Abstract
The ability to efficiently visualize protein targets in cells is a fundamental goal in biological research. Recently, quantum dots (QDots) have emerged as a powerful class of fluorescent probes for labeling membrane proteins in living cells due to breakthrough advances in QDot surface chemistry and biofunctionalization strategies. This review discusses the increasing use of QDots for fluorescence imaging of neuronal receptors and transporters. The readers are briefly introduced to QDot structure, photophysical properties, and common synthetic routes towards the generation of water-soluble QDots. The next section highlights several reports of QDot application that seek to unravel molecular aspects of neuronal receptor and transporter regulation and trafficking. We close with a prospectus of the future of derivatized QDots in neurobiological and pharmacological research.
INTRODUCTION
Since the pioneering neuroanatomical work of Santiago Ramón y Cajal in the early 20th century, the need to visualize dynamic communication within neuronal networks has been a challenge for modern neuroscience.1 Synaptic transmission, and hence central nervous system (CNS) excitability, are modulated in a tightly regulated manner by a myriad of neuronal membrane receptors and transporters that rely on specific ligand binding and transport and the initiation of intracellular signaling cascades. Despite extensive biochemical and genetic analyses, mechanisms that regulate synaptic receptor and transporter activity, trafficking, and localization continue to challenge neuroscientists.2, 3 These considerations have driven our desire to combine recently developed receptor and transporter labeling methods with advanced fluorescence imaging tools to dissect mechanisms of synaptic protein trafficking and signaling at the single-molecule level.
Fluorescent labeling techniques commonly used to interrogate cellular processes can be classified into two broad categories: (1) the construction and expression of fluorescent fusion proteins such as GFP4, 5 and (2) chemical methods of fluorescent labeling.6 With the rapid evolution of fluorescent protein technology, GFP-based labeling methods have quickly found widespread use as the proteins produced bypass many time-consuming probe preparation steps and assure that labeling is restricted to the desired proteins. Unfortunately, there are certain limitations associated with the GFP fusion approach: (1)incompatibility of GFP construct ttransfection with endogenous expression systems; (2) failure of GFP-tagged proteins to localize properly; (3) differences in the activity and signaling of GFP-tagged proteins compared to their wildtype counterparts. As a result, alternative fluorescence labeling strategies have emerged for the visualization of cellular proteins, particularly those systems that are incompatible with the GFP-based methodology. However, since chemical labeling techniques typically utilize conventional fluorescent dyes, live-cell imaging is often hampered by the low photostability and brightness, narrow Stokes shift, and relatively broad emission spectra of these labels. To address these shortcomings, researchers have developed fluorescent nanomaterials with significantly improved optical properties for biological imaging.
Semiconductor nanocrystals, also known as quantum dots (QDots) are one such class of fluorescent nanomaterials that overcome instability issues associated with conventional fluorescent dyes and support versatile protein labeling via their surface functionalization with various targeting probes.7–9 More importantly, QDots are much better suited to advanced fluorescence imaging approaches that pursue live-cell, multicolor detection, in vivo animal imaging,10–12 and single molecule tracking.13, 14 Here we provide an overview of recent advances in QDot synthesis and the conjugation of biological probes to the QDot surface. In addition, we highlight several reports of QDot application that seek to unravel molecular aspects of neuronal receptor and transporter regulation and trafficking. We close with a prospectus of the future of derivatized QDots in neurobiological and pharmacological research.
WATER-SOLUBLE QUANTUM DOTS
What is a quantum dot?
QDots are nanometer-sized crystals that are composed of several hundreds to thousands of atoms of periodic group II/VI (CdSe, CdTe, CdS), IV/VI (PbS, PbSe), III/V (InP), or ternary (InGaP) semiconductor materials, with CdSe being one of the most commonly encountered materials.7, 15–19 Typically, the first step of QDot preparation involves high-temperature pyrolysis of organometallic precursors in organic solvents, such as trioctylphosphine (TOP), trioctylphosphine oxide (TOPO), and hexadecylamine (HDA).17, 18 As a result, the as-synthesized QDot is composed of a CdSe core capped with a layer of hydrophobic ligands. These “bare”, water-insoluble QDots cannot be immediately used in an aqueous, biological environment; moreover, these QDots are susceptible to degradation/aggregation and suffer from relatively low fluorescence quantum yields, as a result of the nonradiative recombination processes occurring at the core surface. To eliminate these problems, the CdSe core is commonly passivated with a shell of a wider bandgap inorganic material (ZnS) to increase its stability and optimize optical properties.16, 18 Thus, the term “quantum dot” usually refers to a CdSe/ZnS core/shell structure. Figure 1A shows an atomic number-contrast scanning transmission electron microscopy (Z-STEM) image of a single CdSe/CdS/ZnS core/shell QDot commercially available from Invitrogen.7
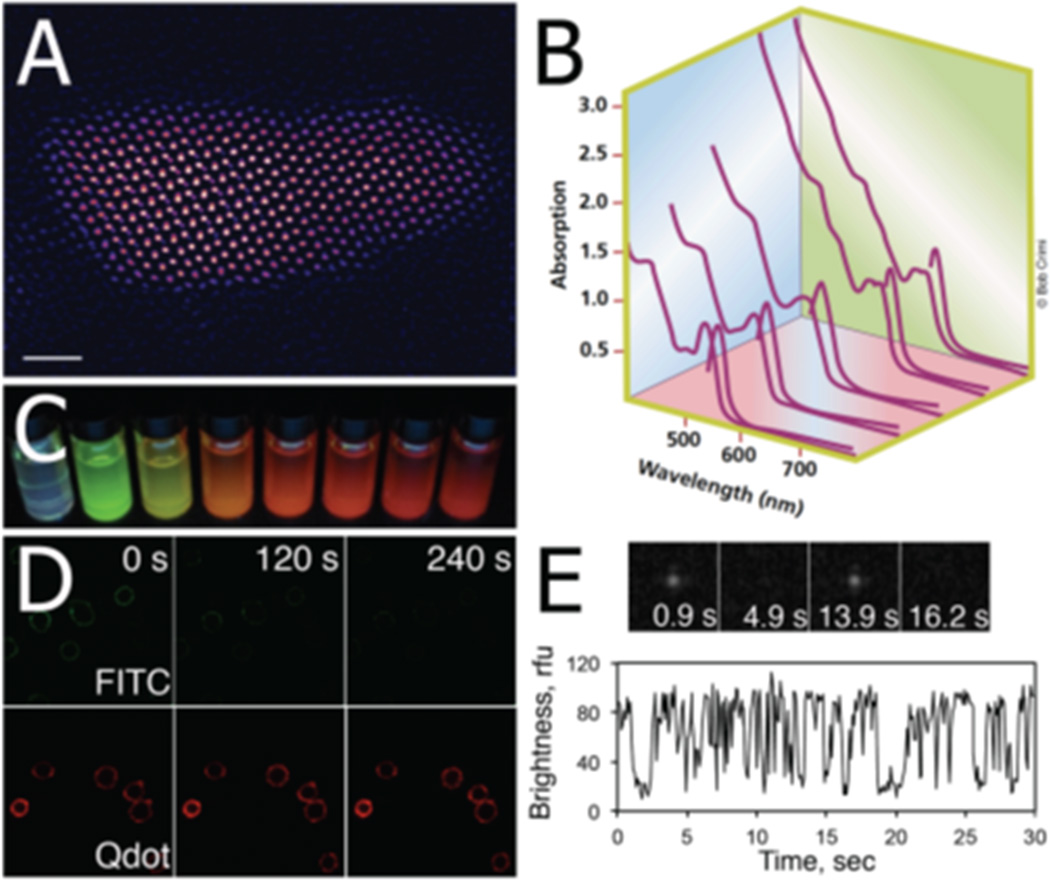
Figure 1.
Photophysical properties of quantum dots. (A) Aberration-corrected Z-STEM image of a commercial core/shell QD655 (Invitrogen). It can be seen that the core/shell QD is actually an elongated, bullet-shaped 3D object rather than a sphere. (Reprinted with permission from Ref 7. Copyright 2007 Elsevier B.V.) (B) Absorption and emission spectra of a series of CdSe nanocrystals. The size of the QD core determines the absorption and emission spectra of QDs. (Reprinted with permission from Ref 14. Copyright 2001 Nature Publishing Group) (C) A series of UV-illuminated CdSe nanocrystals ranging in size from ~ 2 nm to ~6 nm. (D) Time-lapse image series of FITC- and QD-labeled HEK-293 cells. In contrast to traditional fluorophores, QDs are characterized by excellent photostability which enables long-term monitoring of biological processes. (Reprinted with permission from Ref 48. Copyright 2011 American Chemical Society) (E) Fluorescence intermittency or “blinking” in the emission spectrum of a single QD. This QD property can be used as a criterion to distinguish single nanocrystals from aggregates.
Quantum Dot Properties
The properly passivated CdSe core gives rise to a unique combination of photophysical properties of QDots. QDots are characterized by remarkable brightness that is a product of their high molar extinction coefficients (100,000 – 1,000,000 M−1cm−1) and near-unity quantum yields.7, 15, 20 Also, their broad absorption spectra and narrow, Gaussian emission spectra considerably simplify multicolor experiments, allowing visualization of several fluorophores using a single, inexpensive excitation source with minimal cross-talk (Figure 1B).21–25 The dependence of the emission wavelength upon the size of the QDot core is a key property of QDots (Figure 1C), providing for easy diversification of probe identity through relatively simple alterations in chemical synthesis. The inorganic nature of QDots and passivation of the QDot core render QDots extremely resistant to photobleaching.17, 22, 26 This optical stability is an importantadvantage over organic dyes and fluorescent proteins; superior QDot photostability allows for long-term visualization of dynamic biological processes (Figure 1D). An interesting and pivotal feature inherent in the emission spectra of single QDots is the alternation of “dark” (off) and “bright” (on) states, termed “blinking” (Figure 1E).27–31 Although such fluorescence intermittency can be used as a criterion to assure the observation of a single QDot, this also remains problematic when QDot tracking experiments are performed. Finally, another key advantage of QDots over organic dyes and fluorescent proteins is the versatility of QDot surface modification, a feature we will discuss in greater detail below.
