Abstract
Type 2 CXC chemokine receptor CXCR2 plays roles in development, tumorigenesis and inflammation. CXCR2 also promotes demyelination and decreases remyelination by actions toward hematopoietic cells and non-hematopoietic cells. Germline CXCR2 deficient (Cxcr2−/−) mice reported in 1994 revealed the complexity of CXCR2 function and its differential expression in varied cell-types. Here, we describe Cxcr2fl/fl mice for which the targeting construct was generated by recombineering based on homologous recombination in E. coli. Without recombination Cxcr2fl/fl mice have CXCR2 expression on neutrophils in peripheral blood, bone marrow and spleen. Cxcr2fl/fl mice were crossed to Mx-Cre mice in which Cre recombinase is induced by type I interferons, elicited by injection with polyinosinic-polycytidylic acid (poly(I:C)). CXCR2-deficient neutrophils were observed in poly(I:C) treated Cxcr2fl/fl::Mx-Cre+ (Cxcr2-CKO) mice, but not in poly(I:C) treated Cxcr2f//+::Mx-Cre+ mice. CXCR2 deletion was mainly observed peripherally but not in the CNS. Cxcr2-CKO mice showed impaired neutrophil migration in sterile peritonitis. Cxcr2-CKO mice reported here will provide a genetic reagent to dissect roles of CXCR2 in the neutrophil granulocyte lineage. Furthermore Cxcr2fl/fl mice will provide useful genetic models to evaluate CXCR2 function in varied cell populations.
Keywords: CXCR2, chemokine, chemokine receptor, conditional KO mice, neutrophil
Introduction
CXCR2 was cloned in 1991(Holmes et al. 1991; Murphy & Tiffany 1991) and is expressed on myeloid cells in the periphery as well as on oligodendrocyte progenitor cells (OPCs) in the central nervous system (CNS). With its seven differentially-regulated ligands, CXCR2 shows multiple additional functions beyond chemoattraction for myeloid cell trafficking (Cacalano et al. 1994). On OPCs in the developing rodent spinal cord, CXCR2 interacts with CXCL1, arresting migrating OPCs during development, and promoting the OPC proliferative response to PDGFA (Tsai et al. 2002). CXCR2 also plays a role in wound healing (Devalaraja et al. 2000), acetaminophen hepatotoxicity (Hu & Colletti 2010), bone mineralization, intramembranous bone formation (Bischoff et al. 2011), spontaneous tumorigenesis (Jamieson et al. 2012), cancer metastasis and chemoresistance (Acharyya et al. 2012). Recently we and others found that CXCR2 function on neutrophils plays a role in both autoimmune and toxic demyelination (Liu et al. 2010a; Carlson et al. 2008) as well as myelin repair (Liu et al. 2010b). Importantly, CXCR2 plays orthologous roles in humans and rodents (Mihara et al. 2005).
CXCR2 deficient mice (Cxcr2−/−) (Cacalano et al. 1994) are fragile and infertile, which complicates breeding and disease modeling. We generated Cxcr2fl/fl mice to extend research into cell-type specific and inducible deletion of this pleiotropic receptor. In transgenic mice expressing inducible Cre recombinase under control of the Mx1 promoter (Mx-Cre), our induction protocol efficiently deleted the floxed Cxcr2 gene in hematopoietic cells. Cxcr2-CKO mice 4 wks after poly(I:C) injections showed deletion of CXCR2 on peripheral neutrophils and deficient neutrophil migration. Our data indicate that Cxcr2fl/fl mice provide a useful reagent to advance current research into CXCR2 and its chemokine ligands in inflammation, cancer and neurological disease.
Results and Discussion
Generation of Cxcr2 conditional knockout (Cxcr2-CKO) mice
A Cxcr2 conditional targeting construct was generated by recombineering and electroporated into C57BL/6 ES cells, which were screened by Southern blotting (Fig. 1a). Twenty-five candidate ES cell lines were verified both through 3′ screening (Fig. 1a, data not shown) and sequence analysis through the loxP and FRT sites (data not shown). One correct ES clone (clone 1) from seven candidates was re-confirmed by sequence analysis of the loxP and FRT sites (data not shown).
