Abstract
Pilicides prevent pili formation and thereby the development of bacterial biofilms in Escherichia coli. We have performed a comprehensive structure activity relationship (SAR) study of the dihydrothiazolo ring-fused 2-pyridone pilicide central fragment by varying all open positions. Orthogonal projections to latent structures discriminant analysis (OPLS-DA) was used to distinguish active from inactive compounds in which polarity proved to be the most important factor for discrimination. A quantitative SAR (QSAR) partial least squares (PLS) model was calculated on the active compounds for prediction of biofilm inhibition activity. In this model, compounds with high inhibitory activity were generally larger, more lipophilic, more flexible and had a lower HOMO. Overall, this resulted in both highly valuable SAR information and potent inhibitors of type 1 pili dependent biofilm formation. The most potent biofilm inhibitor had an EC50 of 400 nM.
Keywords: Pilicide, Antivirulence, 2-Pyridone, Peptidomimetic, Structure–activity, Biofilm inhibitor
1. Introduction
The discovery of compounds with antimicrobial activity in the 1940s resulted in tremendous advances for humanity. However, the reduced efforts put into antibacterial research in combination with the increasing development of bacterial resistance have resulted in the re-emergence of bacterial diseases as a global health problem. Therefore, the development of new antibacterial targets and strategies, especially those directed towards Gram-negative bacteria, is of great importance.1
Bacterial pathogens have an arsenal of virulence factors that are critical in promoting host–pathogen interactions. One such factor is type 1 pili, assembled by the chaperone/usher pathway (CUP), which constitute a critical virulence factor in uropathogenic Escherichia coli (UPEC).2 Type 1 pili facilitate the attachment to and invasion of the bladder epithelium and is important for IBC formation.3–6 In addition, recent studies concluded that type 1 pili play an important role in human cystitis,7 and it has been reported that type 1 pili fulfill ‘Molecular Koch’s postulates’ of microbial pathogenesis.8
Biofilm formation is a severe complicating factor in bacterial infections due to the increased resistance of bacteria within biofilms to immune- or drug-mediated clearance, leading to persistent, drug-tolerant infections.9–11 Thus, biofilms exacerbate bacterial infections, such as urinary tract infection (UTI), causing significant human morbidity and mortality and billions of dollars in health care expenditures.12
Type 1 pili and other CUP pili are promising targets of anti-virulence therapeutics as they are critical factors for binding and invading the bladder tissue and forming biofilms.13,14
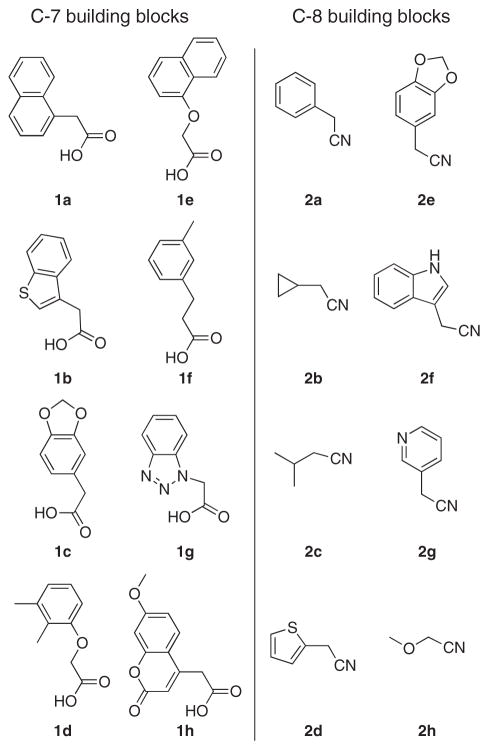
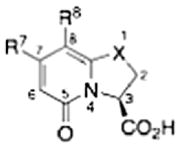
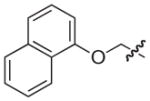
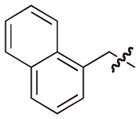
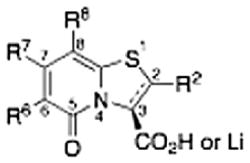
Pilicides are a class of small molecular weight compounds that prevent formation of pili in UPEC.15–17 The assembly proteins of the CUP pilus systems have a high degree of structural conservation and compounds that target pili formation can thus potentially have broad-spectrum activity.18 The pilicide core structure (5) is based on a dihydrothiazolo ring-fused 2-pyridone that can be synthesized via an acyl-ketene imine cyclocondensation of acyl-Meldrum’s acid derivatives (3) and thiazolines (4).19,20 The acyl-Meldrum’s acid derivatives (3) and thiazolines (4) are generally obtained from commercially available carboxylic acids (1) and nitriles (2), respectively. This synthetic procedure introduces substituent diversity in position C-7 and C-8 already in the scaffold formation by the choice of carboxylic acids (1) and nitriles (2). Furthermore, various synthetic methodologies for the introduction of substituents at the C-2 and C-6 positions of the scaffold have been developed (Fig. 1).16,21–24
Figure 1.
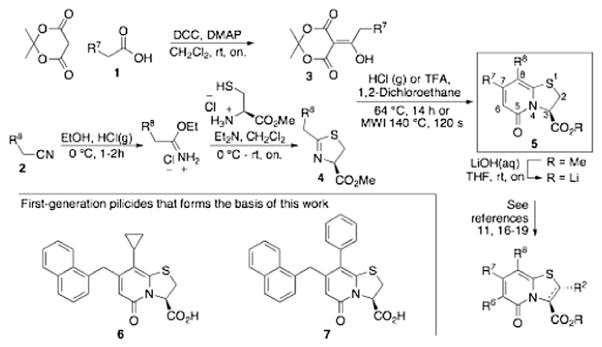
Synthesis of the dihydrothiazolo ring-fused 2-pyridone pilicide scaffold (5) from commercially available carboxylic acids (1) and nitriles (2) via acyl-Meldrum’s acid derivatives (3) and thiazolines (4), respectively. Firs-generation pilicides with a CH2-1-naphthyl substituent in C-7 and a cyclopropyl (6) or phenyl (7) in C-8 that forms the basis of this work.
Rational alteration of the pilicide scaffolds substituents using multivariate design could generate valuable new insights into the intricate relationship between pilicide structure and biological activity and ultimately result in comprehensive structure–activity relationship (SAR) information. Multivariate design can be used to decipher the complex correlation patterns between chemical structures and specific biological responses and properties. In multivariate design, a representative subset of compounds is selected from a more complete virtual set. This considerably reduces the number of compounds to be synthesized and evaluated while retaining much of the chemical information associated with the full compound set. To enable this subset selection, all compounds are described by numerical molecular descriptors. To visualize the data set of different compounds with their corresponding descriptors, the use of principal component analysis (PCA)25 is often applied. From this, a representative selection of a subset can be performed by applying statistical experimental designs, that is, statistical molecular design (SMD).26,27 This results in a balanced subset of compounds that encompass as much as possible of the significant variation from the entire data set.28
Here we present a multivariate design strategy on di-substituted derivatives of the pilicide scaffold from which the most promising compounds could be further substituted to give highly active tri- and tetra-substituted derivatives. Furthermore, the set of di-substituted derivatives could also be used to construct a reliable SAR model that could correctly rank the external validation set. Overall the strategy was rewarding and significantly improved pilicides and valuable SAR information was obtained. The results described herein will guide future studies of these compounds as research tools to study the biological processes they interfere with, which ultimately may lead to novel anti-virulence therapeutics.
2. Results and discussion
2.1. Synthesis and evaluation of di-substituted derivatives
Starting from suitable commercially available carboxylic acids and nitriles, two sets of potential C-7 and C-8 substituents were identified and their chemical diversity was characterized by molecular descriptors. Eight substituents at each position were manually selected from the chemical space generated by PCA. The choice of substituents was based on two known pilicides that both had a CH2-1-naphthyl substituent in C-7 and a phenyl or cyclopropyl in C-8 (Fig. 1).15 Hence, substituents were selected from the chemical space in the vicinity of these substituents with varying sizes, polarities and flexibility (Fig. 2).
Figure 2.
The building blocks used when constructing the library of C-7 and C-8-substituted pilicides.
All combinations of the eight C-7 and eight C-8 substituents, resulted in a virtual set of 64 different pilicides. An SMD using D-optimal design was used to select a balanced subset of 24 compounds from this set, in which all of the substituents were represented three times (Table 1).
Table 1.
Chemical structures and biological evaluation of the balanced set of di-substituted compounds
| |||
|---|---|---|---|
| ID | R8 (C-8) | R7 (C-7) | EC50 (μM)a |
| 8b,c |
|
|
40 |
| 9b,c |
|
|
36 |
| 10b |
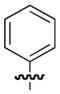
|
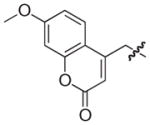
|
NAd |
| 11b |

|
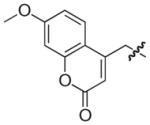
|
NAd |
| 12b |

|
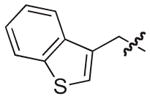
|
NAd |
| 13b |

|
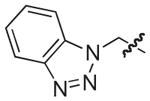
|
NAd |
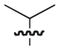
| 14b,c |
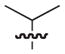
|
|
100 |
| 15b |
|
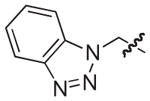
|
NAd |
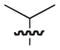
| 16b |
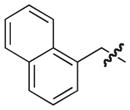
|
|
100 |
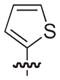
| 17b |
|
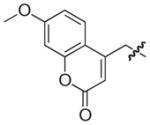
|
NAd |
| 18b |
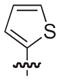
|
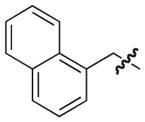
|
39 |
| 19b,c |
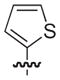
|
|
14 |
| 20b,c |
|
|
27 |
| 21b |
|
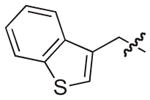
|
67 |
| 22b |
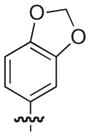
|
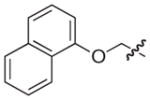
|
21 |
| 23b |
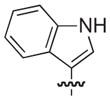
|
|
74 |
| 24b |
|
|
12 |
| 25b |
|
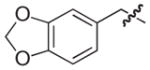
|
39 |
| 26b |
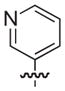
|
|
NAd |
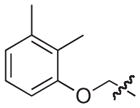
| 27b |
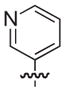
|
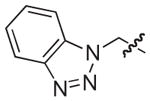
|
NAd |
| 28b |
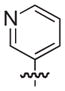
|
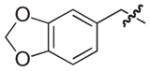
|
NAd |
| 29b |
|
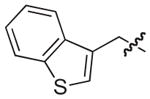
|
NAd |
| 30b |
|
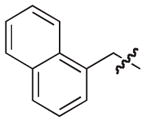
|
NAd |
| 31b,c |
|
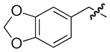
|
NAd |
| 7e |
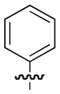
|
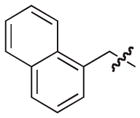
|
40 |
| 6e |
|
|
250 |
Calculated from 16–72 data points per concentration.
Compound is part of the design.
For solubility reasons these compounds were tested as carboxylic acids and not as lithium carboxylates.
Not active.
First generation pilicides that the design was based on.
The 24 derivatives of the pilicide scaffold were next synthesized in a five step synthetic sequence to the target compounds.20 In general, the acyl-ketene imine cyclocondensation produced the desired compounds in good yields (66–93%). However, the polar C-7 substituent in building block 1g proved problematic and resulted in lower yields (15–50%). The C-8 indole building block 2f resulted in both atropisomers, probably due to increased sterical clash with adjacent substituents, and low yields in the acyl-ketene imine cyclocondensation, primarily due to acylation of the indole by the acyl-ketene.
Before biological evaluation could be undertaken, the compounds had to be hydrolyzed to the corresponding lithium carboxylates. This was straightforward and all 24 target compounds were isolated in yields above 90%. The 24 compounds were evaluated using a pili dependent biofilm assay.29 In this assay, blocking the formation of type 1 pili formation in the clinical isolate E. coli strain, UTI89, completely prevents the bacteria’s ability to form biofilm. Thus, the amount of biofilm that is formed in UTI89 grown in the presence of pilicide is related to the potency of the compound in blocking the formation of pili. The results are summarized in Table 1.
We discovered that many of the di-substituted compounds efficiently prevented pilus-dependent biofilm formation. In general, the potency of the compounds turned out to be highly dependent upon the nature of both substituents. The best compound in the set was the C-8 indole and C-7 naphthoxy substituted 24 with an EC50 of 12 μM. Compounds incorporating a thiophene or 3,4-methylenedioxyphenyl group at C-8 (18–22) were also effective, except when combined with a C-7 coumarin substituent as in 17. The C-8 phenyl substituent was also favorable, but pyridine and the smaller methoxy, cyclopropyl, and isopropyl substituents resulted in reduced potency. The most useful C-7 substituents were the 1-naphthylmethyl, naphthoxymethyl, 3-tolylethyl, and 2,3-dimethylphenoxymethyl groups. Exchange of the 1-naphthylmethyl group and extension of the C-7 linker were well tolerated, but heteroaryls were not.
2.2. Multivariate design
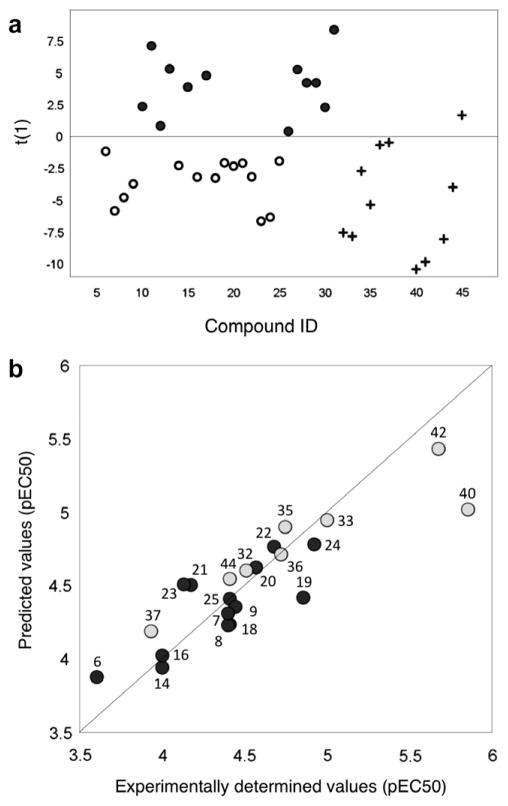
A structure–activity investigation was conducted based on the balanced set of di-substituted compounds and biological responses using a two-step modeling strategy. First, a model to distinguish active from inactive compounds was constructed followed by a QSAR model based on the active compounds for prediction of biofilm inhibition activity. Consequently, all di-substituted compounds (6–31) were characterized with descriptors (see Supplementary data for complete list) and a OPLS-DA model was calculated, in which fourteen of the 26 di-substituted compounds (6–9, 14, 16, 18–25) were classified as active and the remaining in-active. The resulting two-component OPLS-DA model30 ( , Q2= 0.71) discriminated between the two classes of compounds (Fig. 3a) with acceptable accuracy considering the challenge of QSAR modeling when complex whole cell responses are used. The polarity of the compounds appears to be the most important factor for discrimination, where active compounds are more lipophilic than the in-active ones. In the second step, the di-substituted compounds were correlated to their biofilm inhibition activity using PLS.31,32 This resulted in a one-component model ( , Q2 = 0.50) that explained the relationships between the chemical structure of the designed molecules and their ability to prevent biofilm formation (Fig. 3b). The compounds with high inhibitory effects were generally larger, more lipophilic, more flexible and had lower HOMO energies compared to the analogues with lower activity. To follow up on the results of the balanced compound set, and to validate and challenge the generated QSAR model, 14 additional di-substituted derivatives of the pilicide scaffold (32–45; Table 2) were synthesized (for predictions see Table 2 and Fig. 3). Four derivatives of the most potent biofilm inhibitor (24) from the tested compounds were prepared. In the first analogue (32), the C-7 naphthoxy was replaced with the C-7 naphthyl substituent that also had proved promising. The second analogue (33) retained the C-7 naphthoxy group but contained an N-methylated indole at C-8 rather than the free N–H indole found in 24. In the third analogue (34), the indole was similarly methylated and in addition, the carboxylic acid was replaced with a methyl acylsulfonamide. The use of such groups as bioisosteres of carboxylic acids has been reported to enhance pilicide activity.33,34 In the fourth analogue (35), the sulfur atom in the pilicide scaffold was exchanged with oxygen using a one-pot procedure starting from acylated L-serine derivatives.19 Two additional oxygen analogues (36 and 37) of promising compounds from the tested derivatives were prepared using the same procedure. Furthermore, six compounds with presumed high activities that really challenge the model (38–43), one compound predicted to have intermediate activity (44), and one compound that was predicted to be inactive (45) were synthesized. All compounds (32–45) were biologically evaluated using the pili-dependent biofilm inhibition assay (Table 2).
