Summary
In response to DNA damage, eukaryotic cells must rapidly load DNA repair proteins onto damaged chromatin. Chromatin recruitment often entails ubiquitination of a damage-specific DNA repair protein, interaction with a ubiquitin binding factor, assembly of a multisubunit DNA repair complex, and eventually a deubiquitination event once the DNA repair reaction has been completed. This review focuses on the recent discoveries in the Fanconi Anemia (FA) and DNA double strand break (DSB) repair pathways which underscore the importance of regulated chromatin loading in the DNA damage response. Interestingly, these two pathways share several features, suggesting a more general mechanism for DNA repair regulation.
Keywords: Fanconi Anemia, Ubiquitin, Deubiquitination, Chromatin, RNF8, BRCA1
The DNA damage response
When confronted with DNA damage, the eukaryotic cell has several options. It can arrest its own DNA synthesis thus allowing DNA repair to occur. It can continue to replicate its DNA and bypass the damage. Or, it can surrender to the damage and activate its own demise. The cellular decision-making process, known as the DNA Damage Response (DDR), is a complex network of signal transduction pathways, involving a vast array of sensor, transducer, and effector proteins (Harper and Elledge, 2007).
There are several DNA repair pathways in human cells, including base excision repair (BER), mismatch repair (MMR), nucleotide excision repair (NER), homologous recombination (HR), non-homologous end-joining (NHEJ), and translesion synthesis (TLS) (Kennedy and D'Andrea, 2006). These pathways deal with different types of DNA damage. For instance, the HR and NHEJ pathways repair double strand breaks whereas the BER pathway primarily repairs damaged single base residues. The repair pathways also have differential importance, depending on the cell type or the cell cycle phase. For instance, rapidly proliferating cells with a high S phase fraction are more likely to employ HR repair, whereas cells in the G1 phase repair DSBs mainly by NHEJ. A combination of NER, HR, and TLS pathways is required to repair complex DNA lesions, such as DNA crosslinks,. The Fanconi Anemia pathway (see below) is believed to coordinate these three repair pathways (Mirchandani and D'Andrea, 2006).
The cellular reaction to an ionizing radiation (IR)-induced double strand break (DSB) is also complex. Many proteins, including the protein kinase, ataxia-telangiectasia mutated (ATM), histone variant H2AX, mediator of DNA damage checkpoint protein 1 (MDC1), breast cancer antigen 1 (BRCA1), and checkpoint kinases 1 and 2 (Chk1 and Chk2) are involved in the IR-induced DNA damage response, a response that also includes the activation of cell cycle checkpoints (Su, 2006). A DSB can be repaired by either HR or by NHEJ[SC2]. DSBs repaired by HR are initially detected by a complex series[SC3] of events, including the sensory protein ATM and the MRN (MRE11/RAD50/NBS1) complex. The activated ATM kinase next phosphorylates multiple protein substrates, some of which are also assembled at or near the site of the DSB. Eventually, proteins involved in homologous recombination, such as RAD51 and RAD52, are also assembled locally, leading to the repair of the DSB.
A critical event in the DDR, and the focus of this review, is the rapid loading of activated protein complexes onto chromatin at the site of DNA damage. The specific, sequential, and rapid recruitment of these DNA repair proteins is essential for a successful response to the particular DNA damage. Once the relevant proteins are assembled onto chromatin, these complexes provide the checkpoint activity for halting DNA replication or the DNA repair activity for repairing the damaged bases. Accordingly, the eukaryotic cell has established elaborate mechanisms for the regulated loading of proteins onto damaged chromatin; their disruption results in genomic instability and, in many cases, predisposition of the cell to malignant transformation. Of note, many of these mechanisms require protein ubiquitination and deubiquitination.
Interestingly, the importance of ubiquitin-mediated protein loading onto damaged chromatin in the DNA damage response was elucidated largely through the systematic study of human genetic diseases. The major human hereditary breast cancer susceptibility gene product, BRCA1, for example, is critical for the cellular response to ionizing radiation. This protein forms a complex with its structural relative, BARD1, and the heterodimeric complex functions as an E3 ubiquitin ligase. BRCA1 recruitment to damaged chromatin is critical for HR-mediated DNA repair. Also, the genetic disease, Fanconi Anemia (FA), is a chromosome instability syndrome involving defective chromatin loading of DNA repair proteins (see below). Like other DNA damage response pathways, regulated ubiquitination and deubiquitination is critical for an intact FA pathway.
