Abstract
The liver responds to multiple types of injury with an extraordinarily well orchestrated and tightly regulated form of regeneration. The response to partial hepatectomy has been used as a model system to elucidate the molecular basis of this regenerative response. In this study, we used cyclooxygenase (COX)-selective antagonists and -null mice to determine the role of prostaglandin signaling in the response of liver to partial hepatectomy. The results show that liver regeneration is markedly impaired when both COX-1 and COX-2 are inhibited by indocin or by a combination of the COX-1 selective antagonist, SC-560, and the COX-2 selective antagonist, SC-236. Inhibition of COX-2 alone partially inhibits regeneration whereas inhibition of COX-1 alone tends to delay regeneration. Neither the rise in IL-6 nor the activation of signal transducer and activator of transcription-3 (STAT3) that is seen during liver regeneration is inhibited by indocin or the selective COX antagonists. In contrast, indocin treatment prevents the activation of CREB by phosphorylation that occurs during hepatic regeneration. These data indicate that prostaglandin signaling is required during liver regeneration, that COX-2 plays a particularly important role but COX-1 is also involved, and implicate the activation of CREB rather than STAT3 as the mediator of prostaglandin signaling during liver regeneration.
The liver responds to many forms of injury, including traumatic, chemical, metabolic, or infectious injuries, with a proliferative response in the remnant tissue (1–3). Studies using partial hepatectomy (PH) in animal models have indicated that this process is precisely regulated in its initiation, duration, and termination, with the regenerative response proceeding only until the liver to body weight of the animals has been restored (4). Moreover, regeneration occurs while the liver continues to perform its critical functions including glucose homeostasis, protein synthesis, bile secretion, and toxin degradation.
Unlike other regenerating tissues (e.g., skin, gastrointestinal epithelium, and bone marrow) the liver does not require a stem cell population for regeneration. Instead, liver regeneration can proceed by stimulation of existing, normally quiescent, mature cellular populations to re-enter the cell cycle. After proliferation and restructuring, the regenerative response stops and the cells of the liver return to a state of quiescence. The molecular mechanisms that regulate these events include early signaling events such as increased production of hepatocyte growth factor (5), TNFα, and IL-6 (6), followed by induction of a number of immediate early genes (7). Subsequent changes occur in the activity of several transcription factors, including increased activity of NFκB, AP-1, signal transducer and activator of transcription-3 (STAT3), CREB, and CCAAT enhancer binding protein β (C/EBPβ) and decreased activity of C/EBPα (8–13). The transcription factor STAT3 has been shown to be specifically activated after PH by gel shift analysis of hepatic nuclear extracts (8). STAT3 is not activated during the impaired regenerative response seen in the IL-6 null mouse and the TNFα receptor 1-null mouse (14, 15).
Several observations have suggested that prostaglandins, including prostaglandin E2 (PGE2), prostacyclin, and thromboxane, may be involved in growth regulation during liver regeneration (16–20). Prostaglandins are important mediators of normal and abnormal growth control in many other tissues. For example they appear to be involved in the regulatory aspects of angiogenesis (21), early stages of pregnancy (22), and intestinal crypt stem cell survival (23), and they have been implicated in the pathogenesis of several types of cancer (22, 24).
Prostaglandins are also involved in the regulation of, or are regulated by, a number of cytokines and growth factors, including several that have been implicated in liver regeneration. For example, the proinflammatory action of TNFα is in part mediated by its induction of the prostaglandin-synthesizing enzyme cyclooxygenase-2 (COX-2) (25). Furthermore, PGE2 stimulates IL-6 production in macrophages (26).
Prostaglandins are synthesized from arachidonic acid that is released from membrane phospholipid by phospholipase A2. Arachidonic acid is oxidized by COX to generate the precursor PGH2, which is further metabolized by specific synthases to form prostaglandins, thromboxanes, and prostacyclins (27). Two COX isozymes exist. Classically, these have been characterized as the “constitutive form”—i.e., COX-1, which is present in most tissues and mediates the synthesis of prostaglandins required for normal or “housekeeping” functions—and the “regulated form”—i.e., COX-2, which is undetectable in most tissues but is highly inducible (e.g., by inflammatory mediators such as TNFα). Gene disruption of COX-1 or COX-2 in mice gives rise to distinct phenotypes (28–30). COX-1 null mice survive without evidence of gastric pathology and with decreased sensitivity to indocin induced gastric ulceration (28, 29), as well as delayed parturition (31) and mild platelet abnormalities. In contrast, COX-2 null mice develop nephropathy and cardiac fibrosis (30).
The precise nature of the role of prostaglandins in liver regeneration, including whether components of the cytokine-signaling cascade, cAMP-mediated signal transduction, or other signal transduction pathways are prostaglandin-dependent and what is the timing of this dependence, is not well-characterized. Moreover, it is not known which cyclooxygenase isoform (COX-1 or COX-2) is involved. In this study, we used recently developed molecular genetic and pharmacological tools, including mice with targeted gene disruption of COX-1, and enzyme inhibitors that are potent and highly specific for COX-1 [SC-560 (32)] and COX-2 [SC-236 (33, 34)], to undertake detailed analyses of these issues.
Materials and Methods
Animal Husbandry.
