Abstract
Chemotactic and haptotactic cues guide neurite growth toward appropriate targets by eliciting attractive or repulsive responses from the neurite growth cones. Here we present an integrated system allowing both structural and molecular micropatterning in dual hydrogel 3D tissue culture constructs for directing in vitro neuronal growth via structural, immobilized, and soluble guidance cues. These tissue culture constructs were fabricated into specifiable geometries using UV light reflected from a digital micromirror device acting as a dynamic photomask, resulting in dual hydrogel constructs consisting of a cell growth-restrictive polyethylene glycol (PEG) boundary with a cell growth-permissive interior of photolabile α-carboxy-2-nitrobenzyl cysteine agarose (CNBC-A). This CNBC-A was irradiated in discrete areas and subsequently tagged with maleimide-conjugated biomolecules. Fluorescent microscopy showed biomolecule binding only at the sites of irradiation in CNBC-A, and confocal microscopy confirmed 3D binding through the depth of the construct. Neurite outgrowth studies showed contained growth throughout CNBC-A. The diffusion rate of soluble fluorescein-bovine serum albumin through the dual hydrogel construct was controlled by PEG concentration and the distance between the protein source and the agarose interior; the timescale for a transient protein gradient changed with these parameters. These findings suggest the dual hydrogel system is a usefulplatform for manipulating a 3D in vitro microenvironment with patterned structural and molecular guidance cues for modeling neural growth and guidance.
Keywords: digital micromirror device, photolithography, nerve guidance, 3D, polyethylene glycol
1. Introduction
Proper mapping of the nervous system during development requires both chemotactic and haptotactic guidance cues along the growth pathway. These cues orient neurites appropriately and direct their growth toward their targets by eliciting attractive or repulsive responses from the growth cones of neurites (Wilkinson 2001; Yu and Bargmann 2001). Understanding these cues and the neurite growth-permissive environment facilitates the development of better therapies for directed neurite regeneration – specifically, guiding axons to their intended synapse targets. The importance of molecular cues for axonal pathfinding is highlighted in the optic tract, where there is binary divergence of the axon bundle to either hemisphere of the brain at the optic chiasm. During the early stages of development, the optic nerve axon bundle extends from the retina following a concentration gradient of Netrin-1, is maintained along the optic tract by repulsive guidance from semaphorin and slit ligands, and, upon reaching a region of immobilized ephrin-B2, diverges into two pathways toward the contralateral and ipsilateral hemispheres of the brain (Williams et al. 2004). Such neural behavior could potentially be examined and manipulated with in vitro 3D tissue culture models in which the microenvironment may be physically and molecularly tuned to obtain a more physiologically accurate milieu and biomimetic cellular arrangement (Weaver et al. 1997; Abbott 2003; Lutolf and Hubbell 2005). In this report, we present a 3D in vitro tissue culture model incorporating both structural and molecular cues in a dual hydrogel system designed for studying neurite growth and guidance.
A wide spectrum of equipment and procedures previously dedicated to the electronics sector have found new applications in cell biology and tissue engineering, employing techniques like rapid prototyping (Dhariwala et al. 2004; Kim et al. 2010; Melchels et al. 2010), molecular printing (Bernard, et al. 2000; Von Philipsborn et al. 2006; Lehmann et al. 2010), and photolithography (Bryant and Anseth 2003; Luo and Shoichet 2004; Luo and Shoichet 2004; Almany and Seliktar 2005; Hahn et al. 2006; Kloxin et al. 2009) to replicate physiological architecture in three dimensions. A common platform for developing novel and application-specific 3D cultures involves the use of hydrogels – hydrophilic, cross-linked polymer networks (Peppas et al. 2000). Hydrogels utilized for 3D scaffolds exhibit excellent biocompatibility, resemble the extracellular matrix in structure and water content, and can often be modified with functional groups or proteins (Peppas et al. 2000; Lee and Mooney 2001; Zhu 2010). This last quality provides commercially available hydrogels with the flexibility to display new, otherwise unattainable, characteristics (Peppas et al. 2000; Bryant and Anseth 2003; Luo and Shoichet 2004; Moon et al. 2007; Kloxin et al. 2009).
Hydrogels functionalized with proteins can present native protein conformations to cells in the scaffold, while the 3D structure allows cells to present receptors as they would in vivo – a distinct advantage over 2D substrates. The discrepancy between cellular behavior in 2D and 3D cultures has been demonstrated convincingly in tumor cell studies (Bissell and Radisky 2001). In one groundbreaking study, breast cancer cells cultured on 2D substrates showed little response to inhibitory β1-integrin antibodies, but the cancer cells reverted from abnormal growth patterns to a nonmalignant phenotype when presented with the same antibodies in a 3D culture (Weaver et al. 1997; Bissell and Radisky 2001). This study concluded that the extracellular environment may influence cell behavior far more heavily than the cell’s own genotype (Weaver et al. 1997). Since the realization that cells are highly responsive to the structure of their environment, other studies have shown the vast differences in cellular arrangement and response between 2D and 3D cultures (Cukierman et al. 2001; Abbott 2003; Lee et al. 2008). Neuronal studies have observed more physiologically aligned electrophysiological and morphological characteristics in neuronal cells cultured in 3D matrices versus 2D substrates (Desai et al. 2006; Irons et al. 2008). A recent study found that a 3D environment may encourage in vivo growth cone morphology, axonal branching pattern, and polarization features of developing sensory neurons, confirming the importance of 3D tissue culture architecture for cellular studies (Ribeiro et al. 2012).
As mentioned before, functionalized hydrogels are popular candidates for recreating 3D extracellular environments. One particularly useful route for modifying hydrogels, either physically or chemically, utilizes ultraviolet (UV) light-sensitive compounds. The synthetic polymer polyethylene glycol (PEG) can be modified with acrylate groups so that it covalently crosslinks into a hydrogel scaffold when irradiated with UV light in the presence of a photoinitiator (Nguyen and West 2002; Zhu 2010). The PEG gel on its own is resistant to protein adsorption and cell adhesion (Zalipsky and Harris 1997), but many studies have exploited its “blank slate” quality and developed custom gels that are selectively degradable(Kloxin et al. 2009), have adjustable mechanical properties (Bryant and Anseth 2002; Almany and Seliktar 2005; Nemir et al. 2010), contain cell-binding sites (Moon et al. 2007; Lee et al. 2008), or present patterns of proteins for directing cell growth (Hahn et al. 2006). In a similar manner to PEG, the naturally occurring polymer agarose is also extensively used as a 3D hydrogel scaffold due to its minimal cell adhesion properties and capacity for chemical modification (Bellamkonda et al. 1995; Balgude et al. 2001; Luo and Shoichet 2004; Luo and Shoichet 2004). In a study aimed to direct neurite growth through agarose gel, Luo and Shoichet functionalized the free hydroxyl groups of agarose with a modified cysteine that would present free thiol moieties upon UV irradiation (Luo and Shoichet 2004; Luo and Shoichet 2004). To these free thiol moieties, maleimide-activated biomolecules could bind and provide cell-adhesion sites in discrete patterns for directed neurite growth.
By employing some of the photochemical strategies presented above, we have developed an integrated system that allows both structural and molecular 3D micropatterning in dual hydrogel tissue culture constructs. Our dual hydrogel system consists of a photocrosslinkable PEG that surrounds a photolabile agarose gel that can present both immobilized and soluble biomolecules. The PEG gel is commercially available and provides a cell growth-restrictive boundary. The photolabile agarose is modified with α-carboxy-2-nitrobenzyl cysteine, similar to S-2-nitrobenzyl cysteine previously reported to afford photolabile functionality to agarose (Luo and Shoichet 2004). However, the α-carboxy-2-nitrobenzyl leaving group used in the present study has been shown to be less cytotoxic and more water soluble than that of the 2-nitrobenzyl moiety (Pan and Bayley 1997; Walker et al. 1998; Kotzur et al. 2009). A digital micromirror device (DMD) in our photolithography apparatus served as a dynamic mask for irradiating both PEG and the photolabile agarose. The DMD has recently emerged as a potentially high-throughput tool for dynamic patterning of structures and biomolecules on a variety of substrates by projecting UV light to the substrate in distinct, 2D geometries, resulting in a high-resolution extrusion of the mask shape through the depth of the substrate (Lee et al. 2003; Lu et al. 2006; Wang et al. 2009). While this dynamic mask photolithography apparatus can be used in a wide number of applications, in this study it was used to fabricate a dual hydrogel system designed to be a 3D microenvironment prototype as a platform for the study of directed neurite growth in response to patterned structural and molecular guidance cues.