Protein ubiquitination
Proteins are ubiquitinated by an enzymatic cascade which involves an E1 activating enzyme, an E2 conjugating enzyme, and an E3 ubiquitin ligase. If the substrate is polyubiquitinated by K48 ubiquitin linkages, this modification typically leads to proteasome-mediated degradation of the protein. More pertinent to this review, several chromatin-associated proteins (for instance, proteins involved in transcription or DNA repair) are polyubiquitinated through a K63 ubiquitin linkage, which does not target the substrate for proteasome-mediated degradation. Rather, this modification typically leads to changes in the substrate’s activity or location (Pickart and Eddins, 2004). Additionally, [SC4]proteins can be monoubiquitinated. Importantly, mono- and polyubiquitination are often reversible, thereby providing a mechanism to tightly regulate the activity of the modified protein.
Ubiquitination of key proteins in the DNA damage response: specific examples
During the DNA damage response, rapid changes in chromatin compaction and composition must occur. DNA damage causes a local unwinding of chromatin and a rapid entry and assembly of new DNA repair complexes. This assembly is regulated largely by protein ubiquitination events, and several examples of this mode of regulation are known (Huang and D'Andrea, 2006).
UV-light causes the formation of cyclobutane pyrimidine dimers (CPDs). The DDB1/DDB2 complex senses these CPDs as a distortion in the double helix, and activates a reversible polyubiquitination of the XPC protein (Sugasawa et al., 2005). XPC polyubiquitination is a critical upstream event, leading to the chromatin loading of functional[SC5] NER complexes and ultimately to successful excision of the damaged DNA bases (Sugasawa et al., 2005).
DNA damage also triggers the ubiquitination of the DNA processivity factor, PCNA. PCNA can either be monoubiquitinated by the RAD6/RAD18 E2/E3 heterodimer, or polyubiquitinated by the RAD5/UBC13/MMS2 complex, via K63 linkages (Moldovan et al., 2007). These critical modifications can mediate either error prone TLS or possibly error free template switching-mediated TLS, respectively.
Finally, two Fanconi Anemia (FA) pathway proteins, FANCD2 and FANCI, are both monoubiquitinated and loaded onto chromatin in response to DNA damage. Thus, there are many examples of protein ubiquitination that drive selective loading of DNA repair complexes onto chromatin. The importance of ubiquitination in the FA pathway will be discussed in greater detail later.
Role of ubiquitin binding proteins in mediating chromatin loading of DNA repair complexes
Ubiquitin can mediate the DNA damage response by binding a variety of ubiquitin-binding domains (UBDs). Thus, proteins which are monoubiquitinated or K63-linkage polyubiquitinated, as part of the DNA damage response, can load onto chromatin through UBD-containing proteins. Ubiquitin-binding proteins generally have small (20–150 amino acid), independently folded UBDs that can interact directly with monoubiquitin or K63-linked polyubiquitin. UBDs are structurally diverse, and several subgroups have been characterized (Bienko et al., 2005; Hicke et al., 2005).
One example of a rapid ubiquitin-mediated recruitment of a protein to chromatin occurs during translesion DNA synthesis. In response to DNA damage or replication fork arrest, the chromatin bound processivity factor, PCNA, is rapidly monoubiquitinated on Lysine 164. This monoubiquitination enhances the binding of the translesion DNA polymerase, Pol η. Pol η contains a specialized ubiquitin binding domain, the UBZ domain, which binds Ub-PCNA and is essential for the recruitment step (Bienko et al., 2005).
A second example of ubiquitin-mediated recruitment occurs in the cellular response to a double strand break. Following cellular exposure to ionizing radiation, histone H2AX is polyubiquitinated, a modification that triggers RAP80 recruitment and the subsequent recruitment of additional DNA repair proteins. Importantly, RAP80 contains a tandem UIM (ubiquitin interaction motif), which is essential for its recruitment to chromatin (Kim et al., 2007; Sobhian et al., 2007; Wang et al., 2007).[SC6]
The exact mechanism for chromatin recruitment of other ubiquitinated proteins during the DDR remains unknown. For instance, polyubiquitinated CtIP, a BRCA1 substrate, is rapidly recruited to chromatin (Yu et al., 2006). This loading step likely involves still another UBD-containing chromatin-associated protein.
