Abstract
Genome studies suggest that DNA gyrase is the sole type II topoisomerase and likely the unique target of quinolones in Mycobacterium tuberculosis. Despite the emerging importance of quinolones in the treatment of mycobacterial disease, the slow growth and high pathogenicity of M. tuberculosis have precluded direct purification of its gyrase and detailed analysis of quinolone action. To address these issues, we separately overexpressed the M. tuberculosis DNA gyrase GyrA and GyrB subunits as His-tagged proteins in Escherichia coli from pET plasmids carrying gyrA and gyrB genes. The soluble 97-kDa GyrA and 72-kDa GyrB subunits were purified by nickel chelate chromatography and shown to reconstitute an ATP-dependent DNA supercoiling activity. The drug concentration that inhibited DNA supercoiling by 50% (IC50) was measured for 22 different quinolones, and values ranged from 2 to 3 μg/ml (sparfloxacin, sitafloxacin, clinafloxacin, and gatifloxacin) to >1,000 μg/ml (pipemidic acid and nalidixic acid). By comparison, MICs measured against M. tuberculosis ranged from 0.12 μg/ml (for gatifloxacin) to 128 μg/ml (both pipemidic acid and nalidixic acid) and correlated well with the gyrase IC50s (R2 = 0.9). Quinolones promoted gyrase-mediated cleavage of plasmid pBR322 DNA due to stabilization of the cleavage complex, which is thought to be the lethal lesion. Surprisingly, the measured concentrations of drug inducing 50% plasmid linearization correlated less well with the MICs (R2 = 0.7). These findings suggest that the DNA supercoiling inhibition assay may be a useful screening test in identifying quinolones with promising activity against M. tuberculosis. The quinolone structure-activity relationship demonstrated here shows that C-8, the C-7 ring, the C-6 fluorine, and the N-1 cyclopropyl substituents are desirable structural features in targeting M. tuberculosis gyrase.
Fluoroquinolones are active against Mycobacterium tuberculosis and are the first new antimycobacterial drugs to be available since the discovery of rifampin (7, 13, 40). Fluoroquinolones are part of the drug regimens now recommended for treating rifampin-resistant tuberculosis (6, 10). Ofloxacin and ciprofloxacin have a bacteriostatic antimycobacterial activity (13, 20, 40), but several new fluoroquinolones, such as sparfloxacin and moxifloxacin, show high bactericidal activity against M. tuberculosis that compares with that of major antituberculous drugs in animal models (20). By contrast, other new fluoroquinolones, such as gemifloxacin and trovafloxacin, are less active than ofloxacin against M. tuberculosis.
Studies of other bacterial species suggest that the differences in intrinsic activity observed between quinolones are mainly related to differences in quinolone inhibition of the targets. This has been demonstrated for the differences in activities of several compounds against a given bacterial species, e.g., nalidixic acid and ciprofloxacin against Escherichia coli (17), and of a given compound against several species, e.g., ciprofloxacin against E. coli and Staphylococcus aureus (5). In M. tuberculosis, differences in the inhibition of the targets by quinolones might thus explain the differences in quinolone activity. However, efflux pumps (2) and the naturally low permeability of the cell wall (19) could also play a role in determining the antimycobacterial activity of quinolones.
The bacterial targets of quinolones are the type II DNA topoisomerases, DNA gyrase and topoisomerase IV. These ATP-dependent enzymes act by a transient double-stranded DNA break and cooperate to facilitate DNA replication and other key DNA transactions (23). DNA gyrase is unique in catalyzing the negative supercoiling of DNA and is essential for efficient DNA replication, transcription, and recombination, whereas topoisomerase IV has a specialized role in chromosome segregation. DNA gyrase is a tetrameric A2B2 protein. The A subunit (90 to 100 kDa, depending on the bacterial species) carries the breakage-reunion active site, whereas the B subunit (70 to 90 kDa) promotes ATP hydrolysis, needed for energy transduction. M. tuberculosis genes encoding DNA gyrase were identified from the genome analysis as a gyrB-gyrA contig in which gyrA and gyrB encode the A and B subunits, respectively (26). Surprisingly, there is no evidence of the topoisomerase IV parC and parE gene homologs in the genome of M. tuberculosis (8). It appears that DNA gyrase is the sole topoisomerase target for quinolones in M. tuberculosis. Clearly, analysis of quinolone interactions with DNA gyrase will be important in understanding and optimizing the antimycobacterial properties of this class of drugs.
Previous studies of mycobacterial gyrases have largely involved enzymes purified from rapidly growing species by classical methods, i.e., bulk culture, cell disruption, and chromatography on novobiocin-Sepharose. This approach was first applied to gyrase from Mycobacterium smegmatis, a nonpathogenic, rapidly growing (3-day culture) mycobacterium (34). Gyrase was also purified from the low-pathogenicity opportunistic agents M. fortuitum, a rapid grower, and M. avium, a slow grower (14-day culture) (15). However, because M. tuberculosis grows slowly (21-day culture) and is highly pathogenic, the purification of its DNA gyrase requires recombinant methods that do not rely on bulk culture of the organism. In a previous study (28), maltose-binding protein fusions were used to produce M. tuberculosis gyrase subunits and to perform some preliminary assays of gyrase inhibition by quinolones. There are two potential limitations in the earlier work. First, it is known that gyrase subunits expressed as fusion proteins can have very low specific activities, unlike their recombinant His-tagged counterparts (28). Second, only four quinolones were examined, with inhibition of DNA supercoiling as the sole assay.
To investigate the interaction of M. tuberculosis gyrase with quinolones, particularly the newer, highly potent agents, we have developed recombinant plasmid clones that allow the production in E. coli and purification of recombinant M. tuberculosis GyrA and GyrB subunits carrying His tags. This alternative strategy provided M. tuberculosis DNA gyrase subunits safely and in large quantities. The recombinant GyrA and GyrB subunits were stable and reconstituted a functional DNA gyrase activity. We investigated the interaction of the enzyme with a large panel of quinolones with two complementary assays: inhibition of DNA supercoiling and induction of DNA cleavage arising from stabilization of the cleavage complex, which is thought to be the cytotoxic lesion. The results of this systematic study allowed us to establish a quinolone structure-activity relationship in which inhibition of supercoiling activity by 50% (IC50) correlated well (better than DNA cleavage) with inhibition of M. tuberculosis growth (as measured by the MIC). Analysis of drug action on M. tuberculosis gyrase may provide a safe and informative in vitro test for screening new quinolones with putative antituberculosis activity.
MATERIALS AND METHODS
Reagents.
Ofloxacin and levofloxacin (Aventis, Paris, France), gatifloxacin (Grünenthal, Levallois-Perret, France), ciprofloxacin and moxifloxacin (Bayer Pharma, Puteaux, France), sparfloxacin and pefloxacin (Rhône Poulenc Rorer, Vitry sur Seine, France), nalidixic acid, flumequine, novobiocin, oxolinic acid, and pipemidic acid (Sigma-Aldrich Chimie, Saint Quentin Fallavier, France), norfloxacin (Merck Sharp & Dohme Chibret, Paris, France), trovafloxacin (Pfizer, Marnes la Coquette, France), temafloxacin (Abbott, Saint Rémy sur Avre, France), fleroxacin (Roche, Fontenay sous Bois, France), sitafloxacin (Daiichi Pharmaceutical, Co., Ltd., Tokyo, Japan), grepafloxacin (Otsuka Pharmaceutical, Co., Ltd., Tokyo, Japan), garenoxacin (Bristol-Meyers Squibb, Saint Nazaire, France), tosufloxacin (Lederle, Paris La Défense, France), gemifloxacin (GlaxoSmithKline, Marly le Roy, France), and clinafloxacin (Anakena Pharma Marketing, Tokyo, Japan) were provided by the manufacturers. Oligonucleotide primers were synthesized by Eurogentec (Angers, France).
Bacterial strains and plasmids.
gyrA and gyrB genes were amplified from cosmid T776 of the genome bank of M. tuberculosis, which was kindly provided by S. Cole (8). E. coli DH5α was used as the host for cloning purposes, and strain BL21(λDE3)pLysS was used for protein expression. The pMOS Blue plasmid kit (Amersham Biosciences Europe, Orsay, France) was used to clone amplified DNA fragments. Plasmids pET-29a and pET-19b (Novagen, Merck Eurolab, Fontenay Sous Bois, France) were used to construct vectors for overexpression of M. tuberculosis GyrA and GyrB proteins. Supercoiled plasmid pBR322 DNA was provided by Roche Diagnostics, Meylan Cedex, France, and relaxed plasmid pBR322 DNA was obtained from John Innes Enterprises Ltd., Norwich Research Park, Colney, Norwich, United Kingdom.
Drug susceptibility.
M. tuberculosis H37Rv was grown on Löwenstein-Jensen medium. MICs were determined by the proportion method as described previously (14). Briefly, 103 and 105 CFU were inoculated onto 7H11 agar supplemented with 10% oleic acid-albumin-dextrose-catalase and containing serial twofold dilutions of the quinolone. Colonies were enumerated after 21 to 30 days of incubation at 37°C. The MIC was defined as the drug concentration at which the bacterial growth was reduced to 1% or less of that of the drug-free control culture (18). For five quinolones (sitafloxacin, norfloxacin, temafloxacin, fleroxacin, and enoxacin), the MICs were taken from the literature (11, 12, 36, 39, 42).
Construction of GyrA and GyrB expression vectors.
The gyrA and gyrB genes of M. tuberculosis were amplified from cosmid T776, cloned into pMOS Blue, and then inserted into pET-29a and pET-19b, respectively. Into each of the forward primers used for the amplification step, NdeI sites (CA'TATG) were engineered, overlapping the ATG initiation codons of gyrA and gyrB. An XhoI site was engineered before the stop codon for the gyrA primer and after the stop codon for gyrB in each of the reverse primers. The gyrA gene was amplified with forward primer GYRATB1 (5′-GCAAACGAGGAACATATGACAGACAC; the NdeI site overlapping the ATG initiation codon is italic) and reverse primer GYRATB2 (5′-CGAGCCTGATTACTCGAGCGTCTGGT; the XhoI site is italic). The gyrB gene was amplified with the forward primer GYRBTB1 (5′-GGCGCGGTCATATGGGTAAAAACGAG; the NdeI site overlapping the ATG initiation codon is italic) and reverse primer GYRBTB2 (5′-CGAACGCAGGCTCGAGTTAGACATCC; the XhoI site is italic).
Cosmid T776, containing the gyrB and gyrA genes of M. tuberculosis, was the template for amplification with the Taq and Pwo polymerases (Kit Expand Long Template PCR system; Boehringer Mannheim, Meylan, France) in the presence of 5 mM deoxynucleoside triphosphate mix and 2.25 mM MgCl2. Amplification conditions were as follows: after 5 min of denaturation at 94°C, we used 30 amplification cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 68°C for 1 min, with a final extension step for 10 min at 68°C. The PCR products corresponding to the 2.5-kb gyrA and 2.1-kb gyrB fragments were ligated into the pMOS Blue plasmid, transformed into Mos Blue competent cells, and plated on Luria-Bertani (LB) agar containing ampicillin (100 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and isopropylthiogalactopyranoside (IPTG). Recombinant plasmids were recovered from the white colonies and digested with NdeI and XhoI. The DNA fragment corresponding to gyrA was ligated into NdeI- and XhoI-cut pET-29a, and that corresponding to gyrB was ligated into NdeI- and XhoI-cut pET-19b, and both were transformed into E. coli DH5α. Recombinant clones were selected from the resistant colonies selected on plates containing kanamycin (50 μg/ml) for recombinant pET-29a or ampicillin (100 μg/ml) for recombinant pET-19b.
Protein overexpression and purification of GyrA and GyrB subunits.
The recombinant plasmid carrying the gyrA gene of M. tuberculosis (pATB) and that carrying the gyrB gene of M. tuberculosis (pBTB) were separately transformed by electroporation into E. coli BL21(λDE3)pLysS. GyrA and GyrB proteins were purified by the same procedure. Three different BL21 clones were grown separately at 37°C in 4 ml of LB medium containing the selective antibiotic until the optical density at 600 nm reached 0.8 to 1.0. The clone subcultures (1 ml) were then mixed and used to inoculate 50 ml of LB medium containing the selective antibiotic. Cells were grown at 37°C until the optical density at 600 nm reached 0.8 to 1.0. Bacteria were harvested by centrifugation at 3,000 × g for 10 min at 4°C. The pellet was suspended in 20 ml of LB and used to inoculate 500 ml of LB medium containing the selective antibiotic. Cells were grown at 30°C until the optical density at 600 nm reached 0.4 to 0.6 for pATB and 0.4 to 1 for pBTB. IPTG was added to final concentration of 1 mM, and growth was continued for a further 3 h. Bacteria were harvested by centrifugation at 3,000 × g for 15 min at 4°C, and the bacterial pellet was suspended in 12 ml of binding buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, and 5 mM imidazole [Novagen]) prior to storage at −80°C overnight.
The suspension was thawed on ice, lysozyme was added to a final concentration of ≈ 0.1%, and the suspension was centrifuged at 45,000 × g for 60 min. The supernatant was mixed with 2 ml of 50% nickel-nitrilotriacetic acid resin (Qiagen, Courtaboeuf, France) in a sterile precooled tube (Falcon) and gently agitated at 4°C overnight. After the suspension had settled for 1 h, the pellet was washed initially with 30 ml of binding buffer and then four times with 30 ml of wash buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, and 60 mM imidazole [Novagen]). The histidine-tagged GyrA and GyrB proteins were eluted with 2 ml of elution buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, and 250 mM imidazole). The total volume of the elution fraction (2 to 3 ml) was spun at 15,000 × g for 30 min at 4°C and then dialyzed overnight at 4°C against 2.5 liters of 50 mM Tris-HCl (pH 7.9) and 30% glycerol. Dithiothreitol and EDTA were added to final concentrations of 1 mM each. The GyrA and GyrB proteins were then flash frozen in aliquots in liquid nitrogen and stored at −80°C. The protein fractions were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
DNA supercoiling assays.
DNA supercoiling activity was tested with various ratios of purified M. tuberculosis GyrA and GyrB subunits. The reaction mixture (total volume, 30 μl) contained DNA gyrase assay buffer (40 mM Tris-HCl [pH 7.5], 25 mM KCl, 6 mM magnesium acetate, 2 mM spermidine, 4 mM dithiothreitol, 0.1 mg of E. coli tRNA per ml, bovine serum albumin [0.36 mg/ml], 100 mM potassium glutamate, 1 mM ATP) (pH 8.0) and relaxed pBR322 DNA (0.4 μg) as the substrate. Gyrase proteins were added, and the reaction mixtures were incubated at 37°C for 1 h. Reactions were terminated by the addition of 50% glycerol containing 0.25% bromophenol blue, and the total reaction mixture was subjected to electrophoresis in a 1% agarose gel in 0.5× TBE (Tris-borate-EDTA, pH 8.3) buffer. After running for 5.5 h at 50 V, the gel was stained with ethidium bromide (0.7 μg/ml). One unit of enzyme activity was defined as the amount of DNA gyrase that converted 400 ng of relaxed pBR322 to the supercoiled form in 1 h at 37°C.
DNA gyrase of E. coli (John Innes Enterprises Ltd.) was used as a positive control for the assay procedures and buffer. Inhibition of supercoiling activity of the recombinant DNA gyrase was performed by the method described previously (29). In brief, a reaction mixture containing 1 U of purified DNA gyrase and increasing concentrations of quinolones was incubated as described above. The inhibitory effect of quinolones on DNA gyrase was assessed by determining the concentration of drug required to inhibit the supercoiling activity of the enzyme by 50% (IC50). Supercoiling activity was assessed by tracing the brightness of the bands corresponding to the supercoiled pBR322 DNA with Molecular Analyst software (Bio-Rad).
DNA cleavage assays.
DNA cleavage assays were carried out in the same buffer as for DNA supercoiling except that relaxed pBR322 DNA was used instead of supercoiled pBR322 DNA. These assays were performed in the absence and presence of 1 mM ATP (31).
Various DNA gyrase amounts from 0.1 to 10 U were tested to determine the optimal amount of gyrase that had to be incubated with DNA in the presence of increasing concentrations of quinolones for 1 h at 25°C in order to produce the maximum cleaved band. Three microliters of 2% SDS and 3 μl of a 1-mg/ml solution of proteinase K were added, and incubation was continued for 30 min at 37°C. The reactions were stopped as for supercoiling. After electrophoresis for 5.5 h at 50 V, the 1% agarose gel was stained with ethidium bromide (0.7 μg/ml) and photographed under UV transillumination. The extent of DNA cleavage was quantified with the Molecular Analyst software. The concentration of quinolone that induced 50% of the maximum DNA cleavage (CC50) was determined. Plasmid pBR322 linearized by BamHI digestion was used as a marker for cleaved DNA. E. coli gyrase was used as a positive control for the assay procedures and buffer.
Correlation between MICs and IC50s against M. tuberculosis gyrase.
The relationships between the MICs and IC50s or CC50s were assessed by estimating a linear regression between two components, both first translated on the log10 scale. The strength of this relationship was quantified by the R2 coefficient and displayed graphically by the regression line and the two curves defining the 95% confidence interval for this regression.
RESULTS
Construction of expression vectors for M. tuberculosis DNA gyrase genes and purification of recombinant His-tagged GyrA and GyrB proteins.
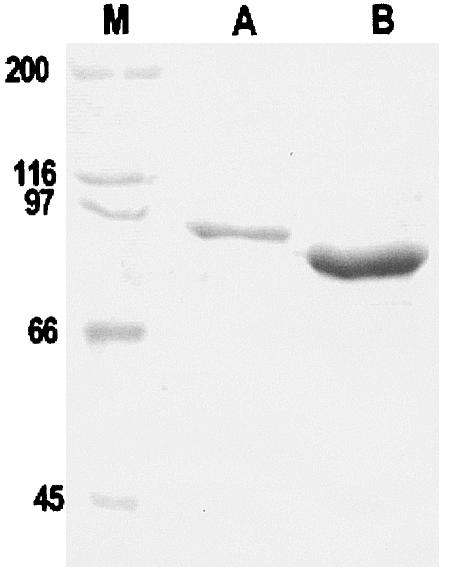
The M. tuberculosis gyrA and gyrB genes were each amplified from cosmid T776, which contains the gyrB-gyrA contig of M. tuberculosis (8). Taq polymerase was mixed with Pwo polymerase to minimize the introduction of polymerase errors, because Pwo polymerase has proofreading activity. The entire amplified gyrA and gyrB genes were inserted separately in-frame downstream of a T7 promoter in pET expression vectors to yield recombinant plasmids pATB (gyrA in pET-29a) and pBTB (gyrB in pET-19b). Expression of the gyrA and gyrB genes in E. coli BL21(λDE3)pLysS by induction with IPTG and subsequent purification by nickel chelate chromatography resulted in 2.5 mg and 10 mg of soluble His-tagged 97-kDa and 72-kDa proteins, respectively, from 500-ml cultures of induced cells (Fig. 1). The recombinant GyrA and GyrB proteins carried hexa- or decahistidine tags, respectively, at the C-terminal and N-terminal ends, respectively.
FIG. 1.
SDS-PAGE analysis of purified M. tuberculosis GyrA and GyrB proteins. The His-tagged proteins were overexpressed in E. coli and purified by nickel resin chromatography, and approximately 16 μl of each protein sample was loaded on an SDS-9% polyacrylamide gel. Following electrophoresis, proteins were revealed by staining with Coomassie blue. Lane M, size markers (sizes are indicated to the left in kilodaltons).
M. tuberculosis DNA gyrase is catalytically efficient for DNA supercoiling.
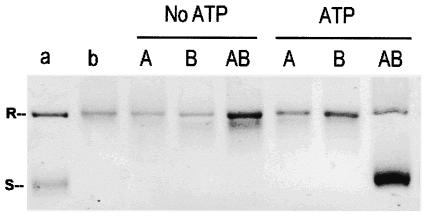
The combination of GyrA and GyrB subunits was tested for DNA supercoiling activity with relaxed pBR322 DNA as a substrate in the absence and presence of ATP (Fig. 2). One unit of GyrA was measured in the presence of excess amounts of GyrB, and conversely, 1 U of GyrB was measured in the presence of excess amounts of GyrA. One unit was defined as the amount that was sufficient to convert 100% of 0.4 μg of relaxed plasmid pBR322 DNA to the supercoiled form. Neither subunit alone induced DNA supercoiling. The combination of GyrA and GyrB led to plasmid supercoiling in the presence of ATP, which demonstrated that they reconstituted a functional DNA gyrase. No supercoiling was observed when ATP was omitted (Fig. 2). Two units of GyrA plus 2 U of GyrB were used subsequently for all the DNA supercoiling experiments. The specific activity of recombinant M. tuberculosis DNA gyrase in the supercoiling assay was 2.6 × 103 U/mg for GyrA and 1.5 × 104 U/mg for GyrB.
FIG. 2.
M. tuberculosis GyrA and GyrB proteins generate an ATP-dependent DNA supercoiling activity. Relaxed pBR322 (0.4 μg) was incubated with DNA gyrase reconstituted from GyrA (1 U) and GyrB (1 U) in the absence and presence of 1 mM ATP. The reactions were stopped, and the DNA products were separated by electrophoresis in a 1% agarose gel. DNA was stained with ethidium bromide and photographed under UV illumination. Lanes: a, supercoiled pBR322 DNA; b, relaxed pBR322 DNA; A, relaxed pBR322 DNA and GyrA (1 U) protein; B, relaxed pBR322 DNA and GyrB (1 U) protein; AB, relaxed pBR322 DNA and both GyrA (1 U) and GyrB (1 U). R and S, relaxed and supercoiled DNA, respectively.
Inhibition of DNA supercoiling by quinolones.
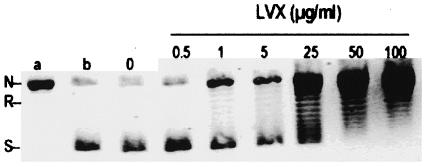
The ability of 22 quinolones to inhibit DNA supercoiling by M. tuberculosis gyrase was compared. An example of representative data is shown for levofloxacin (Fig. 3). The concentration of levofloxacin that inhibited DNA supercoiling by 50% was calculated as 5 μg/ml. Each of the quinolones tested showed dose-dependent inhibition, and their IC50 data are summarized in Table 1. Each experiment was done at least twice with similar results, and representative data are shown in Table 1. IC50 values are shown in rank order of potency from sitafloxacin and sparfloxacin (the most potent) to nalidixic acid (the least potent). The 22 quinolones tested clustered into three groups according to their IC50s. The quinolones in the first group had IC50s of <10 μg/ml and included sitafloxacin, sparfloxacin, clinafloxacin, gatifloxacin, ciprofloxacin, moxifloxacin, and levofloxacin. The quinolones in the second group had IC50s of >10 to <100 μg/ml and included ofloxacin, gemifloxacin, garenoxacin, norfloxacin, trovafloxacin, grepafloxacin fleroxacin, pefloxacin, tosufloxacin, enoxacin, and temafloxacin. The quinolones in the third group had IC50s of >100 μg/ml and included oxolinic acid, flumequine, pipemidic acid, and nalidixic acid.
FIG. 3.
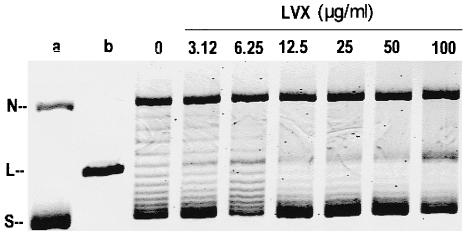
DNA supercoiling activity of wild-type M. tuberculosis DNA gyrase is sensitive to inhibition by levofloxacin (LVX). Relaxed pBR322 (0.4 μg) was incubated with DNA gyrase reconstituted from GyrA (2 U) and GyrB (2 U) in the absence and the presence of levofloxacin. The reactions were stopped, and the DNA products were analyzed by electrophoresis in a 1% agarose gel. Lanes a and b, relaxed and supercoiled pBR322 DNA, respectively. N, R, and S, nicked, relaxed, and supercoiled DNA, respectively.
TABLE 1.
Structural features and concentrations of quinolones that inhibit M. tuberculosis DNA gyrase activity and M. tuberculosis growtha
| Quinolone | Substituent
|
Concn, μg/ml (reference)
|
||||||
|---|---|---|---|---|---|---|---|---|
| R1 | R5 | R6 | R7 | N-8 or C-8 | IC50 | CC50 | MIC | |
| Sparfloxacin | Cyclopropyl | NH2 | F | Piperazine | C-F | 2 | 5 | 0.25 |
| Sitafloxacin | Fluorinated cyclopropyl | H | F | Pyrrolidine | C-Cl | 2.5 | 2 | 0.25 (36) |
| Clinafloxacin | Cyclopropyl | H | F | Pyrrolidine | C-Cl | 2.5 | 5 | 0.5 |
| Gatifloxacin | Cyclopropyl | H | F | Piperazine | C-OCH3 | 3 | 4 | 0.12 |
| Ciprofloxacin | Cyclopropyl | H | F | Piperazine | C-H | 3.5 | 6 | 0.5 |
| Moxifloxacin | Cyclopropyl | H | F | Azabicyclo | C-OCH3 | 4.5 | 4 | 0.5 |
| Levofloxacin | Bridge C1-C8 | H | F | Bridge C1-C8 | C-H | 5 | 12 | 0.5 |
| Ofloxacin | Bridge C1-C8 | H | F | Bridge C1-C8 | C-H | 10 | 20 | 1 |
| Gemifloxacin | Cyclopropyl | H | F | Pyrrolidine | N | 11 | 6 | 4 |
| Garenoxacin | Cyclopropyl | H | H | Azabicyclo | C-OCHF2 | 13 | 15 | 2 |
| Norfloxacin | Ethyl | H | F | Piperazine | C-H | 14 | 40 | 4 |
| Trovafloxacin | Difluorophenyl | H | F | Bicyclique | N | 15 | 25 | 16 |
| Grepafloxacin | Cyclopropyl | CH3 | F | Piperazine | C-H | 16 | 15 | 1 |
| Pefloxacin | Ethyl | H | F | Piperazine | C-H | 37 | 40 | 8 |
| Tosufloxacin | Difluorophenyl | H | F | Pyrrolidine | N | 37 | 25 | 16 |
| Temafloxacin | Difluorophenyl | H | F | Piperazine | C-H | 40 | 35 | 4 (42) |
| Fleroxacin | Fluoroethyl | H | F | Piperazine | C-H | 45 | 50 | 6.25 (39) |
| Enoxacin | Ethyl | H | F | Piperazine | N | 50 | 25 | 8 |
| Oxolinic acid | Ethyl | H | H | Bridge C6-C7 | C-H | 300 | NC | 32 |
| Flumequin | Bridge C1-C8 | H | F | H | Bridge C1-C8 | 500 | NC | 64 |
| Pipemidic acid | Ethyl | H | H | Piperazine | N | 1,000 | NC | 128 |
| Nalidixic acid | Ethyl | H | H | CH3 | N | 1,100 | NC | 128 |
NC, no or weak cleavage observed.
Analysis of the quinolone structure-activity relationship (Table 1) showed that the seven compounds in the first group, i.e., those with the highest activity against M. tuberculosis gyrase, shared several structural features: (i) a carbon at position 8, substituted by F for sparfloxacin, Cl for clinafloxacin, O-methyl for moxifloxacin and gatifloxacin, and an oxygen bridge between C-8 and N-1 for levofloxacin; (ii) a cyclopropyl group at N-1; (iii) a fluorine at C-6; and (iv) a substituent ring at C-7, often piperazinyl. In the other groups, none of the compounds had these four features. Although the compounds in the second group were fluorinated at position 6 except for garenoxacin (which has a difluoromethoxy substituent in C-8), only three quinolones in this group had an N-1 cyclopropyl: gemifloxacin, garenoxacin, and grepafloxacin. These quinolones lacked at least one of the other features, since gemifloxacin is not a C-8 quinolone but has an N-8, garenoxacin is not fluorinated at C-6, and grepafloxacin has an additional group at C-5. Ofloxacin, which is a racemic mixture of an L-isomer, levofloxacin, and an inactive R-isomer, was twofold less active than levofloxacin, as expected. All the compounds in the third group lacked fluorine at the C-6 position except flumequine, which had no substituent at C-7.
Activity of quinolones against M. tuberculosis and comparison with gyrase inhibition.
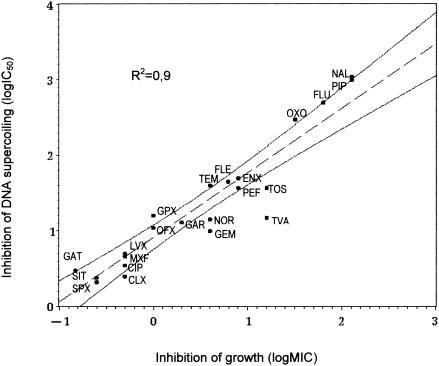
The MICs of the 22 quinolones tested ranged from 0.25 to 128 μg/ml (Table 1). The quinolone IC50 values correlated well with the MICs (R2 = 0.9), as shown in Fig. 4. However for some quinolones, such as trovafloxacin, gemifloxacin, tosufloxacin, norfloxacin, and ciprofloxacin, the MICs were higher than expected from the IC50s, and conversely, for gatifloxacin and grépafloxacine, the MICs were lower than expected from the IC50s.
FIG. 4.
Correlation between quinolone inhibition of M. tuberculosis gyrase (IC50 for DNA supercoiling) and quinolone MICs for M. tuberculosis. Dotted lines represent the confidence interval for 95% of the regression. R2 is the correlation coefficient. CIP, ciprofloxacin; CLX, clinafloxacin; ENX, enoxacin; FLE, fleroxacin; FLU, flumequine; GAR, garenoxacin; GAT, gatifloxacin; GEM, gemifloxacin, GRX, grepafloxacin; LVX, levofloxacin; MXF, moxifloxacin; NAL, nalidixic acid; NOR, norfloxacin, OFX, ofloxacin; OXO, oxolinic acid; PEF, pefloxacin; PIP, pipemidic acid; SIT, sitafloxacin; SPX, sparfloxacin; TEM, temafloxacin; TOS, tosufloxacin; TVA, trovafloxacin.
Cleavable-complex formation by DNA gyrase in the presence of quinolones.
To examine quinolone enhancement of DNA cleavage by M. tuberculosis gyrase, supercoiled pBR322 was incubated with enzyme in the presence of quinolones. After addition of SDS to induce DNA cleavage and proteinase K to remove GyrA protein covalently linked to DNA, DNA products were examined by agarose gel electrophoresis. A representative experiment for levofloxacin is presented in Fig. 5. In the absence of ATP and a quinolone, the DNA gyrase of M. tuberculosis showed a marked DNA relaxation activity. DNA relaxation was inhibited by the quinolones in a dose-dependent fashion. At higher quinolone levels, DNA cleavage led to linearization of the plasmid DNA. However, the relaxation activity hampered visualization of the cleaved band (Fig. 5). When ATP was added to the assay, it inhibited the relaxation activity and allowed better quantitation of the cleaved band.
FIG. 5.
Levofloxacin-mediated DNA cleavage by M. tuberculosis DNA gyrase. Supercoiled pBR322 DNA (0.4 μg) was incubated with M. tuberculosis GyrA (2 U) and GyrB (2 U) proteins in the absence of ATP and in the presence of levofloxacin (LVX) at the concentrations indicated on the figure. After addition of SDS and proteinase K, DNA samples were analyzed by electrophoresis in 1% agarose. Lanes a and b, supercoiled pBR322 DNA and BamHI-linearized pBR322. N, L, and S, nicked, linear, and supercoiled DNA, respectively.
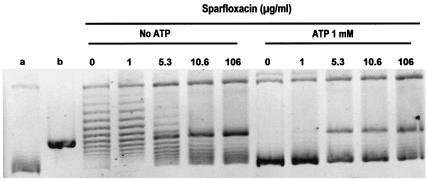
Figure 6 shows the results for sparfloxacin in the absence and presence of ATP. Table 1 summarizes the results obtained for the 22 quinolones. The drug concentration producing 50% of the maximum DNA cleavage (CC50) for sitafloxacin, gatifloxacin sparfloxacin, moxifloxacin, clinafloxacin, ciprofloxacin, and gemifloxacin was below 10 μg/ml; the CC50s of levofloxacin, ofloxacin, garenoxacin, grepafloxacin, trovafloxacin, tosufloxacin, pefloxacin, norfloxacin, temafloxacin, enoxacin, and fleroxacin were between 10 and 100 μg/ml. Little or no DNA cleavage was observed when the assay was performed in the presence of nalidixic acid, oxolinic acid, pipemidic acid, or flumequine.
FIG. 6.
Effect of ATP on the cleavage assay for the DNA gyrase of M. tuberculosis in the presence of quinolones. This figure shows the assay in the presence of sparfloxacin. Supercoiled pBR322 DNA (0.4 μg) was incubated with the M. tuberculosis GyrA (1 U) and GyrB (1 U) proteins in the absence of ATP and in the presence ATP (1 mM) plus increasing concentrations of sparfloxacin at the concentrations indicated in the figure. After addition of SDS and proteinase K, DNA samples were analyzed by electrophoresis in 1% agarose. Lanes a and b, supercoiled pBR322 DNA and BamHI-linearized pBR322, respectively. N, L, and S, nicked, linear, and supercoiled DNA, respectively.