Wild-type (wt) and IL-6 null mice were on a C57BL/6 background (Jackson Laboratory, Bar Harbor, ME). COX-1 null mice were on a mixed C57BL/6 × 129SV background (29). All experiments were performed with 8- to 12-wk-old male mice weighing ≈25–30 g. Mice were kept on 12-h dark/light cycles and maintained on a standard diet of ad libitum mouse chow and water before and after surgery. All experiments were conducted in accordance with the institutional guidelines of Washington University School of Medicine.
Animal Surgery.
Partial hepatectomy, consisting of the removal of the median and left hepatic lobes (≈65%) was performed according to the method of Higgins and Anderson (4) under methoxyflurane anesthesia. Animals typically aroused within 30 min of surgery and were returned to their cages with ad libitum access to food and water. For sham-operated animals, after sedation, the liver was exposed then returned to the abdominal cavity with wound closure.
For experiments with the COX antagonists, gavage dosing was begun the evening before the surgeries and continued twice daily until animal killing. Liver tissue was harvested into fixative or snap frozen in liquid nitrogen and stored at −80°C.
COX Inhibitors.
The COX inhibitors were prepared in PBS with 1% Tween-80. Indocin (Sigma) was used at a dose of 5 mg/kg twice daily. This dose of indocin has previously been shown to suppress both COX-1 and COX-2 activity in a murine model system (35). The COX-1- and COX-2-specific inhibitors, SC-560 (32) and SC-236 (33, 34), were generously provided by Monsanto and were used at a dose of 10 mg/kg twice daily. This dose of SC-560 has been shown to specifically suppress COX-1-derived prostaglandin synthesis in platelets (which only express COX-1) but not COX-2-derived prostaglandin synthesis after lipopolysaccharide injection in the COX-1 null mouse (35). Similarly, this dose of SC-236 efficiently suppresses COX-2-derived prostaglandin synthesis after lipopolysaccharide injection in the COX-1 null mouse, but not COX-1-derived prostaglandin synthesis in platelets (35). Mice given an equal volume of vehicle (PBS with 1% Tween-80) were used as controls.
Histology.
Mice were either injected with 100 μg/g of bromodeoxyuridine (BrdUrd, Sigma) 1 h before killing or had i.p. implantation at the time of surgery of a BrdUrd containing osmotic minipump (Alza, Newark, DE) delivering a flow rate of 1 μl/h (20 mg/ml BrdUrd, 20 μg/h) for 3 days. Livers harvested at killing were fixed for 24 h in 10% neutral buffered-formalin and prepared for histological analysis. Paraffin-embedded sections of liver were stained with hematoxylin and eosin or for BrdUrd by using the BrdUrd Immunohistochemistry kit (Amersham Pharmacia) and hematoxylin as counterstain. For each animal, at least three different random ×400 fields were examined and a total of 300–500 nuclei counted.
Prostaglandin Assays.
Liver was weighed while frozen and then homogenized in 100% ethanol for extraction of prostaglandins. Debris was removed by centrifugation, and each supernatant was assayed in duplicate for PGE2 and the prostacyclin derivative 6-keto-PGF1α by ELISA (Oxford Biomedical Research, Oxford, MI).
IL-6 Assays.
Mice underwent retroorbital phlebotomy at serial time points after surgeries. IL-6 levels were measured by ELISA by using the OptEIA mouse IL-6 kit from PharMingen.
STAT3 and CREB Western Blot Analysis.
Total cellular lysates were made from snap frozen liver by homogenization in 20 mM Tris (pH 8), 1 mM sodium vanadate, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 100 mM NaF, 10% glycerol, 50 mM β-glycerophosphate, and a protease inhibitor mixture (Sigma). Cellular debris was pelleted at 10,000 × g at 4°C. Protein concentration was determined by using the Pierce bicinchoninic acid (BCA) protein assay kit. Total cellular protein (25 μg) was subjected to Western blot analysis for total and phosphorylated STAT3 by using the PhosphoPlus STAT3 (Tyr-705) antibody kit (New England Biolabs), and for total and phosphorylated CREB (p-CREB) by using the PhosphoPlus CREB (Ser-133) antibody kit (Cell Signaling Technology, Beverly, MA). Densitometric analysis was performed with National Institutes of Health image data analysis software.
Statistical Analysis.
Data were analyzed by using sigma plot software (SPSS, Chicago). ANOVA for multiple groups was used to determine statistical significance for the BrdUrd-labeling experiments, serum IL-6 determinations, and densitometric analysis of Western blot studies. χ2 analysis was used to determine statistical significance for mortality and weight data. Data are reported as mean ± SE.
Results
Characterization of Liver Regeneration in wt Mice by Using Two Methods for Assaying Hepatocyte Replication.