Ubiquitin-mediated loading of DNA repair complexes at DNA double strand breaks
It has long been known that the E3 ubiquitin ligase and tumor suppressor protein, BRCA1, accumulates in nuclear foci around a DNA double strand break and that these foci are likely to be sites of BRCA1 effector functions (Scully et al., 1997a; Scully et al., 1997b). BRCA1 foci formation depends on upstream DDR proteins, including ATM, H2AX, and MDC1, among others. Still, the molecular basis of these dependencies (i.e., how BRCA1 is recruited to the site of damage) remains speculative.
Four recent independent reports (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang et al., 2007) have described the mechanism of serial protein recruitment around the site of a DSB[SC7]. ATM-mediated phosphorylation of MDC1 and H2AX, which are early steps in the response to IR, leads to the recruitment of the E3 ligase RNF8. Together with the E2 conjugating enzyme UBC13, RNF8 appears to polyubiquitinate H2AX and perhaps other chromatin associated substrates through K63-linkage. Polyubiquitinated H2AX is now able to serve as a docking site for the UIM-containing protein RAP80, which in turn recruits the repair proteins ABRA1 and BRCA1 to this site on chromatin. In good agreement, a previous report demonstrated the involvement of UBC13 in HR-mediated DSB repair (Zhao et al., 2007).
Although H2AX polyubiquitination has been reported previously (Ikura et al., 2007), the functional relevance of this modification was unknown at the time. These new reports underscore several important features of the ubiquitin pathway and its role in protein complex assembly. For the described RNF8-BRCA1 pathway, polyubiquitination via K63 linkage is used as a chromatin-recruitment tool and not as a degradation tool. The K63-linked ubiquitin chain serves as an anchor for assembling and holding the BRCA1 downstream protein, and its binding partners, in place, in order to conduct additional concerted DSB repair functions.
The protein components of the RNF8-BRCA1-dependent chromatin-loading cascade cooperate in this signaling response to a DSB. Disruption of any of the genes encoding these proteins results in a common cellular outcome, namely, increased sensitivity to ionizing radiation. Still, the degree of radiation sensitivity varies greatly, depending on which gene is disrupted. To date, there is no genetic model system to study the epistatic relationship of the genes in this pathway. For instance, BRCA1 disruption causes a more severe effect than disruption of the other proteins in this pathway, suggesting that BRCA1 might be recruited to chromatin by other mechanisms, or, that it participates in other pathways in addition to the DSB response. Indeed, the BRCA1/BARD1 heterodimer was recently shown to play a role in mitotic spindle assembly (Joukov et al., 2006).
Several questions emerge from these findings. Are RNF8 and BRCA1/BARD1 the only ubiquitin E3 ligases required to complete the task of the DSB response, or do other E3 ligases cooperate with UBC13? Consistent with this notion, UBC13 ablation renders cells more sensitive to IR than does ablation of RNF8 (Huen et al., 2007; Kolas et al., 2007; Zhao et al., 2007). However, it should be noted that one study utilized genetic knockout of UBC13 in chicken DT40 cells whereas the other studies applied siRNA knockdown in HeLa cells.[SC8] Second, is H2AX the only RNF8 substrate involved in the recruitment cascade, or are other proteins K63 polyubiquitinated at the site of the DSB? For instance, the more abundant histone H2A is a possible substrate during this response (Mailand et al., 2007). Third, is RAP80 the only UBD-containing protein that is available for docking on the K63-linkage polyubiquitinated substrate? Fourth, how is the complex of polyubiquitinated proteins disassembled? Is deubiquitinating enzyme activity solely involved, or do some locally polyubiquitinated proteins also undergo degradation after the DSB has been repaired?
Role of deubiquitinating enzymes in regulating chromatin loading of DNA repair complexes
Deubiquitinating enzymes (DUBs) mediate the removal and processing of ubiquitin and are functionally important for regulated chromatin binding during the DDR. There are at least 95 deubiquitinating enzymes in human, and these proteases can be divided into five subfamilies including the USP subfamily (58 members), the UCH subfamily (4 members), the MJD subfamily (5 members), the OTU subfamily (14 members), and the JAMM subfamily (14 members) (Nijman et al., 2005). The DUB enzymes vary greatly in terms of their substrate selection (i.e., monoubiquitinated versus polyubiquitinated substrates), their mechanism of action (i.e, cysteine proteases or metalloproteases, in the case of the JAMM subfamily), their cellular localization, and their specificity for different cellular pathways. Moreover, we recently reported that some human deubiquitinating enzymes require the association of regulatory proteins for their activity (Cohn et al., 2007).