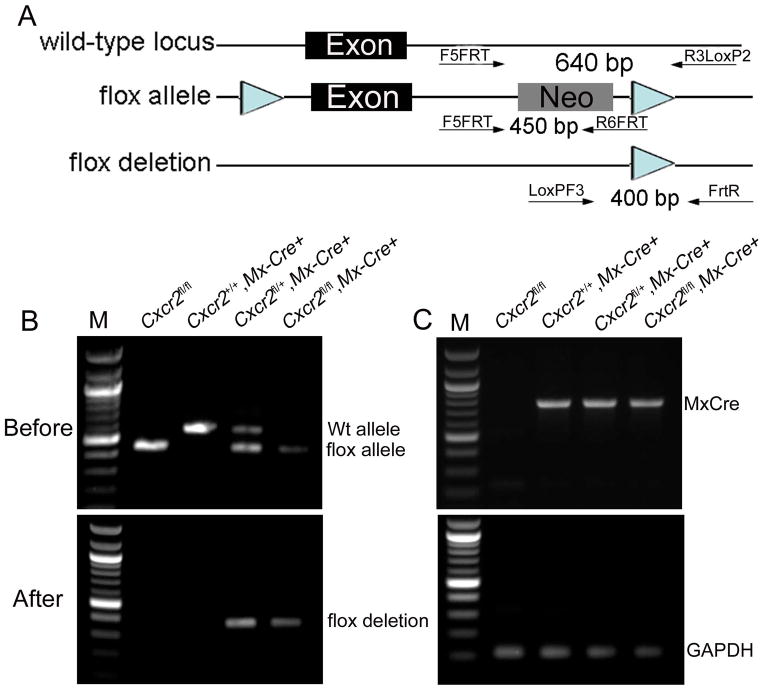
Figure 1. Generation of a conditional Cxcr2 allele and genotype determination of conditional knockouts by PCR.
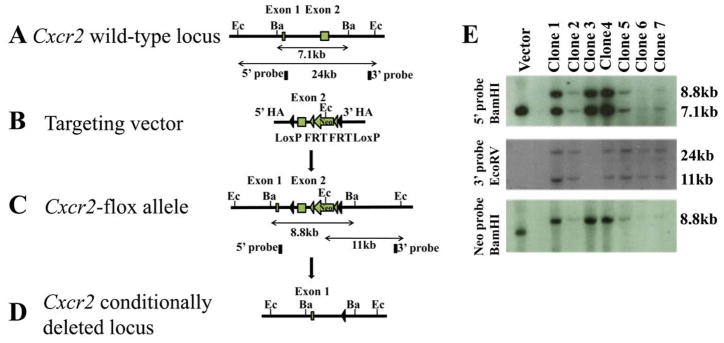
a. The strategy for Cxcr2fl mouse generation. (A). Schematic diagram of the Cxcr2 wild-type genomic locus demonstrating a classic two-exon gene structure that is indicated by two blank boxes. In order to screen for the targeted Cxcr2fl mutant allele, both 5′ and 3′ Southern blot probes (filled boxes) were designed in the genomic regions inside of two homologous arms (HA). Restriction enzymes are in the abbreviated form: Ec, EcoRV and Ba, BamHI. (B). A Cxcr2fl targeting vector was constructed by recombineering. The targeting region includes a 2-kb 5′ HA, a LoxP site that flanks Cxcr2 exon2, a Frt-PGKNeo-Frt cassette that is downstream of Cxcr2 exon2 flanked by another LoxP site and a 3-kb 3′ HA. (C). Schematic demonstration of the targeted Cxcr2fl allele. In Southern blot screening, an 8.8kb-BamHI targeted band (5′ probe) and an 11kb-EcoRV targeted band (3′ probe) are expected. (D). Schematic diagram depicting the conditional deletion of the Cxcr2 gene through Cre-mediated recombination. (E). Seven targeted embryonic stem cells were screened by Southern blot. 7.1kb wild-type and 8.8kb targeted BamHI bands were detected by the 5′ Southern probe. 24kb wild-type and 11kb mutant-type EcoRV bands are detected by the 3′ Southern probe. An 8.8kb targeted BamHI band is detected by using the PGK-neo probe. Targeting vector was used as the control. b. Genotype determination of Cxcr2-CKO mice by PCR. (A) Strategy for designing PCR primers to detect the Wt locus, the flox allele and the deletion of the flox allele. The primers are shown as arrows. (B) PCR amplification of the genomic DNA using primers for the flox gene before injection of poly(I:C); the higher band (~650 bp) indicates the wild-type allele and the lower band (~450 bp) indicates the flox allele (top). PCR amplification of the genomic DNA using primers to detect the deletion of the flox allele (~400 bp) after poly(I:C) induced recombination (bottom). (C) PCR amplification of the genomic DNA using primers to detect the Mx-Cre transgenes (top). The details for designing the primers specific for Mx-Cre are described in supplementary data 1. The bottom figure shows representive PCR results of GAPDH for quality control of the genomic DNA used for (B) and (C).
Chimeric founder mice harboring the Cxcr2fl/+ allele were intercrossed to generate Cxcr2fl/fl mice. To create the Cxcr2-CKO mouse line, Cxcr2fl/fl mice were bred to Mx-Cre mice (Kuhn et al., 1995) and intercrossed to generate Cxcr2fl/+::Mx-Cre+ and Cxcr2fl/fl::Mx-Cre+ genotypes.