2. Materials and Methods
All chemicals were purchased and used as received from Sigma Aldrich unless otherwise noted.
2.1 Synthesis of α-carboxy-2-nitrobenzyl cysteine agarose
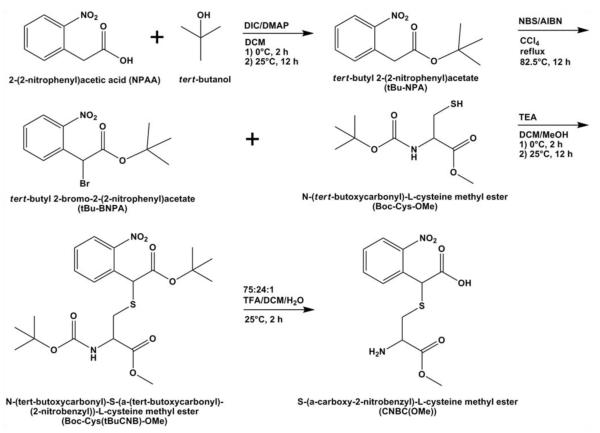
The photolabile agarose was synthesized using a combination of published procedures and inhouse optimization. Figure 1 outlines the synthesis pathway for the photolabile cysteine grafted to agarose. First, the photolabile moiety tert-butyl 2-bromo-2-(2-nitrophenyl)acetate (tBu-BNPA) was synthesized according to a modified protocol (Kotzur et al. 2009). Briefly, 2-(2-nitrophenyl)acetic acid (5.0 g, 27.6 mmol), 4-dimethylaminopyridine (1.85 g, 15.1 mmol), and N,N’diisopropylcarbodiimide (5.2 ml, 33.2 mmol) were dissolved in 50 ml dichloromethane (DCM) and cooled to 0°C. To the reaction mixture, tert-butanol (3.2 ml, 33.7 mmol) was added dropwise, and the reaction mixture was stirred for 2 h at 0°C then kept at room temperature overnight. The solution was washed with 1 M HCl (3×100 ml), 1 M NaOH (3×100 ml), and water (3×100 ml) before filtering over silica gel to obtain the compound tert- butyl 2-(2-nitrophenyl)acetate (tBu-NPA; ~55%; 1H NMR (DMSO-d6): δ 8.1-7.5 (m, 4H, Ar), δ 4.0 (s, 2H, CH2), δ 1.4 (s, 9H, (CH3)3)). The tBu-NPA was dissolved in 100 ml carbon tetrachloride, along with N- bromosuccinimide (5 eq) and azobisisobutyronitrile (0.5 eq). The reaction mixture was purged with nitrogen and refluxed at 82.5°C overnight. The solution was filtered over silica gel to obtain the product tBu-BNPA (~47%; 1H NMR (DMSO-d6): δ 8.1-7.7 (m, 4H, Ar), δ 6.3 (s, 1H, CH), δ 1.4 (s, 9H, (CH3)3)).
Fig. 1.
Synthesis pathway for photoreactive cysteine (CNBC(OMe)). Abbreviations: DIC = N,N’diisopropylcarbodiimide; DMAP = 4-dimethylaminopyridine; DCM = dichloromethane; NBS = N-bromosuccinimide; AIBN = azobisisobutyronitrile; TEA = triethylamine; MeOH = methanol; TFA = trifluoroacetic acid
The tBu-BNPA compound was used to protect the thiol moiety of cysteine using a modified protocol [20]. Briefly, tBu-BNPA was dissolved in 10 ml DCM and N-(tert-butoxycarbonyl)-L-cysteine methyl ester (Boc-Cys-OMe; 2 eq) was dissolved in 30 ml DCM. Both solutions were chilled to 0°C. Separately, triethylamine (1.2 eq) was dissolved in 5 ml DCM. The tBu-BNPA solution was added dropwise to the Boc-Cys-OMe solution, followed by dropwise addition of the triethylamine solution. The reaction was stirred at 0°C for 2 h then kept at room temperature overnight. The solution was filtered over silica gel and the product N-(tert-butoxycarbonyl)-S-(α-(tert-butoxycarbonyl)-(2-nitrobenzyl))-L-cysteine methyl ester (Boc-Cys(tBuCNB)-OMe) was isolated (~45%; 1H NMR (DMSO-d6): δ 8.0-7.6 (m, 4H, Ar), δ 7.4-7.0 (m, 1H, NH), δ 5.2 (d, 1H, ArCH), δ 4.2 (d, 1H, CH), 3.6 δ (t, 3H, OCH3), 3.2-2.9 (m, 2H, CH2), δ 1.4 (s, 18H, 2(CH3)3)). To prepare this modified cysteine for conjugation to agarose, Boc- Cys(tBuCNB)- OMe was dissolved in 75:24:1 trifluoroacetic acid/DCM/water and stirred for 2 h at room temperature to remove the tert-butyl/Boc protective groups. The solvent was removed by reduced pressure and the cysteine residue was dissolved in 5 ml glacial acetic acid prior to lyophilization to yield S-(α-carboxy-2- nitrobenzyl)-L-cysteine methyl ester (CNBC(OMe); >99%; 1H NMR (DMSO-d6): δ 8.1-7.6 (m, 4H, Ar), δ 4.2 (d, 1H, CH), δ 4.0 (d, 1H, ArCH), δ 3.6 (t, 3H, OCH3), δ 2.9-2.8 (m, 2H, CH2)).
The CNBC(OMe) compound was then conjugated to ultra-low gelling temperature (ULGT) agarose according to a published procedure (Luo and Shoichet 2004). For the agarose solution, 0.1 g of ULGT agarose was dissolved in 7 ml DMSO by heating. The coupling agent 1,1′-carbonyldiimidazole (0.04 g, 0.25 mmol) was dissolved in 2 ml DMSO then added dropwise to the agarose solution. The reaction was stirred at room temperature for 1 h. A solution of CNBC(OMe) (0.079 g, 0.25 mmol) dissolved in 2 ml DMSO was added dropwise to the activated agarose solution. This reaction was stirred at room temperature overnight in the dark. The CNBC(OMe)-agarose (CNBC-A) solution was dialyzed against water for 2 d then lyophilized for 3 d. UV-VIS spectrophotometry was used to evaluate the grafting density of CNBC(OMe) to ULGT agarose by measuring the absorbance at 260 nm of 0.1% wt/vol CNBC-A against a standard curve of CNBC(OMe). The grafting density of CNBC(OMe) molecules to ULGT agarose monomers was determined to be 10.7%.
