Abstract
The level of systemic exposure to 2′,3′-dideoxyinosine (ddI) is increased 40 to 300% when it is coadministered with allopurinol (Allo), ganciclovir (GCV), or tenofovir. However, the mechanism for these drug interactions remains undefined. A metabolic route for ddI clearance is its breakdown by purine nucleoside phosphorylase (PNP). Consistent with previous reports, enzymatic inhibition assays showed that acyclic nucleotide analogs can inhibit the phosphorolysis of inosine. It was further established that the mono- and diphosphate forms of tenofovir were inhibitors of PNP-dependent degradation of ddI (Kis, 38 nM and 1.3 μM, respectively). Allo and its metabolites were found to be relatively weak inhibitors of PNP (Kis, >100 μM). Coadministration of tenofovir, GCV, or Allo decreased the amounts of intracellular ddI breakdown products in CEM cells, while they increased the ddI concentrations (twofold increase with each drug at approximately 20 μM). While inhibition of the physiological function of PNP is unlikely due to the ubiquitous presence of high levels of enzymatic activity, phosphorylated metabolites of GCV and tenofovir may cause the increased level of exposure to ddI by direct inhibition of its phosphorolysis by PNP. The discrepancy between the cellular activity of Allo and the weak enzyme inhibition by Allo and its metabolites may be explained by an indirect mechanism of PNP inhibition. This mechanism may be facilitated by the unfavorable equilibrium of PNP and the buildup of one of its products (hypoxanthine) through the inhibition of xanthine oxidase by Allo. These findings support the inhibition of PNP-dependent ddI degradation as the molecular mechanism of these drug interactions.
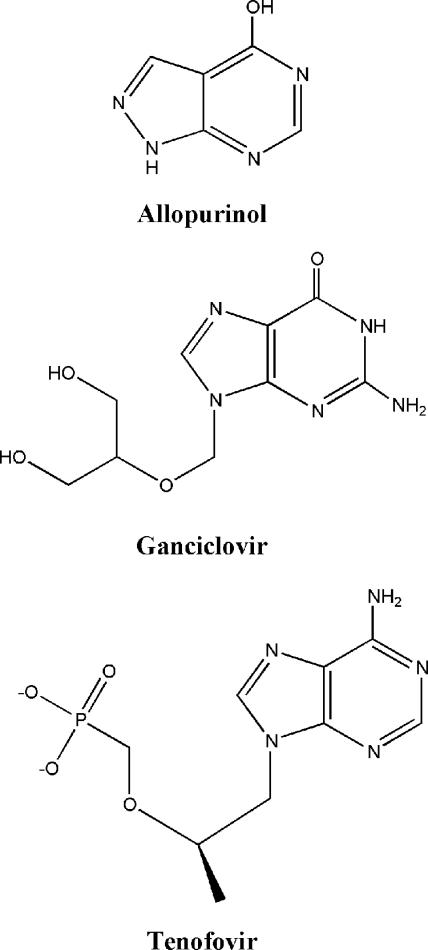
Current treatment regimens for human immunodeficiency virus (HIV) infection call for the use of three or more antiretrovirals of different classes. Other agents are also required for the treatment of opportunistic infections that occur as a result of immunosuppression. The use of multiple treatments increases the potential for drug-drug interactions and, as a result, treatment complications (9). One such interaction is an increase in the level of systemic exposure to the anti-HIV drug 2′,3′-dideoxyinosine (ddI; Videx; Bristol-Myers Squibb) when it is coadministered with allopurinol (Allo) (6; D. Liang, K. Breaux, A. Nornoo, S. Phadungpojna, M. Rodriguez-Barradas, and T. R. Bates, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. A662, 1999; D. Liang, K. Breaux, M. Rodriguez-Barradas, and T. R. Bates, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. A498, 2001); ganciclovir (GCV; Cytovene; Hoffmann-La Roche, Inc.) (16); or tenofovir disoproxil fumarate (TDF; Viread, Gilead Sciences, Inc.), a prodrug of tenofovir (23; J. Flaherty, B. Kearney, J. Wolf, J. Sayre, S. Barriere, S. Wong, M. Wulfsohn, and D. Coakley, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1729, 2001). The structures of the drugs are shown in Fig. 1. Allo, GCV, and TDF are used for the treatment of gout, cytomegalovirus infection, and HIV infection, respectively. Common features of these interactions include (i) a 40 to 300% increase in the level of exposure to ddI through a consistent elevation in its circulating levels over time; (ii) no change in the half-life of ddI; (iii) a proportional increase in the level of renal clearance of ddI, suggesting that the mechanism of the interaction does not involve competition for or impairment of kidney function; and (iv) no changes in the pharmacokinetic profiles of the drugs interacting with ddI. The similarities in the clinically observed effects of these interactions suggest a common mechanism.
FIG. 1.
Structures of drugs known to have drug-drug interactions with ddI. Tenofovir is the dAMP analog found in the blood after treatment with TDF.
The enzyme purine nucleoside phosphorylase (PNP) may be involved in these drug-drug interactions. PNP is a ubiquitous enzyme which functions in the purine nucleoside salvage pathway (reviewed by Bzowska and colleagues [8]). Its physiological function is the phosphorolysis or hydrolysis of inosine and guanosine nucleosides. The highest levels of enzyme activity are found in erythrocytes, peripheral blood lymphocytes, granulocytes, and kidney cells (1). Several observations support the possibility that PNP is the mechanism of ddI clearance: (i) ddI is a substrate for PNP in enzymatic assays (30); (ii) ddI is rapidly degraded in cultured cells, forming products consistent with PNP phosphorolysis (2, 34); (iii) radiolabeled hypoxanthine appears in dogs after treatment with 14C-labeled ddI (17); and (iv) ddI has a short half-life in humans (14). The large proportion of blood volume taken up by erythrocytes, coupled with their high PNP levels, makes the erythrocyte a likely site of ddI clearance (3). There is evidence that drugs known to interact with ddI inhibit PNP: (i) the rare clinical manifestation of PNP deficiency during prolonged high-dose Allo treatment (21, 22), (ii) potent inhibition by acyclovir and GCV metabolites in enzymatic assays (29, 33), and (iii) inhibition by acyclic phosphonate analogs structurally similar to tenofovir in enzymatic assays (4, 18, 35). These observations are consistent with the possibility that PNP inhibition is the mechanism of the clinically observed drug-drug interactions.
The goal of the research described here was to determine if PNP inhibition is in fact the basis for the drug-drug interactions observed with ddI. Studies were designed to gain an understanding of the effects of interacting drugs and their metabolites on PNP-dependent degradation of ddI in both enzymatic and cellular systems. The mechanism of ddI permeation was also addressed, as increased absorption of ddI across the gut wall in response to interacting drugs could serve as an alternate hypothesis.
(This work was presented in part as poster 35 at the 16th International Conference on Antiviral Research, 27 April to 1 May 2003, Savannah, Ga. [A. S. Ray, L. Olson, and A. Fridland, Abstr. 16th Int. Conf. Antivir. Res., abstr. 35, p. A50, 2003].)
MATERIALS AND METHODS
Chemicals.
Cell culture supplies were purchased from Irvine Scientific (Santa Ana, Calif.). [2′,3′-3H(N)]ddI was obtained from Moravek Biochemicals, Inc. (Brea, Calif.). Sugar-labeled ddI appeared to be contaminated with base label (see Results). GCV, Allo, oxypurinol, 7-methylguanosine (m7Guo), and unlabeled ddI were purchased from Sigma-Aldrich, Inc. (St. Louis, Mo.). Tenofovir and tenofovir diphosphate (tenofovir-DP) were provided by Gilead Sciences, Inc. (Foster City, Calif.). Tenofovir monophosphate (tenofovir-MP) was kindly synthesized by Ivan Rosenberg, Institute of Organic Chemistry and Biochemistry, Czech Academy of Sciences, Prague, Czech Republic. The purities of the nucleotides were verified by reverse-phase, ion-pairing high-pressure liquid chromatography (HPLC). All other chemicals used were the highest grade available from Sigma-Aldrich.
Caco-2 cell permeability model.
Caco-2 cell permeability assays were done essentially as described previously (24). In short, Caco-2 cells were seeded at 37,500 per cm2 in a 24-well transwell plate (Corning, Inc., Life Sciences, Acton, Mass.) and cultured for 10 days before the assay was initiated. Cells were pretreated with 50 μM TDF, GCV, or Allo for 30 min. ddI (100 μM) was then added and was allowed to permeate the cells for 1 h. Lucifer yellow (100 μM) was used as a paracellular marker. The ddI in the donor and the receiver wells was extracted by using Speedisk octyldecyl silane columns from J. T. Baker (Phillipsburg, N.J.). Analyses were done by HPLC through a J Sphere C18 fully end-capped column (50 by 2 mm), purchased from Waters (Milford, Mass.), by using an isocratic gradient of 0.2% formate and 5.75% acetonitrile at a flow rate of 0.5 ml/min. ddI was detected with an Aqa single-quad mass spectrometer, purchased from Thermo-Finnigan (San Jose, Calif.), running in the positive electrospray ionization mode.
Enzymatic assays.
In substrate and inhibition assays with ddI and inosine, xanthine oxidase (XOD; Sigma-Aldrich) was used to convert the hypoxanthine liberated by the PNP reaction to uric acid (10). The XOD reaction can be coupled to the cleavage of 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyltetrazolium chloride (INT; Sigma-Aldrich) to a highly colored formazan dye absorbing at 490 nm (12). Enzymatic studies were done essentially as described by Erion and colleagues (12). Briefly, reactions were done in a 96-well plate assay in the presence or absence of inhibitor with various concentrations of substrate, a fixed concentration of cosubstrate, 2 to 20 nM calf or human PNP (Sigma-Aldrich), 35 mU of XOD, 0.075% Triton X-100, 1 mM INT, and 100 mM HEPES (pH 7.6). A second assay was required for analogs that are known to inhibit XOD. The fluorescent substrate m7Guo is an irreversible substrate of PNP, and its phosphorolysis can be conveniently measured by monitoring the decrease in fluorescence at 390 nm when the sample is excited at a wavelength of 290 nm (19). These reactions were done in the same way as the XOD-coupled reactions, except that XOD, Triton X-100, and INT were omitted from the reaction mixture.
Data analyses.
Data from kinetic assays were fitted by using the program KaleidaGraph (Synergy Software, Reading, Pa.). Colorimetric and fluorescent changes were correlated to the substrate concentration by complete phosphorolysis of a sample of known concentration. These values closely matched those reported previously (12, 19). The velocities that were determined (expressed in micromolar per second) were then divided by the concentration of calf or human PNP present in the reaction mixture (2 or 20 nM), yielding the rate in units of inverse seconds. The maximum steady-state rate (kcat) and binding constant (Km) were then obtained by plotting the observed rates (kobsd) versus the substrate concentration and fitting of the result to a hyperbolic curve: kobsd = (kcat[substrate])/([substrate] + Km). To determine the mechanism of inhibition, hyperbolic curves were fitted to the dependence of the rate on the substrate concentration for different substrates in the absence or presence of inhibitor. A significant increase in Km or a significant decrease in kcat compared to the values on the curves generated in the absence of inhibitor indicated a competitive or a noncompetitive mechanism of inhibition, respectively. Consistency in the mechanism of inhibition determined was observed for a specific inhibitor at different concentrations. Ki values were determined by fitting the data to the binding equation for competitive (com) inhibition (kobsd = (kcat[substrate])/{[substrate] + Km(1 + [I]/Kicom)}) or noncompetitive (non) inhibition (kobsd = (kcat[substrate])/(1+[I]/Kinon))/{Km + [substrate]}). Brackets denote concentration, and [I] equals the concentration of inhibitor with which the binding curve was determined.
ddI metabolism in CEM cells.
Human T-leukemic CCRF-CEM lymphoblasts were obtained from the American Type Culture Collection (Manassas, Va.) and were maintained in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum, 100 U of penicillin G per ml, and 100 μg of streptomycin sulfate per ml. The cells were cultured under standard cell culture conditions. The cells were seeded at 106 cells/ml and were grown for 24 h in the presence or the absence of unlabeled compounds known to have an interaction with ddI. After the 24-h pretreatment, 10 μM [2′,3′-3H(N)]ddI (500 dpm/pmol) was added and cells were incubated for an additional 2 h to allow ddI metabolism to take place. Ten milliliters of the cell suspension was then pelleted by spinning at 300 × g for 10 min, and all but 500 μl of the medium was removed. The cells were resuspended in the remaining 500 μl of medium and spun through 200 μl of Nyosil M25 oil (TAI Lubricants, Inc., Hockessin, Del.) at 15,000 × g for 30 s. The medium was then removed from above the oil layer, and the top of the oil layer was washed with 1 ml of phosphate-buffered saline (PBS). The oil and PBS were removed, and the cells were resuspended in 250 μl of 70% methanol buffered with 10 mM Tris (pH 7.6). After 15 min of extraction on ice, the cellular debris was pelleted and the supernatant was dried with a Speed Vac dryer. The dried material was resuspended in 10 mM phosphate buffer (pH 7.6) and analyzed for ddI and its breakdown products by HPLC. Sample stability was tested by HPLC analysis before and after 2 h of incubation at 37°C.
HPLC separation of ddI from PNP-dependent degradation products.
HPLC analyses were done with a Prodigy 5-μm octyldecyl silane (2) reverse-phase C18 column (150 by 4.6 mm; Phenomenex, Torrence, CA) and mobile phase A (4% acetonitrile, 25 mM phosphate [pH 6.0], 5 mM hexyl triethylamine [Q6] ion-pairing reagent [Regis Technologies, Inc., Morton Grove, Ill.]) and mobile phase B (60% acetonitrile, 25 mM phosphate [pH 6.0], 2 mM Q6 ion-pairing reagent). The gradient was as follows: (i) 5 min of an isocratic gradient with 100% mobile phase A at 1.2 ml/min, (ii) 35 min of a linear gradient from 0 to 42% mobile phase B at 1.2 ml/min, (iii) washing for 2 min with 100% mobile phase B at 2 ml/min, and (iv) 8 min of reequilibration in 100% mobile phase A at 1.2 ml/min. This method gave retention times of 3.5 min for 2′,3′-dideoxyribose-1-OH (ddR-1-OH), 4.5 min for adenosine, 6.5 min for ddI, and 12 min for 2′,3′-dideoxyribose-1-phosphate (ddR-1-P). Radioactivity was detected by fraction collection and scintillation counting. Standards were purchased from Sigma-Aldrich or were generated by the PNP-catalyzed degradation of radiolabeled ddI.
RESULTS
Effects of interacting drugs on ddI permeation.
ddI absorption was studied in a Caco-2 cell system. The expression of multiple transporters (32) allows the potential detection of interactions occurring as a result of transport. After treatment with 50 μM TDF, GCV, or Allo (the structures are shown in Fig. 1), no significant increase in ddI permeation, expressed as either apparent permeation or percent flux, was detected (Table 1). Consistent with previous reports (28), data for ddI flux were inferred to be largely paracellular because of the low permeation of ddI and some correlation between that and the flux of the paracellular marker (lucifer yellow) (data not shown).
TABLE 1.
Effects of interacting drugs on ddI permeation in Caco-2 cell model system
| Treatment | ddI permeation
|
|
|---|---|---|
| Papp (cm/s [10−6])a | % Fluxb | |
| None | 2.13 ± 0.37 | 1.15 |
| TDF | 1.83 ± 0.28 | 0.99 |
| GCV | 1.73 ± 0.24 | 0.94 |
| Allo | 1.67 ± 0.24 | 0.90 |
Papp, apparent permeation. Values represent the means ± standard deviation of three independent experiments done in triplicate.
Percent flux by the masses present in donor and receiver wells. The mass balance of the donor and receiver solutions was ≈90%.
Substrate specificity of PNP.
Enzymatic studies were done to determine the interaction of calf PNP with phosphate (Pi), inosine, ddI, and m7Guo. Similar to previous findings, ddI was found to be a poor substrate for PNP (30). The efficiency of phosphorolysis by ddI was approximately 3 orders of magnitude less than that by inosine or m7Guo. This decrease in efficiency was mostly reflected in a 230-fold increase in the Km of ddI relative to that of inosine (Table 2). m7Guo and inosine were found to be similar substrates in the presence of 1 mM Pi. A difference was, however, noted in the binding of Pi in the presence of m7Guo, in which the Km was found to be eightfold lower than that when inosine was used as the cosubstrate.
TABLE 2.