Expression of CXCR2 in Cxcr2fl/fl mice
The Cxcr2fl allele was detected with F5FRT and R6FRT primers (Figure 1b-A). CXCR2 was detected by flow cytometry on Ly6G+ neutrophils of Cxcr2fl/fl mice, at similar frequency and mean fluorescence intensity to Cxcr2+/+ mice (Figure 2). Spleen and bone marrow from Cxcr2fl/fl and Cxcr2+/+ mice showed similar CXCR2 expression patterns as peripheral blood (Supplementary 2, data not shown).
Figure 2. Expression of CXCR2 in Cxcr2fl/fl mice.
Peripheral blood cells from Cxcr2fl/+ and Cxcr2fl/fl mice (bottom) stained with Ly6G and CXCR2 antibodies were analyzed by flow cytometry. Cells from Cxcr2+/+ and Cxcr2−/− mice (top) were used as positive and negative controls respectively. These data represent at least three independent experiments.
CXCR2 is stably deleted by poly(I:C) in Mx-Cre::Cxcr2 CKO mice
Intraperitoneal poly(I:C) injections induced Cre recombinase in Cxcr2fl/fl::Mx-Cre+ and Cxcr2fl/+::Mx-Cre+ mice. Analysis of genomic DNA after poly(I:C) injection showed the Cxcr2 deletion product in samples from Cxcr2fl/fl::Mx-Cre+ and Cxcr2fl/+::Mx-Cre+ but not Cxcr2+/+::Mx-Cre+ or Cxcr2fl/fl mice (Figure 1b).
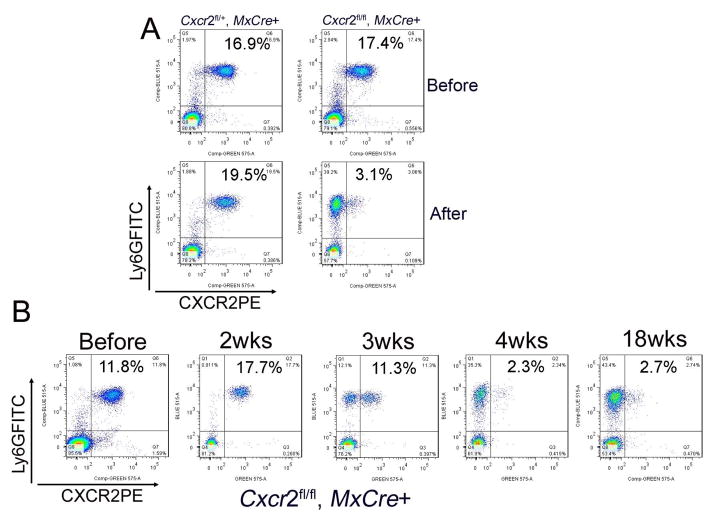
We analyzed CXCR2 protein product on blood leukocytes weekly after poly(I:C) induction. In Cxcr2-CKO mice, CXCR2 deletion on blood neutrophils was time-dependent. To our surprise, deletion of CXCR2 was first observed at 3wks post injection (pi) (Figure 3B). At 4 wks pi, most Cxcr2-CKO mice showed >90% of blood neutrophils were CXCR2-negative (calculated by the ratio of CXCR2 negative cells in total Ly6G+ cells) (Figure 3B, supplementary 2). Cells from the spleen and bone marrow showed equivalent CXCR2 deletion in the neutrophil lineage (Supplementary 2). Monitoring until 18 wks pi, showed stable deletion of CXCR2 (Figure 3B). Cxcr2-CKO mice showed considerable delay in the appearance of CXCR2-negative circulating neutrophils, as compared to previous studies targeting other cell surface molecules where target-deficient cells are detected within a few days (Tiedt et al. 2008; Ulyanova et al. 2007; Yan, 2008). Before appearance of CXCR2-deficient leukocytes in the circulation, necessary events include activating the Mx1 promoter with poly(I:C)-induced type I IFN, producing Cre recombinase, and recombining the floxed gene. Thereafter, turn-over of the targeted protein or cells expressing that protein must occur before target-negative cells predominate (Nagy, 2000). It remains plausible that retention of CXCR2-deficient neutrophil progenitors in bone marrow (Köhler et al. 2011) accounts for the failure of CXCR2-negative neutrophils to accumulate in the bloodstream. In particular, neutrophil progenitors remaining CXCR2+ will be privileged for bone marrow exit and that population must be exhausted before CXCR2-deficient cells will appear in the periphery. Cxcr2fl/fl::Mx-Cre+ mice were born at expected Mendelian ratios and showed normal weight, behavior, fertility and life span. Cxcr2-CKO mice lost weight compared to littermates after efficient CXCR2 deletion on neutrophils (data not shown), reminiscent of the failure-to-thrive phenotype in germline Cxcr2−/− mice.