2.2 Dual hydrogel system
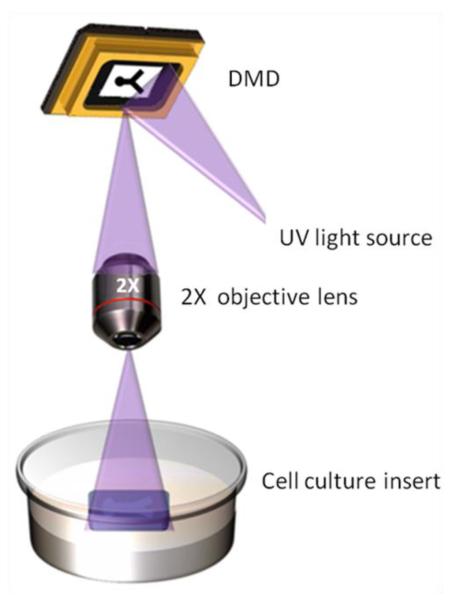
Both hydrogel and molecular micropatterning were performed with a dynamic mask projection photolithography apparatus consisting of a UV light source (OmniCure 1000 with 320-500 nm filter, EXFO, Quebec, Canada) with collimating adapter (EXFO), a DMD as a dynamic photomask (Discovery™ 3000, Texas Instruments, Dallas, TX), and a 2X objective lens (Plan Fluor, Nikon Instruments, Tokyo, Japan) (Fig. 2). For all experiments, the plane of focus of the UV light was the surface of the cell culture insert membrane, resulting in bulk irradiation through the depth of the photoreactive material. The intensity of the UV light at this plane of focus is 181 mW/cm2, as measured by a radiometer (306 UV Powermeter, Optical Associates, San Jose, CA). The 3D dual hydrogel system consisted of a photolabile agarose gel bound by a PEG gel construct on permeable cell culture inserts and fabricated using a published procedure (Curley et al. 2011; Curley and Moore 2011). Briefly, the walls of 6-well polyester cell culture inserts with a diameter of 24 mm and pore size of either 0.4 μm (Transwell®, Corning Inc., Corning, NY) or 1.0 μm (Falcon®, Becton Dickinson Labware, Franklin Lakes, NJ) were treated with Rain-X (SOPUS Products, Houston, TX) to minimize meniscus formation. A photocrosslinkable solution of 10% w/v PEG-diacrylate (MW=1000; Polysciences Inc., Warrington, PA) in PBS with 0.5% Irgacure 2959 photoinitiator (Ciba Specialty Chemicals, Basel, Switzerland) was added to these treated inserts (500 μl per insert). The solution was irradiated with UV light by the photolithography apparatus for 55 sec per construct. Uncrosslinked PEG solution was washed away with PBS.
Fig. 2.
Dynamic mask projection photolithography. The apparatus consists of a digital micromirror device (DMD), a collimated UV light source, and a 2X objective lens. UV light is reflected from the DMD and projected through the objective lens to the substrate, such as a cell culture insert
For each 6-well cell culture insert containing PEG constructs, 1% w/v CNBC-A in PBS was dissolved by heating and sonicating for 30 min and then approximately 1 μl CNBC-A solution was injected into the void of each PEG construct via micropipette. The inserts were placed in 6-well plates and 1.5 ml PBS was added to each well. The plates were chilled at 4°C for 1 h to allow for CNBC-A gelation. Exposure of the photolabile agarose gel to ambient light was minimized by keeping the plates covered in aluminum foil during all steps.
2.3 Biomolecule immobilization within CNBC-A
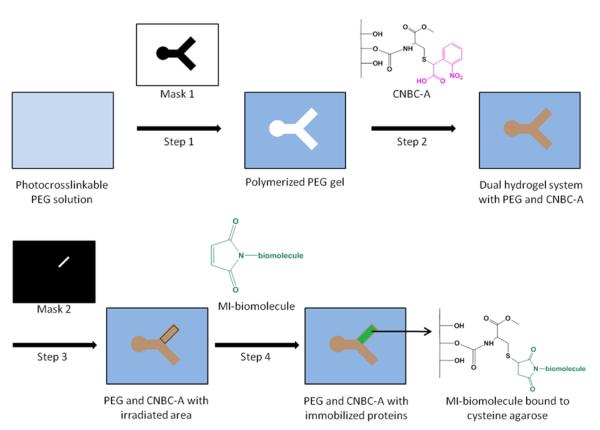
Biomolecule immobilization within select regions of the dual hydrogel constructs was accomplished by irradiating with a second photomask followed by a series of incubation and washing steps. Figure 3 provides an overview of the fabrication process for the dual hydrogel system with immobilized proteins. First, discrete areas of CNBC-A within the PEG construct were irradiated with a second set of photomasks for 180 s per photomask. After irradiation, the gels were washed for 1 h with 1% bovine serum albumin (BSA) in PBS to block nonspecific protein adsorption. The gels were then incubated in a solution of either maleimide-conjugated NeutrAvidin or maleimide-conjugated biotin, and each subsequently tagged with its respective fluorescent complement. For maleimide-biotin immobilization, gels were incubated in 1.5 ml of a 9.5 mM solution of EZ-link Maleimide-PEG2-Biotin (MI-biotin; Thermo Scientific, Rockford, IL) in 5 mM EDTA buffer for 4 h at room temperature. The gels were washed thoroughly with 1% BSA/0.1% Tween 20 in PBS at room temperature, incubated in 1.5 ml of a 13 μM solution of fluorescein-conjugated NeutrAvidin (FITC-NeutrAvidin; Thermo Scientific) in 5 mM EDTA buffer for 4 h at room temperature, and washed thoroughly again with 1% BSA/0.1% Tween 20 solution at room temperature. For maleimide-NeutrAvidin immobilization to CNBC-A, the same procedure for binding MI-biotin was carried out, with the substitution of 50 μM solution of maleimide-activated NeutrAvidin (MI-NeutrAvidin; Thermo Scientific) for MI-biotin, and 0.3 mM solution of the biotin analog Oregon Green 488 biocytin (Molecular Probes) for FITC-NeutrAvidin. For dual labeling of the CNBC-A, this MI-NeutrAvidin binding scheme was repeated in the same construct and tagged with Alexa Fluor 594 biocytin (Molecular Probes). Gradients were produced using sequential masks, either in-focus at the insert membrane or out-of-focus, and irradiation times of 1, 2, 4, 8, 16, and 32 s. The same protocol for labeling the irradiated regions with MI-NeutrAvidin followed by Oregon Green 488 biocytin was followed. The concentrations of immobilized biocytin in the gradient resulting from the out-of-focus masks were quantified against a standard curve of biocytin in PBS in wells of PEG constructs of the same thickness as the dual hydrogel constructs. The relative fluorescence of a 160-pixel long line drawn across the gradient was binned into 10-pixel bins, resulting in 16 total sample points of average fluorescence along the irradiated region.
Fig. 3.
General scheme for fabricating the dual hydrogel system and subsequent protein immobilization. Step 1: A cell culture insert is filled with photocrosslinkable PEG solution. Using Mask 1, PEG solution is crosslinked via UV irradiation and uncrosslinked PEG is removed. Step 2: The void left by Mask 1 is filled with CNBC-A solution. CNBC-A is gelled by cooling. The photocleavable moiety on CNBC-A (pink) cages the thiol on the cysteine. Step 3: After gelation by cooling, CNBC-A is irradiated with UV using Mask 2. Irradiation uncages thiol moieties on CNBC-A to which maleimide-activated (MI) biomolecules can bind. The black rectangular outline indicates the irradiated region. Step 4: The dual hydrogel system is washed with MI-biomolecule solution and tagged with fluorescent markers, resulting in a region of MI-biomolecules bound to cysteine agarose
To test the resolution limits of the dynamic mask projection photolithography apparatus for biomolecule binding, small-sized photomasks were used for irradiating bulk CNBC-A gel. Rain-X–treated 6-well cell culture inserts (1.0 μm pore size) were filled with 500 μl of 1% w/v CNBC-A solution in PBS then incubated at 4°C for 1 h. The bulk CNBC-A gel was irradiated for 180 s per photomask using 10- pixel, 8-pixel, 6-pixel, and 4-pixel diameter circular photomasks (henceforth named 10-pixel mask, etc). The gel was washed in 1% BSA for 1 h at room temperature in the dark. To tag the uncaged thiols, approximately 0.5 mg of Texas Red C2 maleimide (MI-Texas Red; Molecular Probes) was dissolved in 1.5 ml of 5 mM EDTA buffer. The gel was incubated in this MI-Texas Red solution for 4 h at room temperature then washed thoroughly with 1% BSA/0.1% Tween 20 in PBS at room temperature.