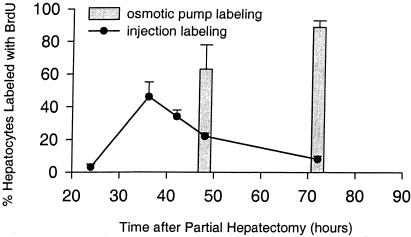
Two methods of BrdUrd delivery were used to characterize the regenerative response in wt mice after partial hepatectomy. In the first, the BrdUrd injection technique, animals were subjected to PH, allowed to recover, and then injected with BrdUrd 1 h before killing at serial time points after surgery. Liver tissue was harvested, fixed, and analyzed by immunohistochemistry for BrdUrd. In this method, only cells in S phase between the time of injection and the time of killing incorporate BrdUrd. Fig. 1 illustrates the BrdUrd-labeling profile observed in hepatocytes of wt animals analyzed in this way. BrdUrd labeling of hepatocytes remains low 24 h after PH, rising to 46 ± 9% at 36 h, and steadily declining to 8 ± 2% at 72 h after PH. This temporal pattern is comparable to that previously described (14, 15). A second method to quantify regenerative response, the BrdUrd sustained-release technique, was developed to provide corroborative data for the first method as well as to generate an integrated measure of hepatocellular entry into S phase (BrdUrd incorporation) spanning the peak of BrdUrd incorporation. This method ensures that the peak of the proliferative response is not missed, even if it is slightly affected by experimental manipulation. In this method, animals were subjected to PH as described, but with implantation of a BrdUrd-containing sustained-release osmotic pump. Again animals were allowed to recover and killed at serial time points after surgery, followed by harvest and fixation of liver tissue and BrdUrd immunohistochemistry. The results in Fig. 1 show that 63 ± 15% of hepatocytes are labeled with BrdUrd at 48 h after surgery, whereas 89 ± 4% are labeled after 72 h. There was no increase in hepatocyte BrdUrd labeling after sham surgery (data not shown).
Figure 1.
Hepatocyte BrdUrd labeling after PH. Percentage of hepatocytes that label with BrdUrd by the injection method is plotted in line form and by the osmotic pump method in bar form. Three to six animals were studied for each time point and treatment group.
Indocin Inhibits Liver Regeneration.
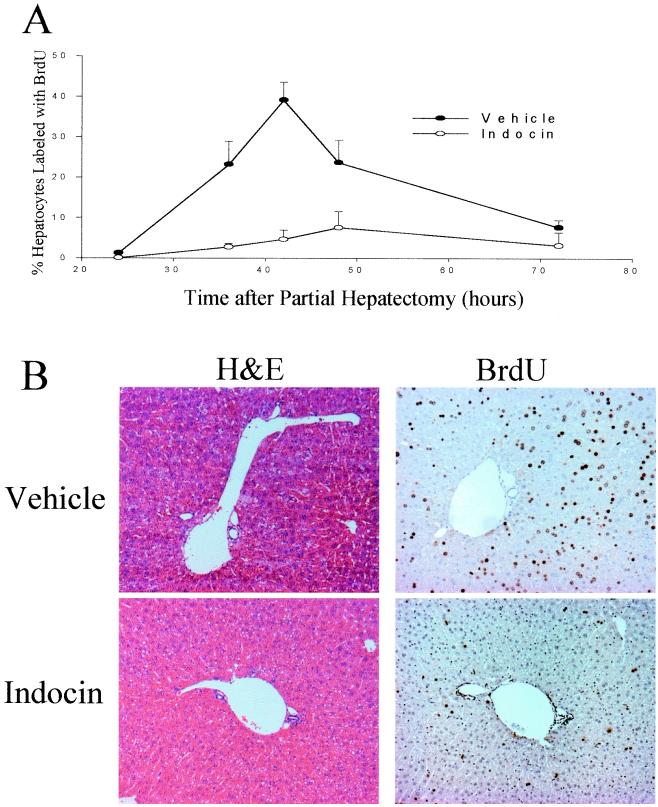
The wt mice were treated by gavage with indocin, which inhibits both COX-1 and COX-2, or with vehicle control and then subjected to PH. Animals treated with vehicle exhibited a pattern of hepatocyte BrdUrd incorporation after PH very similar to that seen in untreated animals (Fig. 2A), with a peak of incorporation of 39 ± 4% at 42 h after PH, after which BrdUrd incorporation steadily declined to 8 ± 3% by 72 h after PH. In contrast, hepatocyte BrdUrd incorporation in the livers of indocin-treated animals exhibited marked suppression compared to the vehicle control with 3 ± 1%, 5 ± 2%, and 8 ± 4% of hepatocytes labeled at 36 h (P = 0.009), 42 h (P = 0.002), and 48 h (P = 0.09), respectively (Fig. 2A). The histological appearances as well as the immunohistochemical patterns of BrdUrd labeling from control- and indocin-treated animals at the peak of BrdUrd incorporation (42 h after PH) are illustrated in Fig. 2B. For the BrdUrd-labeling experiments, nuclei from proliferating cells stained brown and nuclei from quiescent cells counterstained blue with hematoxylin. The results show abundant proliferating hepatocytes in mice treated with vehicle and a markedly reduced number of proliferating cells in mice treated with indocin. There was no evidence for hepatic injury or other changes in liver histology after indocin treatment. There were no statistically significant differences in mortality (vehicle 10%; indocin 16%) or postoperative body weight at 72 h after surgery (data not shown).
Figure 2.
Effect of indocin on hepatocyte BrdUrd labeling and liver histology. (A) Percentage of hepatocytes that label with BrdUrd by the injection method after vehicle or indocin treatment. Three to eight animals were studied for each time point and treatment group. Significant differences were found at 42 h (P < 0.002). (B) Liver sections from vehicle- and indocin-treated animals 42 h after PH stained with hematoxylin/eosin or BrdUrd.
The COX-2-Specific Antagonist Inhibits Liver Regeneration, Whereas the COX-1-Specific Antagonist Exhibits a Tendency to Delay Liver Regeneration.