Figure 3. CXCR2 is stably deleted by poly(I:C) in Mx-Cre::Cxcr2 CKO mice.
(A) Efficient deletion of CXCR2 after poly(I:C) injection. Peripheral blood cells from Cxcr2fl/+::Mx-Cre+ and Cxcr2fl/fl::Mx-Cre+ mice before injection (figure 3A, top) and 4 wks after injection (figure 3A, bottom), were stained with Ly6GFITC and CXCR2PE antibodies and were analyzed by flow cytometry. These data represent at least three independent experiments. (B) CXCR2 deletion is time dependent and irreversible. Peripheral blood cells collected from Cxcr2fl/fl::Mx-Cre+ mice before injection, at 2 wks, 3 wks, 4 wks, and 18 wks after poly(I:C) injection, were stained with Ly6GFITC and CXCR2PE antibodies and were analyzed by flow cytometry. The percentage indicated in the figures is CXCR2 positive neutrophils in the total peripheral blood cells. These data represent at least three independent experiments.
CXCR2-deficient neutrophils show defective ligand scavenging
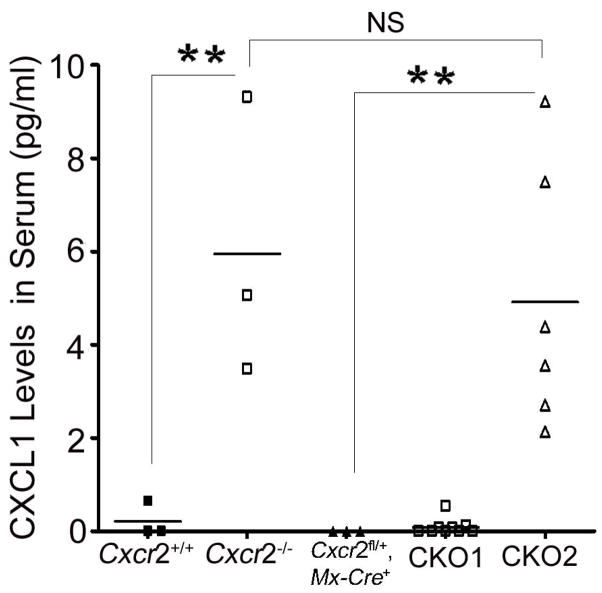
Signaling chemokine receptors such as CXCR2 scavenge their ligands (Cardona et al. 2008). CXCR2 ligand CXCL1 was undetectable in plasma of Cxcr2fl/fl or Cxcr2+/+ mice, while being present at high levels in plasma from positive control Cxcr2−/− animals showing that the Cxcr2fl/fl targeted allele scavenged efficiently (Figure 4, data not shown). Cxcr2-CKO mice with more than 90% CXCR2 deletion on neutrophils showed dramatically increased plasma CXCL1 in the serum comparable to those in Cxcr2−/− mice (Figure 4). However, Cxcr2-CKO mice with less than 90% CXCR2 deletion on neutrophils showed comparable levels of plasma CXCL1 to control mice (Figure 4). These results indicate that near-complete induction of CXCR2 deficiency on peripheral neutrophils is required to abrogate the scavenging of CXCL1.
Figure 4. CXCR2-deficient neutrophils show defective ligand scavenging.
ELISA assays for CXCL1 were performed on sera from Cxcr2+/+, Cxcr2−/−, Cxcr2fl/fl mice (data not shown), Cxcr2fl/+::Mx-Cre+ mice, CKO1 mice: CXCR2 deletion on neutrophils of Cxcr2-CKO mice is less than 90%, and CKO2 mice: CXCR2 deletion on neutrophils of Cxcr2-CKO mice is more than 90%. There is undetectable CXCL1 in the serum of Cxcr2+/+ Cxcr2fl/fl mice (data not shown), Cxcr2fl/+::Mx-Cre+ mice and CKO1 mice. However, there is detectable CXCL1 in Cxcr2−/− mice and CKO2 mice. N is indicated by spots. Each spot represent one mouse on each group; **P<0.01, significant difference. NS: no significant difference. These data represent two independent experiments.