Increasing evidence supports a central role of deubiquitinating enzymes in controlling the DNA damage response. Several DUBs are themselves phosphorylated substrates of the ATM and ATR kinases, suggesting that they play a role in the cellular response to DNA damage (Matsuoka et al., 2007). USP28 is required for the stabilization of Chk2 and 53BP1 (Zhang et al., 2006), and USP1 is required for the deubiquitination of FANCD2 and FANCI (see below). Furthermore, USP1 disruption results in elevated Ub-PCNA levels and increased point mutagenesis (Huang et al., 2006).
The emerging view is that, in response to DNA damage, some DNA damage response proteins are 1) rapidly ubiquitinated, 2) translocated to, or retained in, chromatin and stabilized there by a ubiquitin binding factor and 3) deubiquitinated locally at the site of damaged chromatin. Thus, coupled ubiquitination and deubiquitination appears to be essential for some types of DNA damage responses. Analogously, recent studies indicate that coupled monoubiquitination and deubiquitination also is essential for normal transcriptional regulation (Emre and Berger, 2004; Weake and Workman, 2008).
Fanconi Anemia - a human genetic disease associated with defective loading of DNA repair proteins onto chromatin
A well-described human DNA repair pathway, the Fanconi Anemia Pathway (FA), also depends on sequential ubiquitination and deubiquitination events for its function. FA is a rare human autosomal recessive disease characterized by developmental abnormalities, cancer susceptibility, and cellular hypersensitivity to DNA crosslinking agents, such as Mitomycin C (MMC) (Wang, 2007). There are at least thirteen distinct complementation groups for Fanconi Anemia, and the genes corresponding to all thirteen subtypes have been identified (Wang, 2007). The thirteen Fanconi Anemia proteins cooperate in a common cellular pathway, resulting in the ubiquitin-mediated chromatin loading of protein complexes and ultimately in DNA crosslink repair (Figure 1). The exact mechanism of the repair reaction is not fully understood. However, it is currently believed that the process utilizes elements from the NER, TLS, and HR pathways, in addition to the 13 FA proteins (Thompson et al., 2005; Wang, 2007). [SC9]Interestingly, the FA pathway is activated by numerous types of genotoxic stress in addition to DNA crosslinks, including Hydroxy Urea-mediated replication fork arrest, ultraviolet light (UV) and ionizing radiation (IR), underscoring the complexity of the pathway. Disruption of this pathway leads to genomic instability and to the characteristic developmental abnormalities observed in Fanconi Anemia patients.
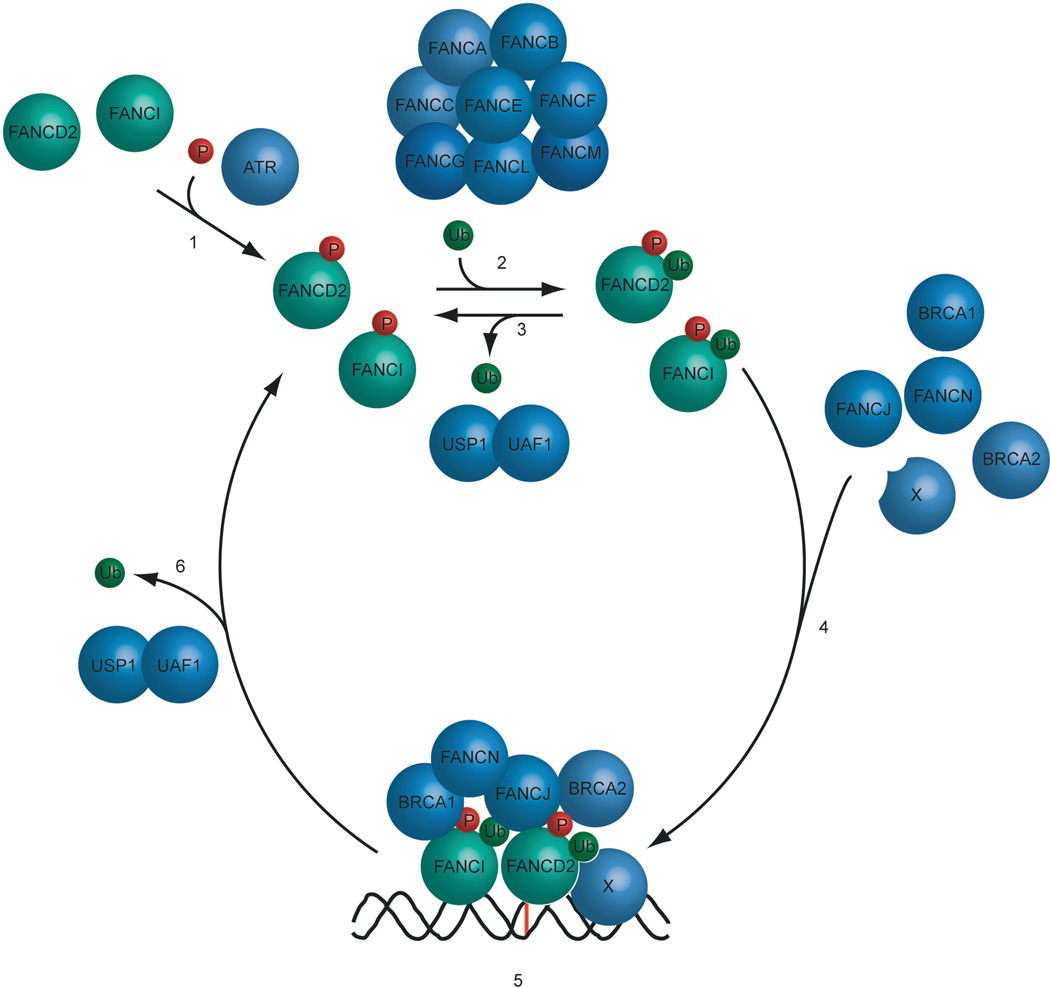
Figure 1. The Fanconi Anemia pathway regulates the loading of DNA repair complexes.