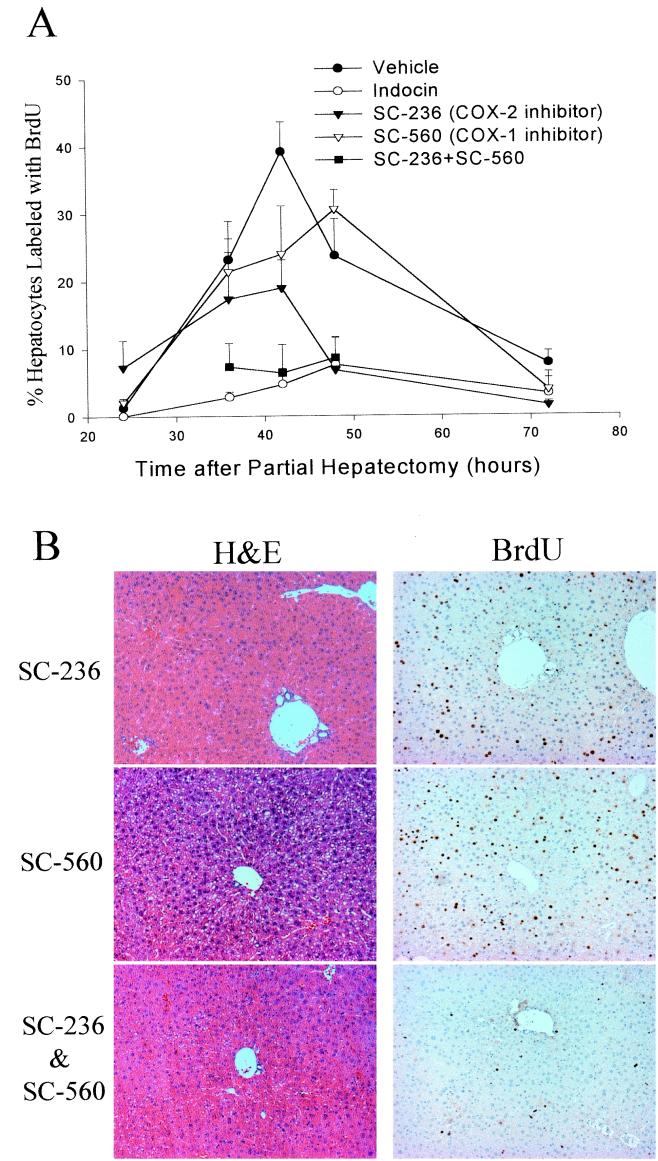
Mice were treated with a COX-1-specific inhibitor (SC-560), a COX-2-specific inhibitor (SC-236), or both and then subjected to PH. Analysis of hepatocyte BrdUrd incorporation in the livers of animals treated with the COX-1-specific inhibitor (SC-560) showed a tendency toward delayed proliferation, with peak values at 48 h instead of 42 h. In animals treated with SC-560, 21 ± 5% of hepatocytes labeled with BrdUrd at 36 h, rising to 24 ± 7% at 42 h and 31 ± 3% at 48 h after PH (Fig. 3A). However the differences between these values and the values for BrdUrd incorporation in mice given vehicle alone did not reach statistical significance (P = 0.11 at 42 h and P = 0.40 at 48 h). Treatment with the COX-2-specific inhibitor (SC-236) resulted in a more significant reduction in BrdUrd incorporation, with peak labeling at 42 h of 19 ± 4% compared to 39 ± 4% in the gavage control treatment group (P = 0.015, Fig. 3A). When animals were treated with a mixture of SC-560 and SC-236 the hepatocyte BrdUrd-labeling profile closely resembled that seen with indocin, with 7 ± 4% of cells labeled at 36 h, 6 ± 4% at 42 h, and 9 ± 3% at 48 h (Fig. 3A). The histological appearances as well as the immunohistochemical patterns of BrdUrd labeling of the regenerating livers from animals treated with SC-560, SC-236, or both recovered at the peak of BrdUrd incorporation (42 h after PH) are illustrated in Fig. 3B. The results show that in liver sections from animals treated with the COX-1-specific antagonist there is a slight decrease in BrdUrd labeled (brown stained) nuclei. In sections from animals treated with the COX-2-specific antagonist, there is a more significant decrease in these BrdUrd-labeled nuclei. Liver sections from animals treated with the mixture of specific inhibitors contain significantly decreased numbers of hepatocytes labeled with BrdUrd, with the results comparable to those seen with indocin treatment. Treatment with either the COX-1-specific inhibitor, the COX-2-specific inhibitor, or the mixture did not result in any apparent histological injury. There was no statistically significant difference between the vehicle treated animals and those treated with the COX-specific inhibitors in mortality (vehicle 10%, SC-560 16%, SC-236 8%, mixture 0%) or postoperative body weight (data not shown). Taken together these data show that inhibition of COX-2 results in a significant reduction of the hepatic regenerative response whereas inhibition of COX-1 tends to slightly delay regeneration. Furthermore, inhibition of COX-1 and COX-2 with a mixture of the specific inhibitors results in marked inhibition of hepatic regeneration, comparable to that seen with indocin. These results argue against the possibility that the inhibitory effect of indocin is because of a nonspecific or toxic side effect.
Figure 3.
Effect of COX-1- and COX-2-specific inhibitors on hepatocyte BrdUrd labeling and liver histology. (A) Percentage of hepatocytes that label with BrdUrd by the injection method is plotted for vehicle-, indocin-, SC-236-, SC-560-, and SC-236 + SC-560-treated animals. Three to eight animals were studied for each time point and treatment group. Significant differences between the vehicle control and the indocin, SC-236, and SC-236 + SC-560 treatment groups were found at 42 h (P < 0.002). (B) Liver sections from SC-236-, SC-560-, and SC-236 + SC-560-treated animals 42 h after PH stained with hematoxylin/eosin or BrdUrd.
To provide further evidence for the role of prostaglandins in liver regeneration and the relative involvement of COX-1 and COX-2, we used a different approach to analyze regenerative activity, the sustained-release BrdUrd-containing osmotic pump, and a different animal model, the COX-1 null mouse. The pumps were placed in the abdomen of wt and COX-1 null mice at the time of hepatectomy. Forty-eight hours later, the mice were killed and their livers evaluated for BrdUrd incorporation. The results show that BrdUrd incorporation into hepatocytes is identical in COX-1 null mice (63 ± 11%) to that seen in wt mice (63 ± 15%) but is markedly decreased in COX-1 null mice treated with the COX-2-specific inhibitor, SC-236 (10 ± 7%, P < 0.04). These data provide further corroborating evidence for the important role of COX-2 in liver regeneration, particularly in the absence of COX-1.
Partial Hepatectomy-Induced Hepatic Prostaglandin Synthesis Is Inhibited by Indocin Treatment.
To investigate which prostaglandins are involved in the hepatic regenerative response to PH, the levels of specific prostaglandins in regenerating liver were measured, including PGE2 and 6-keto-PGF1α (a metabolite of prostacyclin). Hepatic levels of PGE2 and 6-keto-PGF1α were increased after PH with peak levels occurring 2 h after the surgery (3- to 5-fold over baseline). Next, we examined the magnitude of the effect of COX inhibition on peak PH-induced hepatic prostaglandin production of PGE2 and 6-keto-PGF1α, by using the dose of indocin shown to impair the hepatic regenerative response (5 mg/kg bid). Hepatic levels of PGE2 and 6-keto-PGF1α were measured from the livers of animals treated with vehicle or indocin and harvested 2 h after PH. PGE2 levels were significantly suppressed by indocin as compared to vehicle (34 ± 14 vs. 92 ± 12 ng/g liver, P < 0.02). Levels of 6-keto-PGF1α were similarly suppressed (11 ± 2 vs. 48 ± 11 ng/g, P < 0.01). These data indicate that indocin, at a dose that impairs liver regeneration, inhibits prostaglandin synthesis.
Relationship Between Prostaglandin Metabolism and Cytokine Signaling Pathways in Liver Regeneration.
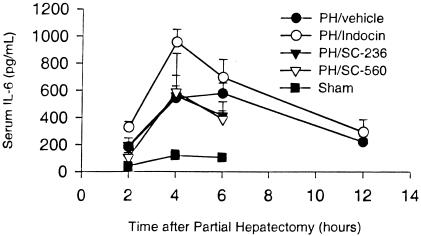
Previous studies have shown that the cytokine-signaling pathway initiated by TNFα and IL-6 and mediated by the transcription factor STAT3 plays an important role in liver regeneration (3, 13, 14). To determine whether prostaglandins function as part of this signaling pathway during liver regeneration, we used two different experimental strategies. First, we examined the effect of COX inhibitors on plasma IL-6 levels during liver regeneration. Animals treated with vehicle exhibited a rise in serum IL-6 to 542 ± 90 and 578 ± 76 pg/ml at 4 and 6 h after PH, respectively (Fig. 4). This rise was significantly increased over that of sham-operated mice at all time points (P < 0.02). Indocin treatment did not inhibit the IL-6 response after PH, with a rise in serum IL-6 to 956 ± 93 pg/ml noted 4 h after PH, declining to 695 ± 134 pg/ml 6 h after PH (Fig. 4). Animals treated with the COX-1- or COX-2-specific inhibitors exhibited serum IL-6 values comparable to those observed in the vehicle control group (Fig. 4). These data show that inhibition of COX-1 and COX-2 with indocin at doses sufficient to impair liver regeneration does not inhibit the associated rise in IL-6. In fact, the serum IL-6 levels noted in indocin-treated animals were significantly greater than those seen in vehicle control animals 4 h after PH (P = 0.02).
Figure 4.
Effect of COX inhibitors on serum IL-6 after PH. Serum IL-6 determination is plotted for vehicle-, indocin-, SC-236-, and SC-560-treated animals, and for sham-operated animals. Three to four animals were studied for each time point and treatment group.
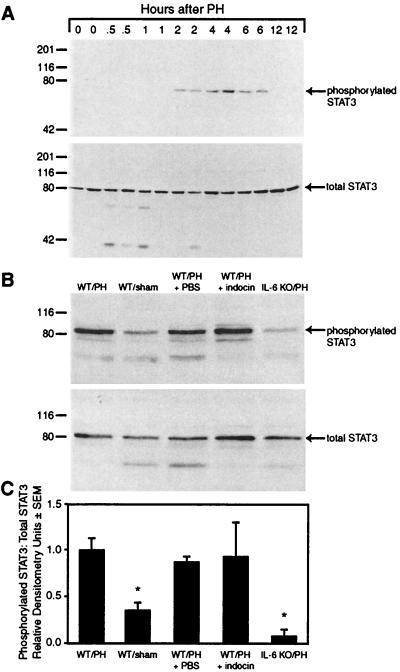
Next, we determined the effect of COX inhibition on STAT3 activation during liver regeneration by Western blot analysis of total STAT3 and phosphorylated STAT3. First, we used this assay to examine the time course of STAT3 activation. The results show the appearance of an 80-kDa band, corresponding to phosphorylated STAT3 between 2 and 6 h after PH (Fig. 5A Upper). The activation of STAT3 was specific and not because of differences in protein loading as there was no change in the amount of total STAT3 noted in these lysates from 0 to 12 h (Fig. 5A Lower). This time course of activation of STAT3 parallels that described in previous reports using electrophoretic mobility gel shift assay (8).
