rejection.
Tolerant indicates off immunosuppression for at least 1 year, with a normal transplant biopsy.
Native kidney disease diagnosis was unknown (biopsies inconclusive), but possibly immune mediated, with the 18-month post-transplant biopsy showing dense deposit disease on electron microscopy.
Disease recurrence; preoperatively undocumented.
Intent to treat without further immunosuppression withdrawal because exclusion criteria were previously unrecognized.
Table 2.
Absolute copy numbers of mRNA in urinary cells per 1 μg total RNA
| Group, Patient No. | CD3 | CD20 | CD25 | CD103 | CXCR3 | IP10 | MIG | PERF | GB | PI9 | IL2 | IFN-r | IL4 | IL10 | TGFB1 | FOXP3 | CTLA4 | TLR5 | BK Virus VP1 | 18S rRNA Copies per 1 μg Total RNA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tolerant | ||||||||||||||||||||
| 2 | 47,997 | 746 | 798 | 653 | 4497 | 1308 | 1255 | 1904 | 1853 | 19,767 | 193 | 333 | 0 | 1028 | 99,225 | 640 | 3460 | 5475 | 0 | 5.18E+08 |
| 5 | 28,250 | 569 | 1452 | 2301 | 10,863 | 6317 | 20,495 | 4800 | 2818 | 3727 | 0 | 386 | 0 | 356 | 25,107 | 1153 | 91 | 1195 | 0 | 2.14E+08 |
| 6 | 3003 | 398 | 201 | 664 | 3706 | 157 | 451 | 854 | 842 | 3087 | 0 | 0 | 0 | 479 | 15,963 | 188 | 29 | 1121 | 0 | 1.22E+08 |
| 8 | 6798 | 5136 | 190 | 144 | 2192 | 1518 | 12,002 | 1261 | 441 | 3604 | 13 | 95 | 13 | 102 | 4414 | 97 | 283 | 3092 | 0 | 2.57E+09 |
| 9 | 24,546 | 1561 | 604 | 304 | 7775 | 2387 | 7244 | 3826 | 1371 | 3105 | 0 | 42 | 0 | 551 | 22,760 | 1898 | 256 | 2178 | 0 | 4.29E+08 |
| Mean | 22,119 | 1682 | 649 | 813 | 5807 | 2337 | 8289 | 2529 | 1465 | 6658 | 41 | 171 | 3 | 503 | 33,494 | 795 | 824 | 2612 | 0 | 7.72E+08 |
| SEM | 8103 | 886 | 232 | 385 | 1559 | 1057 | 3707 | 763 | 414 | 3280 | 38 | 79 | 3 | 152 | 16,821 | 334 | 661 | 802 | 0 | 4.56E+08 |
| Nontolerant | ||||||||||||||||||||
| 1 | 64,091 | 12,963 | 5837 | 784 | 10,692 | 2438 | 8020 | 7430 | 2825 | 58,721 | 90 | 350 | 230 | 5004 | 107,235 | 2994 | 3198 | 6892 | 0 | 7.26E+08 |
| 3 | 5961 | 3889 | 2255 | 88 | 522 | 64 | 458 | 1421 | 863 | 2983 | 60 | 13 | 13 | 179 | 8924 | 613 | 169 | 1536 | 0 | 1.36E+09 |
| 4 | 67,001 | 13,913 | 3134 | 1269 | 16,416 | 24,374 | 99,032 | 10,946 | 12,848 | 37,487 | 341 | 651 | 0 | 3534 | 98,861 | 2041 | 2903 | 5811 | 0 | 1.09E+09 |
| 7 | 32,040 | 2849 | 1625 | 555 | 5749 | 2098 | 4513 | 3522 | 4871 | 9855 | 101 | 36 | 0 | 4473 | 70,882 | 1060 | 1051 | 5141 | 0 | 4.78E+08 |
| 10 | 49,386 | 751 | 4518 | 1919 | 10,858 | 6848 | 26,386 | 9566 | 8784 | 54,651 | 190 | 258 | 0 | 16,656 | 230,633 | 943 | 2348 | 13,304 | 0 | 2.40E+09 |
| Mean | 43,696 | 6873 | 3474 | 923 | 8847 | 7164 | 27,682 | 6577 | 6038 | 32,739 | 156 | 262 | 49 | 5969 | 103,307 | 1530 | 1934 | 6537 | 0 | 1.21E+09 |
| SEM | 11,294 | 2732 | 765 | 313 | 2680 | 4442 | 18,381 | 1798 | 2149 | 11,373 | 51 | 117 | 45 | 2800 | 36,199 | 437 | 575 | 1916 | 0 | 3.33E+08 |
| Mann–Whitney | ||||||||||||||||||||
| P value | 0.15 | 0.10 | 0.01 | 0.80 | 0.53 | 0.41 | 0.53 | 0.15 | 0.056 | 0.22 | 0.10 | 0.80 | 0.72 | 0.10 | 0.22 | 0.31 | 0.31 | 0.10 | 0.22 |
Levels of mRNA were measured with the use of preamplification enhanced real-time quantitative PCR assays using gene-specific oligonucleotide pairs and TaqMan probes. Mean and SEM of the mean copy number of each mRNA measure are shown. Patients are grouped by phenotypic designation as tolerant and nontolerant because of the biopsy findings (see text and Table 1). All tolerant patients had been withdrawn from immunosuppression for ≥1 years at the time of urinary mRNA sampling (between 36 and 48 months post-transplantation), whereas all nontolerant recipients, requiring either continuing immunosuppression (patients 7 and 10) or immunosuppressive reinstitution (patients 1, 3, and 4), had samples obtained during this same interval.