Dose-dependent recombination induced by poly(I:C) in periphery but not CNS of Cxcr2-CKO mice
After 4 injections at 2.5mg/kg poly(I:C), we observed 30% deletion of CXCR2 on neutrophils while 15mg/kg poly(I:C) induced >90% deletion on neutrophils at 4 wks pi, 6 wks pi and 8 wks pi (Figure 5A). Concerned with effects of the 15mg/kg poly(I:C), including inflammatory cytokine (TNF-α, IL-6, or IFNγ) production and sickness behavior (Cunningham et al. 2007) which could confound data analysis for neuroinflammatory disease models, we examined the timing of poly(I:C) injections at lower doses of poly(I:C). At 5mg/kg poly(I:C) with assay 4 wks pi, we observed partial CXCR2 deletion from circulating neutrophils of mice injected at ages >8wks. Using the same dose and timing of assay, we observed >90% CXCR2 deletion on neutrophils of most mice injected at 4wks of age (Figure 5B). Compared to 5 mg/kg, high-doses (15 mg/kg) of poly(I:C) once every other day for 4 injections did not alter the extent or kinetics for generating CXCR2-negative blood neutrophils in young mice (~3–4 wks old). However, high-dose poly(I:C) caused more-efficient CXCR2 deletion on neutrophils in older mice (>8 wks old). We concluded that, 5 mg/kg was an appropriate dose of poly(I:C) for the CXCR2 deletion in young Cxcr2-CKO mice avoiding the neurotoxic effects caused by high-dose poly(I:C).
Figure 5. Dose-dependent recombination induced by poly(I:C) in periphery, not CNS, of Cxcr2-CKO mice.
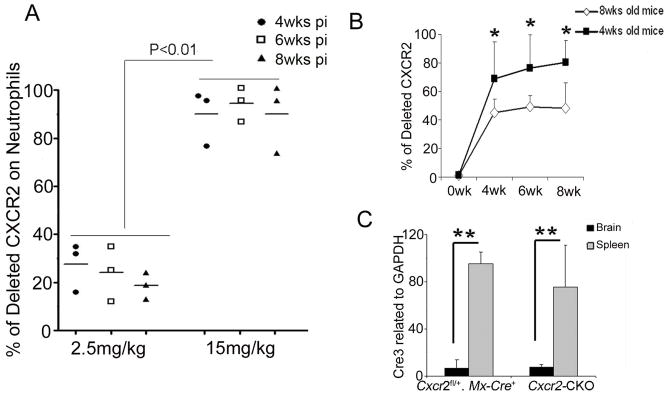
(A) 4 wk old Cxcr2fl/fl::Mx-Cre+ mice were given the indicated doses of poly(I:C) and CXCR2 deletion on neutrophils was examined by flow cytometry at 4 wks, 6 wks, and 8 wks post injection (pi). (B) 4 wks or 8 wks old mice were injected with 5 mg/kg of polyI:C. The deletion of CXCR2 was measured before injection, 4 wks, 6 wks and 8 wks after injection. N= 3 or 4 each group as indicated. The data represent two independent experiments. *P<0.05 (C) Cre recombinase expression in the spleen and brain 4 wks after poly(I:C) injection. Total RNA of brain and spleen tissues was extracted from Cxcr2-CKO and Cxcr2fl/+::Mx-Cre+mice. Real time PCR for Cre was performed on cDNA synthesized from 1ug of RNA. N=3 each group. These data represent two independent experiments. **P<0.01;
To determine whether the deletion of CXCR2 differed in the periphery from the CNS, we performed qPCR to examine Cre expression. Cre recombinase induction in spleen (200-fold) was far greater than that in the brain (six-fold) (Figure 5C). To determine if low-level induction of Cre recombinase mediated recombination in CNS parenchyma, we crossed Mx-Cre mice to reporter ROSA26mTmG mice (Muzumdar et al. 2007), and monitored recombination as conversion from tomato red to GFP labeled cells in the CNS. There were no green neuroepithelial parenchymal CD45 negative cells in the CNS 4wks after injection of 5 mg/kg poly(I:C) by flow cytometry (data not shown), which is consistent with a previous finding (Kuhn et al., 1995).