Figure 3.
(a) A OPLS-DA score plot showing active compounds (circles), inactive compounds (filled circles), and the validation set (plus signs). (b) Observed versus predicted plot of the PLS model showing calculated and predicted values of the model set (black) and the validation set (grey) versus experimentally determined values. The chemical structures can be seen in Tables 1 and 2.
Table 2.
Chemical structures and biofilm inhibition activities (predicted and experimentally determined) for the compounds synthesized to validate the QSAR model that was generated based on the balanced set of 24 di-substituted compounds shown in Table 1
| |||||
|---|---|---|---|---|---|
| ID | R8 (C-8) | R7 (C-7) | Pred. | ||
|
| |||||
| X | EC50 (μM) | EC50 (μM)a | |||
| 32 |
|
|
S | 25 | 31 |
| 33 |
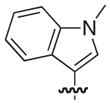
|
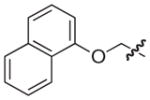
|
S | 11 | 10 |
| 34b |
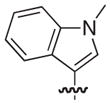
|
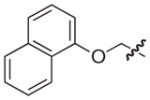
|
S | –c | 9 |
| 35 |
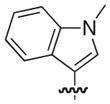
|
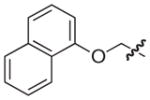
|
O | 13 | 18 |
| 36 |
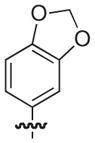
|
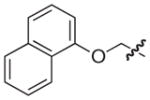
|
O | 19 | 19 |
| 37 |
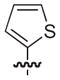
|
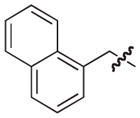
|
O | 65 | 117 |
| 38 |
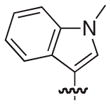
|
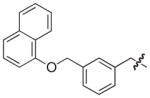
|
S | –c | 1.1 |
| 39 |
|
|
S | –c | 1.9 |
| 40 |
|
|
S | 9.6 | 1.4 |
| 41 |
|
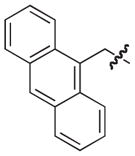
|
S | –c | 3 |
| 42 |
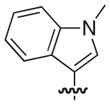
|
|
S | 3.7 | 2.1 |
| 43 |
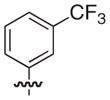
|
|
S | –c | 4.3 |
| 44 |
|
|
S | 28 | 39 |
| 45 |
|
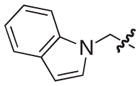
|
S | NAd | NAd |
| 24 |
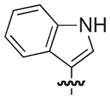
|
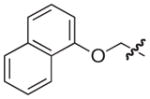
|
S | 17 | 12 |
| 22 |
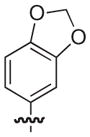
|
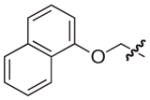
|
S | 17 | 21 |
| 18 |
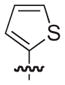
|
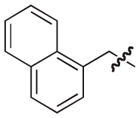
|
S | 58 | 39 |
| – | – | – | – | – | – |
Estimated from 16 to 72 data points per concentration.
In this derivative, the C-3 carboxylic acid was replaced with a methyl acylsulfonamide carboxylic acid isostere.
A reliable predicted EC50 value could not be determined because the compounds were too chemically different from the set of di-substituted compounds the model is based upon (see Section 4 for details).
Not active.
As predicted, several of the compounds based on the SAR data from the first set of di-substituted compounds proved to be highly potent inhibitors of pili-dependent biofilm formation. Considering the differences between the first training set of compounds and the validation set of compounds, the predicted activity correlates well to the experimentally determined activity (Fig. 3b). The biofilm evaluations suggest that the introduction of sterically demanding substituents in position C-7 can greatly enhance biofilm inhibition capacity. Moreover, N-methylation of the C-8 indole substituent in 24 to give 33 does not have a negative effect on the biological response, which greatly facilitates the synthesis of such compounds (Table 2). The use of a naphthoxy group as the C-7 substituent seems to be superior to the C-7 naphthyl substituent (Table 2, 24 and 32). The introduction of the methyl acylsulfonamide carboxylic acid isostere as in 34 was well tolerated, with an EC50 value of 9 μM. The oxygen analogues retain much of the biological activity of the parent sulfur derivatives, which is encouraging in case metabolic sulfur oxidation would emerge as a problem. The oxygen analogue 36 retains all biological activity whereas 35, and 37 result in a two and threefold decrease in activity, respectively as compared to compound 33 and 18 (Table 2, and 35–37 vs 18, 22, and 33).
2.3. Synthesis and evaluation of tri- and tetra-substituted derivatives
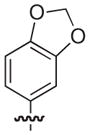
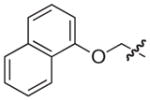
To generate more comprehensive SAR of the pilicide central fragment, the effect of substitution at the C-2 and C-6 positions, respectively, were evaluated by a new SMD (Table 3). The C-7 and C-8 di-substituted compounds 33, 22, and 18 were selected as starting points for exploring C-2 and C-6 variations based on their low molecular weight and moderate to high potency (EC50 values of 10, 21, 39 μM, respectively). The choice of substituents in position C-2 or C-6 on the pilicide scaffold was based on both previous studies of these positions as well as the size of the substituent.16,17,24 Position C-2 was thus substituted with either a phenyl or methoxy substituent and C-6 with a morpholinomethyl or a hydroxymethyl substituent. The C-2 phenyl and methoxy substituents were introduced using a two-step process starting with C-2/C-3 oxidation of 33, 22, and 18 followed by conjugate additions to the corresponding α,β-unsaturated derivatives to give 46–51 as the pure trans stereoisomers.22
Table 3.
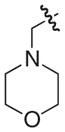
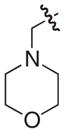
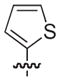
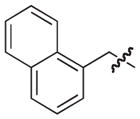
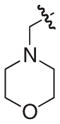
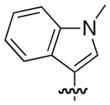
The chemical structures and biological evaluation of tri-substituted derivatives included in the designed set to explore the C-2 and C-6 positions
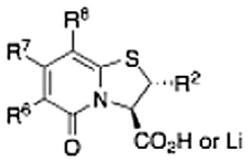
| |||||
|---|---|---|---|---|---|
| ID | R8 (C-8) | R7 (C-7) | R6 (C-6) | R2 (C-2) | EC50 (μM)a |
| 46 |
|
|
–H |
|
1.0 |
| 47 |
|
|
–H |
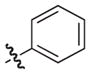
|
1.2 |
| 48 |
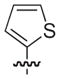
|
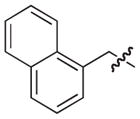
|
–H |
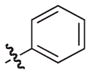
|
0.4 |
| 49 |
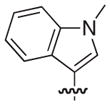
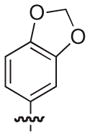
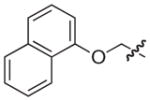
|
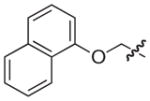
|
–H |
|
2.5 |
| 50 |
|
|
–H |
|
30 |
| 51 |
|
|
–H |
|
92 |
| 52 |
|
|
|
–H | 18 |
| 53 |
|
|
|
–H | 32 |
| 54 |
|
|
|
–H | 88 |
| 55 |
|
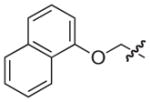
|
|
–H | 16 |
| 56 |
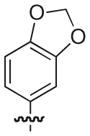
|
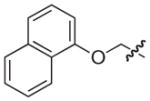
|
|
–H | 30 |
| 57 |
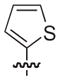
|
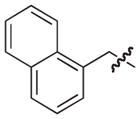
|
|
–H | 250 |
| 33 |
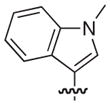
|
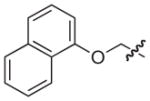
|
–H | –H | 10 |
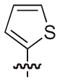
| 22 |
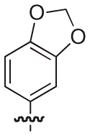
|
|
–H | –H | 21 |
| 18 |
|
|
–H | –H | 39 |
| – | – | – | – | – | – |
Estimated from 16 to 72 data points per concentration.
Addition of a morpholinomethyl or a hydroxymethyl substituent in position C-6 was performed by following previously published protocols.24 Formylation of 33, 22, and 18 gave the corresponding aldehydes from which reduction or reductive amination gave the desired morpholinomethyl or hydroxymethyl substituted compounds 52–57. After hydrolysis the 12 tri-substituted derivatives 46–57 were evaluated using the pili-dependent biofilm formation assay and the results are summarized in Table 3.
The C-2 phenyl substituent was the most effective in terms of increasing the pilicides potency. The most potent of the tri-substituted compounds was the C-2 phenyl substituted 48 with an EC50 of 400 nM, which is almost a 100-fold more potent than the C-2 unsubstituted 18 (Table 3). The incorporation of C-2 phenyl substituents in 46 and 47 resulted in almost 10- and 20-fold increased activities, respectively, relative to 33 and 22. The C-2 methoxy substitution increased the potency of 49 from 33 although in both 50 and 51 the same substitution reduced potency compared to their parent compounds. Both C-6 substituents (morpholinomethyl and hydroxymethyl) reduced the potencies of the compounds. However, 52–57 retained the majority of their potency with C-6 substituents, which could be valuable to improve the compounds solubility properties.
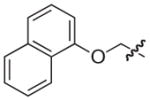
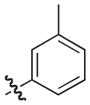
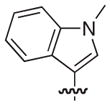
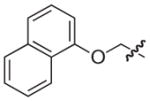
The synthesis and biological evaluation of the di-substituted compounds showed that the incorporation of indole or 3,4-methylenedioxyphenyl substituents at the C-8 position and a naphthoxymethyl substituent at the C-7 position of the pilicide scaffold results in compounds with enhanced potency. Moreover, the introduction of an appropriate C-2 substituent can further increase the compounds potency, while substituents can be incorporated at the C-6 position to improve the pilicides solubility. In using this SAR data to synthesize tetra-substituted pilicide derivatives, 33 and 22 were used as substrates for the introduction of substituents at both C-2 and C-6. Based on a recent study,16 we incorporated a 3-tolyl substituent at the C-2 position of the oxidized scaffold. Our previously developed one-pot oxidation–bromination procedure followed by Suzuki–Miyaura couplings was performed on substrates 33 and 22 to obtain the C-2 substituted intermediates.16 To increase the compounds solubility properties the morpholinomethyl substituent was introduced in position C-6 as described above. After subsequent hydrolysis the tetra-substituted derivatives 58 and 59 and the tri-substituted intermediates 60 and 61 were biologically evaluated for their ability to prevent pili-dependent biofilm formation. The results are shown in Table 4.
Table 4.
The chemical structure and biological evaluation of tri- and tetra-substituted derivatives of the pilicide scaffold
| |||||
|---|---|---|---|---|---|
| ID | R8 (C-8) | R7 (C-7) | R6 (C-6) | R2 (C-2) | EC50 (μM)a |
| 58 |
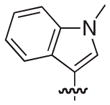
|
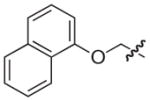
|
|
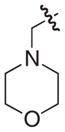
|
5 |
| 59 |
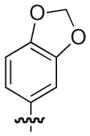
|
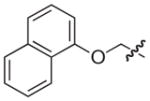
|
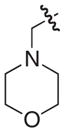
|
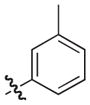
|
5 |
| 60 |
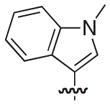
|
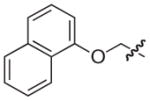
|
–H |
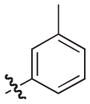
|
86 |
| 61 |
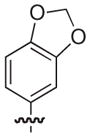
|
|
–H |
|
20 |
| 33 |
|
|
–H | –H | 10 |
| 22 |
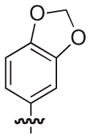
|
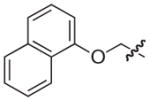
|
–H | –H | 21 |
Estimated from 16 to 72 data points per concentration.
We have previously described that C2/C3 oxidized pilicides bearing a 3-tolyl C-2 substituent were more active than saturated pilicides bearing a phenyl group at the C-2 position.16 However, the incorporation of a 3-tolyl substituent at the C-2 positions of 60 and 61 resulted in decreased activities compared to the analogous unsubstituted (33 and 22) and C-2 phenyl substituted (46 and 47) compounds (Tables 3 and 4). Interestingly, the introduction of a C-6 morpholinomethyl substituent, which previously was found to decrease activity (Table 3), resulted in 17- and 4-fold increases in the respective activities of the tetrasubstituted derivatives 58 and 59 relative to their C-6 unsubstituted counterparts 60 and 61 and also increased their water solubility.
3. Conclusion
Various synthetic methods for the introduction of substituents onto the basic pilicide scaffold were used to prepare a series of 51 new and highly-substituted analogues to generate more comprehensive pilicide SAR information. Biological testing of the 24 di-substituted compounds produced in the initial step of the design process revealed several compounds with improved potency and important QSAR information. Three efficient di-substituted inhibitors of biofilm formation were used to study the effects of further substituting the C-2 and C-6 positions. A set of 14 trisubstituted and two tetra-substituted compounds was synthesized. This showed that substitution of the scaffold’s C-2 position can increase pilicide potency and both the C-2 and C-6 positions can be used to optimize the compounds solubility properties with retained activity. However, the effect of C-2 and C-6 substitution is dependent on the other substituents on the scaffold and is not simply an additive effect. Interestingly, the most potent tri-substituted compound 48 (EC50 400 nM) was obtained from the least potent of the three di-substituted compounds from the initial set that were chosen for further substitution. Compounds in which the sulfur atom in the pilicide scaffold was exchanged for oxygen retained much of the potency of the parent compounds and will thus be useful complements for use in oxidative environments. The use of a methyl acyl sulfonamide as a carboxylic acid isostere was tolerated but did not substantially increase activity.
In addition to generating potent pilicides and valuable SAR, the generated data from the set of di-substituted compounds could be used to construct a multivariate model. The multivariate model could correctly rank the external validation set consisting of 16 compounds with different substituents and substitution pattern. The herein generated comprehensive SAR information can be used to optimize biological activity and properties of these antivirulence compounds, which will be highly useful both from a future therapeutic perspective but also as research tools to study the complex biological processes involved in pili and biofilm formation.
4. Experimental section
4.1. Multivariate design
ChemFinderACX2002Prod35 was used to search for carboxylic acids and nitriles with molecular weights below 250. These building blocks were imported to MOE36 and molecular descriptors were calculated. The nitrile data set was manually filtered to remove undesired substituents mainly based on chemical compatibility giving 531 nitriles. The carboxylic acid data set was imported to SIMCA-P37 and a PCA model was calculated with six components having an eigenvalue over two. The principal components were normalized to unit variance and centered. Building blocks with a Euclidean distance over 1.5 from CH2-1-naphthyl carboxylic acid were discarded in all principal components resulting in a set of 822 carboxylic acids. The carboxylic acid and the cyanomethyl fragments were removed from the respective building blocks, as they are not part of the target compounds. The data sets were washed and molecular descriptors were calculated MOE.36 The filtered datasets were imported to SIMCA-P37 and PCA models were created. The carboxylic acids gave a model with five components with an eigenvalue over two (R2: 0.82) and the nitriles similarly gave a model with five components with an eigenvalue over two (R2: 0.84). From these models, eight C-7 and eight C-8 substituents were manually selected with respect to the properties of 1a and 2a,b from the lead compounds 6 and 7. Combining these substituents in position C-7 and C-8, respectively on the pilicide scaffold resulted in a set of 64 derivatives for which molecular descriptors were calculated (see Supplementary data). The principal component data from the PCA model of the 64 derivatives was imported to Modde38 and a D-optimal design was calculated to select 24 compounds (all substituents represented three times). To discriminate active from inactive compounds, an orthogonal partial least square discriminant analysis (OPLS-DA) model was calculated. Fourteen of the 26 di-substituted compounds (6–9, 14, 16, 18–25) were classified as active and the rest as inactive, resulting in a two-component OPLS-DA model ( , Q2 = 0.71). A PLS model was created with the active di-substituted compounds using the pEC50 values as response giving a one-component model ( , Q2 = 0.50). Compounds from the validation set (32–45) with a DModX below three were used as a prediction set to validate the models.
4.2. Synthesis
4.2.1. General remarks
All reactions were carried out under inert atmosphere with dry solvents under anhydrous conditions, unless otherwise stated. THF was freshly distilled from potassium, CH2Cl2 was distilled from calcium hydride, MeCN and MeOH was dried over activated 3 Å molecular sieves. TLC was performed on Silica Gel 60 F254 (Merck) with detection by UV light (254 nm). Flash column chromatography (eluents given in brackets) was performed on silica gel (Matrex, 60 Å, 35–70 μm, Grace Amicon). Parallel flash chromatography was performed on a Gradmaster parallel, Jones Chromatography using silica gel (Matrex, 60 Å, 35–70 μm, Grace Amicon). 1H and 13C NMR spectra were recorded on a Bruker DRX-400 or Bruker DRX-360 in CDCl3 [residual CHCl3 (δH 7.26 ppm) or CDCl3 (δC 77.0 ppm) as internal standard], CD3OD [residual CD3OD (δH 3.31 ppm) or CD3OD (δC 49.0 ppm) as internal standard], DMSO-d6 [residual DMSO (δH 2.50 ppm) or DMSO-d6 (δC 40.0 ppm) as internal standard] or CDCl3/CD3OD mixtures using CD3OD (see above) as internal standard at 298 K. Microwave reactions were carried out using a monomode reactor (Smith Creator, Biotage AB) in Teflon septa capped 0.5–2 ml or 2–5 ml Smith TM process vials with stirring. Purities of key compounds were >95% as determined by 1H NMR and HPLC.
The synthesis was performed by following previously reported procedures.14,16,19,22,24 The acyl-ketene imine cyclocondensations were typically performed in 0.5 mmol scale. Hydrolysis was generally performed in 0.15 mmol scale.
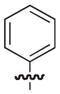
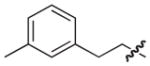
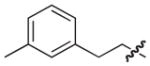
4.2.2. (3R)-7-(3-Methylphenethyl)-5-oxo-8-phenyl-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (8)
Isolated in 76% overall yield as a light grey foam. [α]D −16 (c 0.5, CH2Cl2/MeOH 9:1); 1H NMR (400 MHz, CDCl3) δ 2.20 (s, 3H), 2.44–2.62 (m, 4H), 3.43–3.51 (m, 1H), 3.76–3.85 (m, 1H), 5.49 (dd J1 = 1.38 Hz, J2 = 8.95 Hz, 1H), 6.16 (s, 1H), 6.67–6.76 (m, 2H), 6.91–6.97 (m, 1H), 7.04–7.11 (m, 1H), 7.21–7.56 (m, 4H). 13C NMR (100 MHz, MeOD) δ 20.9, 31.3, 34.7, 34.8, 63.4, 113.1, 114.3, 125.0, 126.5, 128.06, 128.10, 128.7, 128.8 (2C), 129.98, 130.04,136.4, 137.2, 140.8, 147.6,154.0, 160.2, 169.6. HRMS (electrospray ionization) calcd for [M–H] C23H20NO3S 390.1164, obsd 390.1161.
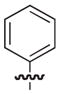
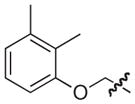
4.2.3. (3R)-7-((2,3-Dimethylphenoxy)methyl)-5-oxo-8-phenyl-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (9)
Isolated in 77% overall yield as a light grey foam. [α]D −48 (c 0.5, DMSO); 1H NMR (400 MHz, MeOD) δ 2.15 (s, 3H), 2.22 (s, 3H), 3.56 (dd J1 = 1.73 Hz, J2= 11.83 Hz, 1H), 3.76 (dd J1 = 8.79 Hz, J2= 11.84 Hz, 1H), 4.56–4.68 (m, 2H), 5.66 (dd J1 = 1.68 Hz, J2 = 8.73 Hz, 1H), 6.40 (d, J = 8.15 Hz, 1H), 6.64 (s, 1H), 6.72 (d, J = 7.55 Hz, 1H), 6.90 (t, J = 7.88 Hz, 1H), 7.24–7.33 (m, 2H), 7.36–7.46 (m, 3H). 13C NMR (100 MHz, MeOD) δ 12.0, 20.2, 32.3, 64.6, 67.5, 109.7, 112.3, 115.5, 123.6, 125.8, 126.4, 129.4, 129.8 (2C), 130.0, 130.4, 135.6, 138.7, 149.0, 152.4, 156.4, 163.1, 170.2. HRMS (electrospray ionization) calcd for [M–H] C23H20NO4S 406.1113, obsd 406.1110.
4.2.4. (3R)-7-((7-Methoxy-2-oxo-2H-chromen-4-yl)methyl)-5-oxo-8-phenyl-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (10)
Isolated in 70% overall yield as a light yellow non-crystalline solid. [α]D −17 (c 0.25, CHCl3); 1H NMR (400 MHz, DMSO-d6) δ 3.49 (dd, J1 = 1.79 Hz, J2 = 11.82 Hz, 1H), 3.72–3.88 (m, 6H), 5.48 (dd, J1 = 1.63 Hz, J2 = 9.01 Hz, 1H), 5.93 (s, 1H), 6.01 (s, 1H), 6.88 (dd, J1 = 2.50 Hz, J2 = 8.83 Hz, 1H), 6.96 (d, J = 2.49, 1H), 7.20–7.28 (m, 1H), 7.29–7.36 (m, 2H), 7.36–7.44 (m, 3H); 13C NMR (100 MHz, DMSO-d6) δ 32.4, 35.7, 56.8, 64.6, 101.9, 112.8, 113.1, 115.0, 115.3 (2C), 127.0, 129.2, 129.8 (2C), 130.8 (broad, 2C), 136.8, 149.6, 150.8, 154.0, 155.8, 160.79, 160.83, 163.3, 170.4. HRMS (electrospray ionization) calcd for [M+H] C25H20NO6S 462.1011, obsd 462.1018.
4.2.5. (3R)-8-Cyclopropyl-7-((7-methoxy-2-oxo-2H-chromen-4-yl)methyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (11)
Isolated in 65% overall yield as a light yellow non-crystalline solid. [α]D −58 (c 0.20, CHCl3); 1H NMR (400 MHz, DMSO-d6) δ 0.51–0.71 (m, 2H) 0.76–0.88 (m, 2H), 1.56–1.66 (m, 1H), 3.52 (dd, J1 = 1.76 Hz, J2= 11.86 Hz, 1H), 3.82 (dd, J1 = 9.08 Hz, J2= 11.87 Hz, 1H), 3.86 (s, 3H), 4.10–4.27 (m, 2H), 5.40 (dd, J1 = 1.75 Hz, J2 = 9.08 Hz, 1H), 5.79 (s, 1H), 6.03 (s, 1H), 6.97 (dd, J1 = 2.55 Hz, J2 = 8.87 Hz, 1H), 7.03 (d, J = 2.51 Hz, 1H), 7.66 (d, J = 8.88 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) δ 7.6, 7.8, 11.2, 32.2, 34.9, 56.5, 63.8, 101.6, 111.8, 112.4, 112.7, 112.9, 114.5, 127.0, 149.6, 152.9, 154.3, 155.6, 160.4, 160.6, 163.0, 170.1. HRMS (electrospray ionization) calcd for [M+H] C22H20NO6S 426.1011, obsd 426.1016.
4.2.6. (3R)-7-(Benzo[b]thiophen-3-ylmethyl)-8-cyclopropyl-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (12)
Isolated in 57% overall yield as a light yellow non-crystalline solid. [α]D −99 (c 0.5, DMSO); 1H NMR (400 MHz, DMSO-d6) δ 0.53–0.73 (m, 2H), 0.81–0.97 (m, 2H), 1.56–1.67 (m, 1H), 3.50 (dd J1 = 1.81 Hz, J2 = 11.93 Hz, 1H), 3.78 (dd J1 = 9.12 Hz, J2 = 11.91 Hz, 1H), 4.15–4.30 (m, 2H), 5.37 (dd J1 = 1.78 Hz, J2 = 9.10 Hz, 1H), 5.61 (s, 1H), 7.35–7.43 (m, 2H), 7.47 (s, 1H), 7.71–7.77 (m, 1H), 7.97–8.04 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 7.9, 8.4, 12.0, 33.0, 34.0, 67.2, 114.8, 115.8, 122.9, 123.8, 125.1, 125.1, 125.4, 134.5, 140.0, 141.9, 151.2, 157.2, 163.7, 174.0. HRMS (electrospray ionization) calcd for [M–Li] C20H16NO3S2 382.0572, obsd 382.0578.
4.2.7. (3R)-7-((1H-Benzo[d][1,2,3]triazol-1-yl)methyl)-8-cyclopropyl-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (13)
Isolated in 46% overall yield as a light pink non-crystalline solid. [α]D −33 (c 0.5, CH2Cl2/MeOH 9:1); 1H NMR (400 MHz, MeOD) δ 0.77–0.89 (m, 2H), 0.91–1.10 (m, 2H), 1.59–1.70 (m, 1H), 3.59 (dd J1 = 1.31 Hz, J2= 11.34 Hz, 1H), 3.73 (dd J1 = 8.64 Hz, J2 = 11.30 Hz, 1H), 5.34 (s, 1H), 5.37 (dd J1 = 1.36 Hz, J2 = 8.61 Hz, 1H), 5.98–6.16 (m, 2H), 7.43–7.51 (m, 1H), 7.54–7.62 (m, 1H), 7.68–7.76 (m, 1H), 8.01–8.07 (m, 1H). 13C NMR (100 MHz, MeOD) δ 8.0, 8.5, 11.3, 34.1, 50.0, 67.3, 111.6, 112.2, 113.9, 120.1, 125.9, 129.2, 134.7, 146.8, 152.46, 152.50, 163.3, 173.8. HRMS (electrospray ionization) calcd for [M–Li] C18H15N4O3S 367.0865, obsd 367.0868.
4.2.8. (3R)-8-Isopropyl-7-(3-methylphenethyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (14)
Isolated in 84% overall yield as a light grey foam. [α]D −21 (c 0.09, CH2Cl2/MeOH 9:1); 1H NMR (400 MHz, CD3OD) δ 1.31 (m, 6H), 2.31 (s, 3H), 2.78–2.83 (m, 4H), 3.07–3.22 (m, 1H), 3.62 (dd J1 = 1.54 Hz, J2 = 11.27 Hz, 1H), 3.72 (dd J1 = 8.45 Hz, J2 = 11.27 Hz, 1H), 5.41 (dd J1 = 1.55 Hz, J2 = 8.42 Hz, 1H), 6.12 (s, 1H), 6.97–7.06 (m, 3H), 7.13–7.19 (m, 1H). 13C NMR (100 MHz, CD3OD) δ 20.0, 21.2, 21.5, 29.7, 34.2, 36.8, 37.4, 66.6, 115.1, 121.6, 126.4, 128.0, 129.5, 130.1, 139.2, 142.2, 147.5, 156.9, 163.5, 173.9. HRMS (electrospray ionization) calcd for [M–H] C20H22NO3S 356.1320, obsd 356.1315.
4.2.9. (3R)-7-((1H-Benzo[d][1,2,3]triazol-1-yl)methyl)-8-isopropyl-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (15)
Isolated in 13% overall yield as a light pink non-crystalline solid. 1H NMR (400 MHz, CD3OD) δ 1.94–1.24 (m, 3H), 1.25–1.31 (m, 3H), 3.22–3.30 (m, 1H), 3.61–3.67 (m, 1H), 3.79 (dd J1 = 8.66 Hz, J2 = 11.52 Hz, 1H), 5.48 (d, J = 8.77 Hz, 1H), 5.63 (br s, 1H), 5.90–5.98 (m, 2H), 7.44–7.50 (m, 1H), 7.54–7.61 (m, 1H), 7.67–7.73 (m, 1H), 8.01–8.07 (m, 1H). 13C NMR (90 MHz, CD3OD) δ 19.6, 20.9, 29.6, 34.3, 50.8, 66.7, 111.7, 114.1, 120.0, 120.2, 125.9, 129.3, 134.7, 146.8, 149.1, 149.7, 162.9, 173.5. HRMS (electrospray ionization) calcd for [M–Li] C18H17N4O3S 369.1021, obsd 369.1019.
4.2.10. (3R)-8-Isopropyl-7-(naphthalen-1-ylmethyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (16)
Isolated in 56% overall yield as a light grey non-crystalline solid. [α]D −9 (c 0.5, CH2Cl2/MeOH 9:1); 1H NMR (400 MHz, CD3OD) δ 1.20–1.32 (m, 6H), 3.01–3.17 (m, 1H), 3.63 (dd J1 = 1.22 Hz, J2 = 11.29 Hz, 1H), 3.74 (dd J1 = 8.51 Hz, J2 = 11.28 Hz, 1H), 4.34 (s, 2H), 5.39 (dd J1 = 1.13 Hz, J2 = 8.33 Hz, 1H), 5.81 (s, 1H), 7.22–7.27 (m, 1H), 7.38–7.44 (m, 1H), 7.46–7.54 (m, 2H), 7.76–7.83 (m, 1H), 7.85–7.97 (m, 2H). 13C NMR (100 MHz, CD3OD) δ 19.7, 20.9, 29.9, 34.3, 37.5, 66.6, 116.3, 121.8, 124.6, 126.6, 126.8, 127.3, 128.2, 128.6, 129.8, 133.1, 135.4, 135.7, 147.5, 155.8, 163.3, 173.9. HRMS (electrospray ionization) calcd for [M–Li] C22H20NO3S 378.1164, obsd 378.1166.
4.2.11. (3R)-7-((7-Methoxy-2-oxo-2H-chromen-4-yl)methyl)-5-oxo-8-(thiophen-2-yl)-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (17)
Isolated in 72% overall yield as a light yellow non-crystalline solid. [α]D −17 (c 0.69, CHCl3); 1H NMR (400 MHz, DMSO-d6) δ 3.51 (dd, J1 = 1.77 Hz, J2 = 11.61 Hz, 1H), 3.78–3.83 (m, 1H), 3.85 (s, 2H), 5.43 (dd, J1 = 1.52 Hz, J2 = 9.16 Hz, 1H), 5.91 (s, 1H), 6.04 (s, 1H), 6.91 (dd, J1 = 2.56 Hz, J2 = 8.88 Hz, 1H), 6.98 (d, J = 2.54 Hz, 1H), 7.03–7.07 (m, 2H), 7.48 (d, J = 8.88 Hz, 1H), 7.56–7.60 (m, 1H) 13C NMR (100 MHz, DMSO-d6) δ 32.6, 35.8, 56.9, 65.1, 101.9, 107.1, 112.78, 112.81, 113.1, 115.5, 126.9, 128.4, 128.9, 130.4, 136.8, 151.5, 152.4, 154.2, 155.8, 160.7, 160.9, 163.3, 170.4. HRMS (electrospray ionization) calcd for [M+H] C23H18NO6S2 468.0576, obsd 468.0587.
4.2.12. (3R)-7-(Naphthalen-1-ylmethyl)-5-oxo-8-(thiophen-2-yl)-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (18)
Isolated in 81% overall yield as a light yellow non-crystalline solid. [α]D33 (c0.05, CH2Cl2/MeOH 9:1); 1H NMR (400 MHz, MeOD) δ 3.50–3.56 (m, 1H), 3.63–3.72 (m, 1H), 4.01–4.16 (m, 2H), 5.36–5.42 (m, 1H), 5.70 (s, 1H), 7.04–7.09 (m, 2H), 7.23–7.28 (m, 1H), 7.34–7.45 (m, 3H), 7.46–7.50 (m, 1H), 7.66–7.77 (m, 2H), 7.79–7.85 (m, 1H). 13C NMR (100 MHz, MeOD) δ 34.0, 37.6, 68.2, 110.2, 114.7, 124.9, 126.5, 126.7, 127.2, 128.38, 128.41, 128.6, 128.8, 129.7, 130.5, 133.1, 135.35, 135.43, 137.9, 153.1, 156.9, 163.7, 173.8. HRMS (electrospray ionization) calcd for [M–Li] C23H16NO3S2 418.0572, obsd 418.0575.
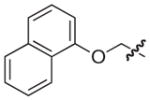
4.2.13. (3R)-7-((Naphthalen-1-yloxy)methyl)-5-oxo-8-(thiophen-2-yl)-3,5-dihydro-2H–thiazolo[3,2-a]pyridine-3-carboxylic acid (19)
Isolated in 87% overall yield as a light yellow foam. [α]D −23 (c 0.36, CH2Cl2/MeOH 9:1); 1H NMR (400 MHz, MeOD) δ 3.55 (dd J1 = 1.55 Hz, J2 = 11.90 Hz, 1H), 3.87 (dd J1 = 9.14 Hz, J2 = 11.89 Hz, 1H), 4.94–5.03 (m, 2H), 5.55 (dd J1 = 1.51 Hz, J2 = 9.05 Hz, 1H), 6.47 (s, 1H), 6.79 (d, J = 7.70 Hz, 1H), 7.11–7.15 (m, 1H), 7.18–7.20 (m, 1H), 7.34–7.40 (m, 1H), 7.47–7.57 (m, 3H), 7.64–7.67 (m, 1H), 7.84–7.90 (m, 1H), 8.13–8.18 (m, 1H). 13C NMR (100 MHz, MeOD) δ 31.4, 63.7, 66.4, 104.8, 105.4, 112.2, 120.6, 121.4, 124.7, 125.6, 126.0, 126.6, 127.5, 127.6, 127.9, 129.2, 134.0, 135.0, 149.8, 151.0, 152.9, 159.9, 169.4. HRMS (electrospray ionization) calcd for [M–H] C23H16NO4S2 434.0521, obsd 434. 0530.
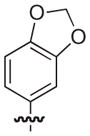
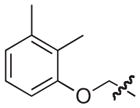
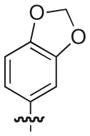
4.2.14. (3R)-8-(Benzo[d][1,3]dioxol-5-yl)-7-((2,3-dimethylphenoxy)methyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (20)
Isolated in 84% overall yield as a light grey foam. [α]D −9 (c 0.36, DMSO); 1H NMR (400 MHz, CDCl3) δ 2.18 (s, 3H), 2.26 (s, 3H), 3.62–3.72 (m, 1H), 3.76–3.83 (m, 1H), 4.57–4.71 (m, 2H), 5.77–5.82 (m, 1H), 6.03 (s, 2H), 6.43–6.48 (s, 1H), 6.68–6.90 (m, 5H), 6.93–6.99 (m, 1H), 8.95 (s, 1H). 13C NMR (100 MHz, DMSO-d6) δ 11.9, 20.1, 30.8, 64.8, 66.7, 101.5, 109.0, 109.2, 110.0, 112.0, 115.5, 122.8, 123.1, 123.5, 125.5, 125.7, 127.9, 138.3, 148.1, 148.6, 152.6, 155.7, 162.9, 168.5. HRMS (electrospray ionization) calcd for [M–H] C24H20NO6S 450.1011, obsd 450.1006.
4.2.15. (3R)-7-(Benzo[b]thiophen-3-ylmethyl)-8-(benzo[d][1,3]dioxol-5-yl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (21)
Isolated in 72% overall yield as a light yellow non-crystalline solid. [α]D −5 (c0.1, DMSO); 1H NMR (400 MHz, MeOD) δ 3.49–3.57 (m, 1H), 3.68–3.78 (m, 1H), 3.80–3.92 (m, 2H), 5.42–5.49 (m, 1H), 5.84–5.97 (m, 2H), 6.07 (s, 1H), 6.64–6.81 (m, 3H), 7.14–7.19 (m, 1H), 7.24–7.34 (m, 2H), 7.45–7.53 (m, 1H), 7.78–7.86 (m, 1H). 13C NMR (100 MHz, MeOD) δ 32.2, 34.0, 68.1 (split, 68.12, 68.07), 102.6, 109.4 (split, 109.34, 109.48), 111.4 (split, 111.08, 111.73), 114.7 (split, 114.65, 114.70), 117.8, 122.7, 123.6, 124.8 (split, 124.55, 124.09), 125.0, 125.3 (2C), 131.2 (split, 131.23, 131.25), 134.1, 139.7, 141.6, 148.9 (split, 148.86, 148.95), 149.2, 151.0, 155.1 (split, 155.05, 155.09), 163.8, 174.0 (split, 173.97, 174.04). HRMS (electrospray ionization) calcd for [M–Li] C24H16NO5S2 462.0470, obsd 462.0480.
4.2.16. (3R)-8-(Benzo[d][1,3]dioxol-5-yl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (22)
Isolated in 78% overall yield as a light grey non-crystalline solid. [α]D −2 (c 0.65, DMSO); 1H NMR (400 MHz, MeOD) δ 3.55–3.60 (m, 1H), 3.77 (dd J1 = 8.70 Hz, J2 = 11.31 Hz, 1H), 4.88–4.99 (m, 2H), 5.49–5.53 (m, 1H), 5.97 (d, J = 13.38 Hz, 1H), 6.64 (s, 1H), 6.68 (d, J = 7.64 Hz, 1H), 6.83–6.94 (m, 3H), 7.27–7.33 (m, 1H), 7.39–7.43 (m, 1H), 7.44–7.51 (m, 2H), 7.76–7.81 (m, 1H), 8.21–8.27 (m, 1H). 13C NMR (100 MHz, MeOD) δ 34.1, 68.0, 68.1, 102.7, 106.3, 109.6 (split), 111.3 (split, 110.9, 111.6), 112.5, 115.7, 121.8, 122.8, 124.7 (split, 124.4, 125.0), 126.3, 126.82, 126.84, 127.5, 128.5, 130.3, 136.1, 149.2 (split), 149.5, 151.3, 151.9, 154.9, 163.9, 173.9 (split). HRMS (electrospray ionization) calcd for [M–Li] C26H18NO6S 472.0855, obsd 472.0845.
4.2.17. (3R)-8-(1H-Indol-3-yl)-7-(3-methylphenethyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (23)
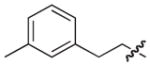
Isolated in 59% overall yield as a light pink non-crystalline solid. As a 1:1.8 mixture of atropisomers. [α]D –29 (c 0.8, DMSO); 1H NMR (400 MHz, MeOD) δ 2.11–2.16 (m, 3H), 2.47–2.65 (m, 4H), 3.45–3.52 (m, 1H), 3.63–3.72 (m, 1H), 5.46–5.52 (m, 1H), 6.21–6.26 (m, 1H), 6.42–6.58 (m, 2H), 6.80–6.87 (m, 1H), 6.90–7.07 (m, 2H), 7.12–7.29 (m, 3H), 7.40–7.48 (m, 1H). 13C NMR (100 MHz, MeOD) δ 21.31 (maj), 21.33 (min), 33.68 (maj), 33.74 (min), 36.9 (maj), 37.0 (min), 37.3 (min), 37.4 (maj), 68.4 (maj), 68.5 (min), 111.18 (maj), 111.24 (min), 111.6 (min), 112.1 (maj), 112.5 (min), 112.7 (maj), 113.66 (maj), 113.69 (min), 120.1 (maj), 120.4 (min), 120.5 (maj), 120.6 (min), 122.8 (min), 122.9 (maj), 125.5 (min), 126.16 (maj), 126.19 (min), 126.6 (maj), 127.4 (maj), 127.5 (min), 127.7 (maj), 128.4 (min), 129.0, 129.88 (maj), 129.92 (min), 137.8 (min), 137.9 (maj), 138.8, 142.5, 152.2 (maj), 152.3 (min), 158.8 (min), 158.9 (maj), 164.2, 174.1 (min), 174.2 (maj). HRMS (electrospray ionization) calcd for [M–Li] C25H21N2O3S 429.1273, obsd 429.1282.
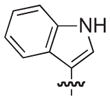
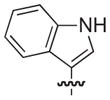
4.2.18. (3R)-8-(1H-Indol-3-yl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (24)
Isolated in 41% overall yield as a light pink non-crystalline solid. As a 1:1.1 mixture of atropisomers. [α]D −52 (c 1, DMSO); 1H NMR (400 MHz, DMSO-d6) δ 3.45–3.53 (m, 1H), 3.77–3.90 (m, 1H), 4.80–5.12 (m, 2H), 5.54–5.62 (m, 1H), 6.47 (s, 1H, maj), 6.48 (s, 1H, min), 6.61 (d, J =7.67 Hz, 1H, maj), 6.70 (d, J =7.68 Hz, 1H, min), 7.00–7.08 (m, 1H), 7.11–7.18 (m, 1H), 7.24–7.33 (m, 1H), 7.39–7.56 (m, 6H), 7.82–7.88 (m, 1H), 8.11–8.19 (m, 1H), 11.39 (s, 1H). 13C NMR (100 MHz, DMSO-d6) δ 31.0 (maj), 31.2 (min), 63.6 (min), 63.7 (maj), 66.5 (maj), 66.7 (min), 105.0 (min), 105.3 (maj) 105.3 (maj), 105.4 (min), 108.0 (min), 108.5 (maj), 111.1 (maj), 111.3 (min), 112.0 (maj/min), 118.8 (min), 118.9 (maj), 119.1 (min), 119.3 (maj), 120.5, 121.30 (min), 121.33 (maj), 121.5 (min), 121.6 (maj), 124.7 (maj), 125.1 (min), 125.5–125.6 (2C, maj/min), 125.8 (min), 125.9 (maj), 126.0 (maj), 126.1 (min), 126.53 (maj), 126.55 (min), 127.5, 134.0, 135.9 (min), 136.0 (maj), 149.7 (maj), 149.9 (min), 150.8 (maj), 151.1 (min), 153.0, 160.2, 169.61 (maj), 169.64 (min). HRMS (electrospray ionization) calcd for [M–Li] C27H19N2O4S 467.1066, obsd 467.1056.
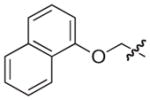
4.2.19. (3R)-7-(Benzo[d][1,3]dioxol-5-ylmethyl)-8-(1H-indol-3-yl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (25)
Isolated in 27% overall yield as a light pink non-crystalline solid. As a 1:1.3 mixture of atropisomers. [α]D −1 (c 0.5, CH2Cl2/MeOH 9:1); 1H NMR (400 MHz, MeOD) δ 3.44–3.62 (m, 3H), 3.64–3.75 (m, 1H), 5.58 (br s, 1H), 5.79–5.85 (m, 2H), 6.07–6.13 (m, 1H), 6.27–6.35 (m, 2H), 6.52–6.57 (m, 1H), 6.97–7.19 (m, 3H), 7.24–7.31 (m, 1H), 7.38–7.45 (m, 1H). 13C NMR (100 MHz, MeOD) δ 35.1 (maj), 35.2 (min), 42.79 (maj), 42.84 (min), 68.6, 104.7, 111.5, 112.89 (min), 112.94 (maj), 113.7 (maj), 114.0 (min), 114.1 (min), 114.3 (maj), 115.2 (min), 115.4 (maj), 117.2, 122.5 (maj), 122.6 (min), 123.1 (min), 123.3 (maj), 125.5 (min), 125.6 (maj), 125.7 (min), 12.8 (maj), 128.3 (min), 129.1 (maj), 130.2 (maj), 130.7 (min), 136.0 (maj), 136.2 (min), 140.3 (min), 140.4 (maj), 150.06 (min), 150.10 (maj), 151.6, 154.3 (maj), 154.5 (min), 162.1 (min), 162.2 (maj), 166.6 (maj), 167.5 (min), 174.2 (broad). HRMS (electrospray ionization) calcd for [M–Li] C24H17N2O5S 445.0858, obsd 445.0863.