1) The FANCD2 and FANCI proteins are phosphorylated on multiple amino acid residues in response to a DNA crosslink. 2) The phosphorylated FANCD2 and FANCI proteins are monoubiquitinated by the FA core complex, containing of the FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL and FANCM protein subunits. FANCL is the E3 ligase subunit. 3) [SC16]In the absence of genotoxic stress FANCD2 and FANCI are deubiquitinated by the USP1/UAF1 complex to prevent the accumulation of monoubiquitinated FANCD2 and FANCI. In response to DNA damage, the level of USP1/UAF1 complex decreases, resulting in the cellular increase in ubiquitinated FANCD2 and FANCI. 4) Monoubiquitinated FANCD2 and FANCI translocate to sites of damage on the chromatin, putatively with the help of a recruitment protein X. On chromatin the proteins assemble around the site of damage together with other FA proteins, including FANCD1(BRCA2), FANCN(PALB2) and FANCJ(BRIP1,BACH1). 5) The assembled FA DNA repair complex repairs the DNA damage in conjunction with other DNA repair proteins. 6) Once the repair process has been completed, FANCD2 and FANCI are deubiquitinated by the USP1/UAF1 complex, resulting in a disassembly of the FA DNA repair complex and dissociation from the chromatin.
Specifically, eight of the FA proteins (A, B, C, E, F, G, L, M) are assembled in a nuclear complex (the FA core complex). This complex itself undergoes regulated chromatin binding. A fraction of the FANCM protein pool is constitutively anchored to chromatin via the FAAP24 protein (Ciccia et al., 2007; Kim et al., 2008). The remaining FA core complex subunits (the ABCEFGL complex) associate with FANCM during S phase of the cell cycle and in particular following DNA damage (Kim et al., 2008). The FANCM protein possesses DNA translocase and ATPase activity (Meetei et al., 2005) as well as DNA branch migration activity (Gari et al., 2008).
The FA core complex contains an E3 ligase (the FANCL subunit), and, in response to DNA damage or replication arrest, this ligase monoubiquitinates two downstream members of the pathway, namely FANCD2 (Meetei et al., 2003) and FANCI (Smogorzewska et al., 2007). Monoubiquitinated FANCD2 and FANCI, a fraction of which forms a heterodimer (the so-called ID complex), are then rapidly loaded onto chromatin. The mechanism of FANCD2/FANCI chromatin loading is not well understood, but it presumably entails the interaction with a protein containing a ubiquitin binding motif (UIM) or another Ub-interaction domain. However, it can not be excluded that the physical properties of the monoubiquitinated proteins enhance their affinities for DNA directly. Indeed, FANCD2 itself possesses some intrinsic DNA-binding activity (Park et al., 2005). More recent evidence suggests that a later deubiquitination of FANCD2 and FANCI by USP1/UAF1 might be required for efficient foci assembly and HR-mediated repair (J. Kim and A. D’Andrea, unpublished observation).
The function of monoubiquitinated FANCD2/FANCI in chromatin is unknown. Indeed, both proteins lack recognizable enzymatic domains. We hypothesize that these modified proteins communicate with the more downstream FA proteins, FANCD1(BRCA2), FANCN(PALB2), and FANCJ([SC10]BRIP1, BACH1) (Wang, 2007). The disruption of FANCD2 (Nakanishi et al., 2005) or FANCI (Smogorzewska et al., 2007) results in a reduction of HR-mediated repair, suggesting that these proteins play a role, either directly or indirectly, in regulating the downstream function of FANCD1(BRCA2).
Comparison of the RNF8-BRCA1 pathway and the FA pathway
Mechanistically, the DSB reponse and the FA pathway share multiple elements. Components of the DSB pathway and the FA pathway are aligned side-by-side in Figure 2. Here we comment on some of the common features between the two pathways, using the FA pathway as a model. In the FA pathway, in response to the DNA crosslinker MMC, the central protein of the pathway, FANCD2, is phosphorylated on several residues by the ATR kinase (Andreassen et al., 2004; Taniguchi et al., 2002). Phosphorylated FANCD2 is subsequently monoubiquitinated by the FA core complex, which contains the E3 ligase activity. Monoubiquitinated FANCD2 is then recruited to and loaded onto chromatin, likely assisted by an unknown factor containing a UBD. Recent evidence indicates that the FANCM(FAAP24) subunit of the FA core complex is required at some stage of this targeted loading event (Gari et al., 2008; Kim et al., 2008). Once the monoubiquitinated FANCD2 protein is correctly localized on sites of DNA damage in the chromatin, it associates with other DNA repair proteins to initiate the DNA repair process. When the DNA crosslink has been repaired, the USP1/UAF1 deubiquitinating enzyme complex deubiquitinates FANCD2, causing disassembly of the DNA repair complex, and completion of the DNA repair process (Cohn et al., 2007; Oestergaard et al., 2007). USP1/UAF1-mediated FANCD2 deubiquitination is critical for DNA crosslink repair. In addition to signaling the completion of the DNA repair reaction, regulated deubiquitination might also allow the recycling of free non-ubiquitinated [SC11]FANCD2 protein. Each [SC12]pathway has missing links, yet the similarity of the pathways allows an important generalization, namely, that DNA damage activated protein assembly and disassembly, mediated by specific E3 ligases, ubiquitin substrates, ubiquitin binding proteins, and DUBs are critical for the successful activation and completion of the different DNA repair pathways.
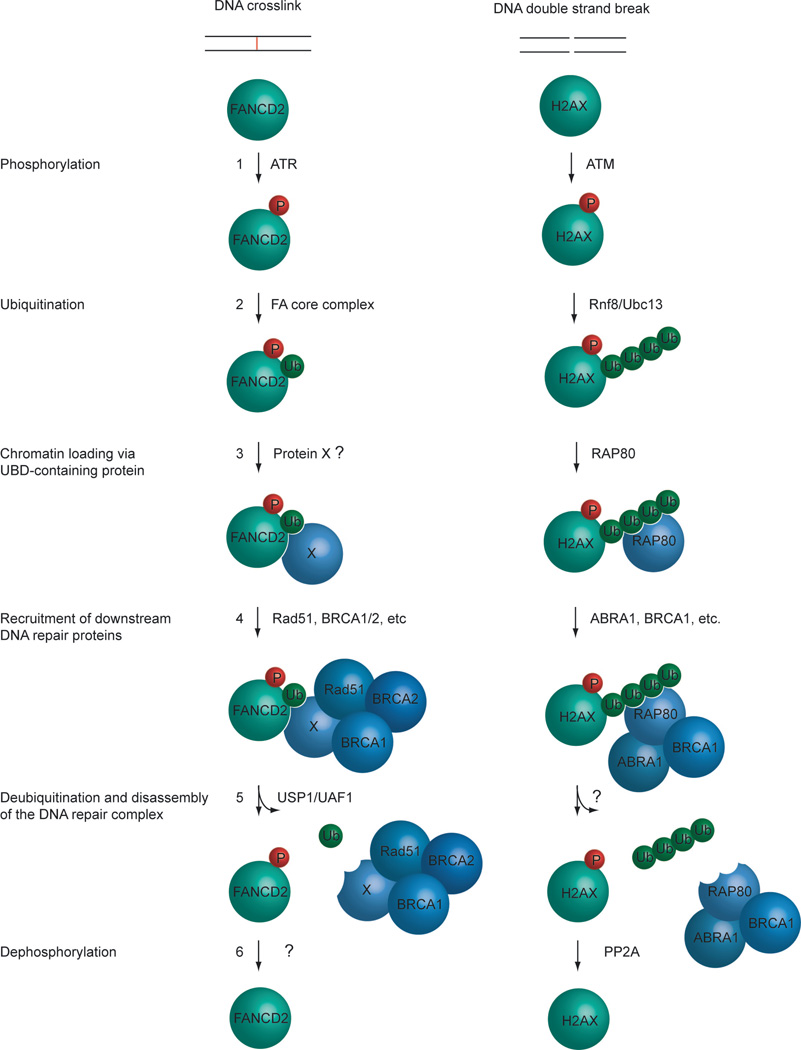
Figure 2. Conserved elements of the Fanconi Anemia and DSB repair pathways.
1) In response to either a DNA crosslink or double strand break, ATR or ATM phosphorylates FANCD2 or H2AX, respectively. 2) The phosphorylated proteins are now substrates for a ubiquitination event, carried out either by the FA core complex or the RNF8/UBC13 complex. 3) Monoubiquitinated FANCD2 protein can now be recruited to sites of damage on the chromatin by a recruitment factor, Protein X. Polyubiquitinated H2AX recruits RAP80 to sites of damage on the chromatin. 4) The properly localized FANCD2 protein associates with other DNA repair proteins and the DNA damage is repaired. In addition to RAP80, other DNA repair proteins are recruited to H2AX, and the DNA damage is repaired. 5) Once the DNA repair process has been completed, the recruited protein complex must be disassembled. This is achieved by deubiquitinating the FANCD2 protein by the USP1/UAF1 complex, and H2AX by an unknown deubiquitinating enzyme. 6) After the DNA repair process has been completed, FANCD2 is dephosphorylated by an unknown phosphatase, and H2AX is dephosphorylated by PP2A (Chowdhury et al., 2005). For simplicity, this model only shows a subset of the proteins known to participate in ICL and DSB repair. For instance, only FANCD2 is shown, although the paralog protein, FANCI, is believed to act in a similar fashion.
Chromatin accessibility and DNA damage and repair
The serial recruitment and disassembly of protein complexes at sites of damaged DNA occurs in the context of chromatin. The basic unit of chromatin is the nucleosome, consisting of a histone octamer composed of two of each of the core histone proteins, H2A, H2B, H3 and H4, wrapped in 146 nucleotides of DNA (Luger et al., 1997). The core histones are rich in positively charged basic amino acids, which, owing to their electrostatic interaction with the negatively charged DNA, result in the stable nucleosomal protein-DNA complexes. A DNA linker region between each nucleosome interacts with the histone H1 protein. The basic structure of repeating nucleosomes, termed the 10 nm fiber, can be additionally compacted into a higher order structure, the 30 nm fiber. The chromatin can be further divided into two principally different structures, euchromatin and heterochromatin, based on the local compaction, accessibility, and transcriptional activity. Euchromatin is considered more open and more transcriptional active than the more condensed heterochromatin (Trojer and Reinberg, 2007).
The degree of chromatin compaction appears to determine, at least in part, the extent of DNA damage and the likelihood of DNA repair. For instance, studies in which histones were gradually dissociated from the chromatin by increasing the salt concentration showed that looser compaction of the chromatin increases the frequency of DSBs following ionizing radiation in vitro (Sak et al., 2000).
The measurement of DNA repair efficiency for different chromatin templates historically has been delayed owing to the lack of reconstituted DNA repair systems. One DNA repair pathway that has been successfully reconstituted in vitro is Nucleotide Excision Repair (NER), [SC13]which typically removes monoadducts such as CPDs from the DNA. An in vitro NER assay showed that chromatinized templates are repaired more slowly than naked DNA (Gong et al., 2005). Furthermore, nucleosome remodeling complexes, such as the SWI/SNF complex, that can reposition nucleosomes in an ATP-dependent mechanism, accelerate NER in vitro (Gong et al., 2005).
Taken together, local chromatin structure appears to partially determine both the likelihood of a DNA sequence to be assaulted by damage and the feasibility of DNA repair. Most DNA repair pathways, including the FA pathway, are likely to be affected by the chromatin structure surrounding the DNA damage.