Decreased migration of CXCR2-deficient neutrophils in sterile peritonitis
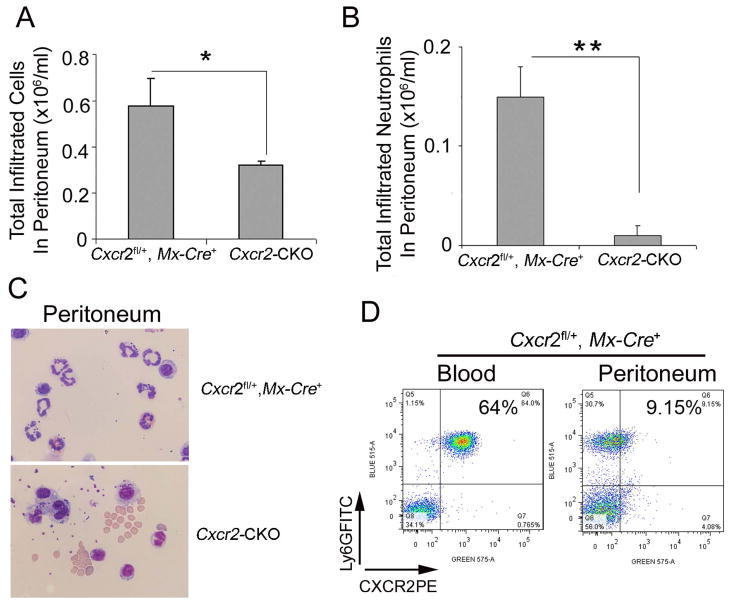
To assess neutrophil migration toward an inflammatory stimulus in Cxcr2-CKO mice, the sterile irritant thioglycollate was administered i.p.. Total peritoneal cell number was reduced by about 50% in Cxcr2-CKO mice (0.32±0.02×106/mL) as compared with the Cxcr2fl/+::Mx-Cre+ group (0.59±0.10×106/mL, P=0.022) due to the virtual absence of recruited neutrophils in Cxcr2-CKO mice (Figure 6A and B). Giemsa-stained cytospin preparations from the peritoneal cavity showed more macrophage-like cells in Cxcr2-CKO mice than in Cxcr2fl/+::Mx-Cre+ mice (Figure 6C). In Cxcr2fl/+::Mx-Cre+ mice, CXCR2 expression was downregulated on infiltrated peritoneal neutrophils (Figure 6D) suggesting receptor engagement during migration.
Figure 6. Decreased migration of CXCR2-deficient neutrophils in sterile peritonitis.
Cxcr2-CKO and Cxcr2fl/+::Mx-Cre+ mice 4 wks after poly(I:C) injection were injected with 4% aged TG 2 hours before analysis. (A) Total cells were collected from the peritoneum and counted on a hemocytometer. (B) Total infiltrated neutrophils in peritoneum were calculated by total cells collected from the peritoneum times the percentage of neutrophils in total cells determined by the staining of peritoneal cells with Ly6G and CD45 antibodies (data not shown). (C) Wright-Giemsa staining of peritoneal cells collected by cytospin showed neutrophils with multiple lobulated nuclei in the peritoneal exudate of Cxcr2fl/+::Mx-Cre+mice (top) and mononuclear and kidney-shaped nuclei consistent with monocytes in the peritoneum of Cxcr2-CKO mice (bottom). (D) Peritoneal exudate cells and blood cells from Cxcr2fl/+::Mx-Cre+ mice were collected and stained with Ly6G and CXCR2 antibodies, then analyzed by flow cytometry. These data represent two independent experiments. Each experiment included 3 mice per group. **P<0.01; *P<0.05;
Our current data indicate that the floxed Cxcr2 gene was deleted by induction of Cre recombinase in Cxcr2fl/fl mice, providing opportunities to elucidate the functions of CXCR2 and its many chemokine ligands in murine models of human disorders such as neurodegenerative disease and cancer.
Methods
Mice
The Cxcr2 targeting construct for generation of a conditional Cxcr2 allele was built by recombineering (Liu et al. 2003), based on homologous recombination in E. coli and applied to modifying BACs. The Cxcr2 targeting construct DNA was electroporated into C57BL/6 ES cells. Twenty-five positive candidates from the total 192 ES clones were screened and verified by both Southern hybridization and genomic DNA sequencing. One targeted ES clone was injected into 129/SvEv blastocysts to create chimeric mice. Chimeric founder mice (C57BL/6×129/SvEv) were backcrossed with C57BL/6 mice. Germline transmission of the Cxcr2flox (Cxcr2fl) allele was confirmed by Southern hybridization and PCR genotyping. To establish the Cxcr2-CKO mouse line, Cxcr2fl/fl mice were further bred to Mx-Cre mice (Stock Number: 003556; Strain name: B6.Cg-Tg (Mx1-Cre)1Cgn/J from the Jackson Laboratory). Conditional knockout mice include the Cxcr2-CKO mice. Control mice used for conditional deletion studies included Cxcr2fl/+::Mx-Cre+ or Cxcr2fl/fl genotypes. ROSA26mTmG mice were obtained from The Jackson Laboratory. All mouse studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the Cleveland Clinic (Cleveland, OH).