4.2.20. (3R)-7-((2,3-Dimethylphenoxy)methyl)-5-oxo-8-(pyridin-3-yl)-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (26)
Isolated in 53% overall yield as a light pink non-crystalline solid. [α]D −11 (c 1.1, DMSO); 1H NMR (400 MHz, MeOD) δ 2.09 (s, 3H), 2.22 (s, 3H), 3.60 (dd J1 = 1.17 Hz, J2 = 11.34 Hz, 1H), 3.80 (dd J1 = 8.74 Hz, J2 = 11.25 Hz, 1H), 4.68 (s, 2H), 5.51 (dd J1 = 1.07 Hz, J2 = 8.52 Hz, 1H), 6.50 (d, J = 8.18 Hz, 1H), 6.56 (s, 1H), 6.72 (d, J = 7.53 Hz, 1H), 6.88–6.94 (m, 1H), 7.46–7.53 (m, 1H), 7.87–7.93 (m, 1H), 7.51–7.60 (m, 2H). 13C NMR (100 MHz, MeOD) δ 11.9, 20.1, 34.3, 68.1, 68.2, 110.2, 112.0, 113.5, 123.9, 125.4, 125.9, 126.9, 134.0, 139.0, 140.1, 149.9, 151.2, 151.68, 151.75, 157.1, 163.9, 173.7. HRMS (electrospray ionization) calcd for [M–Li] C22H19N2O4S 407.1066, obsd 407.1057.
4.2.21. (3R)-7-((1H-Benzo[d][1,2,3]triazol-1-yl)methyl)-5-oxo-8-(pyridin-3-yl)-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (27)
Isolated in 33% overall yield as a light pink non-crystalline solid. [α]D0 (c 0.36, CH2Cl2/MeOH 9:1); 1H NMR (400 MHz, MeOD) δ 3.58 (dd J1 = 1.12 Hz, J2 = 11.42 Hz, 1H), 3.74–3.84 (m, 1H), 5.48 (dd J1 = 1.21 Hz, J2 = 8.56 Hz, 1H), 5.58–5.75 (m, 2H), 5.98 (s, 1H), 7.30–7.44 (m, 2H), 7.46–7.54 (m, 2H), 7.58–7.74 (m, 1H), 7.91–7.96 (m, 1H), 8.28–8.46 (m, 2H). 13C NMR (100 MHz, MeOD) δ 34.4, 49.9, 68.2, 111.4, 111.9, 114.7, 120.0, 125.4, 125.7, 129.1, 133.4, 134.4, 139.9 (split, 139.7, 140.2), 146.5, 149.0, 149.9 (split, 149.8,150.0), 151.0 (split, 150.9,151.1), 153.1, 163.3, 173.4. HRMS (electrospray ionization) calcd for [M–Li] C20H14N5O3S 404.0817, obsd 404.0814.
4.2.22. (3R)-7-(Benzo[d][1,3]dioxol-5-ylmethyl)-5-oxo-8-(pyridin-3-yl)-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (28)
Isolated in 40% overall yield as a light pink non-crystalline solid. [α]D −2 (c 0.9, DMSO); 1H NMR (400 MHz, MeOD) δ 3.52–3.60 (m, 3H), 3.71–3.82 (m 1H), 5.45–5.50 (m, 1H), 5.86 (s, 2H), 6.18 (s, 1H), 6.29 (d J = 7.80 Hz, 1H), 6.37 (s, 1H), 6.59 (d J = 7.92 Hz, 1H), 7.40–7.47 (m, 1H), 7.62–7.70 (m, 1H), 8.21–8.31 (m, 1H), 8.46–8.51 (m, 1H). 13C NMR (100 MHz, MeOD) δ 34.2, 40.1, 67.9, 102.1, 108.9, 110.0, 113.8, 115.6, 122.9, 125.1, 132.5, 134.6, 140.2 (split, 140.0, 140.4), 147.5, 149.0, 149.3 (split, 149.2, 149.4), 151.2, 151.3, 155.5, 163.7, 173.6. HRMS (electrospray ionization) calcd for [M–Li] C21H15N2O5S 407.0702, obsd 407.0709.
4.2.23. (3R)-7-(Benzo[b]thiophen-3-ylmethyl)-8-methoxy-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (29)
Isolated in 63% overall yield as a light yellow non-crystalline solid. [α]D −8 (c 0.5, DMSO); 1H NMR (400 MHz, MeOD) δ 3.67 (dd J1 = 1.48 Hz, J2 = 11.38 Hz, 1H), 3.69 (s, 3H), 3.82 (dd J1 = 8.52 Hz, J2= 11.35 Hz, 1H), 4.06–4.17 (m, 2H), 5.37 (dd J1 = 1.45 Hz, J2 = 8.47 Hz, 1H), 5.89 (s, 1H), 7.31–7.40 (m, 3H), 7.72–7.77 (m, 1H), 7.85–7.90 (m, 1H). 13C NMR (100 MHz, MeOD) δ 29.4, 34.7, 61.0, 67.8, 114.5, 122.9, 123.8, 125.2, 125.4, 125.5, 133.8, 138.8, 139.9,141.8,142.8,151.7,162.6,173.6. HRMS (electrospray ionization) calcd for [M–Li] C18H14NO4S2 372.0364, obsd 372.0374.
4.2.24. (3R)-8-Methoxy-7-(naphthalen-1-ylmethyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (30)
Isolated in 80% overall yield as a light grey non-crystalline solid. [α]D −1 (c 0.36, CH2Cl2/MeOH 9:1); 1H NMR(400 MHz, DMSO-d6) δ 3.56–3.63 (m, 1H), 3.70 (s, 3H), 3.89 (dd J1 = 8.93 Hz,J2 = 11.71 Hz, 1H), 4.24–4.36 (m, 2H), 5.31–5.39 (m, 2H), 7.41–7.58 (m, 4H), 7.85–7.99 (m, 3H). 13C NMR (100 MHz, MeOD) δ 33.4, 34.7, 61.0, 67.8, 114.6, 125.1, 126.6, 126.8, 127.3, 128.7, 128.9, 129.7, 133.3, 135.3, 135.5, 138.7, 142.7, 153.0, 162.6, 173.6. HRMS (electrospray ionization) calcd for [M–Li] C20H16NO4S 366.0800, obsd 366.0803.
4.2.25. (3R)-7-(Benzo[d][1,3]dioxol-5-ylmethyl)-8-methoxy-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (31)
Isolated in 76% overall yield as a light yellow foam. [α]D −1 (c 0.5, CH2Cl2/MeOH 9:1); 1H NMR (400 MHz, DMSO-d6) δ 3.56–3.61 (m, 4H), 3.71 (s, 2H), 3.88 (dd J1 = 8.99 Hz, J2 = 11.70 Hz, 1H), 5.37 (dd J1 = 124 Hz, J2 = 8.79 Hz, 1H), 5.79 (s, 1H), 5.98 (s, 2H), 6.70–6.75 (m, 1H), 6.82–6.87 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 32.0, 34.8, 60.1, 62.9, 100.8, 108.2, 109.4, 113.5, 122.1, 131.8, 135.3, 139.7, 145.8, 147.3, 151.4, 159.1, 169.4. HRMS (electrospray ionization) calcd for [M–H] C17H14NO6S 360.0542, obsd 366.0543.
4.2.26. (3R)-8-(1H-Indol-3-yl)-7-(naphthalen-1-ylmethyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (32)
Isolated in 50% overall yield as a light pink foam. As a 1:1.3 mixture of atropisomers. [α]D −33 (c 0.6, DMSO);1H NMR (400 MHz, MeOD) δ 3.43–3.49 (m, 1H), 3.67–3.77 (m, 1H), 3.95–4.09 (m, 2H), 5.57–5.63 (m, 1H), 5.73 (s, 1H, maj), 5.76 (m, 1H, min), 7.06–7.40 (m, 7H), 7.41–7.49 (m, 2H), 7.54–7.60 (m, 1H), 7.66–7.71 (m, 1H), 7.74–7.79 (m, 1H), 10.77 (s, 1H). 13C NMR (100 MHz, MeOD) δ 32.4 (maj), 32.5 (min), 37.7, 65.7 (maj), 65.8 (min), 111.0 (min), 111.4 (maj), 111.48 (min), 111.51 (maj), 112.8 (min), 112.9 (maj), 114.3, 120.0 (maj), 120.1 (min), 120.7 (min), 120.8 (maj), 123.0 (min), 123.1 (maj), 124.99 (min), 125.01 (maj), 125.7, 126.4 (min), 126.5 (maj), 126.56 (min), 126.62 (maj), 127.00 (maj), 127.02 (min), 127.6 (maj), 128.1 (min), 128.52 (min), 128.55 (maj), 128.8 (min), 128.9 (maj), 129.6, 133.0 (min), 133.1 (maj), 135.3, 135.7 (maj), 135.9 (min), 137.8 (min), 138.0 (maj), 151.6 (maj), 151.7 (min), 159.2 (min), 159.3 (maj), 163.81 (maj), 163.83 (min), 171.2. HRMS (electrospray ionization) calcd for [M–Li] C27H19N2O3S 451.1117, obsd 451.1126.
4.2.27 (3R)-8-(1-Methyl-1H-indol-3-yl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-3,5-dihydro-2H–thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (33)
Isolated in 76% overall yield as a light pink foam. As a 1:1.3 mixture of atropisomers. [α]D −18 (c 0.45, DMSO); 1H NMR (400 MHz, DMSO-d6) δ 3.45–3.52 (m, 1H), 3.78 (s, 3H, min), 3.79 (s, 3H, maj), 3.81–3.89 (m, 1H), 4.81–5.13 (m, 2H), 5.55–5.63 (m, 1H), 6.47–6.50 (m, 1H), 6.60 (d,J = 7.73 Hz, 1H, maj), 6.70 (d,J = 7.67 Hz, 1H, min), 7.03–7.10 (m, 1H), 7.17–7.55 (m, 8H), 7.81–7.86 (m, 1H), 8.11–8.18 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 31.0 (maj), 31.2 (min), 32.58 (maj), 32.62 (min), 63.62 (min), 63.64 (maj), 66.6 (maj), 66.7 (min), 104.7 (maj), 104.9 (min), 105.5 (maj), 105.6 (min), 107.2 (min), 107.7 (maj), 110.26 (maj), 110.31 (min), 111.08 (maj), 111.13 (min), 119.0 (min), 119.2 (maj), 119.4 (min), 119.5 (maj), 120.5, 121.35 (min), 121.37 (maj), 121.6 (min), 121.7 (maj), 124.8, 125.6, 125.94 (maj), 125.97 (min), 125.99 (maj), 126.4 (min), 126.6, 127.5, 129.3 (min), 130.0 (maj), 134.1, 136.5 (min), 136.6 (maj), 149.8 (min), 150.0 (maj), 151.0 (min), 151.2 (maj), 153.0, 160.3, 169.6 (maj), 169.7 (min). HRMS (electrospray ionization) calcd for [M–Li] C28H21N2O4S 481.1222, obsd 481.1217.
4.2.28. (R)-8-(1-Methyl-1H–indol-3-yl)-N-(methylsulfonyl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxamide (34)
Isolated in 51% yield as a light grey foam starting from 0.08 mmol of 33. As a 1:1.9 mixture of atropisomers. [α]D 2 (c 0.5, DMSO); 1H NMR (400 MHz, CDCl3) δ 3.32 (s, 3H), 3.52–3.68 (m, 1H), 3.75 (s, 3H, min), 3.83 (s, 3H, maj), 3.85–3.93 (m, 1H), 4.81–5.06 (m, 2H), 5.77–5.84 (m, 1H), 6.46 (d, J = 7.60 Hz, 1H, maj), 6.50 (d, J = 7.64 Hz, 1H, min), 6.97 (s, 1H, min), 6.99 (s, 1H, maj), 7.06 (s, 1H, min), 7.14 (s, 1H, maj), 7.15–7.54 (m, 9H), 7.74–7.80 (m, 1H), 8.30–8.38 (m, 1H). 13C NMR (100 MHz, CDCl3) δ 28.6 (maj), 29.2 (min), 33.0 (min), 33.1 (maj), 41.36 (min), 41.4 (maj), 65.6 (maj), 65.7 (min), 66.5 (maj), 66.7 (min), 105.06 (maj), 105.11 (min), 107.5 (min), 108.1 (maj), 108.6 (maj), 108.8 (min), 109.7 (min), 110.0 (maj), 111.9 (maj), 112.0 (min), 119.1 (maj), 119.6 (min), 120.26 (min), 120.33 (maj), 121.02 (maj), 121.04 (min), 122.0, 122.5 (min), 122.6 (maj), 125.39 (min, 2C), 125.43 (maj, 2C), 125.57 (maj), 125.60 (min), 126.1 (min), 126.59 (maj), 126.61 (maj), 126.7 (min), 127.4, 128.0 (min), 129.1 (maj), 134.4, 136.8 (min), 137.0 (maj), 149.9, 153.4, 153.6 (min), 153.9 (maj), 162.7 (min), 162.9 (maj), 166.1 (maj), 166.2 (min). HRMS (electrospray ionization) calcd for [M–H] C29H24N3O5S2 558.1158, obsd 558.1173.
4.2.29. (3S)-7-((Naphtalen-1-yloxy)methyl)-5-oxo-8-(1-metyhyl-1H-indol-3-yl)-2,3-dihydro-5H-oxazolo[3,2-a]pyridine-3-carboxylic acid (35)
Isolated in 29% overall yield as a light pink non-crystalline solid. [α]D −23 (c 0.2, CHCl3/CH2Cl2/MeOH 1:1:1); 1H NMR (400 MHz, DMSO-d6) δ 3.77 (s, 3H), 4.69 (m, 1H), 4.86 (m, 1H), 5.07 (m, 3H), 6.28 (s, 1H), 6.71 (d, J =7.52 Hz, 1H), 7.04 (m, 1H), 7.18 (m, 1H), 7.31 (m, 1H), 7.37 (m, 1H), 7.46 (m, 3H), 7.25 (m, 2H), 7.86 (m, 1H), 8.16 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 33.0, 58.2, 67.4, 72.7, 89.0, 104.3, 106.0, 107.2, 110.4, 119.6, 119.8, 120.9, 121.8, 121.8, 125.3, 126.0, 126.5, 127.0, 127.8, 128.0, 130.2, 134.5, 136.9, 153.2, 153.5, 155.7, 158.8, 169.9. HRMS (electrospray ionization) calcd for [M–H] C28H21N2O5 465.1451, obsd 465.1451.
4.2.30. (3S)-7-((Naphtalen-1-yloxy)methyl)-5-oxo-8-(benzo[d][1,3]dioxol-5-yl)-2,3-dihydro-5H-oxazolo[3,2-a]pyridine-3-carboxylic acid (36)
Isolated in 39% overall yield as a light yellow non-crystalline solid. [α]D −37 (c 0.2, CHCl3/MeOH 1:1); 1H NMR (400 MHz, DMSO-d6) δ 4.74 (dd, J1 = 4.01 Hz, J2 = 9.05 Hz, 1H), 4.90 (t, J = 9.31 Hz, 1H), 5.00 (d, J = 14.00 Hz, 1H), 5.10 (d,J = 14.00 Hz, 1H), 5.16 (dd, J1 = 4.01 Hz, J2 = 9.44 Hz, 1H), 6.01 (s, 2H) 6.26 (s, 1H), 6.78 (d, J = 7.65 Hz, 1H), 6.86 (m, 1H), 6.91 (m, 1H), 6.97 (m, 1H), 7.36 (m, 1H), 7.47 (m, 1H), 7.52 (m, 2H), 7.86 (m,1H), 8.11 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 58.1, 67.2, 72.9, 96.7, 101.5, 106.0, 107.8, 108.7, 111.3, 120.9, 121.8, 124.4, 125.3, 125.8, 126.0, 126.5, 127.0, 128.0, 134.5, 147.0, 147.7, 151.8, 153.5, 155.3, 158.6, 167.8. HRMS (electrospray ionization) calcd for [M–H] C25H19NO7 456.1084, obsd 456.1079.
4.2.31. (3S)-7-((Naphthalen-1-yloxy)methyl)-5-oxo-8-(thiophen-2-yl)-2,3-dihydro-5H-oxazolo[3,2-a]pyridine-3-carboxylic acid (37)
Isolated in 34% overall yield as a light grey non-crystalline solid. [α]D −67 (c 0.75, CHCl3); 1H NMR (400 MHz, CDCl3) δ 4.16 (d, J = 17.26 Hz, 1H), 4.23 (d, J = 17.26 Hz, 1H), 4.80 (m, 1H), 5.20 (m, 1H), 5.35 (m, 1H), 5.80 (s, 1H), 7.05 (m, 1H), 7.10 (m, 1H), 7.24 (m, 1H), 7.38–7.48 (m, 4H), 7.62 (m, 1H), 7.78 (m, 1H), 7.85 (m, 1H). 13C NMR (100 MHz, CDCl3) δ 37.1, 59.2, 70.8, 96.0, 109.9, 123.6, 125.5, 125.8, 126.3, 127.4, 127.4, 127.9, 128.0, 128.8, 129.5, 131.5, 131.6, 133.4, 133.9, 155.0, 160.5, 161.0, 167.5. HRMS (electrospray ionization) calcd for [M–H] C23H16NO4S 402.0800, obsd 402.0794.
4.2.32. (3R)-8-(1-Methyl-1H-indol-3-yl)-7-(3-((naphthalen-1-yloxy)methyl)benzyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (38)
Isolated in 40% yield as a light grey foam in a 1:1.3 mixture of atropisomers. [α]D −30 (c 0.82, DMSO); 1H NMR (400 MHz, DMSO-d6) δ 3.47–3.68 (m, 4H), 3.71 (s, 3H, maj), 3.73 (s, 3H, min) 5.05–5.15 (m, 2H), 5.24–5.32 (m, 1H), 5.85–5.90 (m, 1H), 6.88–7.02 (m, 4H), 7.10–7.26 (m, 4H), 7.28–7.33 (m, 1H), 7.36–7.44 (m, 2H), 7.45–7.55 (m, 3H), 7.83–7.89 (m, 1H), 8.11–8.17 (m, 1H); 13C NMR (100 MHz, DMSO-d6) δ 32.88 (maj), 32.93 (min), 33.1 (broad), 39.05 (maj), 39.09 (min), 66.8, 69.82 (min), 69.84 (maj), 106.2, 106.6 (broad and split), 109.7 (min), 110.2 (maj), 110.5, 114.1 (broad and split), 119.5, 119.6 (maj), 119.7 (min), 120.6, 121.8 (min), 121.9 (maj), 122.0,125.5, 125.7 (broad), 125.9, 126.6 (min), 126.7 (maj), 126.9, 127.0 (maj), 127.5 (min), 127.9, 128.4 (min), 128.5 (maj), 128.8, 128.9, 129.6 (min), 130.2 (maj), 134.5, 136.8 (min), 136.9 (maj), 137.5 (split), 139.5 (maj), 139.8 (min), 150.49 (min), 150.52 (maj), 154.17 (min), 154.19 (maj), 154.7 (min), 154.8 (maj), 161.2 (broad and split), 170.1 (broad and split). HRMS (electrospray ionization) calcd for [M–CO2H] C34H27N2O2S 527.1793, obsd 527.1786.
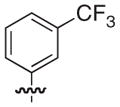
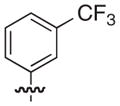
4.2.33. (3R)-7-[3-(Naphthalen-1-yloxymethyl)-benzyl]-5-oxo-8-(3-trifluoromethyl-phenyl)-2,3-dihydro-5H-thiazolo[3,2-a]pyridine-3-carboxylic acid (39)
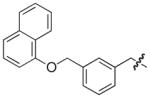
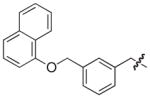
Isolated in 35% overall yield as a colorless foam. [α]D 5 (c 0.25, CHCl3); 1H NMR (400 MHz, DMSO-d6) δ 3.46–3.53 (m, 1H), 3.58–3.71 (m, 2H), 3.76–3.84 (m, 1H), 5.17 (s, 2H), 5.46–5.52 (m, 1H), 6.11 (s, 1H), 6.77–6.84 (m, 1H), 6.97–7.04 (m, 2H), 7.18–7.25 (m, 1H), 7.30–7.60 (m, 8H), 7.63–7.69 (m, 1H), 7.84–7.89 (m, 1H), 8.14–8.19 (m, 1H); 13C NMR (100 MHz, DMSO-d6) δ 31.7, 38.6, 63.6, 69.2, 105.7, 112.8, 114.7, 120.1, 121.5, 123.9 (q, J = 271 Hz, 1C), 124.7 (broad), 125.0, 125.4 (2C), 126.1, 126.4, 126.8 (broad splitted), 127.4, 127.6 (broad), 128.1, 128.4, 129.3 (q, J = 31 Hz, 1C), 129.7, 134.0, 134.4, 137.1, 137.3 (broad), 138.2, 148.5, 153.0 (broad), 153.7, 160.1, 169.5. HRMS (electrospray ionization) calcd for [M–H] C33H23F3NO4S 586.1300, obsd 586.1294.
4.2.34. (3R)-8-Cyclopropyl-7-[3-(naphthalen-1-yloxymethyl)-benzyl]-5-oxo-2,3-dihydro-5H-thiazolo[3,2-a]pyridine-3-carboxylic acid (40)
Isolated in 30% overall yield as a colorless foam. [α]D −17 (c 0.25, CHCl3/MeOH 9:1); 1H NMR (400 MHz, DMSO-d6) δ 0.44–0.62 (m, 2H), 0.69–0.86 (m, 2H), 1.26–1.37 (m, 1H), 3.44–3.53 (m, 1H), 3.67–3.77 (m, 1H), 3.91–4.06 (m, 2H), 5.38 (m, 1H), 5.86 (s, 1H), 7.00–7.06 (m, 1H), 7.19–7.24 (m, 1H), 7.34–7.55 (m, 7H), 7.84–7.89 (m, 1H), 8.17–8.23 (m, 1H);13C NMR (100 MHz, DMSO-d6) δ 7.4, 7.7, 10.9, 31.3, 38.2, 62.8, 69.3, 105.8, 111.8, 114.2, 120.1, 121.5, 125.0, 125.4, 125.5, 126.1, 126.5, 127.5, 128.1, 128.5, 128.7, 134.1, 137.4, 138.9, 148.4, 153.6, 156.2, 160.1, 169.7. HRMS (electrospray ionization) calcd for [M–H] C29H24NO4S 482.1426, obsd 482.1416.
4.2.35. (3R)-7-(Anthracen-9-ylmethyl)-5-oxo-8-(3-(trifluoromethyl)phenyl)-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (41)
Isolated in 57% overall yield as a white non-crystalline solid by following a previously published procedure and the use of naphthalene-1-boronic acid. [α]D −4 (c 0.5, DMSO); 1H NMR (400 MHz, MeOD/CDCl3 (1:1)) δ 3.48–3.56 (m, 1H), 3.63–3.75 (m, 1H), 4.20–4.38 (m, 2H), 5.36 (s, 1H), 5.41–5.50 (m, 1H), 7.32–7.43 (m, 4H), 7.65–7.94 (m, 8H), 8.27 (s, 1H); 13C NMR (100 MHz, MeOD/CDCl3 (3:2)) δ 32.4, 32.6, 65.1 (broad), 114.6, 116.0, 124.4 (2C), 124.7 (q, J = 272.8 Hz), 125.5 (2C), 126.1 (broad and split), 126.9 (2C), 127.6 (split J = 36.5 Hz), 127.84, 129.3, 129.8 (2C), 130.7, 130.9 (2C), 132.1 (2C), 132.3 (q, J = 33.5 Hz), 134.5 (split J = 47.5Hz), 137.9 (split J = 4.3 Hz), 149.3, 155.4, 162.8, 170.6 (broad). HRMS (electrospray ionization) calcd for [M–H] C30H19F3NO3S 530.1038, obsd 530.1038.
4.2.36. (3R)-8-(1-Methyl-1H-indol-3-yl)-5-oxo-7-((3-phenoxyphenoxy)methyl)-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (42)
Isolated in 50% overall yield as a light pink foam in a 1:1.2 mixture of atropisomers. 1H NMR (400 MHz, DMSO-d6) δ 3.41–3.51 (m, 1H), 3.68–3.81(m, 4H), 4.57–4.85 (m, 2H), 5.44–5.51 (m, 1H), 6.23 (s, 1H, maj), 6.25 (s, 1H, min), 6.37 (t, J = 2.25 Hz, 1H, maj), 6.45 (t, J =2.24 Hz, 1H, min), 6.49–6.54 (m, 1H), 6.55–6.62 (m, 1H), 6.93–7.04 (m, 3H), 7.12–7.25 (m, 4H), 7.35–7.41 (m, 2H), 7.43–7.49 (m, 1H); 13C NMR (100 MHz, DMSO-d6) δ 32.0 (maj), 32.2 (min), 33.0 (maj), 33.1 (min), 65.0, 67.0 (maj), 67.1 (min), 104.8 (maj), 105.0 (min), 105.4 (maj), 105.6 (min), 107.7 (min), 108.2 (maj), 110.2 (min), 110.3 (maj), 110.6, 111.3 (min), 111.4 (maj), 111.7 (maj), 111.9 (min), 119.25, 119.31, 119.5, 119.7 (min), 119.9 (maj), 121.95 (min), 122.05 (maj), 124.11 (min), 124.13 (maj), 126.5 (maj), 126.9 (min), 129.7 (min), 130.3 (maj), 130.5 (2C), 131.00 (min), 131.03 (maj), 136.8 (min), 136.9 (maj), 150.4 (min), 150.60 (maj), 150.65 (min), 150.8 (maj), 156.62 (min), 156.63 (maj), 158.2 (maj), 158.3 (min), 159.4 (maj), 159.5 (min), 160.8, 170.3 (maj), 170.4 (min). HRMS (electrospray ionization) calcd for [M–H] C30H23N2O5S 523.1328, obsd 523.1334.
4.2.37. 5-Oxo-7-((3-phenoxy-phenoxy)methyl)-8-(3-trifluoromethyl-phenyl)-2,3-dihydro-5H-thiazolo[3,2-a]pyridine-3-carboxylic acid (43)
Isolated in 54% overall yield as a white non-crystalline solid. [α]D −32 (c 0.5, DMSO); 1H NMR (400 MHz, CDCl3) δ 3.49–3.58 (m, 1H), 379–3.91 (m, 1H), 4.73 (s, 2H), 5.51–5.60 (m, 1H), 6.31–6.41 (m, 2H), 6.48–6.59 (m, 2H), 6.91–7.01 (m, 2H), 7.10–7.25 (m, 2H), 7.33–7.44 (m, 2H), 7.50–7.76 (m, 4H); 13C NMR (100 MHz, CDCl3) δ 32.0, 63.9, 67.0, 105.4, 110.1, 111.5, 111.7, 113.5, 119.3, 124.1, 124.4 (q,J = 277.36 Hz), 125.4 (d, J = 3.43 Hz), 126.8 (d, J = 3.3 Hz), 130.0 (q,J = 31.93 Hz), 130.4, 130.5, 131.0, 134.4, 136.9, 149.1, 149.4, 156.6, 158.2, 159.2, 160.4, 169.9. HRMS (electrospray ionization) calcd for [M–H] C28H19F3NO5S 538.0936, obsd 538.0926.
4.2.38. (3R)-8-Cyclopropyl-7-((3-phenoxy-phenoxy)methyl)-5-oxo-2,3-dihydro-5H-thiazolo[3,2-a]pyridine-3-carboxylic acid (44)
Isolated in 61% overall yield as a grey non-crystalline solid. [α]D −44 (c 0.5, DMSO); 1H NMR (400 MHz, CDCl3) δ 0.49–0,63 (m, 2H), 0.78–0.84 (m, 2H), 1.58–1.64 (m, 1H), 3.50–3.55 (m, 1H), 3.75–3.82 (m, 1H), 5.14 (s, 2H), 5.38–5.43 (m, 1H), 6.14 (s, 1H), 6.55–6.59 (m, 1H), 6.65 (s, 1H), 6.77–6.81 (m, 1H), 7.00–7.05 (m, 2H), 7.13–7.18 (m, 1H), 7.27–7.33 (m, 1H) 7.36–7.42 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 6.7, 6.8, 9.8, 31.4, 62.7, 66.1, 105.1, 109.7, 109.9, 110.8, 111.3, 119.0, 123.7, 130.0, 130.6, 148.5, 151.7, 156.1, 158.0, 159.2, 160.0, 169.6. HRMS (electrospray ionization) calcd for [M–H] C24H20NO5S 434.1062, obsd 434.1051.
4.2.39. 8-Cyclopropyl-7-indol-1-ylmethyl-5-oxo-2,3-dihydro-5H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (45)
Isolated in 21% overall yield as a yellow non-crystalline solid. [α]D −22 (c 0.33, CHCl3/MeOH 9:1); 1H NMR (400 MHz, CDCl3) δ 0.83–0.78 (m, 2H), 1.07–0.91 (m, 2H), 1.75–1.69 (m, 1H), 3,58 (d, J = 11.36 Hz, 1H), 3.75–3.68 (m, 1H), 5.22 (s, 1H), 5.35 (d, J = 8.59 Hz, 1H), 5.49 (s, 2H), 6.53 (d, J = 3.10 Hz, 1H), 7.07–7.01 (m, 1H), 7.16–7.09, (m, 1H), 7.28–7.23 (m, 2H), 7.57 (d, J = 7.88 Hz, 1H); 13C NMR (100 MHz, CDCl3) δ 7.9, 8.4, 11.25, 34.1 (2C), 67.1, 102.8, 110.7, 111.7, 113.9, 120.6, 121.8, 122.8, 129.7, 130.2, 137.8, 151.3, 155.5, 163.7, 173.9. HRMS (electrospray ionization) calcd for [M–Li] C20H17N2O3S 365.0960, obsd 365.0970.
4.2.40. 8-(1-Methyl-1H-indol-3-yl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-2-phenyl-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (46)
Isolated in 54% yield over three steps starting from 0.4 mmol of 33. As a light pink foam in a 1:1.5 mixture of atropisomers. 1H NMR (400 MHz, CDCl3) δ 3.78 (s, 3H, min), 3.80 (s, 3H, maj), 4.85–5.07 (m, 2H), 5.29–5.33 (m, 1H), 5.84–5.90 (m, 1H), 6.49 (d, J = 7.66 Hz, 1H, maj), 6.54 (d, J =7.62 Hz, 1H, min), 7.06–7.54 (m, 15H), 7.75–7.81 (m, 1H), 8.33–8.39 (m, 1H), 8.76 (br s, 1H). 13C NMR (100 MHz, CDCl3) δ 33.0, 48.9 (maj), 49.3 (min), 66.7 (maj), 66.9 (min), 72.5 (min), 72.7 (maj), 105.1 (maj), 105.2 (min), 107.5 (min), 108.0 (min), 108.3 (maj), 108.5 (min), 109.7 (min), 109.9 (maj), 112.0 (maj), 112.3 (min), 119.0 (maj), 119.6 (min), 120.30 (min), 120.31 (maj), 121.0 (maj), 121.1 (min), 121.98 (min), 122.02 (maj), 122.5, 125.4 (min), 125.46 (maj), 125.49 (min), 125.5 (maj, 2C), 126.2 (maj), 126.4, 126.5 (2C), 126.7 (min), 127.41 (maj), 127.45 (min), 128.1, 128.69 (maj), 128.74 (min), 129.1, 129.2 (maj), 129.3 (min), 134.46 (maj), 134.48 (min), 136.8 (min), 137.0 (maj), 139.6 (maj), 139.7 (min), 149.8 (min), 149.9 (maj), 153.47 (maj), 153.50 (min), 153.8 (min), 154.2 (maj), 163.1 (min), 163.3 (maj), 167.99 (min), 168.04 (maj). HRMS (electrospray ionization) calcd for [M–H] C34H25N2O4S 557.1535, obsd 557.1543.
4.2.41. 8-(Benzo[d][1,3]dioxol-5-yl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-2-phenyl-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (47)
Isolated in 62% yield as a grey foam starting from 0.4 mmol of methylester protected 22. 1H NMR (400 MHz, CDCl3) δ 4.76–4.93 (m, 2H), 5.23 (s, 1H), 5.84 (s, 1H), 5.98 (s, 2H), 6.60 (d, J = 7.29 Hz, 1H), 6.67–6.87 (m, 3H), 7.04 (s, 1H), 7.24–7.35 (m, 6H), 7.40–7.53 (m, 3H), 7.75–7.81 (m, 1H), 8.31–8.37 (m, 1H), 11.87 (br s, 1H). 13C NMR (100 MHz, CDCl3) δ 50.0 (split, 50.01, 50.09), 65.6, 71.9 (split, 71.88, 71.93), 101.4, 105.1, 109.0, 109.7 (split, 109.4, 110.1), 112.5, 114.9, 121.1, 122.0, 123.2 (split, 122.9, 123.6), 125.4, 125.5, 125.6, 126.4, 126.5, 126.6, 127.4, 127.8, 128.7, 129.2 (2C), 134.5, 139.3 (split, 139.3, 139.4), 148.0, 148.1 (split, 148.1, 148.2), 148.4, 151.9, 153.5, 162.7, 168.6. HRMS (electrospray ionization) calcd for [M–H] C32H22NO6S 548.1168, obsd 548.1170.
4.2.42. 7-((Naphthalen-1-yloxy)methyl)-5-oxo-2-phenyl-8-(thiophen-2-yl)-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (48)
Isolated in 65% yield as a light yellow foam starting from 0.4 mmol of methylester protected 18. 1H NMR (400 MHz, CDCl3) δ 4.10–4.24 (m, 2H), 5.27 (d, J = 2.07 Hz, 1H), 5.66 (d, J = 2.19 Hz, 1H), 5.96 (s, 1H), 7.06–7.10 (m, 2H), 7.23–7.36 (m, 6H), 7.39–7.50 (m, 4H), 7.67–7.72 (m, 1H), 7.76–7.80 (m, 1H), 7.83–7.88 (m, 1H), 9.04 (br s, 1H). 13C NMR (100 MHz, CDCl3) δ 36.9, 49.4, 72.5, 110.4, 114.8, 123.7, 125.5, 125.8, 126.4, 126.6 (2C), 127.6, 127.7, 128.0 (2C), 128.8 (2C), 129.29 (2C), 129.33, 131.7, 133.2, 133.9, 135.6, 139.1, 150.2, 157.4, 162.7, 167.8. HRMS (electrospray ionization) calcd for [M–H] C29H20NO3S2 494.0885, obsd 494.0892.
4.2.43. 2-Methoxy-8-(1-methyl-1H-indol-3-yl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (49)
Isolated in 65% yield starting from 0.4 mmol of methylester protected 33. As a light pink foam in a 1:1.5 mixture of atropisomers. 1H NMR (400 MHz, CDCl3) δ 3.34 (s, 3H, maj), 3.38 (s, 3H, min), 3.81 (s, 3H, min), 3.83 (s, 3H, maj), 4.84–5.06 (m, 2H), 5.70 (s, 1H, maj), 5.71 (s, 1H, min), 5.89 (s, 1H, maj), 5.92 (s, 1H, min), 6.46 (d, J = 7.64 Hz, 1H, maj), 6.52 (d, J = 7.73 Hz, 1H, maj), 7.03–7.24 (m, 4H), 7.28–7.53 (m, 6H), 7.75–7.81 (m, 1H), 8.30–8.36 (m, 1H), 9.11 (br s, 1H). 13C NMR (100 MHz, CDCl3/MeOD (70:30)) δ 33.07 (maj), 33.09 (min), 56.4 (maj), 56.6 (min), 67.1 (maj), 67.3 (min), 71.8, 87.9 (maj), 88.3 (min), 105.48 (maj), 105.51 (min), 107.89 (min), 107.93 (maj), 108.1 (min), 108.5 (maj), 110.06 (min), 110.14 (maj), 112.2 (maj), 112.6 (min), 119.7, 120.2 (min), 120.3 (maj), 121.09 (maj), 121.13 (min), 122.1, 122.56 (min), 122.63 (maj), 125.62 (maj), 125.64 (min), 125.7, 125.8, 126.66 (maj), 126.73 (maj), 126.74 (min), 127.2 (min), 127.7, 128.7 (min), 129.5 (maj), 134.8, 137.1 (min), 137.3 (maj), 148.0 (maj), 148.1 (min), 153.0 (min), 153.3 (maj), 153.77 (min), 153.79 (maj), 162.7, 167.5. HRMS (electrospray ionization) calcd for [M–H] C29H23N2O5S 511.1328, obsd 511.1335.
4.2.44. 8-(Benzo[d][1,3]dioxol-5-yl)-2-methoxy-7-((naphthalen-1-yloxy)methyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (50)
Isolated in 65% yield as a light grey foam starting from 0.4 mmol of methylester protected 22. 1H NMR (400 MHz, CDCl3) δ 3.36 (s, 3H), 4.73–4.91 (m, 2H), 5.63 (s, 1H), 5.89 (s, 1H), 5.98–6.04 (m, 2H), 6.54–6.60 (m, 1H), 6.64–6.89 (m, 2H), 6.99 (s, 1H), 7.24–7.30 (m, 1H), 7.39–7.51 (m, 3H), 7.74–7.80 (m, 1H), 8.26–8.33 (m, 1H), 11.81 (br s, 1H). 13C NMR (100 MHz, CDCl3) δ 56.6, 66.6, 71.9, 87.9, 101.4, 105.1, 109.0 (split, 108.9, 109.0), 109.8 (split, 109.4, 110.1), 112.6, 115.8, 121.1, 122.0, 123.3 (split, 123.0, 123.6), 125.4, 125.5, 125.6, 126.6, 127.4, 127.9 (split, 127.8, 127.9), 134.5, 146.9, 148.0 (split), 148.2 (split), 151.8, 153.5, 162.8, 166.7. HRMS (electrospray ionization) calcd for [M–H] C27H20NO7S 502.0961, obsd 502.0948.
4.2.45. 2-Methoxy-7-((naphthalen-1-yloxy)methyl)-5-oxo-8-(thiophen-2-yl)-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylic acid (51)
Isolated in 65% yield as a light grey foam starting from 0.4 mmol of methylester protected 18. 1H NMR (400 MHz, DMSO-d6) δ 3.36 (s, 3H), 4.01–4.22 (m, 2H), 5.60 (s, 1H), 5.74 (s, 1H), 5.93 (s, 1H), 7.01–7.10 (m, 2H), 7.19–7.23 (m, 1H), 7.35–7.49 (m, 4H), 7.62–7.68 (m, 1H), 7.73–7.78 (m, 1H), 7.81–7.87 (m, 1H). 13C NMR (100 MHz, MeOD) δ 37.7, 56.8, 72.8, 89.6, 111.2, 115.3, 124.8, 126.5, 126.8, 127.3, 128.6, 128.7, 128.79, 128.83, 129.7, 130.7, 133.0, 135.1, 135.4, 137.2, 150.3, 157.9, 163.4, 168.2. HRMS (electrospray ionization) calcd for [M–H] C24H18NO4S2 448.0677, obsd 448.0691.
4.2.46. (3R)-8-(1-Methyl-1H-indol-3-yl)-6-(morpholinomethyl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (52)
Isolated in 65% yield starting from 0.4 mmol of methylester protected 33. As a pink non-crystalline solid as a 1:1.4 mixture of atropisomers. [α]D −58 (c 0.27, DMSO); 1H NMR (400 MHz, MeOD) δ 2.36–2.54 (m, 4H), 3.37–3.56 (m, 8H), 3.59–3.74 (m, 3H), 4.80–5.32 (m, 2H), 5.56–5.62 (m, 1H), 6.51 (d, J = 7.62 Hz, 1H maj), 6.61 (d, J = 7.65 Hz, 1H, min), 6.83–6.51 (m, 9H), 7.71–7.78 (m, 1H), 8.04–8.09 (m, 1H). 13C NMR (100 MHz, MeOD) δ 32.6 (min), 32.7 (maj), 33.4 (maj), 33.7 (min), 53.9 (maj), 54.1 (min), 54.67 (2C, min), 54.70 (2C, maj), 65.9 (min), 66.0 (maj), 67.95 (2C, min), 67.99 (2C, maj), 69.1 (min), 69.2 (maj), 105.7 (maj), 105.9 (min), 110.3 (min), 110.4 (maj), 110.5 (min), 110.8 (maj), 111.1 (min), 111.3 (maj), 120.3 (min), 120.4 (maj), 120.5 (maj), 121.2, 121.3 (min), 122.8, 122.85 (maj), 122.87 (min), 122.92 (maj), 122.94 (min), 126.0 (maj), 126.2 (min), 126.71 (maj), 126.75 (min), 126.81 (maj), 126.9 (min), 127.27 (maj), 127.35 (min), 128.38 (min), 128.42 (maj), 128.5 (maj), 128.6 (min), 130.1 (min), 131.0 (maj), 135.88 (maj), 135.92 (min), 138.0 (min), 138.1 (maj), 150.4 (min), 150.7 (maj), 151.5 (min), 151.7 (maj), 155.6 (min), 155.7 (maj), 163.99 (maj), 164.03 (min), 173.9 (min), 174.0 (maj). HRMS (electrospray ionization) calcd for [M–Li] C33H30N3O5S 580.1906, obsd 580.1929.
4.2.47. (3R)-8-(Benzo[d][1,3]dioxol-5-yl)-6-(morpholinomethyl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (53)
Isolated in 56% yield as a light pink non-crystalline solid starting from 0.4 mmol of methylester protected 22. [α]D −29 (c 0.22, DMSO); 1H NMR (400 MHz, DMSO-d6) δ 2.23 (br s, 4H), 3.35 (br s, 4H), 3.46–3.60 (m, 3H), 3.78–3.89 (m, 1H), 4.97–5.17 (m, 2H), 5.53–5.60 (m, 1H), 5.82–5.99 (m, 2H), 6.70–6.92 (m, 4H), 7.31–7.37 (m, 1H), 7.40–7.53 (m, 3H), 7.80–7.86 (m, 1H), 7.94–8.00 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 31.0, 52.1, 52.9 (2C), 64.0, 64.5, 65.8 (2C), 100.9, 105.0, 108.0 (split, 108.0, 108.1), 109.9 (split, 109.8, 110.0), 140.1, 120.0, 121.3, 121.7, 123.3, 124.7, 125.0, 125.8, 126.2, 127.1, 129.9, 133.8, 146.4, 146.6, 147.0, 147.5, 153.7, 160.7, 169.2. HRMS (electrospray ionization) calcd for [M–Li] C31H27N2O7S 571.1539, obsd 571.1535.
4.2.48. (3R)-6-(Morpholinomethyl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-8-(thiophen-2-yl)-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (54)
Isolated in 47% yield as a light pink non-crystalline solid starting from 0.4 mmol of methylester protected 18. [α]D −7 (c 0.5, DMSO); 1H NMR (400 MHz, DMSO-d6) δ 2.24 (br s, 4H), 3.19–3.33 (m, 6H), 3.52 (dd J1 = 1.71 Hz, J2 = 11.78 Hz, 1H), 3.86 (dd J1 = 9.17Hz, J2= 11.76 Hz, 1H), 4.21–4.49 (m, 2H), 5.57 (dd J1 = 1.65 Hz, J2 = 9.17Hz, 1H), 6.79–6.87 (m, 2H), 6.96 (d, J = 6.99 Hz, 1H), 7.37–7.43 (m, 2H), 7.46–7.53 (m, 2H), 7.73–7.78 (m, 1H), 7.87–7.98 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 31.1, 31.7, 52.6, 52.9 (2C), 64.7, 65.7 (2C), 107.4, 120.5, 122.7, 123.9, 125.2, 125.5, 125.9, 126.3, 126.7, 127.2, 128.3, 128.7, 131.1, 133.0, 135.0, 136.8, 149.6, 152.2, 160.8, 169.3. HRMS (electrospray ionization) calcd for [M–Li] C28H25N2O4S2 517.1256, obsd 517.1270.
4.2.49. (3R)-6-(Hydroxymethyl)-8-(1-methyl-1H-indol-3-yl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (55)
Isolated in 62% yield starting from 0.4 mmol of methylester protected 33. As a pink non-crystalline solid in a 1:1.2 mixture of atropisomers. [α]D −21 (c 0.5, DMSO); 1H NMR (400 MHz, DMSO-d6) δ 3.44–3.50 (m, 1H), 3.56 (s, 3H, min), 3.62 (s, 3H, maj), 3.74–3.80 (m, 1H), 4.54–4.68 (m, 2H), 4.88–5.18 (m, 2H), 5.51–5.58 (m, 1H), 6.63 (d,J = 7.69 Hz, maj), 6.72 (d, J = 7.65 Hz, min), 6.87–7.00 (m, 1H), 7.07–7.15 (m, 1H), 7.18–7.27 (m, 2H), 7.31–7.51 (m, 5H), 7.77–7.82 (m, 1H), 7.93–8.02 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 31.0 (maj), 31.2 (min), 32.3, 54.91 (min), 54.93 (maj), 64.5, 64.57 (min), 64.62 (maj), 105.3 (maj), 105.5 (min), 106.9 (maj), 107.0 (min), 108.6 (min), 109.2 (maj), 109.9 (maj), 110.1 (min), 119.1 (2C), 120.18 (maj), 120.2 (min), 121.5 (2C), 124.8, 125.27 (min), 125.29 (maj), 125.8 (min), 125.89 (maj), 125.93, 126.37, 126.41 (maj), 126.7 (min), 127.3, 129.0 (min), 129.9 (maj), 133.9, 136.15 (min), 136.24 (maj), 146.5 (min), 146.6 (maj), 148.6 (min), 148.7 (maj), 153.65 (min), 153.70 (maj), 160.50 (min), 160.53 (maj), 169.6. HRMS (electro-spray ionization) calcd for [M–Li] C29H23N2O5S 511.1328, obsd 511.1335.
4.2.50. (3R)-8-(Benzo[d][1,3]dioxol-5-yl)-6-(hydroxymethyl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (56)
Isolated in 50% yield as a yellow non-crystalline solid starting from 0.4 mmol of methylester protected 22. [α]D −10 (c 0.45, DMSO); 1H NMR (400 MHz, DMSO-d6) δ 3.50 (dd J1 = 2.08 Hz, J2 = 11.80 Hz, 1H), 3.84 (dd J1 = 9.11 Hz, J2 = 11.78 Hz, 1H), 4.51–4.59 (m, 2H), 4.96–5.06 (m, 2H), 5.57 (dd J1 = 2.02 Hz, J2 = 9.04 Hz, 1H), 5.80–6.00 (m, 2H), 6.70–6.90 (m, 4H), 7.30–7.37 (m, 1H), 7.42–7.53 (m, 3H), 7.80–7.86 (m, 1H), 7.96–8.02 (m, 1H). 13C NMR (100 MHz, DMSO-d6) δ 31.3, 54.6, 64.1, 64.3 (split, 64.28, 64.35), 101.1, 105.3, 108.3 (split, 123.4), 124.7, 125.3, 126.0 (2C), 126.4, 127.3, 129.9 (split, 129.8, 129.9), 133.9, 145.3 (split, 145.23, 145.80), 146.8 (split, 146.79, 146.86), 147.2, 147.5, 153.6, 160.3, 169.5. HRMS (electrospray ionization) calcd for [M–Li] C27H20NO7S 502.0961, obsd 502.0964.
4.2.51. (3R)-6-(Hydroxymethyl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-8-(thiophen-2-yl)-3,5-dihydro-2H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (57)
Isolated in 53% yield as a yellow non-crystalline solid starting from 0.4 mmol of methylester protected 18. [α]D −3 (c 1.0, DMSO); 1H NMR (400 MHz, MeOD) δ 3.59 (dd J1 = 1.45 Hz, J2 = 11.36 Hz, 1H), 3.77 (dd J1 = 8.83 Hz, J2 = 11.40 Hz, 1H), 4.28–4.48 (m, 4H), 5.59 (dd J1 = 1.41 Hz, J2 = 8.70 Hz, 1H), 6.68–6.77 (m, 2H), 7.05–7.09 (m, 1H), 7.18–7.21 (m, 1H), 7.32–7.37 (m, 1H), 7.41–7.47 (m, 2H), 7.67–7.73 (m, 1H), 7.80–7.89 (m, 2H). 13C NMR (100 MHz, MeOD) δ 33.5, 33.9, 57.8, 68.9, 110.9, 123.8, 125.1, 126.0, 126.5, 126.7, 127.1, 127.7, 127.9, 128.0, 129.6, 130.2, 132,9, 135.1, 136.1, 138.3, 152.8, 153.4, 163.5, 174.0. HRMS (electrospray ionization) calcd for [M–Li] C24H18NO4S2 448.0677, obsd 448.0673.
4.2.52. 8-(1-Methyl-1H-indol-3-yl)-6-(morpholinomethyl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-2-m-tolyl-5H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (58)
Isolated in 34% yield as a yellow non-crystalline solid starting from 0.5 mmol of methylester protected 33. 1H NMR (400 MHz, MeOD) δ 2.30 (s, 3H), 2.48 (br s, 4H), 3.48–3.54 (m, 7H), 3.79 (s, 2H), 5.07–5.40 (m, 2H), 6.61 (d, J = 7.71 Hz, 1H), 6.93–6.98 (m, 1H), 7.11–7.24 (m, 5H), 7.28–7.49 (m, 6H), 7.53 (s, 1H), 7.72–7.76 (m, 1H), 7.99–8.04 (m, 1H). 13C NMR (100 MHz, MeOD) δ 21.3, 32.8, 54.3, 54.7 (2C), 66.2, 68.0 (2C), 106.1, 109.7, 110.3, 110.8, 118.6, 120.3, 120.7, 121.4, 122.8, 123.2, 125.2, 126.1, 126.5, 126.8, 126.9, 127.3, 128.0, 128.5, 129.79, 129.84, 130.6, 130.8, 131.4, 135.7, 135.9, 138.4, 139.9, 147.0, 149.9, 155.7, 161.4, 167.9. HRMS (electrospray ionization) calcd for [M–CO2Li] C39H34N3O3S 624.2321, obsd 624.2307.
4.2.53. 8-(Benzo[d][1,3]dioxol-5-yl)-6-(morpholinomethyl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-2-m-tolyl-5H-thiazolo[3,2-a]pyridine-3-carboxylate lithium salt (59)
Isolated in 42% yield as a yellow non-crystalline solid starting from 0.5 mmol of methylester protected 22. 1H NMR (400 MHz, MeOD) δ 2.34 (s, 3H), 2.45 (s, 4H), 3.48 (s, 4H), 3.72 (s, 2H), 5.19–5.29 (m, 2H), 5.85 (d, J = 36.04 Hz, 2H), 6.72–6.76 (m, 1H), 6.78–6.82 (m, 1H), 6.85–6.92 (m, 2H), 7.17–7.21 (m, 1H), 7.24–7.32 (m, 2H), 7.37–7.48 (m, 3H), 7.50–7.55 (m, 1H), 7.58 (s, 1H), 7.75–7.80 (m, 1H), 8.00–8.06 (m, 1H). 13C NMR (100 MHz, MeOD) δ 21.4, 54.2, 54.6 (2C), 65.8, 68.0 (2C), 102.7, 106.0, 109.8, 111.4, 116.8, 118.8, 121.6, 122.8, 124.9, 125.0, 126.2, 126.6, 126.8, 126.9, 127.4, 128.5, 129.87, 129.94, 130.96, 130.98, 131.3, 135.7, 136.0, 140.0, 145.6, 148.9, 149.3, 149.6, 155.7, 161.2, 167.7. HRMS (electrospray ionization) calcd for [M–CO2Li] C37H31N2O5S 615.1954, obsd 615.1969.
4.2.54. 8-(1-Methyl-1H-indol-3-yl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-2-m-tolyl-5H-thiazolo[3,2-a]pyridine-3-carboxylic acid (60)
Isolated in 66% yield as a yellow foam starting from 0.5 mmol of methylester protected 33. 1H NMR (400 MHz, DMSO-d6) δ 2.26 (s, 3H), 3.83 (s, 3H), 4.96–5.18 (m, 2H), 6.55 (s, 1H), 6.66 (d, J = 7.61 Hz, 1H), 7.02–7.09 (m, 1H), 7.14–7.18 (m, 1H), 7.19–7.30 (m, 3H), 7.36–7.56 (m, 7H), 7.62 (s, 1H), 7.82–7.87 (m, 1H), 8.12–8.18 (m, 1H). 13C NMR (100 MHz, CDCl3/MeOD (9:20)) δ 21.4, 33.2, 67.3, 105.8, 107.4, 107.9, 108.3, 110.6, 119.6, 120.7, 121.4, 122.1, 123.0, 125.95, 125.99, 126.1, 126.2, 126.4, 126.7, 127.2, 128.0, 129.35, 129.42, 129.44 (2C), 130.0, 130.8, 135.2, 137.8, 139.4,149.0,150.1,154.1,160.7,167.5. HRMS (electrospray ionization) calcd for [M–CO2H] C34H25N2O2S 525.1637, obsd 525.1630.
4.2.55. 8-(Benzo[d][1,3]dioxol-5-yl)-7-((naphthalen-1-yloxy)methyl)-5-oxo-2-m-tolyl-5H-thiazolo[3,2-a]pyridine-3-carboxylic acid (61)
Isolated in 67% yield as a yellow foam starting from 0.5 mmol of methylester protected 22. 1H NMR (400 MHz, DMSO-d6) δ 2.31 (s, 3H), 5.11 (s, 2H), 6.08 (s, 2H), 6.54 (s, 1H), 6.81 (d, J = 7.59 Hz, 1H), 6.99–7.06 (m, 2H), 7.13 (s, 1H), 7.22–7.28 (m, 1H), 7.30–7.39 (m, 2H), 7.40–7.57 (m, 5H), 7.84–7.90 (m, 1H), 8.12–8.18 (m, 1H). 13C NMR (100 MHz, CDCl3/MeOD (9:20)) δ 21.4, 40.4, 67.3, 102.3, 105.9, 108.7, 110.0, 110.3, 114.2, 121.6, 122.3, 123.9, 125.9, 126.1, 126.2, 126.2, 127.1, 128.0, 128.4, 129.0, 129.6, 129.7, 129.8, 131.5, 135.3, 139.7, 148.0, 148.4, 149.1, 149.4, 154.1, 160.3, 163.5. HRMS (electrospray ionization) calcd for [M–CO2H] C32H22NO4S 516.1270, obsd 516.1263.
Supplementary Material
Acknowledgments
F.A. was supported by the Swedish Research Council (Grant 621-2010-4730), the Knut and Alice Wallenberg Foundation, JC Kempe Foundation (SJCKMS), S.H. was supported by NIH grants AI048689, AI049950, and AI029549, and A.L. was supported by the Swedish Research Council (Grant 2008-3672).
Abbreviations
- SAR
structure activity relationship
- QSAR
quantitative structure activity relationship
- PLS
partial least squares
- OPLS-DA
orthogonal projections to latent structures discriminant analysis
- UTI
urinary tract infection
- SMD
statistical molecular design
- PCA
principal component analysis
- MRSA
methicillin-resistant Staphylococcus aureus
- UPEC
uropathogenic Escherichia coli
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bmc.2012.01.048.
References and notes
- 1.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Clin Infect Dis. 2009;48:1. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 2.Wiles TJ, Kulesus RR, Mulvey MA. Exp Mol Pathol. 2008;85:11. doi: 10.1016/j.yexmp.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. Science. 2003;301:105. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 4.Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. Science. 1998;282:1494. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 5.Mulvey MA, Schilling JD, Hultgren SJ. Infect Immun. 2001;69:4572. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schilling JD, Lorenz RG, Hultgren SJ. Infect Immun. 2002;70:7042. doi: 10.1128/IAI.70.12.7042-7049.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norinder BS, Köves B, Yadav M, Brauner A, Svanborg C. Microb Pathog. 2012;52:10. doi: 10.1016/j.micpath.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Snyder JA, Lloyd AL, Lockatell CV, Johnson DE, Mobley HLT. Infect Immun. 2006;74:1387. doi: 10.1128/IAI.74.2.1387-1393.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costerton JW, Stewart PS, Greenberg EP. Science. 1999;284:1318. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 10.Davies D. Nat Rev Drug Disc. 2003;2:114. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 11.Xu KD, McFeters GA, Stewart PS. Microbiology UK. 2000;146:547. doi: 10.1099/00221287-146-3-547. [DOI] [PubMed] [Google Scholar]
- 12.Litwin MS, Saigal CS, Yano EM, Avila C, Geschwind SA, Hanley JM, Joyce GF, Madison R, Pace J, Polich SM, Wang M. J Urol. 2005;173:933. doi: 10.1097/01.ju.0000152365.43125.3b. [DOI] [PubMed] [Google Scholar]
- 13.Pratt LA, Kolter R. Mol Microbiol. 1998;30:285. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 14.Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Aberg V, Walker JN, Seed PC, Almqvist F, Chapman MR, Hultgren SJ. Nat Chem Biol. 2009;5:913. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aberg V, Almqvist F. Org Biomol Chem. 2007;5:1827. doi: 10.1039/b702397a. [DOI] [PubMed] [Google Scholar]
- 16.Chorell E, Pinkner JS, Phan G, Edvinsson S, Buelens F, Remaut H, Waksman G, Hultgren SJ, Almqvist F. J Med Chem. 2010;53:5690. doi: 10.1021/jm100470t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinkner JS, Remaut H, Buelens F, Miller E, Åberg V, Pemberton N, Hedenström M, Larsson A, Seed P, Waksman G, Hultgren SJ, Almqvist F. Proc Natl Acad Sci USA. 2006;103:17897. doi: 10.1073/pnas.0606795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nuccio SP, Baumler AJ. Microbiol Mol Biol Rev. 2007;71:551. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chorell E, Edvinsson S, Almqvist F. Tetrahedron Lett. 2010;51:2461. [Google Scholar]
- 20.Emtenäs H, Alderin L, Almqvist F. J Org Chem. 2001;66:6756. doi: 10.1021/jo015794u. [DOI] [PubMed] [Google Scholar]
- 21.Aberg V, Sellstedt M, Hedenstrom M, Pinkner JS, Hultgren SJ, Almqvist F. Bioorg Med Chem. 2006;14:7563. doi: 10.1016/j.bmc.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 22.Chorell E, Das P, Almqvist F. J Org Chem. 2007;72:4917. doi: 10.1021/jo0704053. [DOI] [PubMed] [Google Scholar]
- 23.Pemberton N, Aberg V, Almstedt T, Westermark A, Almqvist F. J Org Chem. 2004;69:7830. doi: 10.1021/jo048554y. [DOI] [PubMed] [Google Scholar]
- 24.Pemberton N, Pinkner JS, Jones JM, Jakobsson L, Hultgren SJ, Almqvist F. Tetrahedron Lett. 2007;48:4543. [Google Scholar]
- 25.Wold S, Esbensen K, Geladi P. Chemom Intell Lab Syst. 1987;2:37. [Google Scholar]
- 26.Eriksson L, Johansson E, Kettaneh-Wold N, Wikstrom C, Wold S. Design of Experiments, Principles and Applications. Umetrics Academy; 2000. [Google Scholar]
- 27.St John RC, Draper NR. Technometrics. 1975;17:15. [Google Scholar]
- 28.Linusson A, Elofsson M, Andersson IE, Dahlgren MK. Curr Med Chem. 2001. p. 17. [Google Scholar]
- 29.O’Toole GA, Kolter R. Mol Microbiol. 1998;28:449. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 30.Bylesjo M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J. J Chemometr. 2006;20:341. [Google Scholar]
- 31.Wold S, Ruhe A, Wold H, Dunn WJ. SIAM J Sci Stat Comp. 1984;5:735. [Google Scholar]
- 32.Wold S, Sjostrom M, Eriksson L. Chemom Intell Lab Syst. 2001;58:109. [Google Scholar]
- 33.Aberg V, Das P, Chorell E, Hedenstrom M, Pinkner JS, Hultgren SJ, Almqvist F. Bioorg Med Chem Lett. 2008;18:3536. doi: 10.1016/j.bmcl.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aberg V, Hedenstrom M, Pinkner JS, Hultgren SJ, Almqvist F. Org Biomol Chem. 2005;3:3886. doi: 10.1039/b509376g. [DOI] [PubMed] [Google Scholar]
- 35.ChemACX2002, CamebridgeSoft. Cambridge, MA: 2002. [Google Scholar]
- 36.MOE. Chemical Computing Group Inc; Montreal, Quebec, Cananda: 2004. [Google Scholar]
- 37.SIMCA-P+ 12.0. Umetrics AB; Box 7960, Umeå, Sweden: 2008. [Google Scholar]
- 38.MODDE 7.0. Umetrics AB; Box 7960, Umeå, Sweden: [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.