Solubilization
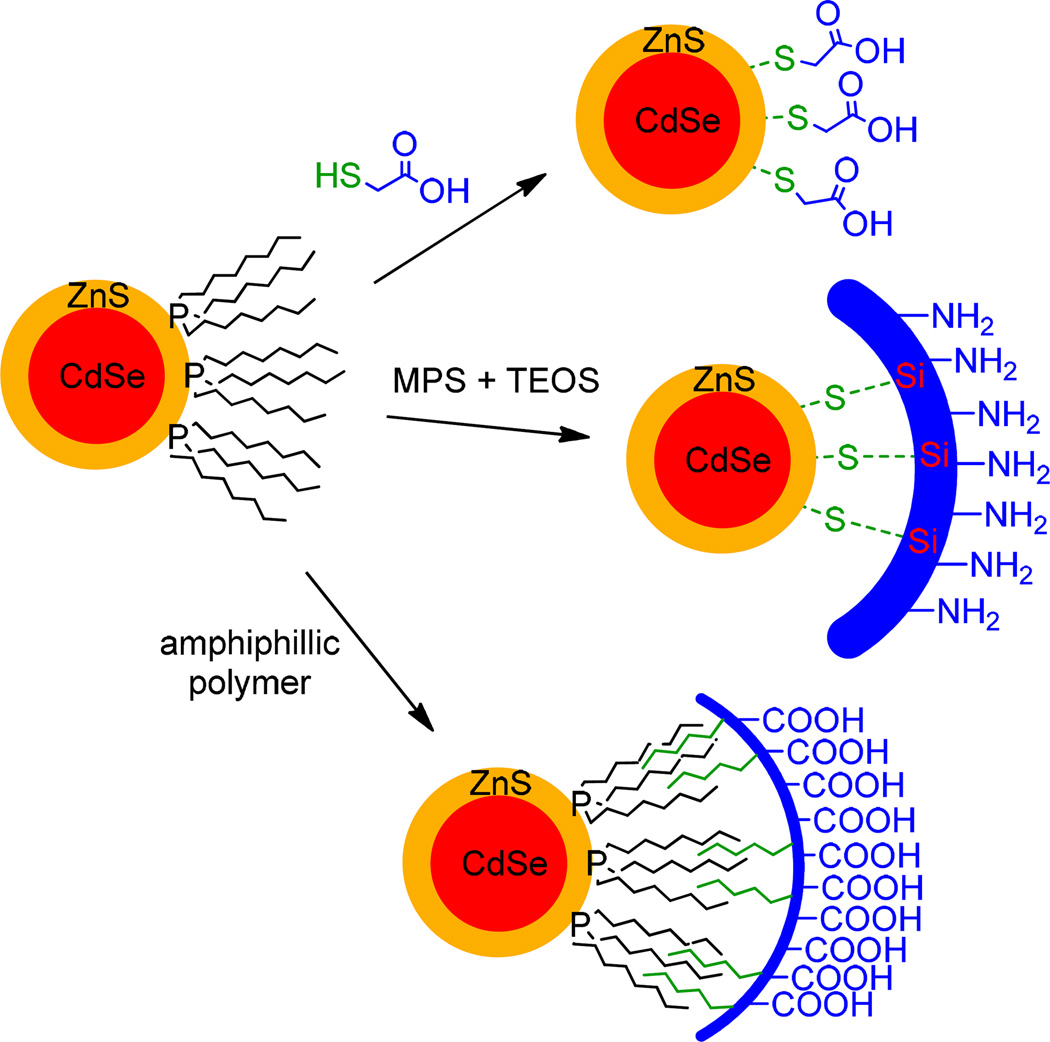
As mentioned above, as-synthesized core/shell QDots are capped with hydrophobic organic ligands and are thus incompatible with an aqueous biological environment. To render QDots biocompatible, one must first make them soluble in water while preserving their photophysical properties. Several strategies are used to achieve this: (1) ligand exchange, (2) polymer encapsulation, and (3) silica shell encapsulation (Figure 2). Before these approaches are discussed, it should be noted that any solubilization route must lead to a QDot that is stable under physiologically relevant conditions, that displays minimal nonspecific interactions and possesses functional surface elements necessary for the introduction of target specificity. In the ligand-exchange approach, bifunctional ligands, which contain a point of attachment to the QDot surface on one end and a hydrophilic moiety on the other, displace native organic ligands (Figure 2, Top).32 In polymer encapsulation, native organic ligands are retained and instead tightly associate with the inner aliphatic chains of an added amphiphilic polymer shell.11, 33, 34 The exterior hydrophilic backbone of the polymer then aids in aqueous dispersion (Figure 2, Middle). With silica encapsulation, as-synthesized QDots are encapsulated within an inert silica shell (Figure 2, Bottom).35–37 The silica encapsulation strategy is not yet in wide use due to the tendency of silica-coated QDots to agglomerate during synthesis as well as lack of control over the growth of silica spheres around QDots. Depending upon the experimental requirements, each of the above solubilization strategies may be the optimal method. Amphiphilic polymer encapsulation has thus far been the preferred strategy since polymer-coated QDots retain native surface-capping ligands and do not exhibit significant decrease in performance, e.g. quantum yield. In addition, polymer-coated QDots are characterized by superior colloidal stability in physiologically relevant conditions. However, the added polymer shell significantly increases the hydrodynamic diameter of QDots, limiting their use in size-sensitive applications. With ligand exchange, it is possible to maintain QDot size below the typical hydrodynamic diameter values of 30–40 nm after polymer encapsulation while preserving the original photophysical properties. It is true that in some cases the exchange of native surface-capping ligands leads to significant decrease of quantum yield and shelf life; however, the use of compact, polydentate ligands (dihydrolipoic acid, DHLA) is a potential solution to these problems.38, 39
Figure 2.
Quantum dot solubilization approaches. The as-synthesized QDs can be rendered soluble in water via ligand exchange (Top), wherein native hydrophobic surfactants are replaced with bifunctional hydrophobic capping ligands; via encapsulation in an insert silica shell (Middle); or via amphiphilic polymer encapsulation (Bottom), wherein native surfactants are retained and integrated into the polymer shell.
To summarize, the goal of solubilization is to modify the QDot surface in such a way that the resulting QDots are water-soluble and stable at physiologically relevant pH and ionic strength ranges. One must carefully ensure that such post-preparative surface modification is achieved with modified QDots that retain their original physical and optical properties. Finally, water-soluble QDots must contain functional reactivity handles to allow subsequent conjugation of biospecific molecules to the QDot surface.
CONJUGATION OF BIOPROBES TO QUANTUM DOTS
Once QDots have been made water-soluble, the next step is to introduce biological specificity for a specific target receptor or transporter. QDots present a very attractive photoluminescent scaffold with a high surface area to volume ratio, enabling conjugation of multiple copies of various reactivity handles and biospecific recognition elements to the QDot surface. In addition to a consideration of the role of bioprobe-binding valency to QDots, which we will describe in detail later, a prerequisite for success in QDot labeling is the choice of an appropriate bioprobe. Bioprobes that have been introduced in QDot conjugation include small organic ligands, peptides, adhesive proteins (i.e. Streptavidin), and antibodies. Each of these probes presents particular advantages and disadvantages (see Table 1 for comparison). QDot-antibody nanoconjugates have thus far been the preferred method of surface protein detection, as they provide excellent specificity, and commercial antibody conjugation kits are readily available. However, QDot-antibody conjugates typically possess a large hydrodynamic diameter, which may complicate the use of such probes in crowded cellular locations. In addition, the long-term stability of the antibody-protein interaction may be a concern, as antibody probes are prone to proteolytic digestion and degradation. The potential for cross-linking of proteins, which serves as an activation signal for some receptors, is another problem, though the use of Fab fragments can overcome this issue. Peptide-based QDots and antibody-conjugated QDots share similar features in that probes are also readily available and they are both prone to proteolytic digestion. A significant advantage with peptide-conjugated QDots is that several neuropeptide receptors still lack small-molecule ligands and specific antibodies for targeting. Biological recognition can also be incorporated in the QDot architecture through the use of biologically active small-molecule ligands. These ligand-conjugated QDots rely on ligand-receptor interactions to provide specificity. Additionally, a ligand-based targeting approach can be designed to elicit either an agonistic or antagonistic physiological response upon QDot probe binding to the target protein.. While the ligand-conjugated Qdots may potentially address the concerns regarding poor specificity associated with the peptide-based QDot conjugates, fairly sophisticated organic chemistry is required to synthesize small-molecule ligand probes for conjugation to the QDot surface. Also, rigorous ligand characterization and purification steps must be involved. Each class of bioprobes that can be conjugated to QDots is associated with distinct advantages and disadvantages, and bioprobe selection will ultimately be dependent upon experimental conditions and goals.
Table 1.
Advantages and weaknesses of major classes of functional probes that can be conjugated to QDots for biological labeling and imaging.
| Targeting Probe |
Advantages | Disadvantages |
|---|---|---|
| Organic Ligand |
|
|
| Peptide |
|
|
| Antibody |
|
|
To take advantage of the unique photophysical properties of QDots for live cell, real-time imaging of surface proteins, the choice of conjugation methodology should be considered as important as the choice of the bioprobe. Several methodologies have been devised to functionalize QDots with target-specific bioprobes and can be grouped into two categories: direct and indirect conjugation. These QDot conjugation approaches are discussed below along with specific examples of their use in targeting neuronal receptors and transporters.
Direct Conjugation Approach
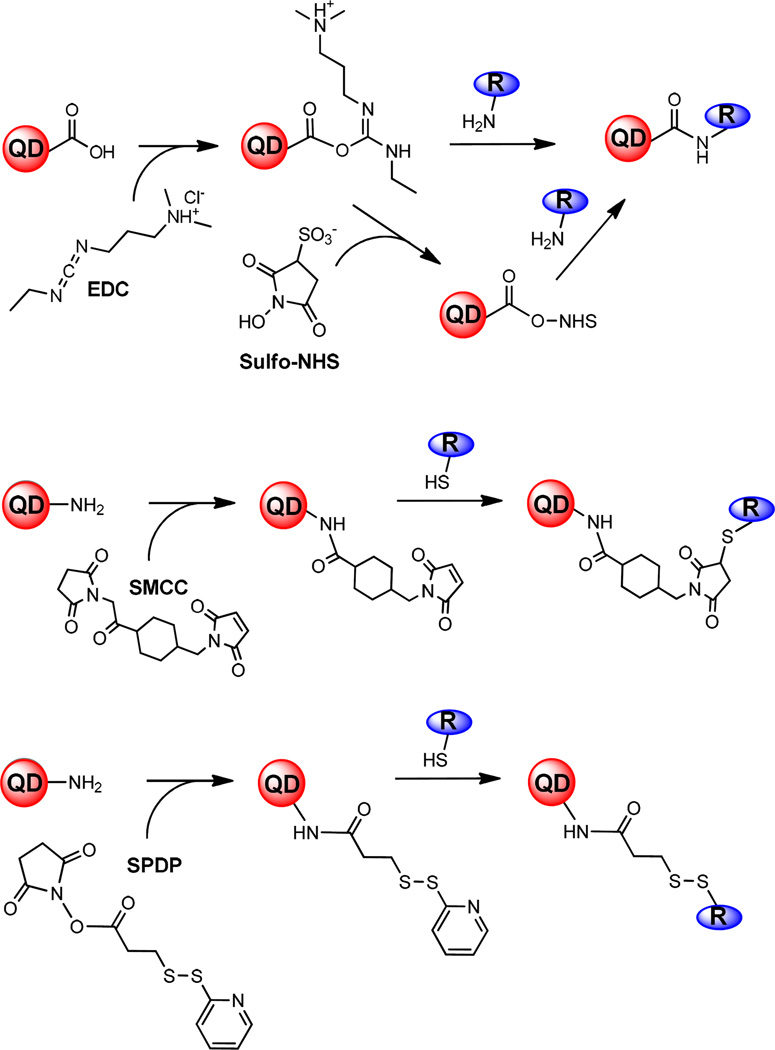
In the direct conjugation approach, a reactive group at the QDot surface (−COOH, −NH2, −SH) is conjugated to a biospecific molecule that has a compatible reactivity handle. This is achieved through the use of heterobifunctional cross-linker molecules (Figure 3), such as 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), succinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (SMCC), and N-succinimidyl 3-(2-pyridyldithio)propionate (SPDP). In EDC coupling, QDot surface carboxyl groups (−COOH) are first reacted with EDC reagent to form an unstable, amine-reactive O-acylisourea intermediate that readily reacts with an amino-terminated (−NH2) biomolecule to form an amide bond. Typically, the unstable amine-reactive intermediate is stabilized using N-hydroxysuccinimide; the NHS-stabilized intermediate then quickly reacts with an amino-terminated biomolecule to form an amide bond. In SMCC coupling, SMCC or its water-soluble analog sulfo-SMCC reacts with QDot surface amino groups to yield maleimide-activated QDots. Then the maleimide-activated QDots are reacted with a biomolecule that contains a sulfhydryl (-SH) group to produce QDot conjugates. Similar to SMCC coupling, SPDP coupling allows conjugation of amino-terminated QDots to sulfhydryl-containing biomolecules, or vice versa. Amino-terminated QDots first react with SPDP to give 2-pyridilthio-terminated QDots, with the 2-pyridil groups readily displaced by sulfhydryls under the neutral pH conditions. The EDC/NHS methodology is typically used to covalently conjugate small molecules, polyethylene glycol (PEG) chains, or high-affinity proteins (avidin) to the surface of water-soluble QDots, whereas the SMCC/SPDP methodology is commonly used to conjugate antibodies or antigen-binding fragments (Fab) to QDots. To do so, the hinge disulfide bonds of an antibody or a Fab fragment must first be reduced with dithiothreitol (DTT) to yield free sulfhydryl groups.40 Another way to confer specificity to the QDot surface is to combine solubilization and subsequent functionalization steps into a one-step ligand-exchange procedure.8 Such a strategy involves the exchange of native capping ligands with heterobifunctional, amphiphilic ligands that contain a chemical group which may be directly conjugated to the bare nanocrystal (e.g. -SH) as well as a functional element at the opposite ligand end that facilitates binding to a neuronal receptor or transporter.41
Figure 3.
Direct conjugation methodology. Schematic diagram of the chemical reactions that occur during covalent coupling of bioprobes to the QD surface by using EDC/NHS (Top), SMCC (Middle), and SPDP (Bottom) reagents.
Indirect Conjugation Approach
In the indirect conjugation approach, a biospecific molecule is not directly conjugated to the surface of the QDot. Instead, this approach takes advantage of high-affinity non-covalent interactions between high-affinity binding partners, such as avidin family proteins that bind biotin, and biotin-tagged molecules. Although other partner molecules exist for such use (e.g. maltose and maltose-binding protein), avidin-biotin noncovalent assembly is the most popular approach to achieve stable, target specific QDot labeling and thus will be discussed in more detail below.
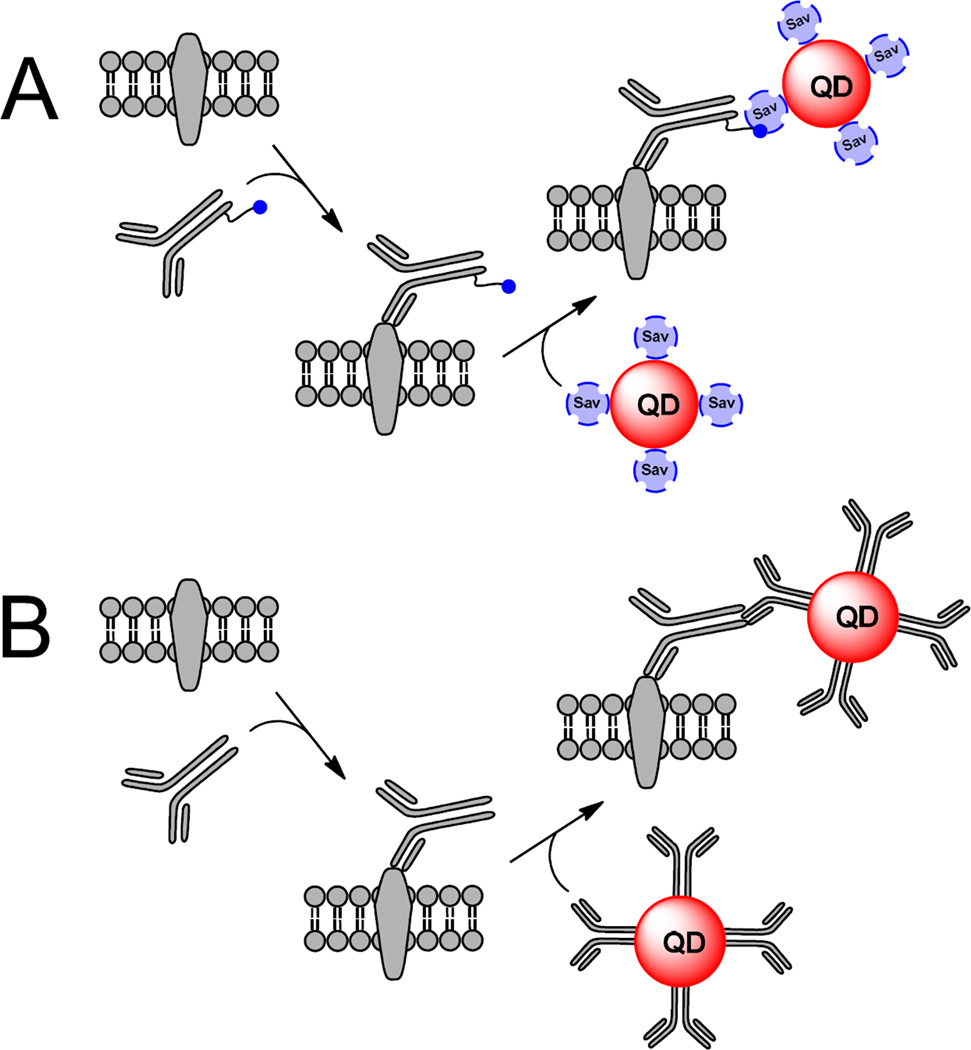
The use of avidin-biotin assembly in a QDot-based assay was first documented in 2002 when Goldman et al. used avidin as a natural bridge between biotinylated antibody and QDots in a fluoroimmunoassay.42 The avidin family proteins are capable of forming tight complexes with biotin, a small organic molecule, and includes Streptavidin and similar, evolutionary unrelated, biotin-binding avidin and neutravidin.43 Streptavidin is a homotetrameric protein with a molecular mass of ~52.8 kDa; each Streptavidin subunit is capable of binding one biotin molecule, resulting in one of the strongest and most stable noncovalent interactions in nature (dissociation constant, Kd ~ 10−15 M; half-life, greater than several days).44, 45 Together with high binding specificity and minimal background, such properties have enabled the Streptavidin-biotin assembly to become a tool of first choice for the QDot visualization of biological targets. In a typical experiment, live cells expressing the membrane protein of interest are first incubated with a biotinylated primary antibody specific for the extracellular epitope of that protein. Then, the unbound, biotinylated probe is washed away, and the cells are incubated with QDots coated with several covalently attached Streptavidin molecules (Sav-QDots). After excess Sav-QDots are washed away, the cells can be immediately visualized (Figure 4A). Alternatively, one can use an “antibody sandwich” method wherein a target-bound primary antibody is recognized by a biotinylated secondary antibody that is then captured by Sav-QDots. Also, one can simply use QDots covalently coupled to secondary antibodies via SPDP/SMCC methodology to recognize the target-bound primary antibody (Figure 4B). Since the use of several antibodies can add substantial bulk to the resulting QDot-antibody probe bound to the target protein, it is not uncommon for antibodies to be digested to yield smaller Fab fragments that retain the antigen-binding site; this becomes particularly important in size-sensitive experiments. Some experiments require that the biotinylated probe be conjugated to Sav-QDots and applied in a one-step labeling protocol; the formation of such a conjugate eliminates unwanted effects due to cellular stimulation with the biotinylated probe, or can allow the preparation of a monovalent QDot probe. Although antibody-based QDot labeling of neuronal receptors and transporters is popular in the field, Sav-QDots can also be used in conjunction with biotinylated peptides46 and organic ligands.47, 48
Figure 4.
Indirect Qdot labeling methodology. Indirect Qdot labeling typically involves either a Streptavidin-biotin interaction between biotinylated probes and Streptavidin-conjugated Qdots (A), or a noncovalent recognition of a target-bound primary antibody with secondary antibody-conjugated Qdots (B).
Each QDot conjugation approach described above is associated with distinct advantages and drawbacks; the suitability of each approach depends on the material/probe availability and experimental requirements. The direct conjugation approach usually results in smaller QDot conjugates and enables multicolor experiments wherein several targets may be monitored with several sizes of QDots preconjugated to distinct biospecific molecules. On the other hand, direct conjugation involves additional preparation, purification, and characterization steps; however, often such steps are easily accomplished through the use of commercially available QDot conjugation kits (Invitrogen). Also, direct conjugation usually results in the presence of several copies of probe at the QDot surface, with the conjugation stoichiometry being problematic to control. Depending upon the application, such multivalent probe presentation at the QDot surface may lead to either cooperative binding and increased avidity or unwanted protein cross-linking due to multivalent binding.49 In the indirect conjugation approach, the use of Streptavidin significantly increases the final size of the QDot conjugates, an undesirable effect when it comes to size-sensitive QDot applications, such as fluorescence resonance energy transfer (FRET) or in vivo imaging studies. Another issue with the use of the Streptavidin-biotin assembly is the possibility of multivalent binding; for example, a commercially available Sav-QDot with a 655-nm emission maximum (Invitrogen) has approximately 5–10 Streptavidin molecules at its surface. Each Streptavidin has 4 biotin-binding sites, which results in a total of 20–40 biotin-binding sites per QDot. To address this issue, Howarth et al. genetically engineered a monovalent Streptavidin protein with a single femtomolar affinity binding site.50 Subsequently, a single copy of the monovalent Streptavidin was conjugated to the QDot surface to yield monovalent QDots capable of binding only a single copy of the biotinylated probe.51 The advantages of the Streptavidin-biotin approach include highly specific recognition of the biotinylated probe, ultra-low nonspecific binding and avoidance of time-consuming preparation steps, as Sav-QDots and biotinylated probes (antibodies) are readily available.24
QUANTUM DOTS FOR BIOLOGICAL IMAGING OF NEURONAL RECEPTORS AND TRANSPORTERS
In the previous section, we discussed the selection of bioprobes and conjugation methods for the preparation of QDot nanoconjugates. In this section, we present specific examples that demonstrate how QDot nanoconjugates have been employed in recent molecular neuroscience studies. We have also compiled a comprehensive list of the instances where QDots were used to target neuronal receptors and transporters, and classified each example based on the targeting probe and QDot conjugation strategy used (Table 2).
Table 2.
Selected examples of quantum dot applications in the studies of neuronal receptors and transporters
| Protein of interest | Targeting probe |
QDot Conjugation strategy |
Cellular expression model | Reference |
|---|---|---|---|---|
| Serotonin transporter | Organic ligand | Ligand exchange | Transfected HEK293 cells | 41 |
| Serotonin transporter | Organic ligand | Biotin-Sav binding | mRNA-microinjected Xenopus | 47 |
| Serotonin transporter | Organic ligand | Biotin-Sav binding | Serotonergic RN46A cells | 67 |
| Dopamine transporter | Organic ligand | Biotin-Sav binding | Transfected flp-In 293 cells | 48, 66 |
| GABAc receptor | Organic ligand | EDC coupling | mRNA-microinjected Xenopus oocytes | 54 |
| GABAA receptor (with GFP tag) | Anti-GFP antibody | 2nd antibody coupled to QDot | Hippocampal neurons | 72 |
| Glycine receptor | Antibody | Biotin-Sav binding | Primary rat spinal cord neurons | 56, 57 |
| Glycine receptor | Antibody | SPDP/SMCC coupling | Transfected HeLa cells, primary neurons | 73 |
| Glial fibrillary acidic protein | Antibody | Biotin-Sav binding | Primary neurons and glia | 70 |
| TrkA and and P75 NGF receptors | NGF peptide | Biotin-Sav binding | PC12 cells | 71, 74 |
| 5-HT1A receptor (with HA tag) | Anti-HA antibody | Biotin-Sav binding | N2a cells | 64 |
| AMPA receptor | Antibody | 2nd antibody coupled to QDot | Primary rat cortical neurons | 58, 75 |
| AMPA receptor | Peptide | 2nd antibody coupled to QDot | Primary rat hippocampal neurons | 62 |
| Cannabinoid type 1 receptor | Antibody | 2nd antibody coupled to QDot | Primary rat hippocampal neurons | 76 |
| NMDA receptor | Antibody | 2nd antibody coupled to QDot | Primary rat hippocampal neurons | 77 |
| metabotropic glutamate receptors (mGluR5) | Antibody | Biotin-Sav binding | Primary rat hippocampal neurons | 78 |
| Nicotinic acetylcholine receptor (nAChR) | Protein/toxin | Biotin-Sav binding | Neuromuscular synapses in the mouse diaphragm | 46 |
| Nicotinic acetylcholine receptor (nAChR) | Antibody; Protein/toxin | Biotin-Sav binding | Chick CG neurons | 79 |
| Angiotensin II receptor type 1 | Peptide | EDC coupling | Transfected CHO cells | 80 |
| Presynaptic L-type calcium channel | Antibody | Biotin-Sav binding | Synapses of the tiger salamander retina | 81 |
| P2 purinergic receptors | Organic ligand | EDC coupling | PC12 cells | 82 |
| D2 dopamine receptor | Organic ligand | EDC coupling | Transfected A9 cells | 55 |
QDot Labeling of Neuronal Receptors and Transporters Using the Direct Conjugation Approach
We mentioned above that QDots are typically synthesized in organic solvents and passivated with hydrophobic surfactants such as trioctylphosphine oxide (TOPO) and trioctylphosphine (TOP). Thus, one of the first methods used to generate ligand functionalized QDots for biological labeling was naturally the exchange of QDot hydrophobic surfactants with thiol (−SH) functionalized hydrophilic ligands.8 In 2002, our group first employed the ligand exchange method in generating organic ligand functionalized QDots for neuronal transporter labeling (Figure 5).41 In this study, a PEGylated, thiol-terminated serotonin ligand (5HT-PEG-SH) was exchanged with surface TOPO to yield water-soluble QDot-PEG-5HT conjugates, which were then used to image membrane serotonin transporters (SERTs) in living cells. Furthermore, these same QDot conjugates were employed in EC50 measurements and electrophysiological studies, demonstrating the potential utility of ligand-conjugated QDots in pharmacological and physiological applications.
Figure 5.

Ligand-conjugated QDots for SERT labeling in living cells. (A) Fluorescence images of SERT expressing HEK cells labeled with serotonin ligand functionalized QDots. Fluorescence labeling of SERT in the membrane is clearly visible (left) while cells pre-incubated with high affinity SERT inhibitor paroxetine prior to the labeling shows no sign of fluorescence (right), indicating the QDot labeling specificity. (B) Electrophysiological currents elicited by serotonin or serotonin ligand functionalized QDots. The current induced by serotonin is typical. In contrast, the current induced by serotonin ligand functionalized QDots is characteristic of currents induced by SERT antagonists. (Reprinted with permission from Ref 41. Copyright 2002 American Chemical Society)
Recently, due to the popularity of high-quality, commercially available QDots, more scientists have begun to use these commercially available carboxyl- or amine-functionalized quantum dots to generate nanoconjugates for biological labeling. Although direct conjugation of biological probes to the these QDots involves additional “wet” chemistry techniques (see previous section), this preparation provides an additional degree of freedom in controlling how nanoconjugates interact with biological targets (e.g., adding a PEG chain to further decrease the cytotoxicity and non-specific binding;52 or manipulating the probe multivalency53). In particular, we covalently conjugated PEGylated muscimol, a GABAC receptor agonist, to the QDot surface via EDC/NHS coupling.54 In the follow-up experiments, it was demonstrated that ligand valency on GABAC affects its binding avidity and affinity.53 Clarke and colleagues used a similar conjugation approach to design a redox-sensitive probe.55 Based on the electron transfer mechanisms, dopamine-conjugated QDots were used to sense an intracellular oxidative state. Under reducing conditions, these QDot probes were only visible in the cell periphery and lysosomes, whereas in mild oxidizing conditions QDot fluorescence appeared in the perinuclear region. Under strongly oxidizing conditions, the QDot probe-associated fluorescence was visible throughout the cell. This experiment demonstrated the potential utility of QDot probes for sensing intracellular environments.
QDot Labeling of Neuronal Receptors and Transporters Using the Indirect Conjugation Approach
Shortly after Goldman and colleagues recognized the potential of the avidin family of proteins to facilitate the preparation of stable QDot-antibody conjugates,42 Dahan and colleagues used the Streptavidin-biotin assembly to detect single endogenous glycine receptor (GlyR) molecules at the surface of cultured rat spinal cord neurons.56 The specific detection of GlyR α1 subunits was achieved through the use of a primary monoclonal antibody, a biotinylated Fab fragment of the secondary antibody, and Sav-QDots. Then QDots were used to visualize the lateral diffusion of single GlyR molecules. Importantly, this elegant study revealed that diffusion dynamics of glycine receptors varies in synaptic, perisynaptic, and extrasynaptic domains of spinal neurons.
In 2008, Lévi et al. established that Ca2+-driven excitatory synaptic transmission significantly restricted GlyR lateral diffusion and led to an increased subsequent clustering of GlyRs within the synaptic domain.57 Furthermore, Charrier et al. in 2010 demonstrated that such a regulation of GlyR lateral diffusion at the excitatory synapses is mediated by β1 and β3 integrins, cell adhesion molecules and signaling receptors that interact via calcium/calmodulin-dependent protein kinase II.58 This progression is an impressive example of the evolution of QDots as fluorescent probes aimed at unraveling the molecular aspects of neuronal receptor regulation.
A similar “sandwich” approach was also employed by Triller and colleagues to label GABA A receptors (GABAAR). The 2 subunit of the GABAAR was sequentially labeled with an anti- 2 antibody, a biotinylated secondary antibody, and then Sav-QDots. This approach was used by Bouzigues et al. to label GABAARs in the growth cones (GCs) of rat spinal cord neurons.59 In the presence of an extracellular GABA gradient, the authors showed that single QDot-labeled GABAA receptors redistribute asymmetrically across the growth cone, located at the axon tip, toward the gradient source in a microtubule- and calcium-dependent manner. In 2009, Bannai et al. relied on a modified approach for GABAAR labeling, in which a biotinylated secondary Fab fragment was used in conjunction with primary antibody and Sav-QDots. In this study, the authors demonstrated that GABAAR diffusion coefficient and confinement domain size increase in response to enhanced excitatory synaptic activity.60
A distinct labeling approach based on Streptavidin-biotin assembly was developed in the Ting Lab, where the authors adapted an enzymatic reaction to specifically biotinylate their proteins of interest. 61 In their design, a fifteen amino acid acceptor peptide sequence (AP) is genetically fused to a C- or N-terminus of the protein, and a bacterial enzyme biotin ligase (BirA) is used to biotinylate a lysine side chain within the AP sequence. Howarth et al. applied this approach to label AP-fused α-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPA) receptors in hippocampal neurons and then study AMPA receptor synaptic localization.62
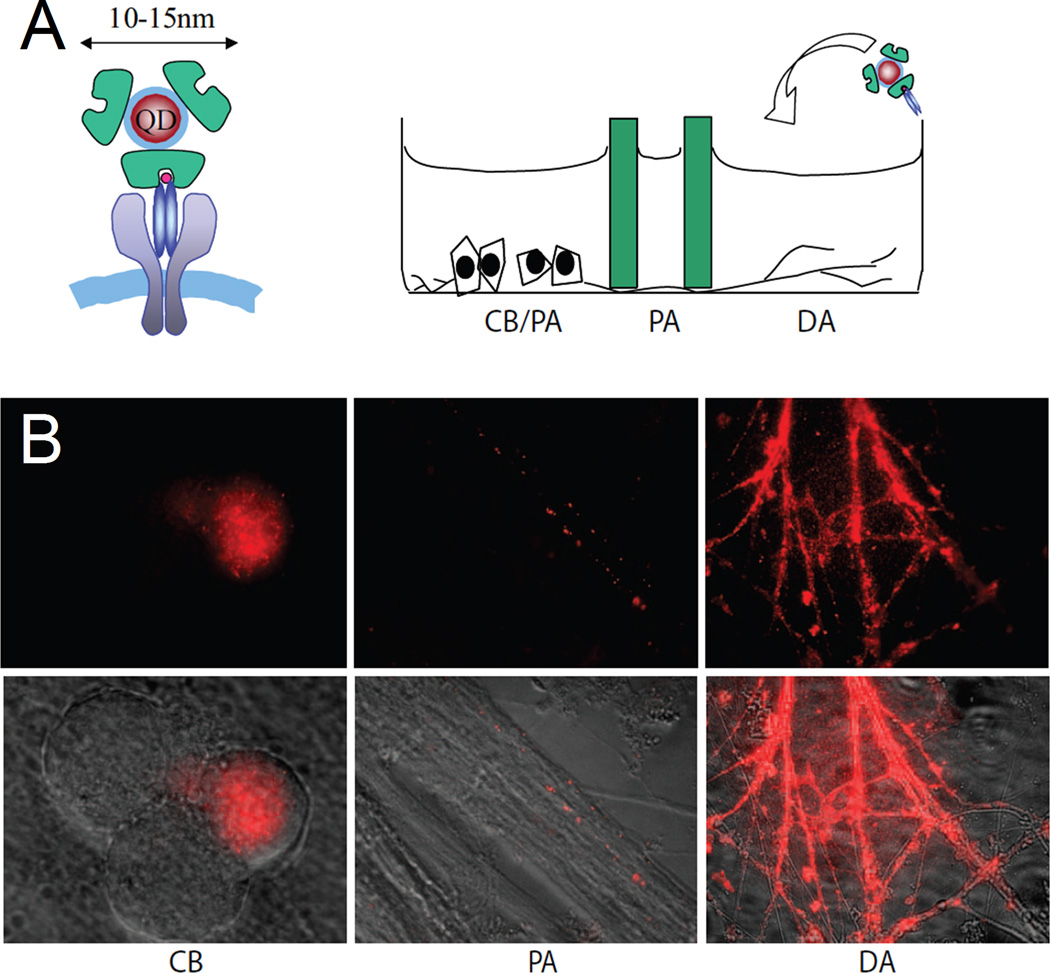
Cui and colleagues also used the biotin-Streptavidin approach to prepare QDot nanoconjugates to label nerve growth factor (NGF) receptors in live PC12 and rat dorsal root ganglion (DRG) neurons (Figure 6).63 In their study, biotinylated NGF peptides were pre-conjugated to Sav-QDots with a stoichiometric ratio of NGF to QDot of 1:1.2 to achieve a monovalent presentation of NGF dimer on the QDot surface. In a clever setup, NGF-QDot conjugates were first added to the microfluidic chamber containing distal axons of DRG neurons and allowed to bind and form complexes with NGF receptors and undergo subsequent internalization into early endosomes. Endosomes containing NGF-QDots were then demonstrated to exhibit “stop-and-go” retrograde transport toward the neuronal cell body with an average speed of 1.31 ± 0.03 µm/s. Similarly to Cui et al.,63 Fichter and coworkers employed the preconjugation strategy to link a biotinylated anti-hemagglutinin (HA) antibody to Sav-QDot. The resulting QDot conjugates were used to label HA-fused serotonin receptor subtype 1A (5-HT1A) and investigate the kinetics of receptor-mediated internalization of QDots.64
Figure 6.
QDot labeling of NGF receptors using Streptavidin-biotin assembly. NGF-conjugated QDs were used to investigate retrograde, vesicular NGF transport in living DRG neurons. (A) Schematic drawing of a QDot-NGF bound to dimerized TrkA receptors (Left) and addition of QDot-NGF to the DA compartment of the three-chamber DRG neuron culture (Right). DA, distal axon; PA, proximal axon; CB, cell body. (B) Representative live fluorescence images of DRG neuron axons or cell bodies 2 h after the addition of 4 nM QDot-NGF to the DA chamber. QDot-NGF seems to bind all axons in distal axon chamber. However, only a small portion of the cell bodies and proximal axons are shown to have QDot fluorescence, reflecting the fact that not all cell bodies extend their axons into the distal axon compartment. (Reprinted with permission from Ref 63. Copyright 2007 National Academy of Sciences, U.S.A.)
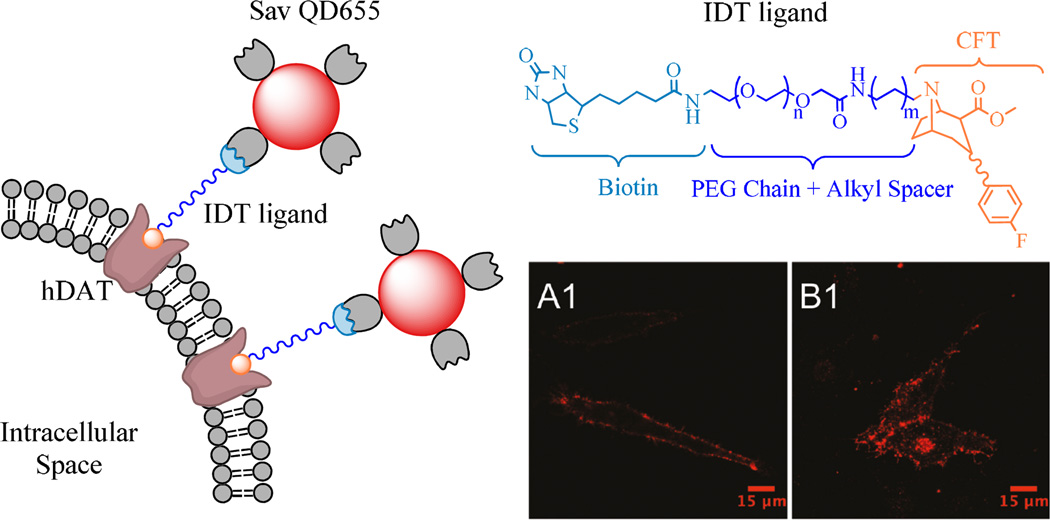
Our group has also previously utilized a biotin-Streptavidin assembly strategy to label cell surface neuronal receptors and transporters with Sav-QDots. In 2008, Orndorff et al. used a biotinylated α-bungarotoxin (α-BTX) and Sav-QDots in a two-step labeling protocol to visualize nicotinic acetylcholine receptors (nAChRs) on the surface of mouse diaphragm neuromuscular junction ex vivo.46 The high affinity and specificity of α-BTX for nAChRs and ultra-low nonspecific binding of Sav-QDots allowed direct assessment of the presence of endogenous nAChRs in native tissue. Such a labeling approach was later used by Fernandes et al. to investigate the lateral diffusion of endogenous nAChRs on the surface of chick ciliary ganglion neurons.65 In a study using the presynaptic dopamine transporter (DAT) expressed in live HeLa and HEK-293 Flp-In cells, Kovtun et al. targeted DAT with a biotinylated ligand, 2-β-carbomethoxy-3-β-(4-fluorophenyl)tropane (IDT444), that can be bound by Sav-QDots (Figure 7).48 The lack of an efficient antibody against the extracellular DAT epitope required that fairly sophisticated organic chemistry be employed to facilitate QDot labeling of DAT, whereby the cocaine analogue β-CFT was conjugated to a biotinylated, flexible linker arm to enable subsequent QDot recognition. QDot-IDT444 probes were demonstrated to possess high affinity for DAT, as evidenced by the IC50 (half-maximal inhibitory concentration) value of roughly 50 nM. The specific interaction of these QDot probes with the DAT primary binding site was completely blocked by high-affinity DAT antagonist, GBR12909. Moreover, the binding interaction was shown to be virtually irreversible, as DAT-bound QDots were not readily displaced after prolonged exposure of QDot-labeled cell cultures to cocaine derivatives and GBR12909. In addition, QDot-labeled membrane DATs were demonstrated to undergo acute internalization in response to protein kinase C (PKC) activation with phorbol 12-myristate 13-acetate (PMA). PKC-mediated phosphorylation of QDot-labeled DATs led to endosomal accumulation of DAT-QDot complexes as evidenced by the appearance of punctate intracellular fluorescence after 30 minutes of PMA incubation (Figure 7-B1). In contrast, the QDot-associated fluorescence remained predominantly on the cell surface in the control group exposed to dimethyl sulfoxide (DMSO/vehicle) only (Figure 7-A1). In a follow-up set of experiments, Kovtun et al. took advantage of excellent specificity, high affinity, and temporally stable binding interaction of these antagonist-conjugated QDots to develop a QDot-based DAT binding assay.66 The use of flow cytometry enabled rapid, multiwell analysis of large QDot-labeled cell populations at the single-cell level. As a proof-of-concept, the QDot-based DAT binding assay was used to accurately measure inhibitory action of known DAT modulators; dose-response curves of median QDot fluorescence intensity per cell were generated for titrations of DAT-expressing HEK293 cells with PMA and GBR12909. Kovtun et al. anticipated that the reported QDot-based binding assay would be of immediate value to the high-throughput screening (HTS) of novel small-molecule modulators of DAT function.
Figure 7.
Labeling of dopamine transporter (DAT) with ligand conjugated-QDots in live HeLa cells. (Left) Streptavidin-conjugated QDots were used to label DATs previously exposed to a biotinylated, PEGylated cocaine analog. (Upright) chemical structure of the DAT ligand used in the study. (A1) QDot labeling of membrane DATs in a live HeLa cell. (B1) QDot-bound DATs underwent acute redistribution from the plasma membrane to intracellular compartments as a result of protein kinase C (PKC) activation. (Reprinted with permission from Ref 48. Copyright 2011 American Chemical Society)
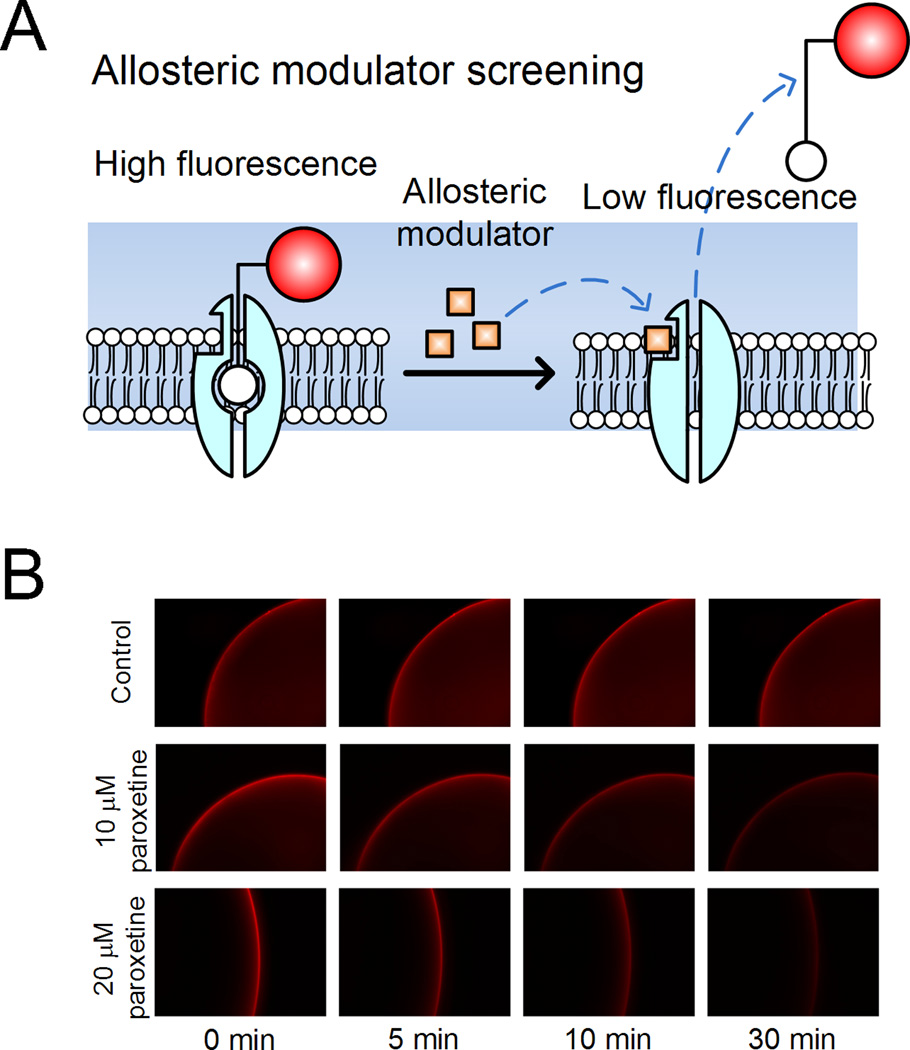
Recently, Chang et al. went a step further to adapt the QDot nanoconjugates for antidepressant drug screening (Figure 8).47 Our approach targeted human SERT (hSERT), the primary target for drugs used to treat major depression. A custom-synthesized, QDot-tagged indoleamine derivative that is structurally similar to 5-HT and that can access the primary neurotransmitter binding site of hSERT was employed to demonstrate that interactions between QDot-tagged ligand and hSERT could be displaced by other antidepressants, providing an opportunity for a screen for allosteric, hSERT-targeted small molecules. Another area of ongoing investigation surrounding SERTs in our group concerns the membrane dynamics of single SERTs in response to their natural signaling stimuli. In a study using serotonergic RN46A cells, Chang et al. implement the ligand-conjugated QDot labeling approach to monitor single SERT proteins.67 Two modes of SERT membrane dynamics were identified in the study, in which the majority of QDot-labeled single SERTs were constrained in cholesterol-rich membrane microdomains (often referred to as lipid rafts), and a small fraction showed relatively free diffusion in the plasma membrane. The diffusion dynamics of single SERTs and the relationship of these dynamics to membrane compartments and regulatory stimuli, including cGMP and IL-1 were also investigated. Notably, single SERTs activated by 8-bromo-cGMP and IL-1 remain co-localized with membrane microdomains, leading to the hypothesis that cholesterol-rich membrane microdomains may have essential roles in SERT regulation. This could be the first direct observation of single SERT membrane dynamics in real-time in a living cell.
Figure 8.
Fluorescence displacement assay based on ligand-conjugated QDots for the drug discovery of allosteric antidepressants. (A) Targeted hSERTs bind to the QDot-tagged ligands, forming complexes that increase fluorescent signal along the membrane. When exposed to a potential drug that induces a conformational change in the binding site, the QDot-tagged ligands are displaced resulting in a decrease in fluorescence intensity. (B) Representative time-lapse fluorescent images show the effect of paroxetine on lhigand-hSERT displacement in the presence of PBS buffer (control), 10 µM, and 20 µM paroxetine. (Reprinted with permission from Ref 62. Copyright 2011 American Chemical Society)
CONCLUSIONS AND FUTURE PERSPECTIVES
In the past two decades, technological advances in QDot synthesis have advanced opportunities for fluorescence-based biological imaging.14 Currently available QDot probes are characterized by minimal cytotoxicity, improved stability in biological environments, and ultra-low non-specific binding. Most importantly, advances in surface chemistry have allowed for the preservation of key QDot optical properties, including high quantum yield, large Stokes shift, and narrow fluorescence emission spectra. These advances have prompted a significant increase in the use of Qdots by molecular and cellular neuroscientists. The most important achievement of QDot technology in molecular neuroscience is likely to be the use of this approach for single-QDot tracking techniques to investigate membrane protein dynamics in neuronal cells at the single-molecule level.68, 69 We mentioned previously that Dahan and coworkers were the first to utilize this tool to interrogate the diffusion dynamics of individual glycine receptors.56 Similar approaches were subsequently employed to investigate various neuronal signaling related targets including nerve growth factor (NGF),63 glial fibrillary acidic protein,70 and gamma-aminobutyric acid A receptor (GABAAR).60 In addition, several solutions have been suggested to overcome the blinking shortcoming for Qdot tracking experiments, including single QDot blinking suppression,28 and synthesis of non-blinking colloidal QDots.30 However, rendering the newly generated QDots biocompatible and readily adaptable to neuroscience research is not a trivial matter, requiring an integration of multiple disciplines, including chemistry, material science, and neurobiology. With this in mind, we expect to witness increasing collaboration among various scientific disciplines towards the pursuit of better QDot reporters for the field of neuroscience.