Genotyping
The Cxcr2fl allele was detected with primers which consist of forward primer F5FRT (AGGGAATAGGGGATATTTGG) and reverse primer R6FRT (GCTTGGCTGGACGTAAACTC). Mice were genotyped by amplification of genomic DNA obtained by tail biopsy as shown by figure 1b. The Cxcr2+ allele was detected with primers which consist of forward primer F5FRT (AGGGAATAGGGGATATTTGG) and reverse primer R3LoxP2 (CGTCTGTGCCTTCTAAGCCT). This PCR reaction yields a 450bp fragment from the Cxcr2fl allele and 640bp fragment from the Cxcr2+ allele. The presence of the deleted allele was detected with LoxPF3 primer (CTACTAGCATGTTTGAGCCC) and FrtR primer (CTTGAATGAGGATGGTTGTT), the amplified fragment measured 400bp (figure 1b). This band was purified with a QIAGEN PCR Purification Kit (Invitrogen) according to the kit manual and the purified product was sequenced by the Molecular Core Facility in the Cleveland Clinic. For Mx-Cre detection, we redesigned specific primers. One primer is located on the Mx1 promoter (Hug et al. 1988) and another one is on the Cre gene. The PCR product from this pair of primers detects Cre genes expressed under the Mx1 promoter (forward primer: MxPF1: TCCCAACCTCAGTACCAAGC, and reverse primer: Cre2: ATTCTCCCACCGTCAGTACG, the DNA product measures 800bp (Supplementary data 1). GAPDH primers were used for positive controls as described before (Liu et al. 2010a).
Injection of poly(I:C) and analysis of CXCR2 deletion
4 or 8 wks old mice were treated with different doses of poly(I:C) (P1530, Sigma, St. Louis, MO) (2.5 mg/kg, 5 mg/kg, 15 mg/kg) as indicated once every other day for a total of 4 injections. The time after poly(I:C) injection was counted from the last day of injection (day 0) as days post injection (pi.).
Deletion of CXCR2 from freshly isolated peripheral leukocytes was determined by flow cytometry. Blood was drawn before the first poly(I:C) injection (time point 0) and once per week for the indicated time. Red blood cells were lysed with red blood cell lysis buffer as decribed previously (Liu et al. 2010a). For the analysis of percentage of deleted CXCR2 on neutrophils, the peripheral leukocytes were stained with antibody mixtures of CD45APC (Clone: 30-F11; Biolegend), CXCR2PE (Clone: 242216; R&D) and Ly6GFITC (Clone: 1A8; Biolegend) and analyzed with flow cytometry as described previously (Liu et al. 2010a). The percentage of CXCR2 deleted neutrophils was determined as the ratio of CXCR2 negative neutrophils to the total Ly6G+ neutrophil population. The percentage of neutrophils in a total blood sample was determined as the ratio of Ly6G+ in total CD45+ cells. Analysis was performed with an LSRII (BD Biosciences) equipped with CellQuest software (BD Biosciences), and 10,000 events per sample, were acquired. Data were analyzed with FlowJo software (Tree Star).
ELISA
At the indicated time-points, blood samples were obtained by submandibular puncture. The level of CXCL1 was detected in the sera by ELISA (Duoset, R&D Systems), as described by the manufacturer.
TG-induced peritonitis
To study the migration of neutrophils to the site of inflammation in the periphery, inflammation in the peritoneal cavity was induced by using 4% TG medium (Sigma-Aldrich, USA) in ddH2O. Autoclaved TG solution was aged in the dark at 4°C for at least one week. Mice were injected i.p. with 4% TG solution (1 mL/20 g mouse). After 2 hours, mice were sacrificed, and peritoneal leukocytes and blood samples were collected. The neutrophil content in the peritoneal cavity and the blood was determined by flow cytometry. Total cell counts from the peritoneum were performed by hemocytometer (Hausser Scientific, Horsham, PA, USA) immediately following peritoneal cell collection. Cells were stained with antibodies as indicated in data and analyzed by flow cytometry. Some of the cells from the peritoneum were cytospun for Wright-Giemsa staining according to the manufacturer’s protocol.
Statistical analysis
Data are expressed as mean ± SD. Multiple comparisons were statistically evaluated by 1-way ANOVA using Prism 4 (GraphPad Software). The Students-t-test was used for the comparisons of cytokine content, the percentage of CXCR2 deletion. A p value <0.05 was considered as significant. *P<0.05, **P<0.01.
Supplementary Material
(A) Strategy for designing PCR primers to detect the Cre expression under the promoter of the Mx1 gene. (B) PCR amplification of the genomic DNA using specific Mx-Cre primers to detect the Cre gene under the promoter of Mx1 in Mx-Cre mice but not in PLP-Cre mice or C57BL/6 (B6) mice. GAPDH primers were used as a quality control for DNA used for the Mx-Cre detection. These data represent at least three independent experiments.