Steady-state kinetic parameters for calf PNP-catalyzed phosphorolysis of inosine, ddI, and m7Guo
| Substrate | Cosubstratea | kcat (s−1)b | Km (μM)b | Efficiency (μM−1 s−1) |
|---|---|---|---|---|
| Inosine | Pi | 3.0 ± 0.5 | 4.3 ± 0.6 | 0.70 |
| ddI | Pi | 0.54 ± 0.12 | 1,000 ± 360 | 0.00054 |
| m7Guo | Pi | 8.8 ± 2.8 | 10 ± 3 | 0.88 |
| Pi | Inosine | 3.1 ± 0.6 | 500 ± 30 | 0.0062 |
| ddI | 0.53 ± 0.15 | 570 ± 230 | 0.00094 | |
| m7Guo | 3.8 ± 1.1 | 69 ± 38 | 0.055 |
Cosubstrate concentrations were held at about the Km for each cosubstrate (1 mM for Pi and ddI and 10 μM for inosine and m7Guo).
Values represent the means ± standard deviations for at least five independent binding curves for inosine, ddI, and m7Guo and two independent binding curves for Pi in the presence of the various cosubstrates.
Enzymatic inhibition of PNP by metabolites of purine base and nucleoside or nucleotide analogs.
To assess the validity of direct PNP inhibition as a mechanism for eliciting the drug interactions observed clinically, enzymatic studies were done with calf PNP and metabolites of Allo, GCV, and tenofovir (Table 3). The relationship of the observed rate to the substrate concentration was determined in the presence of various PNP inhibitors. Since Allo and its 6-hydroxylated metabolite (oxypurinol) are potent inhibitors of XOD, they could not be studied by the XOD-coupled colorimetric assay. Instead, fluorescence assays with the irreversible substrate m7Guo were used for these inhibitors. Studies showed that Allo and oxypurinol were weak inhibitors of PNP. Previous reports have also indicated that allopurinol-1-riboside is a poor inhibitor (21). Taken together, direct inhibition of PNP by Allo and its major metabolites seems unlikely.
TABLE 3.
Inhibition of calf PNP by metabolites of purine base and nucloside or nucleotide analogsa
| Drug | Metabolite |
Ki (μM)
|
|||
|---|---|---|---|---|---|
| Inosine | ddI | m7Guo | Pib | ||
| Allo | Unchanged | 100 ± 20 | 150 ± 20 (m7Guo) | ||
| 6-OH | 85 ± 37 | 210 ± 40 (m7Guo) | |||
| 1-ribo | 277c | ||||
| GCV | Unchanged | 10 ± 1 | 9.5 ± 2.4 | ||
| MP | 6.6d | ||||
| DP | 0.009e | ||||
| TP | 0.0061 ± 0.0007 | 0.018 ± 0.004 | |||
| Tenofovir | Unchanged | 4400 ± 800 | 3100 ± 900 | ||
| MP | 0.031 ± 0.007 | 0.038 ± 0.017 | 0.016 ± 0.007 | ||
| DP | 1.2 ± 0.3 | 3.2 ± 0.9 | 1.5 ± 0.5 | 3.0 ± 0.7 (ddI) | |
Inhibition constants for PNP degradation of inosine ddI, or m7Guo were determined in the presence of 1 mM Pi as the cosubstrate. The inhibition constants represent the means ± standard deviations determined from multiple substrate binding curve studies done in the presence of inhibitor.
Inhibition constants for Pi were determined in the presence of either m7Guo or ddI (indicated in parentheses) at concentrations near their Km values (10 μM and 1 mM, respectively).
The datum was generated with human erythrocyte PNP (21).
The datum is for acyclovir-MP with human erythrocyte PNP (33).
The datum was generated with human erythrocyte PNP (29).
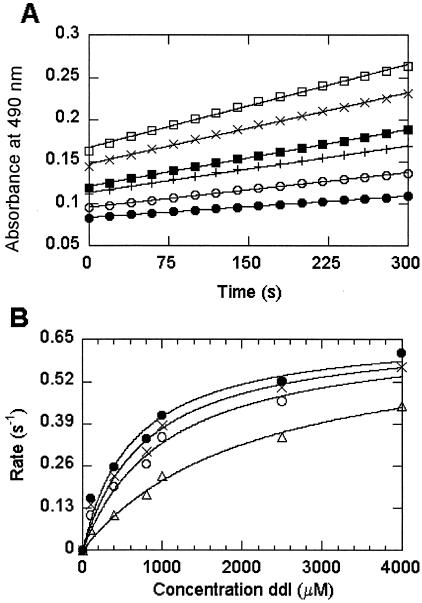
Alternatively, the data generated illustrate that phosphorylated metabolites of GCV and a structurally similar analog, acyclovir, are potent PNP inhibitors (Table 3) (29, 33). While tenofovir was a poor inhibitor, its phosphorylated metabolites showed potent inhibition. Similar to previous results for acyclovir-DP and acyclovir triphosphate (acyclovir-TP) (33), the data summarized in Table 3 show an almost 40-fold decrease in inhibition between tenofovir-MP and tenofovir-DP (Ki values for inosine phosphorolysis, 0.031 and 1.2 μM, respectively). Tenofovir-DP was observed to be competitive with both nucleoside and phosphate substrates. In contrast, inhibition by Allo and oxypurinol was competitive only with nucleoside substrates and was noncompetitive with Pi (data not shown). Although calf and human PNPs have been shown to have similar inhibition profiles (18), we wanted to verify that the inhibition observed with the calf enzyme was representative of that for the human enzyme. Inhibition of human PNP by tenofovir-MP was found to be competitive with ddI, and its inhibition constant was comparable to that found with the calf enzyme (Fig. 2).
FIG. 2.
Inhibition of human PNP phosphorolysis by tenofovir-MP. The rate of PNP-dependent phosphorolysis of various concentrations of ddI in the presence of 1 mM phosphate was studied by a colorimetric XOD-coupled assay (A). The changes in absorbance at 490 nm over time as a result of formazan dye formation at 100 μM (▪), 400 μM (○), 800 μM (+), 1,000 μM (▪), 2,500 μM (×), and 4,000 μM (□) ddI were determined. The rates of ddI phosphorolysis were then plotted against the ddI concentration for experiments done in the presence of 0 nM (•), 30 nM (×), 90 nM (○) and 270 nM (▵) tenofovir-MP and fit to hyperbolic curves (B). The curves show that ddI phosphorolysis by human PNP occurs with a kcat of 0.68 s−1 and a Km of 640 μM, as determined by nonlinear curve fitting of the data in the absence of inhibitor. Tenofovir-MP was observed to be a competitive inhibitor with a Ki of 126 ± 14 nM (mean ± standard deviation of the Ki values determined at each of the three different inhibitor concentrations).
Effects of interacting drugs on intracellular PNP-dependent ddI degradation.
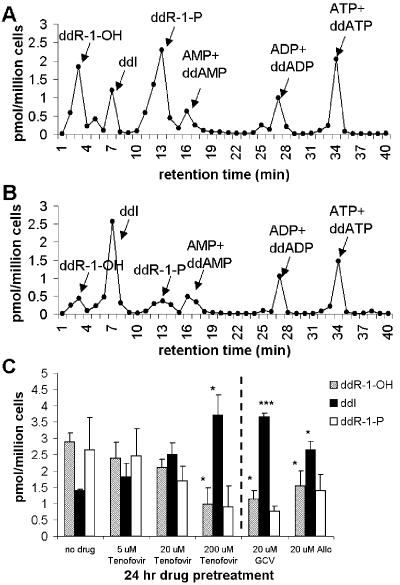
To confirm that PNP-dependent ddI breakdown could be inhibited in a cellular system, studies were done with sugar-labeled [3H]ddI in CEM cells. After pretreatment with interacting drugs, the cells were treated with [3H]ddI. In order to get an accurate assessment of the relative amounts of intracellular ddI and its breakdown products, an oil separation technique was used to separate the cells from extracellular [3H]ddI-containing medium (see Materials and Methods). Samples were then analyzed by HPLC and radioactivity detection. Analyses of the data showed that tenofovir, GCV, or Allo decreased the intracellular level of PNP-dependent ddI breakdown (Fig. 3). Statistically significant decreases in ddR-1-OH levels and increases in ddI levels and the ratio of ddI to its breakdown products (ddR-1-OH and ddR-1-P) were observed with 200 μM tenofovir, 20 μM GCV, and 20 μM Allo (Fig. 3C). While the variability inherent to the experiments caused the change in the ddR-1-P level to only approach statistical significance (P values, 0.09 and 0.15 for 20 μM Allo and 200 μM tenofovir, respectively, by the paired two-tailed Student t test), a marked and consistent decrease in ddR-1-P levels was observed with drug pretreatment in each of three independent experiments. Similar to the results generated with sugar-labeled ddI by other methods (2), the HPLC profiles also revealed radioactively labeled adenosine metabolites, suggesting that [2′,3′-3H(N)]ddI is contaminated with some label on the base. Our HPLC method, which was optimized to separate ddI from its degradation products, could not reproducibly separate ddA phosphorylated metabolites from radiolabeled adenosine nucleotides.
FIG. 3.
Inhibition of intracellular PNP-dependent ddI degradation by tenofovir, GCV, and Allo in CEM cells. Concentrations of metabolites of radiolabeled ddI formed in 2 h following a 24-h pretreatment in the absence (A) or presence (B) of 200 μM tenofovir, as determined by HPLC fractionation and scintillation counting. The concentrations of ddI and its breakdown products were determined by similar methods after 24 h of pretreatment with tenofovir, GCV, or Allo (C). Bar graphs present the means ± standard deviations of three independent experiments. The statistical significance of the changes in individual concentrations relative to the concentrations with no drug treatment were determined by a two-tailed Student t test and are as follows: *, P < 0.05; ***, P < 0.001. Statistically significant (P < 0.05) increases in the ratio of ddI to ddR-1-OH and ddR-1-P relative to no drug pretreatment were observed with all drug concentrations tested.
DISCUSSION
The clinically observed increase in the level of ddI exposure in the absence of a change in its half-life makes an interaction at the level of absorption a potentially plausible explanation for the drug-drug interactions observed. This hypothesis has been presented as a possible reason for the increase in the level of ddI exposure when it is coadministered with Allo or TDF (23; Liang et al. 41st ICAAC). However, we were unable to detect any difference in ddI permeation in response to interacting drugs in a cell-based permeability model. The mostly paracellular mechanism of ddI permeation would argue against a large contribution from efflux pumps, which are dependent on the entry of the drug into cells, in decreasing ddI absorption. Alternatively, ddI absorption could be altered by a change in gut wall integrity or modulation of the gastric contents; however, changes in these properties seem exceedingly unlikely in light of the chemical properties of the interacting drugs. In combination, these findings further support a systemic mechanism for the drug-drug interactions observed, while they do not completely rule out a contribution from changes in ddI absorption.
The observation that GCV triphosphate (GCVTP) is a potent inhibitor of calf PNP coupled with findings from previous studies showing the potent inhibition of human erythrocyte PNP by GCV diphosphate (GCVDP) (29) illustrates that phosphorylated metabolites of GCV are potent PNP inhibitors. Tenofovir-MP was also found to be an inhibitor of human and calf PNPs in this study. The competitive inhibition by tenofovir-DP with both nucleoside and Pi further supports the bisubstrate mode of inhibition previously proposed for acyclic nucleotide analogs (11, 35). Studies have shown that two inhibitors with potencies at millimolar concentrations linked by an optimal linker can result in inhibition at nanomolar concentrations through multiplicative increases in binding energy (27). A similar mechanism can be suggested for tenofovir-MP, because adenine and phosphate bind in the high micromolar to millimolar concentration range; but when they are linked appropriately (as is apparently the case for tenofovir-MP), inhibition at nanomolar concentrations resulted. This binding mode may explain the potent inhibition of PNP by acyclic nucleotide analogs. Their potent inhibition of PNP in enzymatic assays may indicate that acyclic nucleotide analogs (GCVDP, GCVTP, tenofovir-MP, and tenofovir-DP) are responsible for decreasing the level of PNP-dependent ddI breakdown in patients.
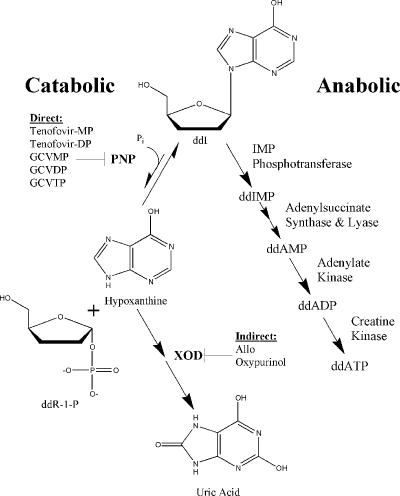
Allo, tenofovir, and GCV all inhibited PNP-dependent ddI breakdown in CEM cells. The consistency of the results between cellular and enzymatic assays suggests that direct inhibition of PNP by metabolites of tenofovir and GCV is responsible for the increased concentrations of ddI observed clinically. However, Allo and its major metabolites, allopurinol-1-riboside and oxypurinol, were poor inhibitors of PNP, suggesting some disconnect between the results of the enzymatic assay and those of the cellular experiments. It is proposed that these discrepancies may be explained by a model in which Allo exerts an indirect effect upon PNP activity. The potent inhibition of XOD by Allo and its metabolites would result in an increase in the levels of hypoxanthine, a product of PNP degradation. In light of the unfavorable equilibrium of PNP (∼50-fold in favor of nucleoside synthesis [13]), increased hypothanthine levels may account for the observed inhibition of PNP activity and the resulting elevation in plasma ddI concentrations. Proposed models for indirect (Allo) and direct (GCV and tenofovir) inhibition of PNP are shown in Fig. 4.
FIG. 4.
Abbreviated model for ddI metabolism showing both its breakdown and its activation to the active antiviral agent ddATP. The metabolites likely to interact with ddI through direct or indirect inhibition of PNP are indicated. Indirect inhibition of PNP by Allo and its metabolite oxypurinol are suggested to occur through potent inhibition of XOD, which causes a buildup of hypoxanthine. The presence of products of PNP may be inhibitory because of the unfavorable equilibrium of PNP. The general mechanisms of the catabolic and anabolic metabolism pathways of ddI are summarized from a previous study (2).
The concentrations of tenofovir necessary to decrease the level of intracellular PNP-dependent ddI breakdown by 50% (between 20 and 200 μM) seem inconsistent with the low micromolar levels of circulating tenofovir observed after oral dosing of TDF. However, tenofovir has limited cellular permeation due to the presence of two negative charges on the phosphonate moiety. Studies show that in antiviral assays tenofovir is approximately 100-fold less potent than TDF due to decreased cellular permeation (25). These studies also demonstrate that the metabolite most potent at inhibiting PNP activity (tenofovir-MP) is the least prevalent. Taken together, these findings may explain the elevated levels of tenofovir necessary to inhibit PNP-dependent ddI degradation in cell culture.
PNP has been explored as a potential drug target for certain types of cancer and immune-related diseases because of the dependence of circulating T cells on PNP activity (8). However, individuals who retain even minimal enzyme activity do not exhibit any T-cell dysfunction, illustrating the considerable difficulty in modulating T cells through PNP inhibition (31). This requirement for nearly complete enzyme inhibition has prevented many potent inhibitors of PNP from showing any clinical effect on T-cell function, and only inhibitors with activities at low picomolar concentrations represent any promise of showing clinical efficacy (20). It is therefore highly unlikely that the low levels of the far less potent metabolites of TDF or GCV have any impact on the physiological function of PNP during antiviral therapy, and clinical trials with TDF have shown favorable increases in CD4-positive T cells (S. Staszewski, J. E. Gallant, A. L. Pozniak, J. M. A. H. Suleiman, E. DeJesus, B. Lu, J. Sayre, and A. Cheng, Abstr. 10th Conf. Retrovir. Opportunist. Infect., abstr. 564b, p. 259, 2003). The weak binding constant (Table 2) and the low levels of circulating ddI make it more susceptible to competitive inhibition of its phosphorolysis, potentially explaining the selective change in its metabolism.
Some of the adverse effects observed with ddI treatment, including pancreatitis, peripheral neuropathy, and lactic acidosis, are thought to be caused by the interaction of its active metabolite, ddATP, with mitochondrial DNA polymerase γ (7, 15). Therefore, increases in circulating ddI levels may cause an elevation in ddI-mediated adverse events. However, cellular studies with acyclic phosphonates and ddI have not shown increases in ddATP levels (26, 34), and initial clinical findings did not show any increase in ddI-mediated adverse events as a result of the drug interaction (Flaherty et al., 41st ICAAC). More recently, this drug interaction has been implicated in adverse events in a limited number of patient case reports (5); in order to reduce any potential risk, dose reduction has successfully been used to normalize circulating ddI levels (23).
In conclusion, we have presented compelling in vitro evidence that the interactions between ddI and GCV, tenofovir, or Allo observed clinically are due to PNP inhibition. A model for the direct and indirect inhibition of PNP based on experimental results potentially explains the cellular mechanism of increased ddI levels. Furthermore, the information in this report should aid in the ability to proactively predict PNP inhibition and the drug-drug interactions that may result.
Acknowledgments
We thank Brian Robbins and John Rodman (St. Jude Research Hospital) and Brenda Hernandez-Santiago (Emory University) for general advice on cellular experimental design, Arash Mahmoudi for aiding with sample analysis, Tomas Cihlar (Gilead Sciences, Inc.) for thoughtful discussion of the data, and Ivan Rosenberg (Czech Academy of Sciences) for synthesizing the tenofovir-MP for the enzymatic inhibition assays.
REFERENCES
- 1.Agarwal, K. C., R. P. Agarwal, J. D. Stoeckler, and R. E. Parks, Jr. 1975. Purine nucleoside phosphorylase. Microheterogeneity and comparison of kinetic behavior of the enzyme from several tissues and species. Biochemistry 14:79-84. [DOI] [PubMed] [Google Scholar]
- 2.Ahluwalia, G., D. A. Cooney, H. Mitsuya, A. Fridland, K. P. Flora, Z. Hao, M. Dalal, S. Broder, and D. G. Johns. 1987. Initial studies on the cellular pharmacology of 2′,3′-dideoxyinosine, an inhibitor of HIV infectivity. Biochem. Pharmacol. 36:3797-3800. [DOI] [PubMed] [Google Scholar]
- 3.Back, D. J., S. Ormesher, J. F. Tjia, and R. Macleod. 1992. Metabolism of 2′,3′-dideoxyinosine (ddI) in human blood. Br. J. Clin. Pharmacol. 33:319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beauchamp, L. M., J. V. Tuttle, M. E. Rodriguez, and M. L. Sznaidman. 1996. Guanine, pyrazolo[3,4-d]pyrimidine, and triazolo[4,5-d]pyrimidine (8-azaguanine) phosphonate acyclic derivatives as inhibitors of purine nucleoside phosphorylase. J. Med. Chem. 39:949-956. [DOI] [PubMed] [Google Scholar]
- 5.Blanchard, J. N., M. Wohlfeiler, A. Canas, K. King, and J. T. Lonergan. 2003. Pancreatitis with didanosine and tenofovir disoproxil fumarate. Clin. Infect. Dis. 37:e57-e62. [DOI] [PubMed] [Google Scholar]
- 6.Boelaert, J. R., G. M. Dom, A. D. Huitema, J. H. Beijnen, and J. M. Lange. 2002. The boosting of didanosine by allopurinol permits a halving of the didanosine dosage. AIDS 16:2221-2223. [DOI] [PubMed] [Google Scholar]
- 7.Brinkman, K., H. J. ter Hofstede, D. M. Burger, J. A. Smeitink, and P. P. Koopmans. 1998. Adverse effects of reverse transcriptase inhibitors: mitochondrial toxicity as common pathway. AIDS 12:1735-1744. [DOI] [PubMed] [Google Scholar]
- 8.Bzowska, A., E. Kulikowska, and D. Shugar. 2000. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol. Ther. 88:349-425. [DOI] [PubMed] [Google Scholar]
- 9.Dasgupta, A., and P. C. Okhuysen. 2001. Pharmacokinetic and other drug interactions in patients with AIDS. Ther. Drug Monit. 23:591-605. [DOI] [PubMed] [Google Scholar]
- 10.de Groot, H., and T. Noll. 1985. Enzymatic determination of inorganic phosphates, organic phosphates and phosphate-liberating enzymes by use of nucleoside phosphorylase-xanthine oxidase (dehydrogenase)-coupled reactions. Biochem. J. 230:255-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ealick, S. E., Y. S. Babu, C. E. Bugg, M. D. Erion, W. C. Guida, J. A. Montgomery, and J. A. Secrist III. 1991. Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors. Proc. Natl. Acad. Sci. USA 88:11540-11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erion, M. D., K. Takabayashi, H. B. Smith, J. Kessi, S. Wagner, S. Honger, S. L. Shames, and S. E. Ealick. 1997. Purine nucleoside phosphorylase. 1. Structure-function studies. Biochemistry 36:11725-11734. [DOI] [PubMed] [Google Scholar]
- 13.Friedkin, M. 1950. Deoxyriboside-1-phosphate. II. The isolation of the crystalline deoxyribose-1-phosphate. J. Biol. Chem. 184:449-459. [PubMed] [Google Scholar]
- 14.Hartman, N. R., R. Yarchoan, J. M. Pluda, R. V. Thomas, K. S. Marczyk, S. Broder, and D. G. Johns. 1990. Pharmacokinetics of 2′,3′-dideoxyadenosine and 2′,3′-dideoxyinosine in patients with severe human immunodeficiency virus infection. Clin. Pharmacol. Ther. 47:647-654. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, A. A., A. S. Ray, J. Hanes, Z. Suo, J. M. Colacino, K. S. Anderson, and K. A. Johnson. 2001. Toxicity of antiviral nucleoside analogs and the human mitochondrial DNA polymerase. J. Biol. Chem. 276:40847-40857. [DOI] [PubMed] [Google Scholar]
- 16.Jung, D., K. Griffy, A. Dorr, R. Raschke, T. L. Tarnowski, J. Hulse, and R. E. Kates. 1998. Effect of high-dose oral ganciclovir on didanosine disposition in human immunodeficiency virus (HIV)-positive patients. J. Clin. Pharmacol. 38:1057-1062. [DOI] [PubMed] [Google Scholar]
- 17.Kaul, S., W. C. Shyu, U. A. Shukla, K. A. Dandekar, and R. H. Barbhaiya. 1993. Absorption, disposition, and metabolism of [14C]didanosine in the beagle dog. Drug Metab. Dispos. 21:447-453. [PubMed] [Google Scholar]
- 18.Kulikowska, E., A. Bzowska, A. Holy, L. Magnowska, and D. Shugar. 1998. Antiviral acyclic nucleoside phosphonate analogues as inhibitors of purine nucleoside phosphorylase. Adv. Exp. Med. Biol. 431:747-752. [DOI] [PubMed] [Google Scholar]
- 19.Kulikowska, E., A. Bzowska, J. Wierzchowski, and D. Shugar. 1986. Properties of two unusual, and fluorescent, substrates of purine-nucleoside phosphorylase: 7-methylguanosine and 7-methylinosine. Biochem. Biophys. Acta 874:355-363. [DOI] [PubMed] [Google Scholar]
- 20.Lewandowicz, A., P. C. Tyler, G. B. Evans, R. H. Furneaux, and V. L. Schramm. 2003. Achieving the ultimate physiological goal in transition state analogue inhibitors for purine nucleoside phosphorylase. J. Biol. Chem. 278:31465-31468. [DOI] [PubMed] [Google Scholar]
- 21.Nishida, Y., N. Kamatani, K. Tanimoto, and I. Akaoka. 1979. Inhibition of purine nucleoside phosphorylase activity and of T-cell function with allopurinol-riboside. Agents Actions 9:549-552. [DOI] [PubMed] [Google Scholar]
- 22.Nishida, Y., N. Kamatani, K. Tanimoto, and I. Akaoka. 1979. Suppression of cellular immunity due to inhibition of purine nucleoside phosphorylase by allopurinol-riboside. Adv. Exp. Med. Biol. 122B:309-313. [DOI] [PubMed] [Google Scholar]
- 23.Pecora Fulco, P., and M. A. Kirian. 2003. Effect of tenofovir on didanosine absorption in patients with HIV. Ann. Pharmacother. 37:1325-1328. [DOI] [PubMed] [Google Scholar]
- 24.Putnam, W. S., L. Pan, K. Tsutsui, L. Takahashi, and L. Z. Benet. 2002. Comparison of bidirectional cephalexin transport across MDCK and Caco-2 cell monolayers: interactions with peptide transporters. Pharm. Res. 19:27-33. [DOI] [PubMed] [Google Scholar]
- 25.Robbins, B. L., R. V. Srinivas, C. Kim, N. Bischofberger, and A. Fridland. 1998. Anti-human immunodeficiency virus activity and cellular metabolism of a potential prodrug of the acyclic nucleoside phosphonate 9-R-(2-phosphonomethoxypropyl)adenine (PMPA), bis(isopropyloxymethylcarbonyl)PMPA. Antimicrob. Agents Chemother. 42:612-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbins, B. L., C. K. Wilcox, A. Fridland, and J. H. Rodman. 2003. Metabolism of tenofovir and didanosine in quiescent or stimulated human peripheral blood mononuclear cells. Pharmacotherapy 23:695-701. [DOI] [PubMed] [Google Scholar]
- 27.Shuker, S. B., P. J. Hajduk, R. P. Meadows, and S. W. Fesik. 1996. Discovering high-affinity ligands for proteins: SAR by NMR. Science 274:1531-1534. [DOI] [PubMed] [Google Scholar]
- 28.Sinko, P. J., P. Hu, A. P. Waclawski, and N. R. Patel. 1995. Oral absorption of anti-AIDS nucleoside analogues. 1. Intestinal transport of didanosine in rat and rabbit preparations. J. Pharm. Sci. 84:959-965. [DOI] [PubMed] [Google Scholar]
- 29.Stein, J. M., J. D. Stoeckler, S. Y. Li, R. L. Tolman, M. MacCoss, A. Chen, J. D. Karkas, W. T. Ashton, and R. E. Parks, Jr. 1987. Inhibition of human purine nucleoside phosphorylase by acyclic nucleosides and nucleotides. Biochem. Pharmacol. 36:1237-1244. [DOI] [PubMed] [Google Scholar]
- 30.Stoeckler, J. D., C. Cambor, and R. E. Parks, Jr. 1980. Human erythrocytic purine nucleoside phosphorylase: reaction with sugar-modified nucleoside substrates. Biochemistry 19:102-107. [DOI] [PubMed] [Google Scholar]
- 31.Stoeckler, J. D., S. E. Ealick, C. E. Bugg, and R. E. Parks, Jr. 1986. Design of purine nucleoside phosphorylase inhibitors. Fed. Proc. 45:2773-2778. [PubMed] [Google Scholar]
- 32.Taipalensuu, J., H. Tornblom, G. Lindberg, C. Einarsson, F. Sjoqvist, H. Melhus, P. Garberg, B. Sjostrom, B. Lundgren, and P. Artursson. 2001. Correlation of gene expression of ten drug efflux proteins of the ATP-binding cassette transporter family in normal human jejunum and in human intestinal epithelial Caco-2 cell monolayers. J. Pharmacol. Exp. Ther. 299:164-170. [PubMed] [Google Scholar]
- 33.Tuttle, J. V., and T. A. Krenitsky. 1984. Effects of acyclovir and its metabolites on purine nucleoside phosphorylase. J. Biol. Chem. 259:4065-4069. [PubMed] [Google Scholar]
- 34.Weibel, M., J. Balzarini, A. Bernhardt, and P. Mamont. 1994. Potentiating effect of (2-[2-[(2-amino-1,6-dihydro-6-oxo-9H-purin-9-yl)methyl]-phenyl]ethenyl)-phosphonic acid (MDL 74,428), a potent inhibitor of purine nucleoside phosphorylase, on the antiretroviral activities of 2′,3′-dideoxyinosine combined with ribavirin in mice. Biochem. Pharmacol. 48:245-252. [DOI] [PubMed] [Google Scholar]
- 35.Wierzchowski, J., E. Kulikowska, A. Bzowska, A. Holy, L. Magnowska, and D. Shugar. 1999. Interactions of calf spleen purine nucleoside phosphorylase with antiviral acyclic nucleoside phosphonate inhibitors: kinetics and emission studies. Nucleosides Nucleotides 18:875-876. [DOI] [PubMed] [Google Scholar]