Figure 5.
Hepatic STAT3 activation after PH in the presence or absence of COX inhibition. (A) Western blot analysis of phosphorylated and total STAT3 in hepatic lysates at serial time points after PH. (B) Western blot analysis of phosphorylated and total STAT3 from hepatic lysates of wt mice subjected to PH, sham surgery (sham), subjected to PH while treated with PBS, subjected to PH while treated with indocin, or IL-6 null mice subjected to PH (IL-6 knockout) is shown. Lysates were prepared from liver harvested 2 h after surgery. (C) Densitometric analysis of the ratio of phosphorylated STAT3 to total STAT3 from three separate experiments.
We next used the 2 h time point to examine the effect of COX inhibition on STAT3 activation (Fig. 5B). The results show the presence of abundant phosphorylated STAT3 in wt mice after PH but much less in wt mice after sham surgery. The wt mice subjected to PH and treated with vehicle or indocin exhibit comparable amounts of phosphorylated STAT3 in hepatic lysates to that seen in the untreated wt control. IL-6 knockout mice were used as a negative control and showed a marked reduction in phosphorylated-STAT3 after PH, consistent with previously reported data using gel shift analysis (14). Comparable amounts of total STAT3 were noted in all samples. Densitometric analysis of three separate experiments (Fig. 5C) shows comparable amounts of phosphorylated STAT3 to total STAT3 from the livers of animals subjected to PH in the absence or presence of vehicle or in the presence of indocin, with significantly reduced activated STAT3 from sham-operated animals (P < 0.02 compared to animal subjected to PH) or IL-6 knockout mice subjected to PH (P < 0.05). These data show that STAT3 remains activated even though regeneration is impaired during COX inhibition.
Relationship Between Prostaglandin Signaling and cAMP-Signaling Pathways in Liver Regeneration.
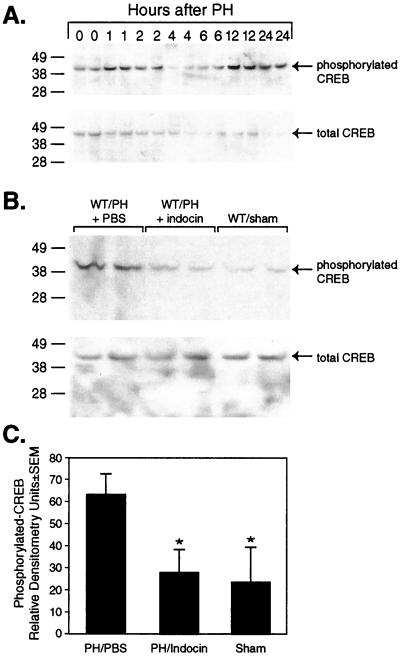
Prostaglandins mediate many of their protean biological activities by binding to G protein coupled transmembrane receptors, which modulate cAMP signaling (36). The action of cAMP is in part mediated by its binding to and activation of a protein kinase which, in turn, phosphorylates and activates the DNA binding protein CREB, the cAMP response element binding protein. To determine whether prostaglandin activity during liver regeneration depends on modulation of this signaling pathway, the effect of COX inhibition on CREB activation during liver regeneration was examined by Western blot analysis of total and phosphorylated-CREB (p-CREB). First, we used this assay to examine the time course of CREB activation. The results show the appearance of a 43-kDa band, corresponding to p-CREB, in a biphasic manner 1–2 h and again 12–24 h after PH. These changes in the level of p-CREB were not associated with corresponding changes in the level of total CREB (Fig. 6A). These observations are consistent with previous studies of cAMP and CREB activation after PH (13).
Figure 6.
Hepatic CREB activation after PH in the presence or absence of COX inhibition. (A) Western blot analysis of phosphorylated and total CREB in hepatic lysates at serial time points after PH. (B) Western blot analysis of phosphorylated and total CREB from hepatic lysates of mice subjected to PH in the presence of vehicle (PBS) or indocin or mice subjected to sham surgery is shown. Lysates were prepared from liver harvested 1 h after surgery. (C) Densitometric analysis of the relative amounts of p-CREB from three separate experiments.
Next the effect of COX inhibition on CREB activation was examined at 1 h and 12 h after PH. The results show the presence of p-CREB in hepatic lysates from wt mice subjected to PH and treated with vehicle at 1 h after PH, with significantly reduced amounts of p-CREB in lysates from animals treated with indocin and subjected to PH or sham-operated animals at the same time point (P < 0.05, Fig. 6B). The differences seen in the amount of activated CREB between the treatment groups was not explained by changes in the levels of total CREB, which were not significantly different between the treatment groups (Fig. 6B). Densitometric analysis of these results (Fig. 6C) shows that activation of CREB after PH is significantly inhibited by COX inhibition. Similar analysis of hepatic lysates harvested 12 h after PH showed there was also a reduction in the amount of p-CREB in indocin- vs. vehicle-treated animals; however this difference did not reach statistical significance (data not shown).
Discussion
The results indicate that prostaglandins are required for liver regeneration to proceed efficiently after PH. The temporal pattern of hepatocyte DNA synthesis (BrdUrd labeling) noted after PH in animals treated with a COX-2-specific inhibitor demonstrated marked impairment in regenerative response and therein indicated an important role for COX-2. Although there was only a slight delay in regeneration in mice treated with the COX-1-specific antagonist and no decrease in overall hepatocyte replication in COX-1 null mice, a role for COX-1 was implicated by the greater effect of indocin and the combination of the COX-1- and COX-2-specific antagonists than for the COX-2-specific antagonist alone, as well as by the marked effect of the COX-2-specific antagonist in COX-1 null mice. Indeed, these data suggest that COX-2 may compensate for chemical or genetic inhibition of COX-1 activity.
Previous studies have implicated TNFα and IL-6 as being important in liver regeneration (14, 15, 37): TNFα receptor 1-deficient mice and IL-6 null mice exhibit impaired regeneration in response to PH (14, 15). IL-6 supplementation at the time of surgery rescues the regenerative response in both of these genetic backgrounds indicating that TNFα is upstream of IL-6 in this important signaling pathway during liver regeneration. The results of our studies show that IL-6 levels remain elevated and STAT3 remains activated during the hepatic regenerative response that is impaired by indocin. These results suggest that prostaglandin signaling is independent of the cytokine-signaling cascade during liver regeneration. Instead, the results implicate cAMP-mediated signaling pathways, including the activation of CREB, in the mechanism by which prostaglandins influence liver regeneration. Interestingly, recent studies have suggested that CREB plays a role in the transcriptional regulation of C/EBPβ in vivo (38). CCAAT enhancer binding protein β (C/EBPβ) is now known to be required for liver regeneration and its effect on the regenerative response also involves a mechanism that is independent of STAT3 activation (12, 14, 15).
The results of our studies also suggest that COX-1 and COX-2 may play distinct roles during liver regeneration. This concept has previously been established in studies of angiogenesis induced by colon cancer cells (21). In those studies, COX-2 modulated the production of angiogenic factors, such as vascular endothelial growth factor, by colon carcinoma cells, whereas COX-1 directly modulated the angiogenic activity of endothelial cells. Future studies will address what specific perturbations in hepatic transcriptional regulation occur during liver regeneration when prostaglandin synthesis is impaired pharmacologically or genetically and whether these events are primarily dependent on COX-1- or COX-2-derived prostaglandins.
Acknowledgments
We thank Peter Isakson of Monsanto Corporation for providing the COX-specific antagonists. The studies were supported by grants from National Institutes of Health, by an American Digestive Health Foundation/American Gastroenterological Association Research Scholar Award (to D.A.R.), and by a Burroughs Wellcome Fund Career Development Award (to L.J.M.). D.A.R. is a Scholar of the Washington University School of Medicine Child Health Research Center of Excellence in Developmental Biology (HD33688).
Abbreviations
- COX
cyclooxygenase
- PH
partial hepatectomy
- wt
wild-type
- BrdUrd
bromodeoxyuridine
- PGE2
prostaglandin E2
- p-CREB
phosphorylated-CREB
- STAT3
signal transducer and activator of transcription-3
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Michalopoulos G K, DeFrances M C. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 2.Diehl A M E, Rai R. In: Schiff's Diseases of the Liver. 8th Ed. Schiff R, Sorrell M C, Maddrey W C, editors. Philadelphia: Lippincott; 1999. pp. 39–54. [Google Scholar]
- 3.Fausto N. J Hepatol. 2000;32,Supp. 1:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 4.Higgins G M, Anderson R M. Arch Pathol. 1931;12:186–202. [Google Scholar]
- 5.Boros P, Miller C M. Lancet. 1995;345:293–295. doi: 10.1016/s0140-6736(95)90279-1. [DOI] [PubMed] [Google Scholar]
- 6.Yamada Y, Webber E M, Kirillova I, Peschon J J, Fausto N. Hepatology. 1998;28:959–970. doi: 10.1002/hep.510280410. [DOI] [PubMed] [Google Scholar]
- 7.Taub R. FASEB J. 1996;10:413–427. [PubMed] [Google Scholar]
- 8.Cressman D E, Diamond R H, Taub R. Hepatology. 1995;21:1443–1449. [PubMed] [Google Scholar]
- 9.Cressman D E, Greenbaum L E, Haber B A, Taub R. J Biol Chem. 1994;269:30429–30435. [PubMed] [Google Scholar]
- 10.Heim M H, Gamboni G, Beglinger C, Gyr K. Eur J Clin Invest. 1997;27:948–955. doi: 10.1046/j.1365-2362.1997.2100770.x. [DOI] [PubMed] [Google Scholar]
- 11.Diehl A M. J Biol Chem. 1998;273:30843–30846. doi: 10.1074/jbc.273.47.30843. [DOI] [PubMed] [Google Scholar]
- 12.Greenbaum L E, Wei L, Cressman D E, Peng Y, Ciliberto G, Poli V, Taub R. J Clin Invest. 1998;102:996–1007. doi: 10.1172/JCI3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Della Fazia M A, Servillo G, Sassone-Corsi P. FEBS Lett. 1997;410:22–24. doi: 10.1016/s0014-5793(97)00445-6. [DOI] [PubMed] [Google Scholar]
- 14.Yamada Y, Kirillova I, Peschon J J, Fausto N. Proc Natl Acad Sci USA. 1997;94:1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cressman D E, Greenbaum L E, DeAngelis R A, Ciliberto G, Furth E E, Poli V, Taub R. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 16.Macmanus J P, Braceland B M. Prostaglandins. 1976;11:609–620. doi: 10.1016/0090-6980(76)90064-2. [DOI] [PubMed] [Google Scholar]
- 17.Tsujii H, Okamoto Y, Kikuchi E, Matsumoto M, Nakano H. Gastroenterology. 1993;105:495–499. doi: 10.1016/0016-5085(93)90725-r. [DOI] [PubMed] [Google Scholar]
- 18.Miura Y, Fukui N. Cell Mol Biol. 1979;25:179–184. [PubMed] [Google Scholar]
- 19.McNeil G E, Chen T S, Leevy C M. Hepatology. 1985;5:43–46. doi: 10.1002/hep.1840050110. [DOI] [PubMed] [Google Scholar]
- 20.Kanzaki Y, Mahmud I, Asanagi M, Fukui N, Miura Y. Cell Mol Biol. 1979;25:147–152. [PubMed] [Google Scholar]
- 21.Tsujii M, Kawano S, Tsuji S, Sawaoka H, Hori M, Dubois R N. Cell. 1998;93:705–716. doi: 10.1016/s0092-8674(00)81433-6. [DOI] [PubMed] [Google Scholar]
- 22.Majerus P W. Curr Biol. 1998;8:87–89. doi: 10.1016/s0960-9822(98)70053-3. [DOI] [PubMed] [Google Scholar]
- 23.Cohn S M, Schloemann S, Tessner T, Seibert K, Stenson W F. J Clin Inv. 1997;99:1367–1379. doi: 10.1172/JCI119296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotha S, Zakim D, Weksler B B, Dannenberg A J. Proc Soc Exp Biol Med. 1997;216:201–210. doi: 10.3181/00379727-216-44170. [DOI] [PubMed] [Google Scholar]
- 25.Jobin C, Morteau O, Han D S, Balfour S R. Immunology. 1998;95:537–543. doi: 10.1046/j.1365-2567.1998.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams J A, Schachter E. J Biol Chem. 1997;272:25693–25699. doi: 10.1074/jbc.272.41.25693. [DOI] [PubMed] [Google Scholar]
- 27.Smith W L, Garavito R M, DeWitt D L. J Biol Chem. 1996;271:33157–33160. doi: 10.1074/jbc.271.52.33157. [DOI] [PubMed] [Google Scholar]
- 28.Langenbach R, Morham S G, Tiano H F, Loftin C D, Ghanayem B I, Chulada P C, Mahler J F, Davis B J, Lee C A. Adv Exp Med Biol. 1997;407:87–92. [PubMed] [Google Scholar]
- 29.Langenbach R, Morham S G, Tiano H F, Loftin C D, Ghanayem B I, Chulada P C, Mahler J F, Lee C A, Goulding E H, Kluckman K D, et al. Cell. 1995;83:483–492. doi: 10.1016/0092-8674(95)90126-4. [DOI] [PubMed] [Google Scholar]
- 30.Morham S G, Langenbach R, Mahler J, Smithies O. Adv Exp Med Biol. 1997;407:131–138. doi: 10.1007/978-1-4899-1813-0_20. [DOI] [PubMed] [Google Scholar]
- 31.Gross G A, Imamura T, Luedke C, Vogt S K, Olson L M, Nelson D M, Sadovsky Y, Muglia L J. Proc Natl Acad Sci USA. 1998;95:11875–11879. doi: 10.1073/pnas.95.20.11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith C J, Zhang Y, Koboldt C M, Muhammad J, Zweifel B S, Shaffer A, Talley J J, Masferrer J L, Seibert K, Isakson P C. Proc Natl Acad Sci USA. 1998;95:13313–13318. doi: 10.1073/pnas.95.22.13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gierse J K, McDonald J J, Hauser S D, Rangwala S H, Koboldt C M, Seibert K. J Biol Chem. 1996;271:15810–15814. doi: 10.1074/jbc.271.26.15810. [DOI] [PubMed] [Google Scholar]
- 34.Penning T D, Talley J J, Bertenshaw S R, Carter J S, Collins P W, Docter S, Graneto M J, Lee L F, Malecha J W, Miyashiro J M, et al. J Med Chem. 1997;40:1347–1365. doi: 10.1021/jm960803q. [DOI] [PubMed] [Google Scholar]
- 35.Gross G, Imamura T, Vogt S K, Nelson D M, Sadovsky Y, Muglia L J. Am J Physiol. 2000;278:R1415–R1423. doi: 10.1152/ajpregu.2000.278.6.R1415. [DOI] [PubMed] [Google Scholar]
- 36.Fennekohl A, Lucas M, Puschel G P. Hepatology. 2000;31:1128–1134. doi: 10.1053/he.2000.7055. [DOI] [PubMed] [Google Scholar]
- 37.Akerman P, Cote P, Yang S Q, McClain C, Nelson S, Bagby G J, Diehl A M. Am J Physiol. 1992;263:G579–G585. doi: 10.1152/ajpgi.1992.263.4.G579. [DOI] [PubMed] [Google Scholar]
- 38.Niehof M, Manns M P, Trautwein C. Mol Cell Biol. 1997;17:3600–3613. doi: 10.1128/mcb.17.7.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]