Peripheral blood cells, bone marrow and spleen cells from Cxcr2fl/fl, Cxcr2fl/+::Mx-Cre+ and Cxcr2fl/fl::Mx-Cre+ mice as indicated stained with Ly6G and CXCR2 antibodies were analyzed by flow cytometry. Cxcr2fl/+::Mx-Cre+ and Cxcr2fl/fl::Mx-Cre+ mice were 8 wks after polyI:C injection. The percentage of CXCR2 negative neutrophils is indicated. Data are showed as gating on Ly6G+ cell population. These data represent at least three independent experiments.
Acknowledgments
This work was supported by National Institutes of Health Grants R01NS032151 (R.M.R) and NS051400 (R.M.R).
We thank the Murine Molecular Constructs Laboratory from the Mouse Biology Program at UC Davis and the Case Transgenic and Targeting Facility at Case Western Reserve University. We thank Dr. Qi Shi (Neuroscience Department, Cleveland Clinic) for help with genotyping the mice; we thank the LRI-Flow Core for providing excellent service. Thank you also to Dr. Christine L. White (Molecular Genetics Department) for providing primers and techniques for checking the activation of interferons. ML is supported by funding from Yunnan Province High-tech Talent Introduction Project and the National Natural Science Foundation of China No.30960091 and No.81271330.
Footnotes
There is no conflict of interest to declare.
References
- Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N, Seshan VE, Norton L, Brogi E, Massagué J. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–171. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff DS, Sakamoto T, Ishida K, Makhijani NS, Gruber HE, Yamaguchi DT. CXC receptor knockout mice: characterization of skeletal features and membranous bone healing in the adult mouse. Bone. 2011;48:267–274. doi: 10.1016/j.bone.2010.09.026. [DOI] [PubMed] [Google Scholar]
- Cacalano G, Lee J, Kikly K, Ryan AM, Pitts-Meek S, Hultgren B, Wood WI, Moore MW. Neutrophil and B cell expansion in mice that lack the murine IL-8 receptor homolog. Science. 1994;265:682–684. doi: 10.1126/science.8036519. [DOI] [PubMed] [Google Scholar]
- Cardona AE, Li M, Liu L, Savarin C, Ransohoff RM. Chemokines in and out of the central nervous system: much more than chemotaxis and inflammation. J Leukoc Biol. 2008;84:587–594. doi: 10.1189/jlb.1107763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson T, Kroenke M, Rao P, Lane TE, Segal B. The Th17-ELR+ CXC chemokine pathway is essential for the development of central nervous system autoimmune disease. J Exp Med. 2008;205:811–823. doi: 10.1084/jem.20072404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C, Campion S, Teeling J, Felton L, Perry VH. The sickness behaviour and CNS inflammatory mediator profile induced by systemic challenge of mice with synthetic double-stranded RNA (poly I:C) Brain Behav Immun. 1997;21:490–502. doi: 10.1016/j.bbi.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Devalaraja RM, Nanney LB, Du J, Qian Q, Yu Y, Devalaraja MN, Richmond A. Delayed wound healing in CXCR2 knockout mice. J Invest Dermatol. 2000;115:234–244. doi: 10.1046/j.1523-1747.2000.00034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes WE, Lee J, Kuang WJ, Rice GC, Wood WI. Structure and functional expression of a human interleukin-8 receptor. Science. 1991;253:1278–1280. doi: 10.1126/science.1840701. [DOI] [PubMed] [Google Scholar]
- Hu B, Colletti LM. CXC receptor-2 knockout genotype increases X-linked inhibitor of apoptosis protein and protects mice from acetaminophen hepatotoxicity. Hepatology. 2010;52:691–702. doi: 10.1002/hep.23715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug H, Costas M, Staeheli P, Aebi M, Weissmann C. Organization of the murine Mx gene and characterization of its interferon- and virus-inducible promoter. Mol Cell Biol. 1988;8:3065–3079. doi: 10.1128/mcb.8.8.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson T, Clarke M, Steele CW, Samuel MS, Neumann J, Jung A, Huels D, Olson MF, Das S, Nibbs RJ, Sansom OJ. Inhibition of CXCR2 profoundly suppresses inflammation-driven and spontaneous tumorigenesis. J Clin Invest. 2012;122:3127–3144. doi: 10.1172/JCI61067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A, De Filippo K, Hasenberg M, van den Brandt C, Nye E, Hosking MP, Lane TE, Männ L, Ransohoff RM, Hauser AE, Winter O, Schraven B, Geiger H, Hogg N, Gunzer M. G-CSF-mediated thrombopoietin release triggers neutrophil motility and mobilization from bone marrow via induction of Cxcr2 ligands. Blood. 2011;117:4349–57. doi: 10.1182/blood-2010-09-308387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Liu L, