Abstract
Introduction
The characteristics and antimicrobial resistance profiles of Staphylococcus aureus differs according to geographical regions and in relation to antibiotic usage. The aim of this study was to determine the biochemical characteristics of the prevalent S. aureus from Ekiti State, Nigeria, and to evaluate three commonly used disk diffusion methods (cefoxitin, oxacillin, and methicillin) for the detection of methicillin resistance in comparison with mecA gene detection by polymerase chain reaction.
Materials and methods
A total of 208 isolates of S. aureus recovered from clinical specimens were included in this study. Standard microbiological procedures were employed in isolating the strains. Susceptibility of each isolate to methicillin (5 μg), oxacillin (1 μg), and cefoxitin (30 μg) was carried out using the modified Kirby–Bauer/Clinical and Laboratory Standard Institute disk diffusion technique. They were also tested against panels of antibiotics including vancomycin. The conventional polymerase chain reaction method was used to detect the presence of the mecA gene.
Results
Phenotypic resistance to methicillin, oxacillin, and cefoxitin were 32.7%, 40.3%, and 46.5%, respectively. The mecA gene was detected in 40 isolates, giving a methicillin-resistant S. aureus (MRSA) prevalence of 19.2%. The S. aureus isolates were resistant to penicillin (82.7%) and tetracycline (65.4%), but largely susceptible to erythromycin (78.8% sensitive), pefloxacin (82.7%), and gentamicin (88.5%). When compared to the mecA gene as the gold standard for MRSA detection, methicillin, oxacillin, and cefoxitin gave sensitivity rates of 70%, 80%, and 100%, and specificity rates of 76.2%, 69.1%, and 78.5% respectively.
Conclusion
When compared with previous studies employing mecA polymerase chain reaction for MRSA detection, the prevalence of 19.2% reported in Ekiti State, Nigeria in this study is an indication of gradual rise in the prevalence of MRSA in Nigeria. A cefoxitin (30 μg) disk diffusion test is recommended above methicillin and oxacillin for the phenotypic detection of MRSA in clinical laboratories.
Keywords: evaluation, disk diffusion, mecA gene, MRSA, Nigeria
Introduction
Staphylococcus aureus is an important pathogen in human infections and is implicated in a wide variety of infections, from mild skin infections to more serious and invasive infections, including sepsis, pneumonia, endocarditis, deep-seated abscesses, and toxinoses including food poisoning and toxic shock syndrome.1
Staphylococci infections are treated with antibiotics. Over the decades, some strains of staphylococci such as methicillin-resistant S. aureus (MRSA) have become resistant to antibiotics that once destroyed it. MRSA strains, though first reported in 1961, did not become a major problem until the late 1970s and early 1980s when outbreaks were reported from many parts of the world.2 MRSAs are resistant to methicillin, amoxicillin, penicillin, oxacillin, and many other antibiotics. They are also resistant clinically to all beta-lactam antibiotics, despite apparent in vitro efficacy. It is also important to note that MRSA are often multidrug-resistant and are resistant to antibiotics such as the macrolides and aminoglycosides.1
Until recently, MRSA was predominantly a nosocomial pathogen that caused hospital-acquired infections, but MRSA strains are now being increasingly isolated from community-acquired infections as well. Vancomycin has been the antibiotic of choice to treat MRSA infections, and the emergence of vancomycin nonsusceptible S. aureus reported in recent years is a cause of great public health concern and has made therapy of MRSA infections even more challenging for clinicians.3 Numerous studies have indicated that S. aureus is among the most frequently encountered microorganisms in medical microbiology laboratories in Nigeria.4–7
Resistance in MRSA is mediated by the presence of penicillin-binding protein (PBP)-2a, encoded by the chromosomal mecA gene.8 PBP-2a is a cell wall enzyme that has low affinity for β-lactam antibiotics, and allows cell wall formation in concentrations of a drug that render the other PBPs inactive.9 The gene responsible for methicillin resistance in S. aureus is mecA,10 which is part of a 21-kb to 60-kb staphylococcal chromosome cassette mec, a mobile genetic element that may also contain genetic structures such as Tn554, pUB110, and pT181, which encode resistance to non-β-lactam antibiotics.11 There are four classes of the mec gene complex, of which only classes A (the original clone) and B have been identified in S. aureus, while classes C and D have been identified in coagulase negative staphylococci. These classes of staphylococcal chromosome cassette mec differ from each other.12
In line with the worldwide spread of MRSA, there are increasing reports of MRSA strains involved in clinical infections in Nigeria.1,12–15 Most of these strains are detected by a phenotypic method using oxacillin and methicillin disks, which may not reflect the true prevalence because of many factors that affect the performance and reliability of the disk diffusion test. The cefoxitin disk was introduced a few years ago to test S. aureus for expression of methicillin resistance, and it was found to be reliable and even better than the oxacillin disk.16 Although detection of the mecA gene by polymerase chain reaction (PCR) is the gold standard test for MRSA confirmation, only a few studies in Nigeria have employed this gold standard for MRSA detection.14 This test is also not available for use in most routine diagnostic laboratories in Nigeria. Therefore, the onus lies on the laboratory to find the most reliable disk diffusion method for MRSA screening.
The objectives of this study are to determine the prevalence rate of methicillin resistance and antibiotic susceptibility patterns of S. aureus clinical isolates in Ekiti State, Nigeria, and to compare the performance of methicillin, oxacillin, and cefoxitin disk diffusion tests in detecting methicillin resistance with the gold standard mecA gene PCR.
Materials and methods
Specimens from various clinical infections were received at the Medical Microbiology Laboratory of University Teaching Hospital, Ado-Ekiti, between June 2010 and December 2010. The specimens were processed according to recommended guidelines by first culturing on Mannitol salt agar (Oxoid Ltd, Cambridge, UK) and incubating at 37°C for 24 hours.17S. aureus isolates were identified by colony morphology and fermentation reaction on Mannitol salt agar, Gram stain reaction, catalase, tube coagulase, and deoxyribonuclease tests.17
Methicillin resistance screening
This was done according to Clinical and Laboratory Standard Institute guidelines.18 Pure cultures of each S. aureus isolate and control strain were used. Five colonies of each isolate and control strain were transferred to 5 mL of nutrient broth and were cultured overnight at 35°C. The overnight cultures were then diluted with sterile saline (0.85% NaCl) in Bijou bottles, and their turbidity was compared to 0.5 McFarland standards. The inocula were spread with a sterile cotton wool swab on Mueller–Hinton agar supplemented with 2% NaCl (Oxoid Ltd). Oxacillin (1 μg), methicillin (5 μg), and cefoxitin (30 μg) disks (Oxoid Ltd) were applied with sterile forceps, and the agar plates were incubated for a full 24 hours at 35°C aerobically. The inhibition zone diameter (ZD) for each isolate was measured and compared with the ZD interpretative standard.18 MRSA National Center for Type Cultures 119040 was used as a control strain.
Antimicrobial susceptibility testing
The susceptibility of all S. aureus isolates to penicillin (ten units), gentamicin (10 μg), erythromycin (15 μg), pefloxacin (5 μg), tetracycline (30 μg), and vancomycin (30 μg) was determined using the modified Kirby–Bauer disk diffusion method.19 The isolates were considered sensitive, intermediately resistant, and resistant based on the Clinical and Laboratory Standard Institute guidelines.18
Molecular detection of methicillin resistance (mecA gene)
DNA extraction
Deoxyribonucleic acid (DNA) extraction from each S. aureus isolate was carried out by modification of the simple crude extraction methods previously described for Salmonella enterica20 and Streptococcus pneumonia.21 Twenty-four-hour-old pure colonies of S. aureus were emulsified in 200 μL of deoxygenated water in Eppendorf tubes (Eppendorf North America, Hauppauge, NY, USA) and sealed with cellophane. The mixture was boiled at 100°C for 10 minutes in a water bath, cooled on ice, and then centrifuged at 15,000 × g for 30 seconds. The supernatant containing the DNA was stored at 4°C before use. Aliquots of 5 μL of template DNA were used for PCR.
PCR for mecA gene
The mecA DNA was amplified with the primers mecA F1 –AAA ATC GAT GGT AAA GGT TGG C, and mecA B1 –AGT TCT GCA GTA CCG GAT TTG C. PCR was carried out in a GeneAmp® PCR System 9700 (Life Technologies, Carlsbad CA, USA) as previously described22 as follows: pre-denaturation at 95°C for 5 minutes, 40 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 1 minute, and final extension at 72°C for 5 minutes. The products were detected by gel electrophoresis using 1.5% w/v agarose gel in 1 × Tris-buffered ethylenediaminetetraacetic acid for 2 hours at 80 V The agarose gel was stained with 1 μg/mL of ethidium bromide (Sigma-Aldrich, St Louis, MO, USA) solution and visualized under ultraviolet illumination and then photographed (SynGene Bioimaging System; Syngene UK, Cambridge, UK). The molecular weight marker band was used to estimate the molecular size of the amplified mecA gene, which was 533 base pairs. MRSA National Center for Type Cultures 119040 served as the positive control in the PCR reaction.
Data analysis
Data were entered into a Windows 7 (Microsoft Corporation, Redmond WA, USA) laptop computer with the Statistical Package for the Social Sciences version 17.0 software package (SPSS; IBM Corporation, Armonk, NY, USA). Frequency tables were generated and analysis was performed using the appropriate statistical tools.
The sensitivity, specificity, positive predictive value and negative predictive value of the cefoxitin, oxacillin, and methicillin disk diffusion test in detecting phenotypic methicillin resistance in the S. aureus isolates using the presence of the mecA gene as “gold standard” were calculated with the formulas:
| (1) |
| (2) |
| (3) |
| (4) |
where TP (true positive) means that the disk showed the isolate to be resistant when the mecA gene is present, TN (true negative) means that the disk showed the isolate to be susceptible when the mecA gene is absent, FP (false positive) means that the disk showed the isolate to be resistant when the mecA gene is absent, and FN (false negative) means that the disk showed the isolate to be susceptible when the mecA gene is present.
Results
A total of 208 S. aureus isolates recovered from various clinical specimens (Table 1) were studied. Clinical data were not included. The antibiotic susceptibility pattern of the 208 isolates showing varying degrees of resistance is shown in Table 2. The highest degree of resistance was to penicillin G (82.7%; 172/208). All the isolates were susceptible to vancomycin (100%; 208/208), and were also highly sensitive to gentamicin (84.6%; 176/208) and sparfloxacin (73.1%; 152/208).
Table 1.
Staphylococcus aureus isolates from clinical specimens in Ekiti State, Nigeria
| Clinical specimens | Number of isolates | mecA genepositive (%) (MRSA) | mecA genenegative (%) (MSSA) |
|---|---|---|---|
| Blood culture | 23 | 0 (0) | 23 (100) |
| Urine | 53 | 8 (15.1) | 45 (84.9) |
| Sputum | 18 | 0 (0) | 18 (100) |
| Anterior nares | 20 | 0 (0) | 20 (100) |
| Pleural fluid | 5 | 0 (0) | 5 (100) |
| Endocervical swab | 11 | 0 (0) | 11 (100) |
| High vaginal swab | 10 | 4 (40) | 6 (60) |
| Semen | 8 | 0 (0) | 8 (100) |
| Wound swab | 46 | 24 (52.2) | 22 (47.8) |
| Ear swab | 14 | 4 (28.6) | 10 (71.4) |
| Total | 208 | 40 (19.2) | 168 (80.8) |
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; Mssa, methicillin-sensitive Staphylococcus aureus.
Table 2.
Antibiotic susceptibility of Staphylococcus aureus isolates from clinical specimens in Ekiti State, Nigeria
| Antibiotic | Susceptible number (%) | Intermediate number (%) | Resistant number (%) |
|---|---|---|---|
| Penicillin G | 36 (17.3) | – | 172 (82.7) |
| Cefoxitin | 112 (53.8) | – | 96 (46.2) |
| Methicillin | 140 (67.3) | – | 68 (32.7) |
| Oxacillin | 124 (59.6) | – | 84 (40.3) |
| Erythromycin | 100 (48.1) | 64 (30.7) | 44 (21.2) |
| Gentamicin | 176 (84.6) | 8 (3.8) | 24 (11.5) |
| Pefloxacin | 124 (59.6) | 48 (23.1) | 36 (17.3) |
| Sparfloxacin | 152 (73.1) | 16 (7.7) | 40 (19.2) |
| Tetracycline | 64 (30.8) | 8 (3.8) | 136 (65.4) |
| Vancomycin | 208 (100) | – | 0 (0) |
Forty of the 208 S. aureus isolates were mecA genepositive on PCR analysis, giving a MRSA prevalence rate of 19.2% while 168 were mecA gene-negative (Table 1). Of the 40 mecA gene-positive isolates, the cefoxitin disk diffusion test detected phenotypic resistance (ZD ≤ 21 mm) in 40 of 40 (100%), the oxacillin disk diffusion test detected resistance (ZD ≤ 10 mm) in 32 of 40 (80%), and the methicillin disk diffusion test detected resistance (ZD ≤ 9 mm) in 28 of 40 (70%) S. aureus isolates. In the 168 mecA-negative S. aureus isolates, the cefoxitin disk detected resistance (false) in 36 (21.4%) isolates, the oxacillin disk detected resistance (false) in 52 (30.9%) isolates, and the methicillin disk detected resistance (false) in 40 (23.8%) isolates (Tables 3–5). Using the mecA gene as the “gold standard” test, the sensitivity of cefoxitin, oxacillin, and methicillin disks for MRSA detection were 100%, 80%, and 70% while the specificity rates were 78.5%, 69.1%, and 76.2%, respectively.
Table 3.
Comparison of cefoxitin disk diffusion test and mecA gene detection
| ZD ≤ 21 mm (R) | ZD ≥ 22 mm (S) | Total | |
|---|---|---|---|
| Cefoxitin ZD (mm) | |||
| + | 40 | 0 | 40 |
| mecA gene | |||
| − | 36 | 132 | 168 |
| Total | 76 | 132 | 208 |
Notes: Sensitivity = 100%; specificity = 78.6%; PPV = 52.6%; NPV = 100%.
Abbreviations: ZD, zone diameter; R, resistant; S, sensitive; PPV, positive predictive value; NPV, negative predictive value.
Table 5.
Comparison of methicillin disk diffusion test and mecA gene detection
| ZD ≤ 9 mm (R) | ZD ≥ 14 mm (S) | Total | |
|---|---|---|---|
| Methicillin ZD (mm) | |||
| + | 28 | 12 | 40 |
| mecA gene | |||
| − | 40 | 128 | 168 |
| Total | 76 | 132 | 208 |
Notes: Sensitivity = 70%; specificity = 76.2%; PPV = 36.8%; NPV = 96.9%.
Abbreviations: ZD, zone diameter; R, resistant; S, sensitive; PPV, positive predictive value; NPV, negative predictive value.
Figure 1 shows the agarose gel electrophoresis of PCR products of representative isolates with the mecA-positive isolate showing band at approximately 533 base pairs.
Figure 1.
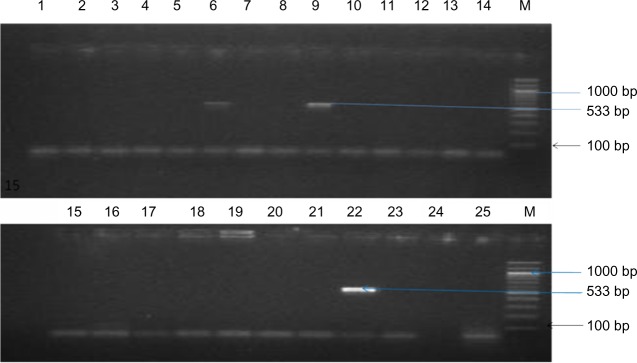
Agarose gel electrophoresis of PCR products of representative isolates.
Notes: The mecA gene is indicated by the amplification of 533 base pair products. Lanes 6 and 9 are positive for mecA, as indicated by the 533 base pair PCR product. Lanes 1–5, 7, 8, 10–21, 24, and 25 were negative for mecA. Lane 22 was the MRSA-positive control (NCTC 119040), lane 23 was the negative control (water), and lane M was the molecular weight size marker.
Abbreviations: PCR, polymerase chain reaction; MRSA, methicillin-resistant Staphylococcus aureus; NCTC, National Center for Type Cultures; M, molecular weight.
Discussion
Antibiotic resistance has been described as one of the paramount microbial threats of the 21st century.23S. aureus has always been a stumbling block for antimicrobial chemotherapy, and the introduction of new classes of antimicrobial agents is usually followed by the emergence of resistant forms of this pathogen.24 Therefore, surveillance of the antimicrobial susceptibility patterns of S. aureus is of utmost importance in understanding new and emerging resistance trends, and in the management of both hospital- and community-acquired infections.
In this study, S. aureus was isolated from various clinical specimens obtained from different infective foci, with the highest being from urine. S. aureus is ubiquitous in its distribution, and although it is a normal fora of the anterior nares, skin, and genital tract of humans, it can cause infections in virtually any organ or system of the body; infections of wounds, skin, soft tissue, blood, and the lower urinary tract are particularly common. In a similar study carried out by Shittu and Lin,25 more than 80% of the total number of isolates recovered were from infected wounds.
The antimicrobial susceptibility pattern of S. aureus isolates varies with place and time. The highest degree of antibiotic resistance in this study was to penicillin G (82.7%) and tetracycline (65.4%). These two drugs are available over the counter and are widely used in Nigeria without medical authorization.25 Resistance to other antibiotics such as erythromycin, gentamicin, and fluoroquinolones appear to be relatively low, while all the isolates were susceptible to vancomycin, which agrees with previous reports.23,25–27 This second category of antibiotics is not readily available for consumption to a majority of the populace because of its high cost. Vancomycin in particular is not available for routine clinical use in Nigeria; hence, most S. aureus isolates, including MRSA, are still susceptible to the drug,1,12–15 although there are few reports of the emergence of vancomycin-intermediate and vancomycin-resistant S. aureus clinical isolates in some centers in Nigeria.28,29
Phenotypic resistance to methicillin, oxacillin, and cefoxitin were 32.7%, 40.3%, and 46.5%, respectively. These three antibiotics are commonly used in the disk diffusion susceptibility test by the clinical diagnostic laboratory in our environment to screen staphylococci isolates for resistance to methicillin. All three antibiotic disks overestimated the prevalence of MRSA, as the mecA gene that encodes resistance to methicillin was detected by PCR in 19.2% of the S. aureus isolates. The implication is that previous reports on MRSA in our environment, which were based on these disk diffusion tests, overestimate MRSA prevalence.1,12,30 The MRSA prevalence rate of 19.2% in this study compared favorably with the rate of 22.2% reported in a recent mecA PCR study of MRSA isolates obtained from three major tertiary health care institutions in southwestern Nigeria.31 When these results are compared with those of previous studies that reported 1.4%32 and 1.5%25 prevalence rates of MRSA using mecA gene PCR, we can conclude that there is a gradual increase in the prevalence of MRSA strains in Nigeria.
When compared to the gold standard mecA gene test for MRSA detection, the cefoxitin disk (30 μg) gave the highest sensitivity (100%) and specificity (78.5%), performing better than oxacillin and methicillin. This finding is in agreement with previous reports.16,33 From the results of our study and those of other studies,16,33 we recommend that the cefoxitin disk should replace the oxacillin disk or methicillin disk as a screening tool for MRSA. One advantage of the cefoxitin disk test is that incubation can take place within 18 hours at 36°C or 37°C, the usual temperature of an aerobic incubator in most microbiology diagnostic laboratories, whereas the optimal incubation temperature for oxacillin or methicillin is 35°C for a full 24 hours.16
Table 4.
Comparison of oxacillin disk diffusion test and mecA gene detection
| ZD ≤ 10 mm (R) | ZD ≥ 13 mm (S) | Total | |
|---|---|---|---|
| Oxacillin ZD (mm) | |||
| + | 32 | 8 | 40 |
| mecA gene | |||
| − | 52 | 116 | 168 |
| Total | 84 | 124 | 208 |
Notes: Sensitivity = 80%; specificity = 69.1%; PPV = 38.1%; NPV = 93.5%.
Abbreviations: ZD, zone diameter; R, resistant; S, sensitive; PPV, positive predictive value; NPV, negative predictive value.
Acknowledgments
The molecular biology works were all carried out in the Molecular Biology Laboratory, Department of Biomedical Sciences, Ladoke Akintola University of Technology, Mercy Land Campus, Osogbo, Nigeria. The assistance of the Management of Ladoke Akintola University of Technology is appreciated. We also wish to thank Mr Oyenike for his technical assistance.
Footnotes
Disclosure
The authors report no conflilcts of interest in this work.
References
- 1.Taiwo SS, Onile BA, Akanbi AA., II Methicillin-resistant Staphylococcus aureus (MRSA) isolates in Ilorin, Nigeria. African Journal of Clinical and Experimental Microbiology. 2004;5(2):189–197. [Google Scholar]
- 2.Grubb WB. Genetics of MRSA. Rev Med Microbiol. 1998;9(3):153–162. [Google Scholar]
- 3.Kim HB, Jang HC, Nam HJ, et al. In vitro activities of 28 antimicrobial agents against Staphylococcus aureus isolates from tertiary-care hospitals in Korea: a nationwide survey. Antimicrob Agents Chemother. 2004;48(4):1124–1127. doi: 10.1128/AAC.48.4.1124-1127.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ako-Nai AK, Lamikanra AB, Onipede AO. Incidence of pathogenic microorganisms in clinical specimens from hospitals in south-western Nigeria. East Afr Med J. 1995;72(7):436–441. [PubMed] [Google Scholar]
- 5.Oliveira DC, Tomasz A, de Lencastre H. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb Drug Resist. 2001;7(4):349–361. doi: 10.1089/10766290152773365. [DOI] [PubMed] [Google Scholar]
- 6.Adejuyigbe EA, Adeodu OO, Ako-Nai KA, Taiwo O, Owa JA. Septicaemia in high risk neonates at a teaching hospital in Ile-Ife, Nigeria. East Afr Med J. 2001;78(10):540–543. doi: 10.4314/eamj.v78i10.8965. [DOI] [PubMed] [Google Scholar]
- 7.Ako-Nai AK, Oluga FA, Onipede AO, Adejuyigbe EA, Amusa YB. The characterization of bacterial isolates from acute otitis media in Ile-Ife, southwestern Nigeria. J Trop Pediatr. 2002;48(1):15–23. doi: 10.1093/tropej/48.1.15. [DOI] [PubMed] [Google Scholar]
- 8.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci U S A. 2002;99(11):7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Zu’bi E, Bdour S, Shehabi AA. Antibiotic resistance patterns of mecA-positive Staphylococcus aureus isolates from clinical specimens and nasal carriage. Microb Drug Resist. 2004;10(4):321–324. doi: 10.1089/mdr.2004.10.321. [DOI] [PubMed] [Google Scholar]
- 10.Tenover FC, Gaynes RP. The epidemiology of Staphylococcus infections. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram-Positive Pathogens. Washington DC; ASM Press; 2000. pp. 414–421. [Google Scholar]
- 11.Holmes A, Ganner M, McGuane S, Pitt TL, Cookson BD, Kearns AM. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. J Clin Microbiol. 2005;43(5):2384–2390. doi: 10.1128/JCM.43.5.2384-2390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okesola AO, Oni AA, Bakare RA. Prevalence and antibiotic sensitivity pattern of methicillin-resistant Staphylococcus aureus in Ibadan, Nigeria. J Hosp Infect. 1999;41(1):74–75. doi: 10.1016/s0195-6701(99)90041-5. [DOI] [PubMed] [Google Scholar]
- 13.Taiwo SS, Bamidele M, Omonigbehin EA, et al. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Ilorin, Nigeria. West Afr J Med. 2005;24(2):100–106. [PubMed] [Google Scholar]
- 14.Ghebremedhin B, Olugbosi MO, Raji AM, et al. Emergence of a community-associated methicillin-resistant Staphylococcus aureus strain with a unique resistance profile in Southwest Nigeria. J Clin Microbiol. 2009;47(9):2975–2980. doi: 10.1128/JCM.00648-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shittu A, Oyedara O, Abegunrin F, et al. Characterization of methicillin-susceptible and -resistant staphylococci in the clinical setting: a multicentre study in Nigeria. BMC Infect Dis. 2012;12:286. doi: 10.1186/1471-2334-12-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broekema NM, Van TT, Monson TA, Marshall SA, Warshauer DM. Comparison of cefoxitin and oxacillin disk diffusion methods for detection of mecA-mediated resistance in Staphylococcus aureus in a large-scale study. J Clin Microbiol. 2009;47(1):217–219. doi: 10.1128/JCM.01506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheesbrough M. District Laboratory Practice in Tropical Countries. 2nd ed. Cambridge, UK: Cambridge University Press; 2000. pp. 64–67. [Google Scholar]
- 18.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing; Sixteenth Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute; 2006. [Google Scholar]
- 19.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493–496. [PubMed] [Google Scholar]
- 20.De Medici D, Croci L, Delibato E, Di Pasquale S, Filetici E, Toti L. Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype enteritidis in poultry. Appl Environ Microbiol. 2003;69(6):3456–3461. doi: 10.1128/AEM.69.6.3456-3461.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayoral C, Noroña M, Baroni MR, Giani R, Zalazar F. Evaluation of a nested-PCR assay for Streptococcus pneumoniae detection in pediatric patients with community-acquired pneumonia. Rev Argent Microbiol. 2005;37(4):184–188. [PubMed] [Google Scholar]
- 22.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Idenitification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29(10):2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shittu AO, Lin J, Morrison D, Kolawole DO. The discovery of a multi-resistant Staphylococcus haemolyticus clone in the hospital and community environment in south western Nigeria. Ostomy Wound Manage. 2005;51(1):67–70. [PubMed] [Google Scholar]
- 24.Kesah C, Ben Redjeb S, Odugbemi TO, et al. Prevalence of methicillin-resistant Staphylococcus aureus in eight African hospitals and Malta. Clin Microbiol Infect. 2003;9(2):153–156. doi: 10.1046/j.1469-0691.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- 25.Shittu AO, Lin J. Antimicrobial susceptibility patterns and characterization of clinical isolates of Staphylococcus aureus in KwaZulu-Natal province, South Africa. BMC Infect Dis. 2006;6:125. doi: 10.1186/1471-2334-6-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fridkin SK, Hageman JC, Morrison M, et al. Active Bacterial Core Surveillance Program of the Emerging Infections Program Network Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med. 2005;352(14):1436–1444. doi: 10.1056/NEJMoa043252. [DOI] [PubMed] [Google Scholar]
- 27.Udo EE, Farook VS, Mokadas EM, Jacob LE, Sanyal SC. Molecular fingerprinting of mupirocin-resistant methicillin-resistant Staphylococcus aureus from a burn unit. Int J Infect Dis. 3(2):1998–1999. 82–87. doi: 10.1016/s1201-9712(99)90014-0. [DOI] [PubMed] [Google Scholar]
- 28.Onolitola OS, Olayinka BO, Salawu MJ, Yakubu SE. Nasal carriage of methicillin resistant Staphylococcus aureus with reduced vancomycin susceptibility (MRSA-RVS) by healthy adults in Zaria, Nigeria. Journal of Tropical Microbiology and Biotechnology. 2007;3(1):19–22. [Google Scholar]
- 29.Taiwo SS, Bamigboye TB, Odaro O, Adefioye OA, Fadiora SO. Vancomycin intermediate and high level resistant Staphylococcus aureus clinical isolates in Osogbo, Nigeria. Microbiol Res. 2011;3(e6):22–25. [Google Scholar]
- 30.Oni AA, Bakare RA, Okesola AO, Ogunlowo HA, Ewete AF. Pattern of bacterial pathogens in surgical wound infections. Afr J Med Med Sci. 1997;26(3–4):139–140. [PubMed] [Google Scholar]
- 31.Terry Alli OA. Distribution of mecA gene amongst Staphylococcus aureus isolates from south western Nigeria. African Journal of Biomedical Research. 2011;14(1):9–16. [Google Scholar]
- 32.Adesida S, Boelens H, Babajide B, et al. Major epidemic clones of Staphylococcus aureus in Nigeria. Microb Drug Resist. 2005;11(2):115–121. doi: 10.1089/mdr.2005.11.115. [DOI] [PubMed] [Google Scholar]
- 33.Skov R, Smyth R, Larsen AR, et al. Phenotypic detection of methicillin resistance in Staphylococcus aureus by disk diffusion testing and Etest on Mueller-Hinton agar. J Clin Microbiol. 2006;44(12):4395–4399. doi: 10.1128/JCM.01411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