Another promising application of QDot nanoconjugates in molecular neuroscience is exertion of specific physiological effects as a result of the interaction with protein targets and neuronal cells and subsequent uncovering of the underlying neuronal signaling mechanisms. However, current QDot applications in neuroscience are mostly limited to revealing the position of the protein target without further extending into the use of QDot nanoconjugates for stimulation of cellular responses/processes. The potential of this approach has recently been demonstrated through the use of QDot nanoconjugates functionalized with biologically active organic ligands and small peptides. Specifically, our serotonin-conjugated QDots displayed target-specific serotonin transporter labeling and, importantly, effectively retained pharmacological and physiological properties of native serotonin.41 Another fine example was demonstrated by Vu et al. wherein NGF-functionalized QDots were employed to stimulate the neuronal differentiation in PC12 cells.71
In conclusion, as with any new technology, there is always a period of time before the technology matures and begins to bear fruit. To fully explore the tremendous potential that QDots offer to the field of neuroscience, collaborations of researchers representing a diverse range of disciplines, including chemistry, material science, neuroscience, pharmacology, and medicine, are required. With the continuous advances in QDot synthesis in parallel with the improvements in QDot bioconjugation protocols, we envision it will not be long before the QDot-based biological labeling techniques are considered routine methods in a standard neuroscience laboratory.
ACKNOWLEDGEMENT
J.C.C. acknowledges a research fellowship from the Vanderbilt Institute of Nanoscale Science and Engineering (VINSE). This work was supported by the US National Institute of Health (EB003728, GM72048, MH07802 and MH094527).
REFERENCES
- 1.Rozental R, Giaume C, Spray DC. Gap junctions in the nervous system. Brain Res Rev. 2000;32:11–15. doi: 10.1016/s0165-0173(99)00095-8. [DOI] [PubMed] [Google Scholar]
- 2.Collingridge GL, Isaac JT, Wang YT. Receptor trafficking and synaptic plasticity. Nat Rev Neurosci. 2004;5:952–962. doi: 10.1038/nrn1556. [DOI] [PubMed] [Google Scholar]
- 3.Blakely RD, Defelice LJ, Galli A. Biogenic amine neurotransmitter transporters: just when you thought you knew them. Physiology. 2005;20:225–231. doi: 10.1152/physiol.00013.2005. [DOI] [PubMed] [Google Scholar]
- 4.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 5.Bastiaens PIH, Pepperkok R. Observing proteins in their natural habitat: the living cell. Trends BiochemSci. 2000;25:631–637. doi: 10.1016/s0968-0004(00)01714-x. [DOI] [PubMed] [Google Scholar]
- 6.Marks KM, Nolan GP. Chemical labeling strategies for cell biology. Nat Methods. 2006;3:591–596. doi: 10.1038/nmeth906. [DOI] [PubMed] [Google Scholar]
- 7.Rosenthal SJ, McBride J, Pennycook SJ, Feldman LC. Synthesis, surface studies, composition and structural characterization of CdSe, core/shell and biologically active nanocrystals. Surf Sci Rep. 2007;62:111–157. doi: 10.1016/j.surfrep.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 9.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 10.Akerman ME, Chan WC, Laakkonen P, Bhatia SN, Ruoslahti E. Nanocrystal targeting in vivo. Proc Natl Acad Sci USA. 2002;99:12617–12621. doi: 10.1073/pnas.152463399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao XH, Cui YY, Levenson RM, Chung LWK, Nie SM. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, et al. Near-infrared fluorescent type II quantum dots for sentinel lymph node mapping. Nat Biotechnol. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenthal SJ, Chang JC, Kovtun O, McBride JR, Tomlinson ID. Biocompatible quantum dots for biological applications. Chem Biol. 2011;18:10–24. doi: 10.1016/j.chembiol.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu WW, Qu LH, Guo WZ, Peng XG. Experimental determination of the extinction coefficient of CdTe, CdSe, and CdS nanocrystals. Chem Mater. 2003;15:2854–2860. [Google Scholar]
- 16.Dabbousi BO, RodriguezViejo J, Mikulec FV, Heine JR, Mattoussi H, Ober R, et al. (CdSe)ZnS core-shell quantum dots: Synthesis and characterization of a size series of highly luminescent nanocrystallites. J Phys Chem B. 1997;101:9463–9475. [Google Scholar]
- 17.Murray CB, Norris DJ, Bawendi MG. Synthesis and characterization of nearly monodisperse CdE (E = sulfur, selenium, tellurium) semiconductor nanocrystallites. J Am Chem Soc. 1993;115:8706–8715. [Google Scholar]
- 18.Talapin DV, Rogach AL, Kornowski A, Haase M, Weller H. Highly luminescent monodisperse CdSe and CdSe/ZnS nanocrystals synthesized in a hexadecylamine-trioctylphosphine oxide-trioctylphospine mixture. Nano Lett. 2001;1:207–211. doi: 10.1021/nl0155126. [DOI] [PubMed] [Google Scholar]
- 19.Bharali DJ, Lucey DW, Jayakumar H, Pudavar HE, Prasad PN. Folate-receptor-mediated delivery of InP quantum dots for bioimaging using confocal and two-photon microscopy. J Am Chem Soc. 2005;127:11364–11371. doi: 10.1021/ja051455x. [DOI] [PubMed] [Google Scholar]
- 20.Kucur E, Boldt FM, Cavaliere-Jaricot S, Ziegler J, Nann T. Quantitative analysis of cadmium selenide nanocrystal concentration by comparative techniques. Anal Chem. 2007;79:8987–8993. doi: 10.1021/ac0715064. [DOI] [PubMed] [Google Scholar]
- 21.Chan WCW, Maxwell DJ, Gao XH, Bailey RE, Han MY, Nie SM. Luminescent quantum dots for multiplexed biological detection and imaging. Curr Opin Biotechnol. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 22.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 23.Stroh M, Zimmer JP, Duda DG, Levchenko TS, Cohen KS, Brown EB, et al. Quantum dots spectrally distinguish multiple species within the tumor milieu in vivo. Nat Med. 2005;11:678–682. doi: 10.1038/nm1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chattopadhyay PK, Price DA, Harper TF, Betts MR, Yu J, Gostick E, et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat Med. 2006;12:972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 25.Veenstra-VanderWeele J, Muller CL, Iwamoto H, Sauer JE, Owens WA, Shah CR, et al. Autism gene variant causes hyperserotonemia, serotonin receptor hypersensitivity, social impairment and repetitive behavior. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1112345109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum dots versus organic dyes as fluorescent labels. Nat Methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 27.Nirmal M, Dabbousi BO, Bawendi MG, Macklin JJ, Trautman JK, Harris TD, et al. Fluorescence intermittency in single cadmium selenide nanocrystals. Nature. 1996;383:802–804. [Google Scholar]
- 28.Frantsuzov PA, Volkan-Kacso S, Janko B. Model of fluorescence intermittency of single colloidal semiconductor quantum dots using multiple recombination centers. Phys Rev Lett. 2009;103:207402. doi: 10.1103/PhysRevLett.103.207402. [DOI] [PubMed] [Google Scholar]
- 29.Mahler B, Spinicelli P, Buil S, Quelin X, Hermier JP, Dubertret B. Towards non-blinking colloidal quantum dots. Nat Mater. 2008;7:659–664. doi: 10.1038/nmat2222. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Ren X, Kahen K, Hahn MA, Rajeswaran M, Maccagnano-Zacher S, et al. Non-blinking semiconductor nanocrystals. Nature. 2009;459:686–689. doi: 10.1038/nature08072. [DOI] [PubMed] [Google Scholar]
- 31.Yao J, Larson DR, Vishwasrao HD, Zipfel WR, Webb WW. Blinking and nonradiant dark fraction of water-soluble quantum dots in aqueous solution. Proc Natl Acad Sci USA. 2005;102:14284–14289. doi: 10.1073/pnas.0506523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 33.Wu XY, Liu HJ, Liu JQ, Haley KN, Treadway JA, Larson JP, et al. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol. 2003;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 34.Pellegrino T, Manna L, Kudera S, Liedl T, Koktysh D, Rogach AL, et al. Hydrophobic nanocrystals coated with an amphiphilic polymer shell: A general route to water soluble nanocrystals. Nano Lett. 2004;4:703–707. [Google Scholar]
- 35.Ow H, Larson DR, Srivastava M, Baird BA, Webb WW, Wiesner U. Bright and stable core-shell fluorescent silica nanoparticles. Nano Lett. 2005;5:113–117. doi: 10.1021/nl0482478. [DOI] [PubMed] [Google Scholar]
- 36.Selvan ST, Tan TT, Ying JY. Robust, non-cytotoxic, silica-coated CdSe quantum dots with efficient photoluminescence. Adv Mater. 2005;17:1620. [Google Scholar]
- 37.Nann T, Mulvaney P. Single quantum dots in spherical silica particles. Angew Chem, Int Ed. 2004;43:5393–5396. doi: 10.1002/anie.200460752. [DOI] [PubMed] [Google Scholar]
- 38.Mattoussi H, Mauro JM, Goldman ER, Anderson GP, Sundar VC, Mikulec FV, et al. Self-assembly of CdSe-ZnS quantum dot bioconjugates using an engineered recombinant protein. J Am Chem Soc. 2000;122:12142–12150. [Google Scholar]
- 39.Kim S, Bawendi MG. Oligomeric ligands for luminescent and stable nanocrystal quantum dots. J Am Chem Soc. 2003;125:14652–14653. doi: 10.1021/ja0368094. [DOI] [PubMed] [Google Scholar]
- 40.Pathak S, Davidson MC, Silva GA. Characterization of the functional binding properties of antibody conjugated quantum dots. Nano Lett. 2007;7:1839–1845. doi: 10.1021/nl062706i. [DOI] [PubMed] [Google Scholar]
- 41.Rosenthal SJ, Tomlinson I, Adkins EM, Schroeter S, Adams S, Swafford L, et al. Targeting cell surface receptors with ligand-conjugated nanocrystals. J Am Chem Soc. 2002;124:4586–4594. doi: 10.1021/ja003486s. [DOI] [PubMed] [Google Scholar]
- 42.Goldman ER, Balighian ED, Mattoussi H, Kuno MK, Mauro JM, Tran PT, et al. Avidin: a natural bridge for quantum dot-antibody conjugates. J Am Chem Soc. 2002;124:6378–6382. doi: 10.1021/ja0125570. [DOI] [PubMed] [Google Scholar]
- 43.Wilchek M, Bayer EA. Introduction to avidin-biotin technology. Method Enzymol. 1990;184:5–13. doi: 10.1016/0076-6879(90)84256-g. [DOI] [PubMed] [Google Scholar]
- 44.Chaiet L, Wolf FJ. The properties of streptavidin, a biotin-binding protein produced by Streptomycetes. Arch Biochem Biophys. 1964;106:1–5. doi: 10.1016/0003-9861(64)90150-x. [DOI] [PubMed] [Google Scholar]
- 45.Weber PC, Ohlendorf DH, Wendoloski JJ, Salemme FR. Structural origins of high-affinity biotin binding to streptavidin. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 46.Orndorff RL, Warnement MR, Mason JN, Blakely RD, Rosenthal SJ. Quantum dot ex vivo labeling of neuromuscular synapses. Nano Lett. 2008;8:780–785. doi: 10.1021/nl072460x. [DOI] [PubMed] [Google Scholar]
- 47.Chang JC, Tomlinson ID, Warnement MR, Iwamoto H, DeFelice LJ, Blakely RD, et al. A fluorescence displacement assay for antidepressant drug discovery based on ligand-conjugated quantum dots. J Am Chem Soc. 2011;133:17528–17531. doi: 10.1021/ja204301g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kovtun O, Tomlinson ID, Sakrikar DS, Chang JC, Blakely RD, Rosenthal SJ. Visualization of the cocaine-sensitive dopamine transporter with ligand-conjugated quantum dots. ACS Chem Neurosci. 2011;2:370–378. doi: 10.1021/cn200032r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaiswal JK, Simon SM. Potentials and pitfalls of fluorescent quantum dots for biological imaging. Trends Cell Biol. 2004;14:497–504. doi: 10.1016/j.tcb.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 50.Howarth M, Chinnapen DJ, Gerrow K, Dorrestein PC, Grandy MR, Kelleher NL, et al. A monovalent streptavidin with a single femtomolar biotin binding site. Nat Methods. 2006;3:267–273. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howarth M, Liu W, Puthenveetil S, Zheng Y, Marshall LF, Schmidt MM, et al. Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nat Methods. 2008;5:397–399. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bentzen EL, Tomlinson ID, Mason J, Gresch P, Warnement MR, Wright D, et al. Surface modification to reduce nonspecific binding of quantum dots in live cell assays. Bioconjug Chem. 2005;16:1488–1494. doi: 10.1021/bc0502006. [DOI] [PubMed] [Google Scholar]
- 53.Gussin HA, Tomlinson ID, Muni NJ, Little DM, Qian H, Rosenthal SJ, et al. GABAC receptor binding of quantum-dot conjugates of variable ligand valency. Bioconjug Chem. 2010;21:1455–1464. doi: 10.1021/bc100050s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gussin HA, Tomlinson ID, Little DM, Warnement MR, Qian HH, Rosenthal SJ, et al. Binding of muscimol-conjugated quantum dots to GABA(c) receptors. J Am Chem Soc. 2006;128:15701–15713. doi: 10.1021/ja064324k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clarke SJ, Hollmann CA, Zhang Z, Suffern D, Bradforth SE, Dimitrijevic NM, et al. Photophysics of dopamine-modified quantum dots and effects on biological systems. Nat Mater. 2006;5:409–417. doi: 10.1038/nmat1631. [DOI] [PubMed] [Google Scholar]
- 56.Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302:442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 57.Levi S, Schweizer C, Bannai H, Pascual O, Charrier C, Triller A. Homeostatic regulation of synaptic GlyR numbers driven by lateral diffusion. Neuron. 2008;59:261–273. doi: 10.1016/j.neuron.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 58.Charrier C, Machado P, Tweedie-Cullen RY, Rutishauser D, Mansuy IM, Triller A. A crosstalk between b1 and b3 integrins controls glycine receptor and gephyrin trafficking at synapses. Nat Neurosci. 2010;13:1388–1395. doi: 10.1038/nn.2645. [DOI] [PubMed] [Google Scholar]
- 59.Bouzigues C, Morel M, Triller A, Dahan M. Asymmetric redistribution of GABA receptors during GABA gradient sensing by nerve growth cones analyzed by single quantum dot imaging. Proc Natl Acad Sci USA. 2007;104:11251–1126. doi: 10.1073/pnas.0702536104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bannai H, Levi S, Schweizer C, Inoue T, Launey T, Racine V, et al. Activity-dependent tuning of inhibitory neurotransmission based on GABAAR diffusion dynamics. Neuron. 2009;62:670–682. doi: 10.1016/j.neuron.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 61.Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 62.Howarth M, Takao K, Hayashi Y, Ting AY. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc Natl Acad Sci USA. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui B, Wu C, Chen L, Ramirez A, Bearer EL, Li W-P, et al. One at a time, live tracking of NGF axonal transport using quantum dots. Proc Natl Acad Sci USA. 2007;104:13666–13671. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fichter KM, Flajolet M, Greengard P, Vu TQ. Kinetics of G-protein-coupled receptor endosomal trafficking pathways revealed by single quantum dots. Proc Natl Acad Sci USA. 2010;107:18658–18663. doi: 10.1073/pnas.1013763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ehrensperger M-V, Hanus C, Vannier C, Triller A, Dahan M. Multiple association states between glycine receptors and gephyrin identified by SPT analysis. Biophys J. 2007;92:3706–3718. doi: 10.1529/biophysj.106.095596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kovtun O, Ross EJ, Tomlinson ID, Rosenthal SJ. A flow cytometry-based dopamine transporter binding assay using antagonist-conjugated quantum dots. Chem Commun. 2012 doi: 10.1039/c2cc31951a. [DOI] [PubMed] [Google Scholar]
- 67.Chang JC, Tomlinson ID, Warnement MR, Ustione A, Carneiro AMD, Piston DW, et al. Single molecule analysis of serotonin transporter regulation using antagonist-conjugated quantum dots reveals restricted, p38 MAPK-dependent mobilization underlying uptake activation. J Neurosci. 2012;32:8919–8929. doi: 10.1523/JNEUROSCI.0048-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pinaud F, Clarke S, Sittner A, Dahan M. Probing cellular events, one quantum dot at a time. Nat Methods. 2010;7:275–285. doi: 10.1038/nmeth.1444. [DOI] [PubMed] [Google Scholar]
- 69.Chang Y-P, Pinaud F, Antelman J, Weiss S. Tracking bio-molecules in live cells using quantum dots. J Biophotonics. 2008;1:287–298. doi: 10.1002/jbio.200810029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pathak S, Cao E, Davidson MC, Jin S, Silva GA. Quantum dot applications to neuroscience: new tools for probing neurons and glia. J Neurosci. 2006;26:1893–1895. doi: 10.1523/JNEUROSCI.3847-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sundara Rajan S, Vu TQ. Quantum dots monitor TrkA receptor dynamics in the interior of neural PC12 cells. Nano Lett. 2006;6:2049–2059. doi: 10.1021/nl0612650. [DOI] [PubMed] [Google Scholar]
- 72.Muir J, Arancibia-Carcamo IL, MacAskill AF, Smith KR, Griffin LD, Kittler JT. NMDA receptors regulate GABAA receptor lateral mobility and clustering at inhibitory synapses through serine 327 on the gamma2 subunit. Proc Natl Acad Sci USA. 2010;107:16679–16684. doi: 10.1073/pnas.1000589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yeow EKL, Clayton AHA. Enumeration of oligomerization states of membrane proteins in living cells by homo-FRET spectroscopy and microscopy: Theory and application. Biophys J. 2007;92:3098–3104. doi: 10.1529/biophysj.106.099424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rajan SS, Liu HY, Vu TQ. Ligand-bound quantum dot probes for studying the molecular scale dynamics of receptor endocytic trafficking in live cells. ACS Nano. 2008;2:1153–1166. doi: 10.1021/nn700399e. [DOI] [PubMed] [Google Scholar]
- 75.Bats C, Groc L, Choquet D. The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron. 2007;53:719–734. doi: 10.1016/j.neuron.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 76.Mikasova L, Groc L, Choquet D, Manzoni OJ. Altered surface trafficking of presynaptic cannabinoid type 1 receptor in and out synaptic terminals parallels receptor desensitization. Proc Natl Acad Sci USA. 2008;105:18596–18601. doi: 10.1073/pnas.0805959105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Michaluk P, Mikasova L, Groc L, Frischknecht R, Choquet D, Kaczmarek L. Matrix metalloproteinase-9 controls NMDA receptor surface diffusion through integrin b1 signaling. J Neurosci. 2009;29:6007–6012. doi: 10.1523/JNEUROSCI.5346-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Renner M, Lacor PN, Velasco PT, Xu J, Contractor A, Klein WL, et al. Deleterious effects of amyloid beta oligomers acting as an extracellular scaffold for mGluR5. Neuron. 2010;66:739–754. doi: 10.1016/j.neuron.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fernandes CC, Berg DK, Gomez-Varela D. Lateral mobility of nicotinic acetylcholine receptors on neurons is determined by receptor composition, local domain, and cell type. J Neurosci. 2010;30:8841–8851. doi: 10.1523/JNEUROSCI.6236-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tomlinson ID, Mason JN, Blakely RD, Rosenthal SJ. Peptide-conjugated quantum dots: imaging the angiotensin type 1 receptor in living cells. Methods Mol Biol. 2005;303:51–60. doi: 10.1385/1-59259-901-X:051. [DOI] [PubMed] [Google Scholar]
- 81.Mercer AJ, Chen M, Thoreson WB. Lateral mobility of presynaptic L-type calcium channels at photoreceptor ribbon synapses. J Neurosci. 2011;31:4397–4406. doi: 10.1523/JNEUROSCI.5921-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang S, Liu AP, Duan HW, Soo J, Chen P. Labeling and tracking P2 purinergic receptors in living cells using ATP-conjugated quantum dots. Adv Funct Mater. 2011;21:2776–2780. [Google Scholar]