2.4 Culturing dorsal root ganglia in dual hydrogel constructs
All procedures involving vertebrate animals were approved by the Institutional Animal Care and Use Committee. Dual hydrogel constructs containing live dorsal root ganglion explants were prepared as described previously (Curley et al. 2011; Curley and Moore 2011). Briefly, collagen-coated PTFE cell culture inserts for 6-well plates were soaked overnight in adhesion medium (Neurobasal medium supplemented with L-glutamine, nerve growth factor (NGF), 10% fetal bovine serum (FBS) and penicillin/streptomycin; Gibco-Invitrogen, Carlsbad, CA). Four dorsal root ganglia (DRG) isolated from embryonic day 15 Long-Evans rat pups (Charles River, Wilmington, MA) in adhesion medium were placed on the surface of each cell culture insert membrane and maintained in growth medium (Neurobasal medium supplemented with B27, L-glutamine, nerve growth factor and penicillin/streptomycin) in an incubator (37°C, 5% CO2).
After allowing 2 h for the DRGs to adhere to each insert membrane, the adhesion medium in the insert was replaced with 500 μl 10% PEG/0.5% Irgacure 2959 in growth medium. The time tissue cultures spent outside of the biosafety cabinet was kept minimal for the following polymerization steps in order to prevent contamination of the samples. By aligning the projected photomasks around the adhered DRG using a visible light source and inverted microscope, the PEG constructs of the dual hydrogel system effectively confined the DRG after UV irradiation for 55 s as described in Section 2.2. Inserts were washed with PBS to remove uncrosslinked PEG solution, and a solution of 1% CNBC-A in growth medium was pipetted in the voids of the PEG constructs. To quickly gel the thermosensitive CNBC-A and minimize exposure of the DRG to low temperatures, the insert was set in pre-chilled (4°C) growth medium for 5 min, then transferred to a 6-well plate with 37°C growth medium. The inserts were washed several times with growth medium after polymerization to help prevent contamination. The DRG explants in the dual hydrogel systems were cultured in an incubator for 7 d, with medium changes every 48 h.
To demonstrate neurite viability following biomolecule immobilization, the same steps for binding MI-NeutrAvidin to CNBC-A and subsequent tagging with Oregon Green 488 biocytin (described in Section 2.3) were carried out under sterile conditions the same day the dual hydrogel systems were fabricated around the DRG explants, with the exception that Tween 20 and EDTA were omitted from the wash solution. Culture conditions remained the same as without biomolecule immobilization.
Neurite outgrowth in the dual hydrogel systems was evaluated using standard immunohistochemical techniques. After fixing the DRG in 4% paraformaldehyde for 2 h, cell nuclei were stained with DAPI nucleic acid stain (Molecular Probes, Eugene, OR) and neurites were tagged with mouse monoclonal [2G10] neuron-specific beta III tubulin primary antibody, followed by fluorescent tagging with Cy2-congugated goat antimouse immunoglobulinG (H+L) secondary antibody (AbCam, Cambridge, MA). The staining and tagging steps were carried out in PBS with 0.1% saponin and 0.2% BSA for 1 h, followed by three 10-min washes in PBS with 0.1% saponin. Fluorescent images were acquired with a Nikon AZ100 stereo zoom microscope, and all images were processed with ImageJ software (National Institutes of Health, Bethesda, MD).
2.5 Diffusion of proteins through dual hydrogel constructs
To explore the establishment of a concentration gradient of a soluble protein within the growth-supporting region of the dual hydrogel constructs, a circular well (diameter = 780 μm) that acted as a protein source was positioned next to the bifurcated void in the PEG construct, and diffusion through the gel constructs was monitored over time. This was done by adding a circle to the photomask used for irradiating the photocrosslinkable PEG solution, allowing for both the PEG construct and the circular well to be fabricated simultaneously. The circular well was placed either 115 μm (close) or 545 μm (far) away from the void. The dual hydrogel constructs were fabricated the same way as previously stated, with the exceptions of the void (not the circular well) being filled with 1% w/v unmodified ULGT agarose in PBS instead of CNBC-A and using both 10% to 20% w/v PEG solutions. Once the agarose in the PEG constructs gelled by incubation at 4°C for 1 h, the circular wells were filled with 0.2 μl of 0.01 mg/ml fluorescein-conjugated BSA (fluorescein-BSA; Molecular Probes) in PBS. Fluorescent images were taken at various time points (0, 15, 30, and 60 min) to monitor fluorescein-BSA diffusion into the agarose gel of the dual hydrogel system. Using ImageJ software, the intensity profile for each fluorescent image was measured using a straight, 270-pixel long line drawn across the length of the agarose gel. Pixels were binned in groups of ten pixels per bin to give 27 points of average relative intensity. This relative intensity was averaged over three experiments to give a mean relative intensity and standard deviation per point. Relative intensity was converted to concentration of fluorescein-BSA (μg/ml) by comparison to a calibration curve obtained from images of known concentrations of fluorescein-BSA contained in circular PEG wells with the same thickness as the dual hydrogel constructs.
To sustain a gradient of fluorescein-BSA in a physiological range in the dual hydrogel constructs, the diffusion of the protein was monitored as before, but the well was refilled once during the experiment. Briefly, dual hydrogel constructs using 10% PEG and ULGT agarose were fabricated with the inclusion of a well (diameter = 1115 μm)in the PEG as described previously. This well, positioned 210 μm away from the bifurcating void, was filled with 0.4 μl of 0.005 mg/ml fluorescein-BSA in PBS, and the diffusion monitored over time. The well was refilled after 60 min, and the diffusion monitored for another 60 min (120 min total.) The relative fluorescence was measured using a 300-pixel line, binned into 30 points of average relative intensity, and then converted into concentration (nM). The decay parameter, used as a measure of the gradient slope, was obtained from best-fit exponential curves for each time point and expressed as percent steepness. The equations of the best-fit exponential curves were also used to determine the absolute magnitude of the concentration at the starting position at various time points.
2.6 Characterization of 3D micropatterning
Biotin/NeutrAvidin binding in the dual hydrogel constructs was verified by fluorescent microscopy (Nikon AZ100). The gel thickness and extent of binding of MI-Texas Red through the depth of the bulk CNBC-A gel were characterized using confocal microscopy. Confocal imaging was performed on a Zeiss LSM 510 Meta microscope (Carl Zeiss, Jena, Germany). Optical slices were taken at 7.87 μm (10- pixel mask), 8.83 μm (8-pixel mask), 10.80 μm (6-pixel mask), and 7.89 μm (4-pixel mask) intervals at a resolution of 1024×1024 pixels. Confocal projection images were taken as summed voxel projections. Both fluorescent and confocal images were processed with ImageJ software. V3D software (Janelia Farm Research Campus, Howard Hughes Medical Institute, Ashburn, VA) allowed 3D visualization of confocal z-stack images.
The average diameter of the fluorescent region in bulk CNBC-A resulting from the projected 4-pixel mask was also quantified. The stack of 60 confocal images was condensed to 20 images by using a summed voxel projection of three consecutive images to create one new image. For each of these summed slices, the intensity profile was measured using a straight line drawn across the center of the fluorescent region. This was done at four angles (0°, 45°, 90°, and 135°), where 0° is the horizontal axis. Since the intensity profile resembled a plateau, the midpoint between the minimum and maximum intensities on either side of the intensity plateau was established, and the diameter of the fluorescent region was determined to be the distance between these two midpoints. The diameter measurements taken at each angle from the 20 summed slices were averaged to give a mean diameter per slice.