The CC50s grouped the quinolones similarly to the IC50s derived from the DNA supercoiling test. Two exceptions were gemifloxacin, which was more effective in DNA cleavage induction than in inhibition of DNA supercoiling, and levofloxacin, which was less effective in cleavage than in supercoiling inhibition. From quinolone structure-activity considerations, the presence of C or N at position 8 (gemifloxacin has N-8) is probably less important for DNA cleavage than the N-1 cyclopropyl, which is not present in levofloxacin. The quinolones that were inefficient in stimulating DNA cleavage, i.e., the classical quinolones, also lacked a fluorine at position 6 or a piperazinyl ring at C-7. Overall, the CC50s for DNA cleavage showed a poorer correlation with the MICs (R2 = 0.7) than the IC50 did (Table 1).
DISCUSSION
The slow growth and high pathogenicity of M. tuberculosis preclude classical purification methods that have yielded native gyrases from the less pathogenic mycobacteria M. smegmatis, M. fortuitum, and M. avium (15, 25). To circumvent these difficulties, M. tuberculosis DNA gyrase was produced as His-tagged A and B subunits. Its interaction with quinolones was evaluated with two assays, DNA supercoiling and quinolone-mediated DNA cleavage (4).
The use of His tags facilitated safe and rapid purification and yielded products that were free of host (E. coli) proteins. Although results with native M. tuberculosis gyrase are not available, it does not seem that the His tags hampered the function of M. tuberculosis DNA gyrase or its interaction with quinolones. Indeed, the data obtained compared with those observed for the native M. smegmatis gyrase (15, 25, 34). Maltose-binding protein fusions with the GyrA and GyrB subunits have been used to obtain recombinant M. tuberculosis DNA gyrase, which has been tested against four quinolones (28). Of the IC50s of these four quinolones measured, only that of sitafloxacin agreed with our data. Although the DNA supercoiling assay was performed under the same conditions, the earlier IC50s for levofloxacin, ciprofloxacin, and sparfloxacin were two- to fourfold higher than those we measured. Maltose-binding protein fusion could result in misfolding compared to the native form. His-tagged gyrases have been shown to display interactions with quinolones comparable to those of the native bacterial gyrases for bacteria other than mycobacteria (5, 29, 35).
The ability to reconstitute M. tuberculosis gyrase activity allowed us to examine and compare the inhibitory effects of a large panel of quinolones, including recently developed agents. Twenty-two quinolones were evaluated for the ability to inhibit DNA supercoiling activity and to stimulate gyrase-mediated DNA cleavage. The quinolones inhibited the DNA supercoiling of M. tuberculosis gyrase in a dose-dependent manner, as observed in bacteria other than mycobacteria (5, 17, 30). The potency of the quinolones was demonstrated in both assays.
From the inhibition results, seven quinolones showed high inhibitory activity against M. tuberculosis DNA gyrase, with IC50s below 10 μg/ml. Analysis of the quinolone structure-activity relationship showed that these seven compounds shared certain structural features (C-8 with or lacking a substitution, N-1 cyclopropyl group, a ring at C-7, and a fluorine at C-6) (see Table 1). Quinolone structure-activity relationship analyses done to date for quinolones and mycobacteria (21, 24, 32, 38) were not based on target inhibition, as in our study, but on MICs, which are the result of target inhibition and many other factors of activity, such as cell wall permeability and efflux (19). Moreover, these analyses were done with nontuberculous mycobacteria (M. smegmatis, M. avium, and M. fortuitum) and not with M. tuberculosis. Although cyclopropyl at N-1 was shown to be a key feature (24, 32, 33), these studies often attributed added importance to the substituent at C-8.
There was no relationship between quinolone activity against M. tuberculosis gyrase and activity against other bacteria classified according to gram-positive and gram-negative status (16). The quinolones that were highly active against M. tuberculosis gyrase included compounds that are highly active against gram-positive bacteria and indeed were developed especially for pneumococci (sitafloxacin, sparfloxacin, clinafloxacin, moxifloxacin, and gatifloxacin). Conversely, four compounds (grepafloxacin, gemifloxacin, trovafloxacin, and the des[6]fluoroquinolone garenoxacin) with high activity against pneumococci showed only moderate activity against M. tuberculosis gyrase. Most of the classical fluoroquinolones developed for their activity against gram-negative bacteria (norfloxacin, pefloxacin, enoxacin, fleroxacin, ofloxacin, temafloxacin, and tosufloxacin) had moderate IC50s except for levofloxacin and ciprofloxacin, which had low IC50s against M. tuberculosis gyrase. Contrary to its effects against pneumococci, the presence of a group at C-5 (27) or a substituent in the 7-piperazinyl ring (1) does not seem to improve gyrase affinity. Moreover, the presence of a naphthyridone core (N-8) in gemifloxacin, which has the lowest MIC against gram-positive bacteria, seems unfavorable for a tight interaction with M. tuberculosis gyrase. Similarly, the naphthyridones, tosufloxacin and enoxacin, were only moderately active (Table 1).
The fact that the quinolone structure-activity relationship against M. tuberculosis does not follow those established for other gram-positive organisms (Staphylococcus aureus and Streptococcus pneumoniae) may arise from two unique characteristics of the M. tuberculosis DNA gyrase: (i) it is the sole type II topoisomerase in its host, and therefore there is no dual activity on DNA gyrase and on topoisomerase IV, and (ii) the peptidic structure of the quinolone resistance-determining region in the A and B subunits is unique, as described previously (14). Ser-83 of E. coli GyrA is the key residue for interaction with quinolones (3) and is conserved in the GyrA proteins of many bacterial species, such as Staphylococcus aureus and Streptococcus pneumoniae. The equivalent residue (position 90) in M. tuberculosis GyrA is an alanine, a difference that may have key importance for quinolone interactions.
The concentrations of quinolones that inhibited 50% of the DNA supercoiling activity of the M. tuberculosis DNA gyrase correlated well with the MICs, i.e., their ability to inhibit the growth of M. tuberculosis. However, the IC50s and MICs were not always proportional; for example, grepafloxacin and trovafloxacin were equipotent in the gyrase assay, and yet the trovafloxacin MIC was about 16-fold higher than that of grepafloxacin against M. tuberculosis. This nonproportionality has been noted by others (41) and presumably reflects basic differences in the cell-permeating properties and accumulation of the different quinolones (19). Penetration of the M. tuberculosis cell wall by quinolones has not been evaluated yet because the study of the mycobacterial cell wall is still a difficult and uncertain task (19). However, penetration of the M. tuberculosis cell wall seems to be at least 100-fold less efficient than that of E. coli (9). In the present study, as in studies on gyrases from other bacteria, it has been shown that the concentration of quinolones required to inhibit DNA supercoiling by gyrase is substantially higher than that required to inhibit growth. This fact has been attributed to the poisoning effect of quinolones interacting with the topoisomerases (22).