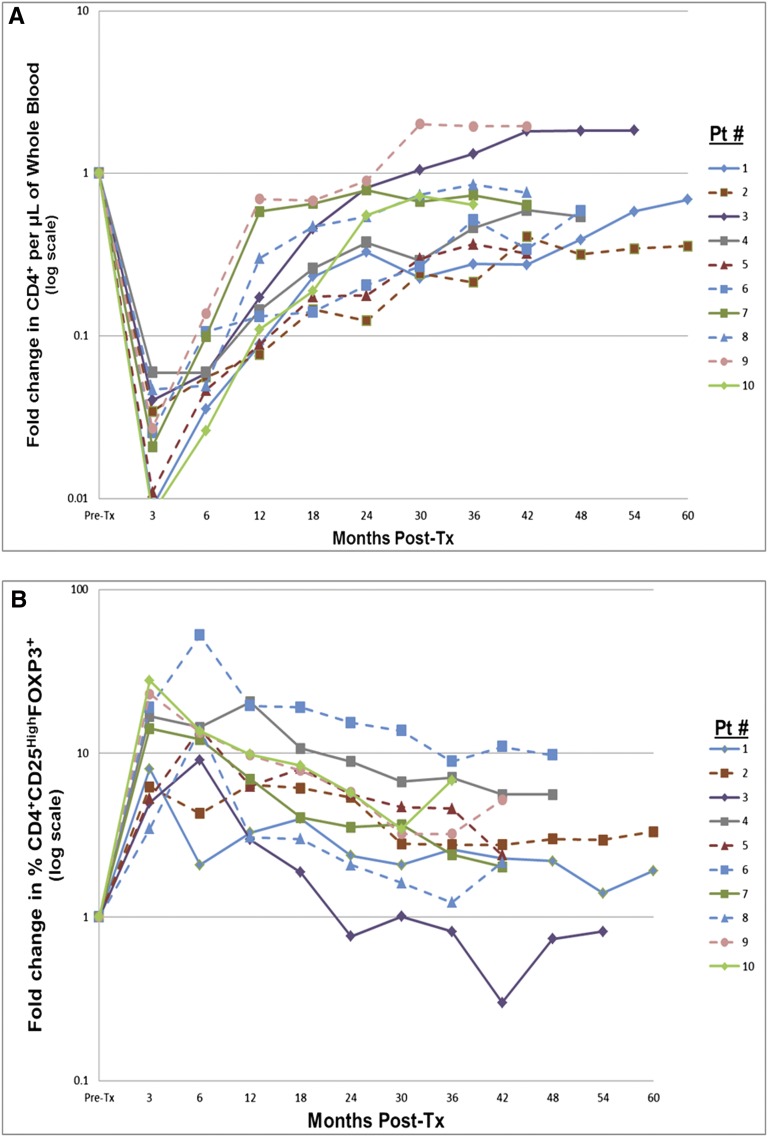
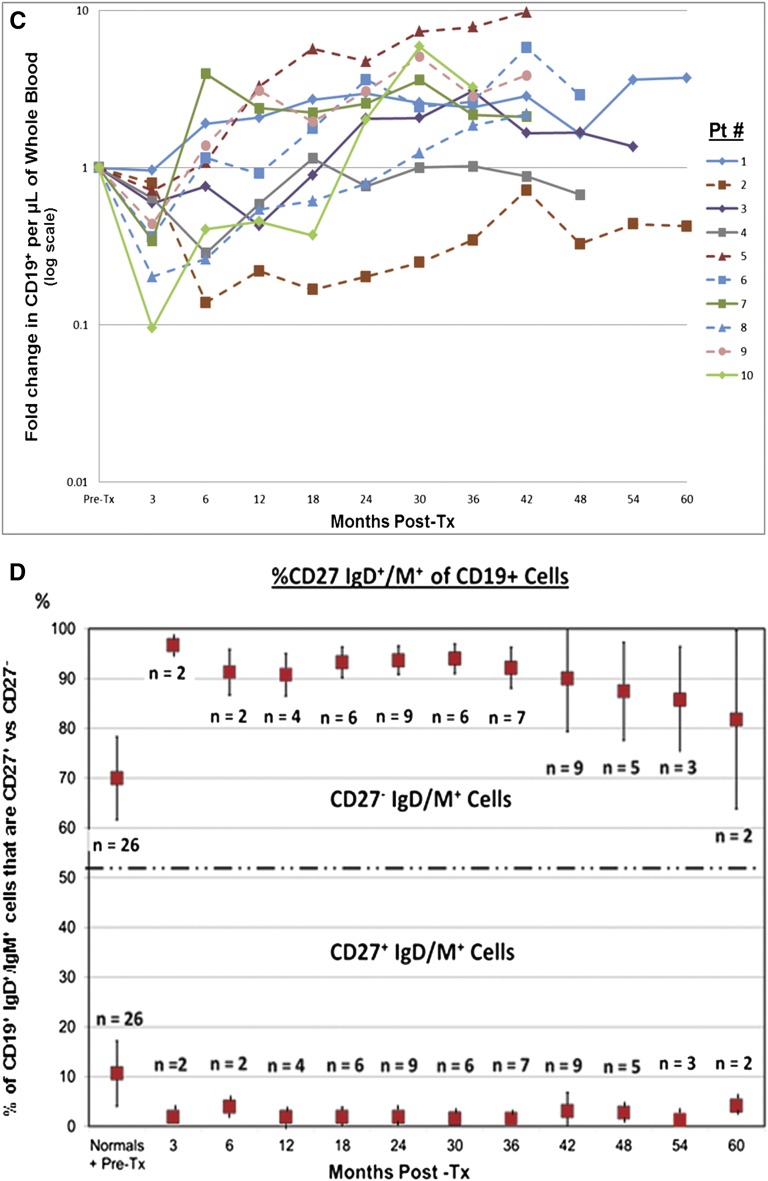
The total number of donor CD34+ purified cells infused ranged between 3.74 and 14.40×106/kg recipient body weight. Only temporary PBMC chimerism was observed during the first year (not above 3%), unrelated to tolerance or nontolerance (Table 1). Prolonged CD4+ T cell depression (P<1.0×10−7; Figure 2A) and parallel CD8+ changes (not shown) persisted through 60 months post-transplantation. Increased percentages of CD4+CD25highCD127−FOXP3+ cells (phenotypic regulatory T cells; Figure 2B), some over 10 times pretransplant values (P=0.01), also persisted through 60 months. CD19+ B cells reached more varied nadirs, but rebounded to prolonged higher levels than before transplantation (P=0.01; Figure 2C). Parallel increases in CD19+IgD/IgM+CD27− (naïve) B cells also persisted through 60 months post-transplantation (Figure 2D), similar to but longer than recently described.7 All of the above changes occurred independent of the tolerance versus nontolerance designation. Other PBMC immunophenotypic changes (CD14, CD56, CD25, CD3/CD16) were also indistinguishable (not shown).
Figure 2.
T and B cell monitoring. (A) Prospective monitoring of recipient CD4+ T cell immunophenotyping for at least 36 months (up to 60 months) postoperatively. Preoperative values have all been normalized to 1. Note that with one exception (outlier) in each group in these first 10 of 20 enrollees, there is a profound and continuing depression in both the nontolerant group (solid lines) and tolerant group (dashed lines), indistinguishable from each other. The vertical axis is logarithmic. The depression actually occurred within 1 week postoperatively, but is recorded here at 3 months and then at 6-monthly intervals. (B) Prospective monitoring of recipient immunophenotypic regulatory T cells (CD4+CD25highCD127−FOXP3+cells) for at least 36 months postoperatively. Preoperative values have all been normalized to 1. Note the prolonged increase in the percentages of this subset lasting up to 60 months postoperatively, indistinguishable between nontolerant (solid lines) and tolerant (dashed lines) groups. Vertical axis is logarithmic. These percentages could only be measured from 3 months onward because of the dearth of CD4+ T cells before that time. (C) Prospective monitoring of recipient CD19+ B cells (pan-B cell epitope) immunophenotyping for at least 36 months (up to 60 months) postoperatively. Preoperative values have all been normalized to 1. Note that the early depression, although similar to the CD4+ subset, is neither as marked nor as prolonged, and that there is an overshoot above preoperative values in both the nontolerant (solid lines) and tolerant (dashed lines) groups that is indistinguishable. Vertical axis is logarithmic in each figure. Although not indicated, these changes appeared earlier as also described in A. (D) The early conversion from preoperative values, reflecting the expected mixture of naïve and activated/memory B cells, to postoperative values favoring naive B cells. Preoperative mean ratios of 7:1 naïve:activated/memory B cells changed to postoperative mean ratios of 20:1. This change persisted from early postoperatively for at least 60 months. Although not indicated in the figure, there is again no difference between the nontolerant and tolerant groups. (This is included as Supplemental Table 1.) This study initiated after the first several enrollees were being followed. It includes patients 19 and 20 so that the earlier preoperative and postoperative time points could be indicated. Preoperative values depicted also include those for 18 normal volunteers and 8 ESRD patients before transplantation, 6 (with their consent) who were not in this trial. There were no differences between these two latter groups. From 12 months onward, the values are only from the first 10 recipients enrolled, as in the remainder of this report. Pt, patient; Post-tx, post-transplantation.
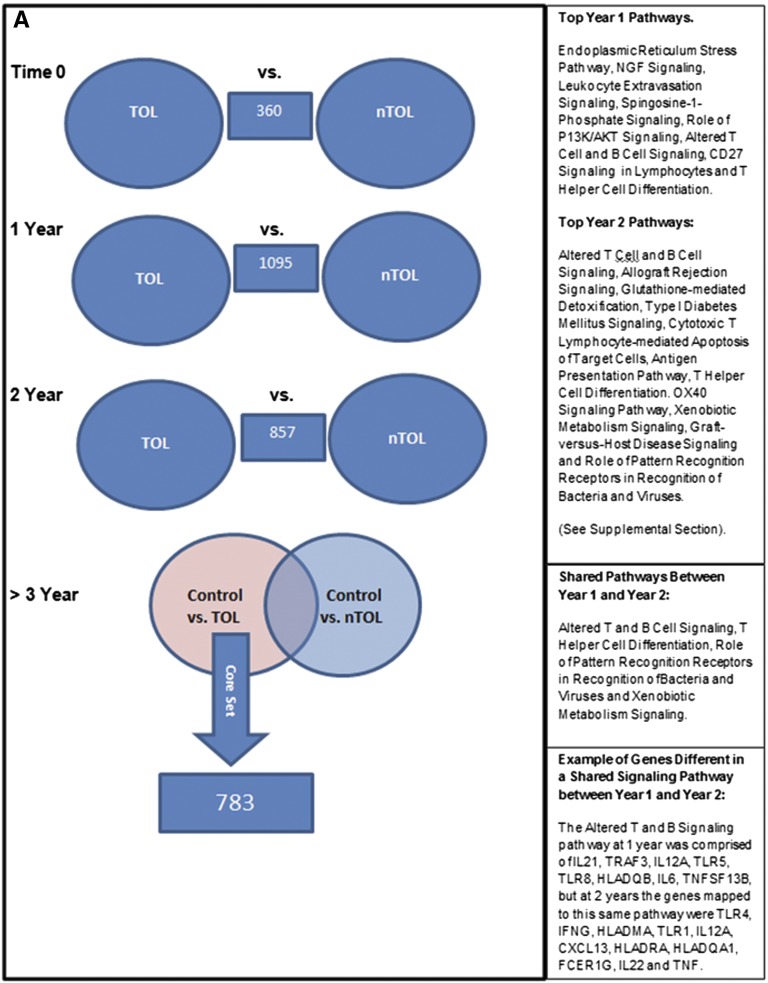
In contrast, PBMC global gene expression profiling and urinary quantitative mRNA PCR correlated with tolerance. PBMC global gene expression studies compared serial samples from five tolerant and four nontolerant patients (Figure 3A). Before transplantation, only 360 genes were differentially expressed at relatively low levels and significance, not mapping to any coherent set of molecular pathways, suggesting that patients were not significantly different pretransplantation. In contrast, 1095 and 857 genes were differentially expressed (P<0.001), 1 year (sirolimus conversion) and 2 years (complete withdrawal) post-transplantation, and mapped significantly (P<0.05) to 35 and 47 molecular functional pathways, respectively (Supplemental Material). Strikingly, differential gene expression for these two time points mapped to different pathways (Supplemental Figure 1). Of the 1095 and 857 differentially expressed genes, 55% and 42% were downregulated in tolerant versus nontolerant patients respectively, specifically for inflammatory genes (65% and 35%, respectively). The better suppression of the 1-year signature in the tolerant group may signal/favor tolerance development and, at this early point post-transplantation, the opportunity to safely continue weaning immunosuppression, whereas the 2-year signature may be more reflective of weaning immunosuppression, but not indicative of failure of tolerance assessed 1 year later. We also identified four functional pathways that were differentially shared between tolerant and nontolerant recipients by comparing differential gene expression between years 1 and 2. However, these were populated with a different set of genes at each time (an example of which is shown in Supplemental Figure 1), consistent with significant differences in immunosuppression protocols at those time points, as called for by the weaning protocol.
Figure 3.
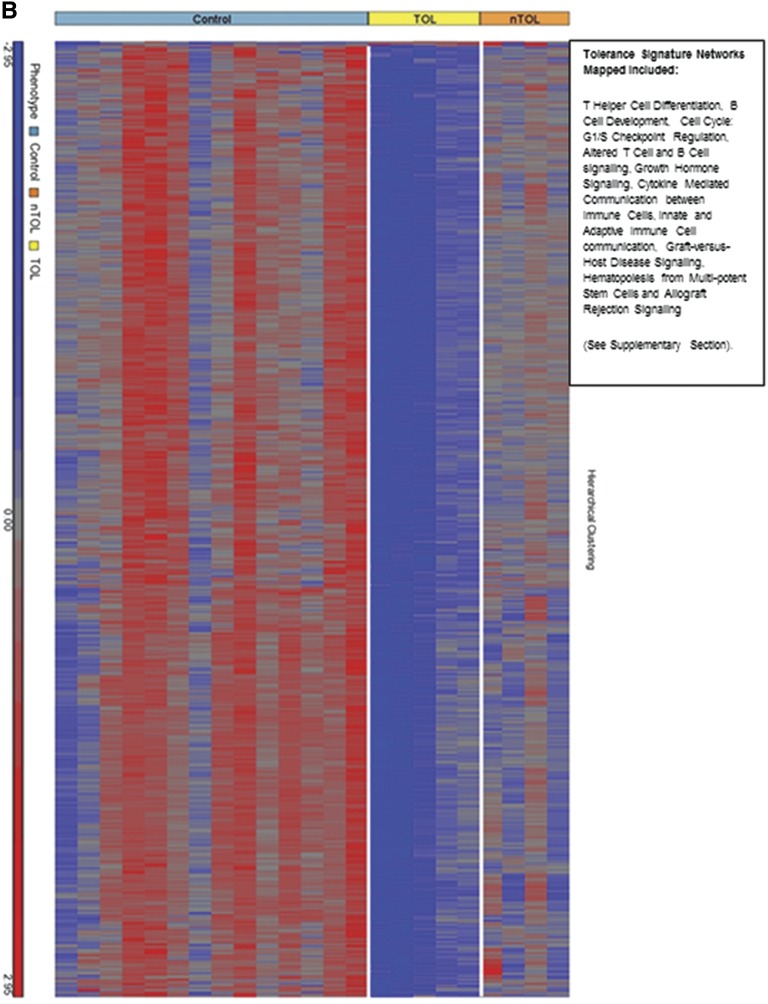
Genomic differences between tolerant and nontolerant recipients. (A) Schematic diagram of the differential gene expression comparisons made between various sample classes in the study. Numbers in the blue boxes represent the differentially expressed genes from the comparisons made in those circles. The right panels enumerate the pathways involved. The numbers in the right lower (overlapping) circle represents the genes “unique” to the control versus the tolerant group. (B) Heat map showing the hierarchical clustering of the 783 differentially expressed genes that were unique in the control versus tolerant comparison at the 3-year time point. Red indicates signals that are greater than the mean signal intensity of the given gene, blue indicates signals that are lower than the mean, and gray indicates signal intensities that do not vary much from the mean intensity. Tol, tolerant; nTol, nontolerant; Control, nonimmunosuppressed normal volunteers.
After 3 years post-transplantation, the tolerant group remained off of immunosuppression, whereas the nontolerant group was still on immunosuppression. Therefore, we compared healthy controls (no immunosuppression) with nontolerant patients (immunosuppression), and subtracted differentially expressed genes from this analysis from differentially expressed genes between healthy (no immunosuppression) and tolerant (no immunosuppression) patients, leaving a tolerance signature of 2169 unique genes. Because some genes differentially expressed in the nontolerant group might not make the conservative P<0.01 cutoff, any differentially expressed genes with a P<0.05 cutoff between healthy and nontolerant participants were subtracted from those differentially expressed between the healthy versus tolerant groups, kept at the more stringent P<0.001, leaving a core set of 783 genes differentially expressed only in tolerance (Figure 3A). Notably, 780 of 783 genes were downregulated in the tolerant versus healthy participants, despite none of the tolerant participants being on immunosuppression. The majority of participants (n=512, 66%) were also downregulated in the tolerant versus the nontolerant (immunosuppression) patients. A heat map of these 783 genes is shown in Figure 3B for each subject by all groups. Mapping of the top functional pathways revealed 16 significant pathways (Figure 3B). Function of these downregulated genes included 66 immune/inflammatory and cytokine genes as well as growth factors, kinases, transcription factors, and G protein–coupled receptors linked to immunity and inflammation.
The urinary cell mRNA levels in the five tolerant patients trended 2- to 3-fold lower (despite not being on any immunosuppression for at least 12 months) than the nontolerant group still on therapy (Table 2), and were not statistically significant, except for CD25 mRNA (P=0.01), and were almost significant for granzyme B mRNA (P=0.06).
Lifetime immunosuppression remains necessary even in HLA-identical renal transplantation,8 despite recent reports that late dose reductions might result in stable function,9,10 or that “spontaneous operational tolerance” may occur11,12 without deliberate attempts to induce it. There are presently three renal transplant tolerance trials that have used “nonmyeloablative” conditioning and DHSC1–4 that have resulted in both transient and permanent multi-lineage chimerism.2,3 One trial showed that in HLA-identical pairs, complete withdrawal was successful in two thirds with stable mixed chimerism in one third.2 In our HLA-identical study, prolonged immunoregulation deliberately induced by DHSC infusions, alemtuzumab induction, and temporary sirolimus allowed planned immunosuppressive withdrawal without chimerism in 5 of the first 10 patients enrolled.
Ex vivo tolerance assays are challenging,1,13,14 especially with HLA identity,15 and high zone chimerism may represent the most predictive biomarker.14,16 However, HLA-identical pairs might be unique for genomic interpretation because these would not be complicated by the noise of MHC-linked immune response polymorphisms (i.e., polymorphic responsiveness against disparate donor HLA antigens being eliminated).17,18 In this respect, no (early) tolerance correlation occurred with individual A, B, C, DR, DQ, or DP HLA loci.
Although this is an interim report, our biomarker data give credence to the notion that genomic signatures may predict tolerance, and reflect tolerance once off immunosuppression. Thus, our sequential 3-year genomic PBMC analysis appears to discriminate between tolerant and nontolerant patients at 1 year (patients on sirolimus, only 1095 genes and 65% of inflammatory genes downregulated in tolerant patients), and at 2 years (when tolerant patients were weaned off immunosuppression—857 genes and 4 pathways), with 69 genes shared between the two time points. These results suggest an evolving molecular signature that may signal both successful drug weaning and a tolerance outcome. We also identified a tolerance signature 1 year off immunosuppression comprising 783 genes, 780 of which were significantly downregulated compared with both normal healthy controls and nontolerant patients despite the latter being on immunosuppression.
These early results suggest two intriguing hypotheses. The first is that the tolerant patients, even in the absence of chimerism, are naturally immunosuppressed by a regulatory activity resembling regulatory T cells and/or regulatory B cells,19–21 not detected by the immunophenotyping thus far described. Alternatively, the tolerant patients may have a form of relative immunodeficiency created by the DHSC infusion protocol, not evident in the nontolerant group. In addition, with tolerance signature genes significantly downregulated compared with the nontolerant patients, this suggests that our current immunosuppressive drugs may not adequately suppress important immune/inflammatory pathways, with further implications regarding long-term allograft dysfunction and chronic rejection. Notably, the signature at 2 years, just after complete immunosuppression withdrawal, is significantly different from the signature 1 year later, with the final state of tolerance speculatively evolving in distinct stages over the 3-year time frame studied.
Consistent with PBMC microarray profiles, urinary cell mRNA levels trended lower in the tolerant (no immunosuppression) versus the nontolerant group (immunosuppression), despite the small number of participants profiled. The CD25 mRNA, a marker of activated T cells, was significantly lower, and several additional mRNAs previously associated with acute cellular rejection (CD3ε, granzyme B, perforin, IP-10, CXCR3, and CD10312) all trended lower. Although levels of mRNA for tolerance markers (FOXP3, CTLA-4, TGF-β1, IL-10, and CD20) were numerically higher in the nontolerant group on immunosuppression,12 these have also been associated with rejection.22,23 Yet, none of the five patients designated nontolerant had graft dysfunction and might easily have been classified as “operationally tolerant,” without protocol-mandated biopsies.
Limitations of this study include the interim nature of the report with only 10 of 20 enrolled patients reaching the phenotypic milestone, and small sample sizes constituting only preliminary discovery sets for functional genomic profiling. Moreover, the two patients with recurrent disease who were considered nontolerant might be tolerant to histocompatibility antigens but still have immunopathology of their original renal disease, especially absent chimerism.24 Despite these shortcomings, we believe that this report is warranted because of the strong suggestion that predictive genomic biomarkers may be able to discriminate tolerant from nontolerant patients, absent either chimerism, or correlation with immunoregulatory phenotyping.
Concise Methods
Informed Consent and Inclusion/Exclusion Criteria
Informed consent was obtained under joint supervision of Northwestern University and the Jesse Brown Veterans Affairs Medical Center Institutional Review Boards, an extramural Data Safety Monitoring Board, the National Institute of Diabetes and Digestive and Kidney Diseases sponsor, and a US Food and Drug Administration–approved investigational new drug application. Inclusion criteria included primary renal transplant recipients aged >18 years, and excluded conditions in which recipient renal disease recurrence was considered likely, and recipients with PRAs >20% by either cytotoxicity or (more commonly) flow cytometry. All had negative crossmatches against their donors.
Immunosuppression and Withdrawal
The immunosuppressive protocol (Figure 1A) consisted of two intravenous doses of 0.3 mg/kg of Campath (alemtuzumab), the first intraoperatively and the second on day 4 postoperatively. Early maintenance therapy consisted of FK506 (tacrolimus) to obtain trough levels of 8–10 ng/ml and 500 mg of concomitant mycophenolate mofetil twice daily dosed to maintain at least a minimal nucleated white blood cell count of >700/µl. At 3 months post-transplantation, tacrolimus was replaced by sirolimus (trough levels of 8–10 ng/ml). Mycophenolate was discontinued between 12 and 18 months and sirolimus was discontinued (complete immunosuppressive withdrawal) by 24 months post-transplantation.
HLA Typing and Antibody Assays
Confirming HLA identity and flow cytometric crossmatching included the use of sequence specific oligonucleatide probe hybridization molecular typing for HLA A, B, C, DR, DQ, and DP alleles. When possible, genotypes were confirmed with parents of the sibling pairs. Solid phase-based PRA testing was performed using Labscreen flow beads (One Lambda Inc., Canoga Park, CA). Donor-specific flow cytometric crossmatching and cytotoxicity PRA screening were performed preoperatively and yearly post-transplantation as previously described.25
Recipient DHSC Infusion Protocol
The DHSC protocol consisted of four infusions. The first DHSC product was obtained intraoperatively by bilateral aspiration of approximately 750 ml of donor iliac crest bone marrow after the donor kidney had been removed and before cessation of anesthesia. From this product, CD34+ selection and immediate cryopreservation were performed. Then DHSC infusions into the recipients were given on postoperative day 5, 1 day after the second alemtuzumab dose (Figure 1). The number of cells infused varied between 0.3 and 1.0×106 cells/kg recipient lean body weight. Three months post-transplantation, the donors underwent two successive daily leukaphereses for CD34+ Neupogen mobilized DHSC immunoselection from peripheral blood. From the first day’s leukapheresis product, immunoselected CD34+ cells were infused into the recipient freshly that day. From the second day’s product, the immunoselected CD34+ cells were cryopreserved in two equal aliquots and then infused at 6 and 9 months postoperatively, respectively. At least 0.7×106 cells/kg recipient body weight were infused each time, containing at least 100-fold fewer CD3+ cell contaminants.
Chimerism Testing
Chimerism analysis was performed by Dr. David Senitzer of the Gift of Hope Medical Center (Duarte, CA) and in the Histocompatibility Laboratory of the Northwestern Comprehensive Transplant Center. This consisted of real-time PCR using tandem repeat sequences of DNA isolated from recipient cryopreserved PBMC and iliac crest bone marrow aspirations obtained sequentially at 3, 6, and 12 months postoperatively using (nonchimeric) donor and recipient PBMC obtained preoperatively as standards. The sensitivity of the assay was 0.001%.
Flow Cytometry Analyses
Recipient heparinized peripheral blood and iliac crest marrow aspirates were labeled with mAbs for membrane epitope detection of PBMC and bone marrow subsets and intracellular detection of FOXP3. The labeled membrane epitopes were for T cells and subsets (CD3, CD4, CD8, and CD127) or activation markers (CD25 and CD28), for B cells and subsets (CD19, IgM/IgD, CD5, and CD27), and for monocytes (CD14) and natural killer cells (CD56), all directly conjugated with one of the following five fluorochromes: FITC, phycoerythrin, electron-couple dye, phycoerythrin-cyanin 5, and phycoerythrin-cyanin 7 (all from Beckman-Coulter, Miami, FL). The FOXP3 monoclonal reagent was obtained from e-Bioscience (San Diego, CA). Red cells were treated with lysing solution (Beckman-Coulter) followed by washings. Then 1×105 cellular events were acquired on a five-color FC 500 flow cytometer. For intracellular FOXP3 detection, Ficoll gradients were first used to isolate mononuclear cells.
Biopsy Processing
Protocol transplant biopsies were obtained preimplantation, at 12, 18, 24, and 36 months postoperatively, or at other times for cause; stained routinely using hematoxylin and eosin, periodic acid–Schiff, and Trichrome; and when indicated, immunostaining and electron microscopy were used.
Peripheral Blood Genomics (mRNA Analyses)
A total of 46 PBMC samples representing blood draws from four time points in the first nine recipients were processed for microarray analysis (The Scripps Research Institute, La Jolla, CA). The analyzed time points were as follows: immediately preoperatively in the absence of immunosuppression (n=9), postoperatively at 1 year (n=8; range, 11–13 months), at 2 years (n=12; range, 18–25 months), and >3 years (n=17; range, 32–48 months). To note, at year 2 and at >3 years, repeated samples were obtained from individual participants; at 1 year, one participant had a technically unsatisfactory sample. To discount the effects of immunosuppression on gene expression, microarray data were included on whole blood from 18 healthy human participants (controls: GSE40586; NCBI Gene Expression Omnibus [GEO] repository). All samples were prepared using standard Affymetrix protocols and analyzed on Hu Gene 1.1 ST microarrays, composed of exon-level probe sets for the entire human transcriptome. CEL files from all samples (n=46 from study participants, n=18 from controls) were normalized using the Robust Multichip Average and the normalized signals were analyzed for significant differential gene expression signatures by ANOVA, including multiple corrections testing using Partek Genomics Suite 6.6. Ingenuity Pathway Analysis software was used to map the genes to functional pathways. The gene expression results presented in this manuscript are available from the GEO repository using accession number GSE45593.
Urinary Cell MRNA Profiling
Urine samples (approximately 50 ml) were collected and cell pellets were prepared with the use of a standard protocol for urine cell sedimentation, stored at −80°C and shipped to the Gene Expression Monitoring (GEM) Core at Weill Cornell Medical College (New York, NY). Total RNA was isolated from the pellets using a commercial kit and the quantity (absorbance at 260 nm) and purity (ratio of the absorbance at 260 and 280 nm) of the isolated RNA were measured using the NanoDrop ND-1000 UV-Vis spectrophotometer (Thermo Scientific). A RNA sample was classified as quality control passed if the 18S rRNA copy number was ≥5×107 and its TGF-β1 mRNA copy number was ≥100 copies in 1 μg of RNA. The total RNA was reverse transcribed to cDNA using the TaqMan RT kit (cat. N808-0234; Applied Biosystems) on the same day. The GEM Core designed gene-specific oligonucleotide primers and TaqMan fluorogenic probes (hydrolysis probes) were used for the measurement of levels of mRNAs, 18S rRNA, and BKV-VP-1 mRNA in the real-time quantitative PCR assays using a two-step process (preamplification step followed by measurement of mRNAs with an ABI Prism 7500 fast detection system; 18S rRNA and BKV-VP-1 mRNA were measured without preamplification with gene-specific primers). Absolute levels of mRNAs were calculated using the standard curve method and transcript abundance was reported as mRNA copies per 1 µg RNA. The GEM Core was blinded to any clinical information and as to tolerant or nontolerant status until PCR assay results were reported to the clinicians.
Statistical Analyses
Data were analyzed as the mean ± SD or SEM. Parametric (paired t tests) and nonparametric (Mann–Whitney U test/Wilcoxon signed-rank tests) were used among compared groups. Significance was established at two-sided α levels of 0.05 using statistical software (SAS Inc., Cary, NC).
Disclosures
None.
Acknowledgments
The authors acknowledge the technical expert participation of Xuemei Huang, Li Chen, Tony Mondala, and Terri Gelbart. We also are very grateful to Sue Benning and Alina Ibrahim for their desktop publishing skills.
J.M.M. and J.M. were supported by National Institutes of Health Grant R01-DK025243 and Veterans Affairs Merit Review Support I01-CX000323. D.R.S., S.M.K., T.M., and T.G. were supported by National Institutes of Health Grants U19-AI063603 and U01-AI084146.
Footnotes
J.R.L. and J.M.M. contributed equally to this work and should both be considered first authors.
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/10.1681/ASN.2013010068/-/DCSupplemental.
References
- 1.Kawai T, Cosimi AB, Spitzer TR, Tolkoff-Rubin N, Suthanthiran M, Saidman SL, Shaffer J, Preffer FI, Ding R, Sharma V, Fishman JA, Dey B, Ko DSC, Hertl M, Goes NB, Wong W, Williams WW, Jr, Colvin RB, Sykes M, Sachs DH: HLA-mismatched renal transplantation without maintenance immunosuppression. N Engl J Med 358: 353–361, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scandling JD, Busque S, Dejbakhsh-Jones S, Benike C, Millan MT, Shizuru JA, Hoppe RT, Lowsky R, Engleman EG, Strober S: Tolerance and chimerism after renal and hematopoietic-cell transplantation. N Engl J Med 358: 362–368, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Leventhal J, Abecassis M, Miller J, Gallon L, Ravindra K, Tollerud DJ, King B, Elliott MJ, Herzig G, Herzig R, Ildstad ST: Chimerism and tolerance without GVHD or engraftment syndrome in HLA-mismatched combined kidney and hematopoietic stem cell transplantation. Sci Transl Med 4: 124ra128, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dave SD, Vanikar A, Trivedi HL, Gumber MR, Patel HV, Shah PR, Kute VB. Stem cells versus donor specific transfusions for tolerance induction in living donor renal transplantation: A single-center experience.. Transplantation 95: 155–160, 2013 [DOI] [PubMed] [Google Scholar]
- 5.Miller J, Leventhal J, Friedewald J, Levitsky J, Charette J, Huang X, Gopalakrishnan MT, Chandrasekaran D, Tambur AR, Mathew JM: Enhanced immunoregulatory profiles in HLA identical renal transplant recipients given donor hematopoetic stem cells alemtuzumab and sirolimus followed by immunosuppression withdrawal [Abstract]. Am J Transplant 11: 92, 2011 [Google Scholar]
- 6.Miller J, Leventhal J, Abecassis M, Chen L, Chandrasekaran D, Huang X, Levitsky J, Tambur A, Friedewald J, Mathew J: Prospectively induced immune tolerance vs. spontaneous operational tolerance in non-chimeric HLA identical renal transplants [Abstract]. Am J Transplant 12: 563, 2012 [Google Scholar]
- 7.Heidt S, Hester J, Shankar S, Friend PJ, Wood KJ: B cell repopulation after alemtuzumab induction-transient increase in transitional B cells and long-term dominance of naïve B cells. Am J Transplant 12: 1784–1792, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seigler HF, Gunnells JC, Jr, Robinson RR, Ward FE, Amos DB, Rowlands DT, Burkholder PM, Klein WJ, Stickel DL: Renal transplantation between HL-A identical donor-recipient pairs. Functional and morphological evaluation. J Clin Invest 51: 3200–3215, 1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerrits JH, van de Wetering J, Weimar W, van Besouw NM: T-cell reactivity during tapering of immunosuppression to low-dose monotherapy prednisolone in HLA-identical living-related renal transplant recipients. Transplantation 87: 907–914, 2009 [DOI] [PubMed] [Google Scholar]
- 10.van de Wetering J, Gerrits JH, van Besouw NM, Ijzermans JNM, Weimar W: Successful tapering of immunosuppression to low-dose monotherapy steroids after living-related human leukocyte antigen-identical renal transplantation. Transplantation 87: 740–744, 2009 [DOI] [PubMed] [Google Scholar]
- 11.Brouard S, Mansfield E, Braud C, Li L, Giral M, Hsieh SC, Baeten D, Zhang M, Ashton-Chess J, Braudeau C, Hsieh F, Dupont A, Pallier A, Moreau A, Louis S, Ruiz C, Salvatierra O, Soulillou J-P, Sarwal M: Identification of a peripheral blood transcriptional biomarker panel associated with operational renal allograft tolerance. Proc Natl Acad Sci U S A 104: 15448–15453, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newell KA, Asare A, Kirk AD, Gisler TD, Bourcier K, Suthanthiran M, Burlingham WJ, Marks WH, Sanz I, Lechler RI, Hernandez-Fuentes MP, Turka LA, Seyfert-Margolis VL, Immune Tolerance Network ST507 Study Group : Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest 120: 1836–1847, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LoCascio SA, Morokata T, Chittenden M, Preffer FI, Dombkowski DM, Andreola G, Crisalli K, Kawai T, Saidman SL, Spitzer TR, Tolkoff-Rubin N, Cosimi AB, Sachs DH, Sykes M: Mixed chimerism, lymphocyte recovery, and evidence for early donor-specific unresponsiveness in patients receiving combined kidney and bone marrow transplantation to induce tolerance. Transplantation 90: 1607–1615, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leventhal J, Abecassis M, Miller J, Gallon L, Tollerud D, Elliott MJ, Bozulic LD, Houston C, Sustento-Reodica N, Ildstad ST: Tolerance induction in HLA disparate living donor kidney transplantation by donor stem cell infusion: Durable chimerism predicts outcome. Transplantation, 95: 169–176, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scandling JD, Busque S, Dejbakhsh-Jones S, Benike C, Sarwal M, Millan MT, Shizuru JA, Lowsky R, Engleman EG, Strober S: Tolerance and withdrawal of immunosuppressive drugs in patients given kidney and hematopoietic cell transplants. Am J Transplant 12: 1133–1145, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spitzer TR, Delmonico F, Tolkoff-Rubin N, McAfee S, Sackstein R, Saidman S, Colby C, Sykes M, Sachs DH, Cosimi AB: Combined histocompatibility leukocyte antigen-matched donor bone marrow and renal transplantation for multiple myeloma with end stage renal disease: The induction of allograft tolerance through mixed lymphohematopoietic chimerism. Transplantation 68: 480–484, 1999 [DOI] [PubMed] [Google Scholar]
- 17.Benacerraf B: Role of major histocompatibility complex in genetic regulation of immunologic responsiveness. Transplant Proc 9: 825–831, 1977 [PubMed] [Google Scholar]
- 18.Guo Z, Hood L, Malkki M, Petersdorf EW: Long-range multilocus haplotype phasing of the MHC. Proc Natl Acad Sci U S A 103: 6964–6969, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M: Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol 155: 1151–1164, 1995 [PubMed] [Google Scholar]
- 20.Bouaziz J-D, Yanaba K, Tedder TF: Regulatory B cells as inhibitors of immune responses and inflammation. Immunol Rev 224: 201–214, 2008 [DOI] [PubMed] [Google Scholar]
- 21.Iwata Y, Matsushita T, Horikawa M, Dilillo DJ, Yanaba K, Venturi GM, Szabolcs PM, Bernstein SH, Magro CM, Williams AD, Hall RP, St Clair EW, Tedder TF: Characterization of a rare IL-10-competent B-cell subset in humans that parallels mouse regulatory B10 cells. Blood 117: 530–541, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muthukumar T, Dadhania D, Ding R, Snopkowski C, Naqvi R, Lee JB, Hartono C, Li B, Sharma VK, Seshan SV, Kapur S, Hancock WW, Schwartz JE, Suthanthiran M: Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N Engl J Med 353: 2342–2351, 2005 [DOI] [PubMed] [Google Scholar]
- 23.Afaneh C, Muthukumar T, Lubetzky M, Ding R, Snopkowski C, Sharma VK, Seshan S, Dadhania D, Schwartz JE, Suthanthiran M: Urinary cell levels of mRNA for OX40, OX40L, PD-1, PD-L1, or PD-L2 and acute rejection of human renal allografts. Transplantation 90: 1381–1387, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naji A, Silvers WK, Bellgrau D, Barker CF: Spontaneous diabetes in rats: Destruction of islets is prevented by immunological tolerance. Science 213: 1390–1392, 1981 [DOI] [PubMed] [Google Scholar]
- 25.Bray RA, Lebeck LK, Gebel HM: The flow cytometric crossmatch. Dual-color analysis of T cell and B cell reactivities. Transplantation 48: 834–840, 1989 [PubMed] [Google Scholar]