3. Results
3.1 Immobilized maleimide-biomolecules
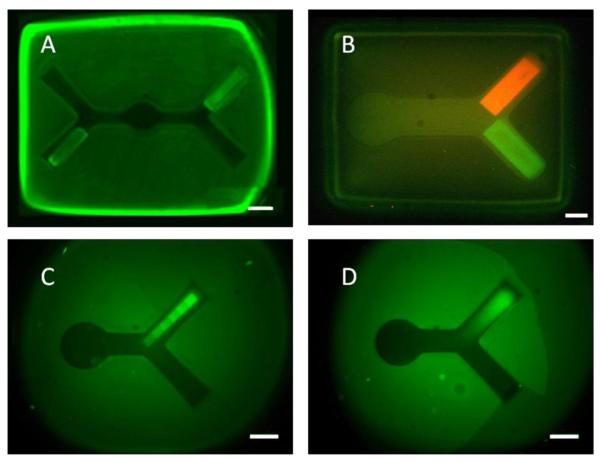
The utility of the photolithography apparatus was demonstrated by fabricating dual hydrogel systems that presented both structural and molecular guidance cues through the use of two different photoreactive hydrogels (Fig. 4, 6). Just as the geometry of the cell growth-restrictive PEG boundary could be specified by a photomask, sections of cell growth-permissive CNBC-A could also be irradiated with a specifiable pattern for biomolecule immobilization. Binding of one or two biomolecules, as well as gradients of biomolecules, was demonstrated. Figure 4 shows fluorescent images of MI-biotin tagged with FITC-NeutrAvidin (Fig. 4A) and MI-NeutrAvidin tagged with fluorescent biocytin (Fig. 4B-D), bound to an irradiated section of CNBC-A. Figure 4B shows both Oregon Green 488 biocytin (green) and Alexa Fluor 594 biocytin (red) bound to MI-NeutrAvidin after two separate irradiation and MI-NeutrAvidin– binding steps. When UV light was focused at the insert membrane, the resulting fluorescent regions had well-defined edges (Fig. 4A-C). However, to obtain the smooth gradient of immobilized biomolecules seen in Figure 4D, the UV light was just out-of-focus from the insert membrane during irradiation. From the standard curve made from the fluorescence of known concentrations of Oregon Green 488 biocytin, the gradient from the out-of-focus masks ranged linearly from 48.9-620 nM biocytin between irradiation times of 1-32 s. The immobilized areas in Figure 4 are 1000 μm in length and 200 μm in width, a scale relevant for cellular studies. For all of the images shown in Figure 4, minimal fluorescence was observed in the PEG constructs or in the non-irradiated parts of CNBC-A
Fig. 4.
Dual hydrogel systems with fluorescent-tagged immobilized MI-biomolecules. A) MI-biotin tagged with FITC-NeutrAvidin. B-D) MI-NeutrAvidin tagged with Oregon Green 488 biocytin B) and Alexa Fluor 594 biocytin (red) after two separate irradiation steps, C) in a gradient pattern using in-focus masks and D) in a gradient pattern using out-of-focus masks (scale bars = 500 μm)
Fig. 6.
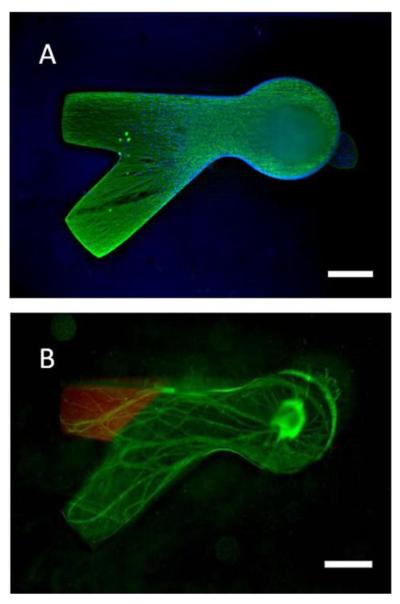
Dorsal root ganglia neurite outgrowth in dual hydrogel system. A) Neurites (green) and cell bodies (blue nuclei) are confined to the CNBC-A gel of the dual hydrogel. B) Neurite growth in the presence of immobilized MI-NeutrAvidin/Oregon Green 488 biocytin (scale bars = 500 μm)
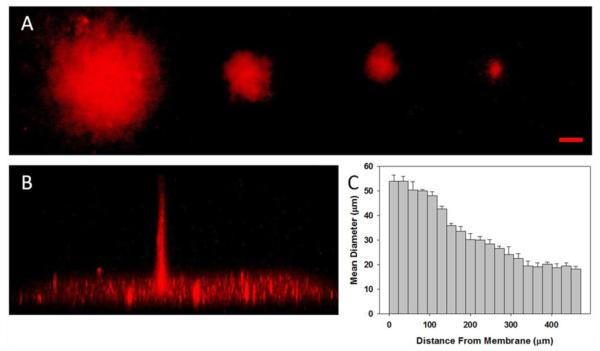
The resolution of protein binding was investigated through the depth of the CNBC-A via confocal microscopy. Using 10-pixel, 8-pixel, 6-pixel, and 4-pixel circular masks, bulk CNBC-A was irradiated for 180 s per mask, and MI-Texas Red was bound to the irradiated regions (Fig. 5). Figure 5A shows confocal z-stack summed voxel projections of MI-Texas Red bound to the irradiated regions resulting from the multiple mask sizes. For all mask sizes, the fluorescent region maintains a distinct circular shape that matched the shape of the original photomask and, as expected, the site of the fluorescent region correlated with the mask size. A side view of the confocal image stack of the 4-pixel mask-irradiated region indicated that binding of MI-Texas Red occurred through the depth of the gel (~470 μm), and the diameter of the column of bound MI-Texas Red decreased as the distance from the membrane increased (Fig. 5B). The bright plane of fluorescence at the bottom of Figure 5B is likely due to autofluoresence of and nonspecific binding to the cell culture support membrane. Because the diameter for the fluorescent region decreased with increasing distance from the insert membrane, the diameter was measured at 24.7 μm intervals (equal to the thickness of 3 optical slices) along the depth of the gel (Fig. 5C). Near the membrane, the diameter was found to be ~50 μm but was only ~30 μm in the center of the gel and ~20 μm at a distance greater than 350 μm from the membrane. This discrepancy in the size of immobilized regions of biomolecules through the gel became less evident as the feature size increased (data not shown.) Even though the resolution varies by 30 μm through the depth of the gel, these feature sizes are in line with the resolution necessary for cell studies. More importantly, binding was evident through the depth of the gel, showing that 3D immobilization of biomolecules is possible with the smallest current resolution obtainable with the photolithography apparatus.
Fig. 5.
Confocal images of MI-Texas Red in bulk CNBC-A following UV irradiation with circular photomasks. A) Top view of summed voxel projection of MI-Texas Red bound through depth of CNBC-A using 10-pixel, 8-pixel, 6-pixel, and 4-pixel diameter circle photomasks, from left to right (scale bar = 50 μm). B) Side view of 3D image stack of MI-Texas Red binding after using the 4-pixel mask, with binding depth of 472 μm. C) Graph of the mean diameter of MI-Texas Red binding through depth of CNBC-A from Part B, where the cell culture insert membrane is at 0 μm and bar width is equivalent to summed slice thickness (error bars represent standard deviation)
3.2 Neurite growth in dual hydrogel constructs
To use the dual hydrogel system as a tissue culture model, tissue explants were cultured on the surface of the cell culture insert, the PEG construct was polymerized around the explants, and liquid CNBC-A was subsequently added to the void of the PEG construct. When using the dual hydrogel constructs for protein immobilization studies, the CNBC-A gelled by cooling to 4°C for 1 h. When incorporating cells into the dual hydrogel constructs, though, the agarose was gelled by submerging the insert in pre-chilled (4°C) growth media. This rapid gelation method minimized the exposure of the cells to cold temperatures while simultaneously gelling the agarose in the PEG void.
Neurite outgrowth from the DRG explants after 7 days spanned the length of the CNBC-A gel, and was entirely contained by the PEG barrier (Fig. 6). As seen in Figure 6A, the neurites (green) were frequently concentrated along the walls of the PEG construct without growing over this barrier. The DAPI-stained cell nuclei (blue) represent both supportive cells and neurons and are similarly contained by the PEG construct. The DRG explant, containing the majority of DAPI-stained nuclei, is out of the plane of focus in the figure. These results confirm that the dual hydrogel system is a suitable construct for 3D tissue culture and growth studies, as neurites are capable of navigating the modified agarose gel while simultaneously confined by the PEG barrier. We have previously confirmed that neurite growth is found throughout the depth of dual hydrogel constructs with a slight preference for the surface of the permeable membrane, as quantified with confocal microscopy (Curley and Moore 2011).
The DRG explants cultured in the presence of immobilized MI-NeutrAvidin/biocytin showed equally robust growth as those cultured without (Fig 6B). This comparison suggests that the wash steps necessary to immobilize biomolecules should not adversely affect cell viability or neurite outgrowth. Immobilizing biomolecules in the dual hydrogel constructs that normally do not induce a response in vivo should not induce a response in the construct, either. Since neurites from DRG do not respond to either NeutrAvidin or biotin in vivo, the neurites extended equally down either channel of the in vitro construct, confirming that the immobilized biomolecules do not alter the structural integrity of the construct enough to influence the neurite growth patterns.
3.3 Protein diffusion
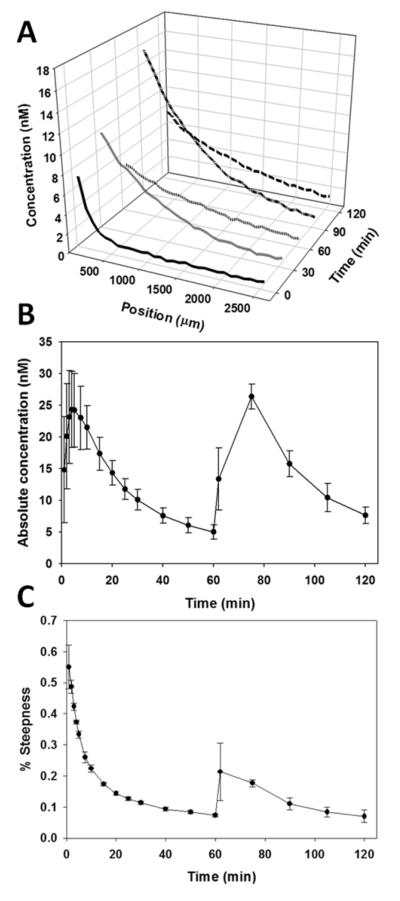
Figure 8 shows that the gradient can be sustained in relevant physiological ranges upon refilling the well with additional protein solution. The 3D plot of the gradient profiles at different time points (Fig. 8A) shows that the diffusion trends between the first and second filling of the well are similar. Though the magnitude of the gradient at time 30 min is smaller than at time 90 min, the gradients have similar steepness. After 60 min since the well was refilled (120 min total), the gradient profile resembled the profile from the 60 min mark. In Figure 8B, the absolute magnitude of the gradient at different time points remains similar between fillings of the well, with peaks in magnitude shortly after the well is filled for both the first and second time. However, the magnitude between time 60 min and time 120 min are nearly the same. The steepness of the gradient (Fig. 8C) is greatest when the well is filled at time 0 min, and quickly resolves to a more linear profile around time 20 min. The gradient steepness increases again when the well is refilled after time 60 min, and is slower to resolve than the first filling of the well. Though the steepness profile is more linear upon refilling the well, the steepness profiles between time 60 min and time 120 min are nearly identical.
Fig. 8.
Diffusion of soluble fluorescein-BSA through dual hydrogel construct with sustained gradient. The well in the construct was filled with fluorescein-BSA at 0 min and refilled after 60 min. A) 3D plot of mean concentration of fluorescein-BSA after 0, 30, 60, 90, and 120 min relative to position in construct. B) Absolute magnitude of the concentration at the starting position over time. C) Graph of gradient steepness over time. Error bars represent standard deviation
4. Discussion
The dynamic mask projection photolithography apparatus described in this report allowed for quick and simple fabrication of micropatterned hydrogels directly on permeable cell culture inserts. To fabricate the dual hydrogel constructs, the DMD in our photolithography apparatus acted as a dynamic mask for irradiating both PEG and the photolabile agarose with UV light in discrete patterns, thereby controlling the spatial distribution of structural and molecular guidance cues. All micropatterning resulted from a 2D image extruded through the depth of the material, giving the user full control over patterning in the horizontal plane. This is advantageous because, for a given photoreactive material, all patterns required the same irradiation time regardless of the size of the pattern. Thus, projecting a 2D image to fabricate a 3D structure rendered irradiation time independent from the size and shape of the pattern – a distinct advantage over other photolithographical techniques like two-photon irradiation. Another advantage of dynamic photolithography over two-photon irradiation is that this setup allows for several experiments to be performed simultaneously, since the constructs can be fabricated in quick succession with minimal time outside of the sterile field. This aspect of dynamic photolithography reduces the amount of time required to run a statistically significant number of trials. Though smaller feature sizes may be attained with other techniques, dynamic photolithography is most useful for fabricating constructs for 3D tissue cultures and studying multiple cells at one time.
With the use of dynamic photolithography, both homogenous and gradient immobilization of molecules could be achieved, with either one or two biomolecules presented in the same construct (Fig. 3). Though only the top view was shown in Figure 3, the depth of binding for all images must be considered. Confocal imaging showed MI-Texas Red to bind through the depth of bulk CNBC-A (Fig. 5B) with decreasing diameter of the immobilized region due to the plane of focus and light attenuation through the depth of the gel, as is common with projection-type photolithographical techniques. Since the irradiation times between Figures 4A-B and Figures 4C-D differed greatly (180 s versus 1-32 s, respectively), the depth of biomolecule binding through the gel is also likely to vary. In Figure 4D, the concentration of bound Oregon Green 488 biocytin ranged linearly from 48.9-620 nM. This change in concentration with irradiation time is likely due to increased depth of binding through the gel with increasing irradiation time.
Because fluorescent biocytin was able to bind to MI-NeutrAvidin, it was evident that maleimide-conjugated proteins retain their functionality after binding to the agarose gel. Thus, the gel itself did not interfere with the conformation of the protein when using a maleimide moiety to bind proteins to the gel. With this dual hydrogel system, multiple biomolecules can be immobilized within the same construct in a variety of patterns, thus providing an additional facet of customization for in vitro tissue cultures. From both the fluorescent and confocal microscopy, it is evident that the immobilized biomolecules were localized to the irradiated sections of CNBC-A. By simply changing the focus of the UV light and irradiation time, the resolution – and thus the intended application – of the region of immobilized biomolecules can be manipulated for specific applications. In order to modify our dual hydrogel system to be an in vitro model for studying the development of the optic nerve, future studies will include immobilized guidance cues such as ephrin-B2, the ligand expressed by glial cells at the optic chiasm that selectively repels retinal neurites presenting the receptor EphB1.
For the resolution of the dual hydrogel system, we obtained structural features and immobilized areas of biomolecules on the order of tens of microns – a scale appropriate for directing neurite growth from tissue explants (Fig. 4-6). In this study, mask geometries were chosen to be representative of nerve bifurcation, such that the geometry provided a choice point for binary divergence. Neurite growth and containment in dual hydrogel constructs have been characterized extensively with a peptide gel in the place of CNBC-A (Curley et al. 2011; Curley and Moore 2011). This study reinforces those findings by demonstrating similarly robust neurite growth in CNBC-A, with and without immobilized biomolecules (Fig. 6). Using biomolecules that do not normally induce a response from neural tissue confirmed that simply irradiating and functionalizing CNBC-A does not affect directional neurite growth.