DNA cleavage assays have been proposed as a more relevant test than supercoiling inhibition to correlate inhibition of gyrase with antibacterial activity (4, 41). Because of the high relaxation activity of M. tuberculosis DNA gyrase, cleavage data needed to be evaluated in the presence of ATP. Unlike the gyrases of E. coli and Streptococcus pneumoniae, M. tuberculosis gyrase exhibited overall a low level of DNA cleavage activity in the presence of quinolones. This has been described previously for gyrA mutants of Streptococcus pneumoniae (31). Moreover, classical quinolones (nalidixic acid, pipemidic acid, oxolinic acid, and flumequine), which lack F-6 or a 7-piperazinyl ring, were very poor inducers of DNA cleavage. Low DNA cleavage stimulation suggests that quinolones may interact less avidly with M. tuberculosis DNA gyrase than with gyrases from other bacterial species (4, 5). This may be due in part to the presence of an alanine at residue 90 of M. tuberculosis GyrA. A Ser83Ala change in E. coli GyrA confers a degree of resistance to quinolones.
In the DNA cleavage assay, the effective quinolone concentrations were slightly different from those inhibiting supercoiling and less correlated with those inhibiting M. tuberculosis growth. The DNA gyrase supercoiling inhibition assay and DNA gyrase cleavable-complex assay are distinct in that the former is a measure of catalytic inhibition, whereas the latter probes an established equilibrium between the ternary DNA-enzyme-drug complexes in which the DNA is either broken or intact (4). For M. tuberculosis DNA gyrase in the presence of low concentrations of quinolones, the balance between the cleaved and rejoined DNA forms of the enzyme complex might favor not the cleaved intermediate but the rejoined one, since the quinolone-stabilized DNA cleavage is known to be reversible and ATP can stimulate the dissociation of the DNA-quinolone-gyrase complex (37). The DNA cleavage results might be better correlated to bactericidal activity. Although the in vitro bactericidal activity of quinolones has not been widely explored for mycobacteria, in animal models moxifloxacin showed much higher bactericidal activity than levofloxacin, which is concordant with the CC50 data (20).
The study of quinolone interaction with M. tuberculosis DNA gyrase represents a crucial step in investigating quinolone structure-activity relationships and in developing compounds with good activity against tubercle bacilli. The inhibition of DNA supercoiling by M. tuberculosis gyrase could be a safe and quick test for screening drugs with promising antituberculosis activity. Quinolones with low IC50s (below 10 μg/ml) will potentially be active against M. tuberculosis and will justify testing in experimental in vivo models of tuberculosis. By contrast, quinolones with high IC50s in the enzyme assay will likely not be suitable for further evaluation as antimycobacterial drugs.
Acknowledgments
We thank Wladimir Sougakoff for helpful discussion and Stephanie Petrella and Sylvie Escolano for help with some of the experiments.
This work was supported by grants from INSERM (EMI 0004), the University of Paris (UPRES 1541), the Association Française Raoul Follereau, and the Association Claude Bernard. X.S.P was funded by project grant C16747 from the Biotechnology and Biological Sciences Research Council, United Kingdom
REFERENCES
- 1.Alovero, F. L., X. S. Pan, J. E. Morris, R. H. Manzo, and L. M. Fisher. 2000. Engineering the specificity of antibacterial fluoroquinolones: benzenesulfonamide modifications at C-7 of ciprofloxacin change its primary target in Streptococcus pneumoniae from topoisomerase IV to gyrase. Antimicrob. Agents Chemother. 44:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee, S. K., K. Bhatt, S. Rana, P. Misra, and P. K. Chakraborti. 1996. Involvement of an efflux system in mediating high level of fluoroquinolone resistance in Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 226:362-368. [DOI] [PubMed] [Google Scholar]
- 3.Barnard, F. M., and A. Maxwell. 2001. Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser83 and Asp87. Antimicrob. Agents Chemother. 45:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett, J. F., J. I. Bernstein, H. M. Krause, J. J. Hilliard, and K. A. Ohemeng. 1993. Testing potential gyrase inhibitors of bacterial DNA gyrase: a comparison of the supercoiling inhibition assay and “cleavable complex” assay. Anal. Biochem. 214:313-317. [DOI] [PubMed] [Google Scholar]
- 5.Blanche, F., B. Cameron, F. X. Bernard, L. Maton, B. Manse, L. Ferrero, N. Ratet, C. Lecoq, A. Goniot, D. Bisch, and J. Crouzet. 1996. Differential behaviors of Staphylococcus aureus and Escherichia coli type II DNA topoisomerases. Antimicrob. Agents Chemother. 40:2714-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumberg, H. M., W. J. Burman, R. E. Chaisson, C. L. Daley, S. C. Etkind, L. N. Friedman, P. Fujiwara, M. Grzemska, P. C. Hopewell, M. D. Iseman, R. M. Jasmer, V. Koppaka, R. I. Menzies, R. J. O'Brien, R. R. Reves, L. B. Reichman, P. M. Simone, J. R. Starke, and A. A. Vernon. 2003. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am. J. Respir. Crit. Care Med. 167:603-662. [DOI] [PubMed] [Google Scholar]
- 7.Cambau, E., W. Sougakoff, M. Besson, C. Truffot-Pernot, J. Grosset, and V. Jarlier. 1994. Selection of a gyrA mutant of Mycobacterium tuberculosis resistant to fluoroquinolones during treatment with ofloxacin. J. Infect. Dis. 170:479-483. [DOI] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 9.Connell, N., and H. Nikaido. 1994. Membrane permeability and transport in Mycobacterium tuberculosis, p. 333-352. In B. R. Bloom (ed.), Tuberculosis: pathogenesis, protection, and control. American Society for Microbiology, Washington, D.C.
- 10.Crofton, J., P. Chaulet, D. Maher, J. Grosset, W. Harris, N. Horne, M. Iseman, and B. Watt. 1997. Guidelines for the management of drug-resistant tuberculosis. World Health Organization, Geneva, Switzerland.
- 11.Fenlon, C. H., and M. H. Cynamon. 1986. Comparative in vitro activities of ciprofloxacin and other 4-quinolones against Mycobacterium tuberculosis and Mycobacterium intracellulare. Antimicrob. Agents Chemother. 29:386-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorzynski, E. A., S. I. Gutman, and W. Allen. 1989. Comparative antimycobacterial activities of difloxacin, temafloxacin, enoxacin, pefloxacin, reference fluoroquinolones, and a new macrolide, clarithromycin. Antimicrob. Agents Chemother. 33:591-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosset, J. H. 1992. Treatment of tuberculosis in HIV infection. Tuberc. Lung. Dis. 73:378-383. [DOI] [PubMed] [Google Scholar]
- 14.Guillemin, I., V. Jarlier, and E. Cambau. 1998. Correlation between quinolone susceptibility patterns and sequences in the A and B subunits of DNA gyrase in mycobacteria. Antimicrob. Agents Chemother. 42:2084-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillemin, I., W. Sougakoff, E. Cambau, V. Revel-Viravau, N. Moreau, and V. Jarlier. 1999. Purification and inhibition by quinolones of DNA gyrases from Mycobacterium avium, Mycobacterium smegmatis and Mycobacterium fortuitum bv. peregrinum. Microbiology 145:2527-2532. [DOI] [PubMed] [Google Scholar]
- 16.Hooper, D. C. 2002. Fluoroquinolone resistance among Gram-positive cocci. Lancet Infect. Dis. 2:530-538. [DOI] [PubMed] [Google Scholar]
- 17.Hooper, D. C., J. S. Wolfson, E. Y. Ng, and M. N. Swartz. 1987. Mechanisms of action of and resistance to ciprofloxacin. Am. J. Med. 82:12-20. [PubMed] [Google Scholar]
- 18.Inderlied, C. B., and K. A. Nash. 1996. Antimicrobial agents: in vitro susceptibility testing, spectra of activity, mechanisms of action and resistance, and assays for activity in biologic fluids, p. 127-175. In M. D. V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. The Williams & Wilkins Co., Baltimore, Md.