Additionally, it is possible that some of the described[SC14] histone modifications that occur in response to DNA damage, for instance H2AX polyubiquitination, in addition to enhancing the recruitment of DNA repair factors, also might affect the local chromatin structure and compaction. An alternation in chromatin compaction could affect the accessibility for DNA repair factors. Indeed, it is well known that acetylation and methylation of various histones affect chromatin structure and accessibility (Trojer and Reinberg, 2007). Furthermore, histone modifications on different nucleosomes might affect the recruitment of protein complexes through cooperative binding.(Ruthenburg et al., 2007). Whether this latter mechanism also affects the DNA repair process remains unknown.
Future challenges
Although studies in recent years have provided significant insight into the mechanisms of DNA repair in general and the FA pathway in particular, a number of questions remain. Monoubiquitination of the central player of the FA pathway, FANCD2, is crucial for its function and for an intact FA pathway. However, the mechanistic consequence of this modification remains unclear. In particular, the faithful monoubiquitination of the FANCD2 protein by the FA core complex has not yet been reconstituted in vitro. We propose that a recruitment factor mediates the translocation of monoubiquitinated FANCD2 to sites of DNA damage in the chromatin; however the identity of this factor remains unknown. As discussed extensively in this review, coordinated chromatin loading of DNA repair factors is essential for most DNA repair pathways. Elucidating the exact timing, order, and mechanism of recruitment of these factors will likely bring us closer to a better understanding of how these DNA repair pathways operate. A complete understanding of the molecular mechanisms underlying DNA repair, however, will likely depend on an in vitro reconstitution of DNA replication and of DNA repair, using purified components.
Acknowledements
We are grateful to Drs. Younghoon Kee, Lucian Moldovan and Vladimir Joukov for critical reading of the manuscript and helpful discussions. We apologize to colleagues whose work was not cited or was only referred to by citing review articles due to space restrictions. This work was supported by NIH grant 1PO1CA092584.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, de Winter JP, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D'Andrea AD. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Emre NC, Berger SL. Histone H2B ubiquitylation and deubiquitylation in genomic regulation. Cold Spring Harb Symp Quant Biol. 2004;69:289–299. doi: 10.1101/sqb.2004.69.289. [DOI] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- Gong F, Kwon Y, Smerdon MJ. Nucleotide excision repair in chromatin and the right of entry. DNA Repair (Amst) 2005;4:884–896. doi: 10.1016/j.dnarep.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Huang TT, D'Andrea AD. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol. 2006;7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov V, Groen AC, Prokhorova T, Gerson R, White E, Rodriguez A, Walter JC, Livingston DM. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell. 2006;127:539–552. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Kennedy RD, D'Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol. 2006;24:3799–3808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- Kim JM, Kee Y, Gurtan A, D'Andrea AD. Cell cycle dependent chromatin loading of the fanconi anemia core complex by FANCM/FAAP24. Blood. 2008 doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirchandani KD, D'Andrea AD. The Fanconi anemia/BRCA pathway: a coordinator of cross-link repair. Exp Cell Res. 2006;312:2647–2653. doi: 10.1016/j.yexcr.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Oestergaard VH, Langevin F, Kuiken HJ, Pace P, Niedzwiedz W, Simpson LJ, Ohzeki M, Takata M, Sale JE, Patel KJ. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell. 2007;28:798–809. doi: 10.1016/j.molcel.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WH, Margossian S, Horwitz AA, Simons AM, D'Andrea AD, Parvin JD. Direct DNA binding activity of the Fanconi anemia D2 protein. J Biol Chem. 2005;280:23593–23598. doi: 10.1074/jbc.M503730200. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sak A, Stuschke M, Wurm R, Budach V. Protection of DNA from radiation-induced double-strand breaks: influence of replication and nuclear proteins. Int J Radiat Biol. 2000;76:749–756. doi: 10.1080/09553000050028896. [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997a;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997b;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, Elledge SJ. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT. Cellular responses to DNA damage: one signal, multiple choices. Annu Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, Hanaoka F. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, Lane WS, Kastan MB, D'Andrea AD. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Hinz JM, Yamada NA, Jones NJ. How Fanconi anemia proteins promote the four Rs: replication, recombination, repair, and recovery. Environ Mol Mutagen. 2005;45:128–142. doi: 10.1002/em.20109. [DOI] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]