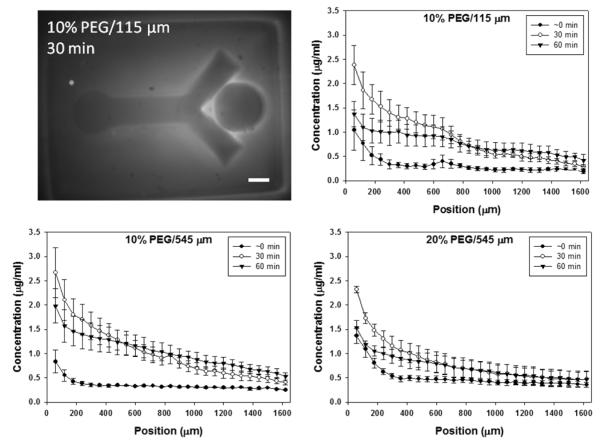
The diffusion experiments demonstrated how the steepness, or strength, of a soluble signal gradient can be tuned to some extent without altering the initial concentration of the soluble factor. Here we used fluorescein-BSA as an analog for netrin, as BSA and netrin have similar molecular weights (Mn ~ 70 kDa). As expected, the steepness of the exponential fluorescein-BSA gradient decreased over time for all three schemes (Fig 7). This setup provided some limited control over the concentration profile, or strength of the soluble signal, along the length of the agarose by varying the concentration of the PEG through which the protein must diffuse. There could also be some diffusion of the protein through the bottom of the cell culture insert membrane that may dominate when lateral diffusion through the gel is sufficiently slowed, as in the 20% far case. The accumulation of fluorescein-BSA along the boundary between the two gels may be due to the diffusivity mismatch or possibly space between the two the gels. However, this protein accumulation would not be expected to affect neurite behavior since neurite growth is contained within the agarose gel. For our purposes, these results and techniques will be applied to modeling the diffusion of netrin through the dual hydrogel constructs. Other biomolecules, such as guidance proteins and growth factors, may be used in this setup, with the change in gradient profiles being dependent upon inherent characteristics of the biomolecule of interest, such as molecular weight, protein radius, functional groups, isoelectric point, etc. Still, the concentration of the PEG and the distance of the well from the agarose can be adjusted to obtain the desired gradient profiles.
Fig. 7.
Diffusion of soluble fluorescein-BSA through the dual hydrogel system. Relative fluorescence measured at 0, 30, and 60 min for three different schemes was converted to concentration (μg/ml). Graphs show changes in fluorescein-BSA concentration over the length of the agarose-filled bifurcating void, where percentage indicates polyethylene glycol (PEG) concentration and 115 μm and 545 μm indicate distance of protein source well from the bifurcating void. Fluorescent image is representative of the 10% PEG/115 μm scheme. Error bars represent standard deviation (scale bar = 500 μm)
Refilling the well demonstrated that a predictable pattern of diffusion through the dual hydrogel construct could be obtained with the current setup. The gradient profiles at corresponding time points after the first and second filling of the well were very similar in steepness and in magnitude (Fig 8). As expected, the magnitude of the gradient was greatest shortly after each filling of the well, and accumulated protein within the agarose may have contributed to the peak magnitude after time 60 min being greater than that of the first filling of the well. After the well was filled a second time with fluorescein-BSA, the steepness profiles over time appeared more linear than after the first filling (Fig 8C). This may be due to a burst release of the protein attributing to the initial gradient steepness between 0-10 min, in addition to some degree of saturation of the PEG and, to a lesser extent, agarose with fluorescein-BSA. The steepness profiles after the second filling suggest that the burst release of a protein solution may only be a factor for the initial filling of the well and not for subsequent refills. If so, then it may account for the slowed decrease in gradient magnitude after filling the well a second time, though the final magnitude at time 120 min is almost the same as that at time 60 min (Fig 8B). Additionally, the protein accumulation at the boundary of the PEG did not appear to affect the steepness profile after time 60 min, as the corresponding time points before and after the refilling of the well have similar steepness profiles. Though the maintenance of a gradient over time scales appropriate for neurite guidance studies is typically a few days, establishment of concentration gradients over longer periods could easily be achieved with successive applications or more complicated geometries of the protein source position, the possibilities of which are nearly limitless with the projection lithography approach. Alternatively, a well could be filled with a suspension of cells that secrete the biomolecule of interest in order to provide a steadier source of soluble cues.
Neurites rely on chemotaxis during development in order to synapse with their targets, and the concentration gradients of both immobilized and soluble guidance cues influence the direction of neurite growth. Some known guidance proteins have been incorporated into in vitro models to determine the ideal concentration and gradient steepness for maximum neurite growth and turning response. One study immobilized Ephrin-A2 in concentrations varying between 0 and 14.8 nM and observed a graded response to the different concentrations of Ephrin-A2 in which low concentrations enhanced neurite growth while high concentrations restricted growth (Hansen et al. 2004). A soluble gradient of netrin varying from 0-200 ng/ml (0-2.8 nM) across a 500 μm space induced axon turning in cortical neurons in one study that utilized a microfluidic chamber to provide a steady soluble gradient (Bhattacharjee et al. 2010). Another study on the effects of soluble netrin gradients determined that a concentration of 0-1 μg/ml (0-14.2 nM) across a 1200 μm space induced the maximum positive turning effect in DRG neurites (Kothapalli et al. 2011). A study on gradients of NGF found that increasing the steepness of the NGF gradient increased the number of neurites guided toward the high end of the gradient and that, at a 0.2% gradient steepness, maximum neurite turning occurred between 1-10 nM of NGF (Rosoff et al. 2004). As seen in Figure 8, at time 60 min and time 120 min we were able to maintain gradients of fluorescein-BSA between 1-7.8 nM across a 2500 μm space, with steepness between 0.1-0.2%, suggesting that our setup and evaluation of experiments with BSA could be used to establish physiologically-relevant netrin gradients. These experimental conditions are attainable with the relatively simple setup we have described, and experiments incorporating multiple cues or more complex configurations are easily imaginable. Just as the soluble gradients of fluorescein-BSA were quantified in this study, the concentrations of immobilized guidance cues can be determined through the use of fluorescent microscopy.
Here we have shown how the dynamic photolithography apparatus afforded control over several aspects of the design and fabrication of a 3D in vitro microenvironment that is initially independent of cell type. Using a single apparatus, we can incorporate structural and molecular cues into a cell culture model that can be tuned for a wide range of applications. The ease at which many facets of the experimental design can be adjusted to suit specific needs is the greatest utility of the dynamic photolithography apparatus and dual hydrogel system. For our purposes, future studies will include characterization of guided neural axon growth in our novel in vitro tissue culture model using both immobilized and soluble cues. Eliciting expected in vivo responses to guidance cues in vitro would validate our model as an appropriate platform for 3D tissue culture. While we presented only a few examples of possible biomolecule guidance methods and in vitro model geometries generated by the dynamic mask photolithography apparatus, these techniques are easily adaptable for manipulating the extracellular environment of many different cell culture models.
5. Conclusions
Using a dynamic mask photolithography apparatus, we have developed a dual hydrogel system with a commercially available PEG gel and a custom photolabile agarose gel. Within this dual hydrogel system, we have demonstrated the immobilization of proteins in discrete locations and the establishment of a concentration gradient of soluble proteins. Because of the nonspecific nature of this in vitro model, it can easily be tailored to enhance 3D cultures of a wide variety of tissues and cells. The binding of maleimide-activated NeutrAvidin and biotin suggests that any maleimide-conjugated biomolecule could bind to the uncaged thiols of the photolabile agarose gel. The concentration profiles for soluble fluorescein-BSA could be adjusted by both the source position and the crosslink density of the PEG hydrogel, and physiologically-relevant gradients can be maintained over time by reapplication. Our results suggest that the dual hydrogel system presented here may be useful as a 3D in vitro microenvironment that can be modified with patterned structural and molecular cues. This approach will be particularly useful for the study of neural growth in response to various guidance cues.
Acknowledgements
This research was funded in part by grants from the Louisiana Board of Regents (LEQSF[2009-10]-RD-A-18), the NIH (NS065374), and an NSF CAREER award to MJM (CBET-1055990). We also thank Chris Rodell for assistance with tBu-BNPA synthesis.