- 19.Jarlier, V., and H. Nikaido. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123:11-18. [DOI] [PubMed] [Google Scholar]
- 20.Ji, B., N. Lounis, C. Maslo, C. Truffot-Pernot, P. Bonnafous, and J. Grosset. 1998. In vitro and in vivo activities of moxifloxacin and clinafloxacin against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:2066-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klopman, G., J. Y. Li, S. Wang, A. J. Pearson, K. Chang, M. R. Jacobs, S. Bajaksouzian, and J. J. Ellner. 1994. In vitro anti-Mycobacterium avium activities of quinolones: predicted active structures and mechanistic considerations. Antimicrob. Agents Chemother. 38:1794-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreuzer, K. N., and N. R. Cozzarelli. 1979. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J. Bacteriol. 140:424-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine, C., H. Hiasa, and K. J. Marians. 1998. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta 1400:29-43. [DOI] [PubMed] [Google Scholar]
- 24.Lu, T., X. Zhao, X. Li, A. Drlica-Wagner, J. Y. Wang, J. Domagala, and K. Drlica. 2001. Enhancement of fluoroquinolone activity by C-8 halogen and methoxy moieties: action against a gyrase resistance mutant of Mycobacterium smegmatis and a gyrase-topoisomerase IV double mutant of Staphylococcus aureus. Antimicrob. Agents Chemother. 45:2703-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madhusudan, K., and V. Nagaraja. 1995. Mycobacterium smegmatis DNA gyrase: cloning and overexpression in Escherichia coli. Microbiology 141:3029-3037. [DOI] [PubMed] [Google Scholar]
- 26.Madhusudan, K., V. Ramesh, and V. Nagaraja. 1994. Molecular cloning of gyrA and gyrB genes of Mycobacterium tuberculosis: analysis of nucleotide sequence. Biochem. Mol. Biol. Int. 33:651-660. [PubMed] [Google Scholar]
- 27.Morris, J. E., X. S. Pan, and L. M. Fisher. 2002. Grepafloxacin, a dimethyl derivative of ciprofloxacin, acts preferentially through gyrase in Streptococcus pneumoniae: role of the C-5 group in target specificity. Antimicrob. Agents Chemother. 46:582-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onodera, Y., M. Tanaka, and K. Sato. 2001. Inhibitory activity of quinolones against DNA gyrase of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 47:447-450. [DOI] [PubMed] [Google Scholar]
- 29.Pan, X. S., and L. M. Fisher. 1999. Streptococcus pneumoniae DNA gyrase and topoisomerase IV: overexpression, purification, and differential inhibition by fluoroquinolones. Antimicrob. Agents Chemother. 43:1129-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan, X. S., and L. M. Fisher. 1997. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob. Agents Chemother. 41:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan, X. S., G. Yague, and L. M. Fisher. 2001. Quinolone resistance mutations in Streptococcus pneumoniae GyrA and ParC proteins: mechanistic insights into quinolone action from enzymatic analysis, intracellular levels, and phenotypes of wild-type and mutant proteins. Antimicrob. Agents Chemother. 45:3140-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renau, T. E., J. W. Gage, J. A. Dever, G. E. Roland, E. T. Joannides, M. A. Shapiro, J. P. Sanchez, S. J. Gracheck, J. M. Domagala, M. R. Jacobs, and R. C. Reynolds. 1996. Structure-activity relationships of quinolone agents against mycobacteria: effect of structural modifications at the 8 position. Antimicrob. Agents Chemother. 40:2363-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renau, T. E., J. P. Sanchez, J. W. Gage, J. A. Dever, M. A. Shapiro, S. J. Gracheck, and J. M. Domagala. 1996. Structure-activity relationships of the quinolone antibacterials against mycobacteria: effect of structural changes at N-1 and C-7. J. Med. Chem. 39:729-735. [DOI] [PubMed] [Google Scholar]
- 34.Revel-Viravau, V., Q. C. Truong, N. Moreau, V. Jarlier, and W. Sougakoff. 1996. Sequence analysis, purification, and study of inhibition by 4-quinolones of the DNA gyrase from Mycobacterium smegmatis. Antimicrob. Agents Chemother. 40:2054-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saiki, A. Y., L. L. Shen, C. M. Chen, J. Baranowski, and C. G. Lerner. 1999. DNA cleavage activities of Staphylococcus aureus gyrase and topoisomerase IV stimulated by quinolones and 2-pyridones. Antimicrob. Agents Chemother. 43:1574-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito, H., H. Tomioka, K. Sato, and S. Dekio. 1994. In vitro and in vivo antimycobacterial activities of a new quinolone, DU-6859a. Antimicrob. Agents Chemother. 38:2877-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheirer, K. E., and N. P. Higgins. 1997. The DNA cleavage reaction of DNA gyrase. Comparison of stable ternary complexes formed with enoxacin and CcdB protein. J. Biol. Chem. 272:27202-27209. [DOI] [PubMed] [Google Scholar]
- 38.Tillotson, G., and J. Blondeau. 1999. Structure-activity-function evaluation of the fluoroquinolones, p. 91-101. In D. Adam and R. Finch (ed.), Moxifloxacin in practice, vol. 2. Bayer, Berlin, Germany. [Google Scholar]
- 39.Tomioka, H., K. Sato, and H. Saito. 1991. Comparative in vitro and in vivo activity of fleroxacin and ofloxacin against various mycobacteria. Tubercle 72:176-180. [DOI] [PubMed] [Google Scholar]
- 40.Tsukamura, M., E. Nakamura, S. Yoshii, and H. Amano. 1985. Therapeutic effect of a new antibacterial substance ofloxacin (DL8280) on pulmonary tuberculosis. Am. Rev. Respir. Dis. 131:352-356. [DOI] [PubMed] [Google Scholar]
- 41.Walton, L., and L. P. Elwell. 1988. In vitro cleavable-complex assay to monitor antimicrobial potency of quinolones. Antimicrob. Agents. Chemother. 32:1086-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yew, W. W., L. J. Piddock, M. S. Li, D. Lyon, C. Y. Chan, and A. F. Cheng. 1994. In-vitro activity of quinolones and macrolides against mycobacteria. J. Antimicrob. Chemother. 34:343-351. [DOI] [PubMed] [Google Scholar]