References
- Abbott A. Nature. 2003;424:870–872. doi: 10.1038/424870a. [DOI] [PubMed] [Google Scholar]
- Almany L, Seliktar D. Biomaterials. 2005;26:2467–77. doi: 10.1016/j.biomaterials.2004.06.047. [DOI] [PubMed] [Google Scholar]
- Balgude AP, Yu X, Szymanski A, Bellamkonda RV. Biomaterials. 2001;22:1077–84. doi: 10.1016/s0142-9612(00)00350-1. [DOI] [PubMed] [Google Scholar]
- Bellamkonda RV, Ranieri JP, Aebischer P. J Neurosci Res. 1995;41:501–509. doi: 10.1002/jnr.490410409. [DOI] [PubMed] [Google Scholar]
- Bernard A, Renault JP, Michel B, Bosshard HR, Delamarche E. Adv Mater. 2000;12:1067–1070. [Google Scholar]
- Bhattacharjee N, Li NZ, Keenan TM, Folch A. Integr Biol. 2010;2:669–679. doi: 10.1039/c0ib00038h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Radisky D. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant SJ, Anseth KS. J Biomed Mater Res. 2002;59:63–72. doi: 10.1002/jbm.1217. [DOI] [PubMed] [Google Scholar]
- Bryant SJ, Anseth KS. J Biomed Mater Res Part A. 2003;64A:70–79. doi: 10.1002/jbm.a.10319. [DOI] [PubMed] [Google Scholar]
- Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- Curley JL, Jennings SR, Moore MJ. J Vis Exp. 2011;48:e2636. doi: 10.3791/2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curley JL, Moore MJ. J Biomed Mater Res Part A. 2011;99A:532–543. doi: 10.1002/jbm.a.33195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Kisaalita WS, Keith C, Wu ZZ. Biosens Bioelectron. 2006;21:1483–1492. doi: 10.1016/j.bios.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Dhariwala B, Hunt E, Boland T. Tissue Eng. 2004;10:1316–1322. doi: 10.1089/ten.2004.10.1316. [DOI] [PubMed] [Google Scholar]
- Hahn MS, Taite LJ, Moon JJ, Rowland MC, Ruffino KA, West JL. Biomaterials. 2006;27:2519–2524. doi: 10.1016/j.biomaterials.2005.11.045. [DOI] [PubMed] [Google Scholar]
- Hansen MJ, Dallal GE, Flanagan JG. Neuron. 2004;42:717–730. doi: 10.1016/j.neuron.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Irons HR, Cullen DK, Shapiro NP, Lambert NA, Lee RH, Laplaca MC. J Neural Eng. 2008;5:333–41. doi: 10.1088/1741-2560/5/3/006. [DOI] [PubMed] [Google Scholar]
- Kim K, Yeatts A, Dean D, Fisher JP. Tissue Eng Part B-Rev. 2010;16:523–539. doi: 10.1089/ten.teb.2010.0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothapalli CR, van Veen E, de Valence S, Chung S, Zervantonakis IK, Gertler FB, Kamm RD. Lab on a Chip. 2011;11:497–507. doi: 10.1039/c0lc00240b. [DOI] [PubMed] [Google Scholar]
- Kotzur N, Briand B, Beyerman M, Hagan V. Chem Comm. 2009:3255–3257. doi: 10.1039/b900865a. [DOI] [PubMed] [Google Scholar]
- Kotzur N, Briand B, Beyerman M, Hagan V. J Amer Chem Soc. 2009;131:16927–16931. doi: 10.1021/ja907287n. [DOI] [PubMed] [Google Scholar]
- Lee J, Cuddihy MJ, Kotov NA. Tissue Eng Part B-Rev. 2008;14:61–86. doi: 10.1089/teb.2007.0150. [DOI] [PubMed] [Google Scholar]
- Lee KN, Shin DS, Lee YS, Kim YK. J Micromech Microeng. 2003;13:18–25. [Google Scholar]
- Lee KY, Mooney DJ. Chem Rev. 2001;101:1869–1879. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- Lee SH, Moon JJ, West JL. Biomaterials. 2008;29:2962–2968. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K, Herklotz M, Espig M, Paumer T, Nitschke M, Werner C, Pompe T. Biomaterials. 2010;31:8802–8809. doi: 10.1016/j.biomaterials.2010.07.071. [DOI] [PubMed] [Google Scholar]
- Lu Y, Mapili G, Suhali G, Chen SC, Roy K. J Biomed Mater Res Part A. 2006;77A:396–405. doi: 10.1002/jbm.a.30601. [DOI] [PubMed] [Google Scholar]
- Luo Y, Shoichet MS. Biomacromolecules. 2004;5:2315–2323. doi: 10.1021/bm0495811. [DOI] [PubMed] [Google Scholar]
- Luo Y, Shoichet MS. Nat Mater. 2004;3:249–253. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- Melchels FPW, Bertoldi K, Gabbrielli R, Velders AH, Feijen J, Grijpma DW. Biomaterials. 2010;31:6909–6916. doi: 10.1016/j.biomaterials.2010.05.068. [DOI] [PubMed] [Google Scholar]
- Moon JJ, Lee SH, West JL. Biomacromolecules. 2007;8:42–49. doi: 10.1021/bm060452p. [DOI] [PubMed] [Google Scholar]
- Nemir S, Hayenga HN, West JL. Biotechnol Bioeng. 2010;105:636–644. doi: 10.1002/bit.22574. [DOI] [PubMed] [Google Scholar]
- Nguyen KT, West JL. Biomaterials. 2002;23:4307–4314. doi: 10.1016/s0142-9612(02)00175-8. [DOI] [PubMed] [Google Scholar]
- Pan P, Bayley H. FEBS Lett. 1997;405:81–85. doi: 10.1016/s0014-5793(97)00165-8. [DOI] [PubMed] [Google Scholar]
- Peppas NA, Huang Y, Torres-Lugo M, Ward JH, Zhang J. Annu Rev Biomed Eng. 2000;2:9–29. doi: 10.1146/annurev.bioeng.2.1.9. [DOI] [PubMed] [Google Scholar]
- Ribeiro A, Vargo S, Powell EM, Leach JB. Tissue Eng Part A. 2012;18:93–102. doi: 10.1089/ten.tea.2011.0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosoff WJ, Urbach JS, McAllister RG, Richards LJ, Goodhill GJ. Nat Neurosci. 2004;7:678–682. doi: 10.1038/nn1259. [DOI] [PubMed] [Google Scholar]
- Von Philipsborn AC, Lang S, Bernard A, Loeschinger J, David C, Lehnert D, Bastmeyer M, Bonhoeffer F. Nat Protoc. 2006;1:1322–1328. doi: 10.1038/nprot.2006.251. [DOI] [PubMed] [Google Scholar]
- Walker JW, Gilbert SH, Drummond RM, Yamada M, Sreekumar R, Carraway RE, Ikebe M, Fay FS. Proc Nat Acad Sci USA. 1998;95:1568–1573. doi: 10.1073/pnas.95.4.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Foo CWP, Warrier A, Poo MM, Heilshorn SC, Zhang X. Biomed Microdev. 2009;11:1127–1134. doi: 10.1007/s10544-009-9329-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG. Nat Rev Neurosci. 2001;2:155–164. doi: 10.1038/35058515. [DOI] [PubMed] [Google Scholar]
- Williams SE, Mason CA, Herrera E. Curr Opin Neurobiol. 2004;14:51–60. doi: 10.1016/j.conb.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Yu TW, Bargmann CI. Nat Neurosci. 2001;4:1169–76. doi: 10.1038/nn748. [DOI] [PubMed] [Google Scholar]
- Zalipsky S, Harris JM. In: Poly(Ethylene Glycol) Harris JM, Zalipsky S, editors. Washington, DC; American Chemical Society: 1997. pp. 1–13. [Google Scholar]
- Zhu JM. Biomaterials. 2010;31:4639–4656. doi: 10.1016/j.biomaterials.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]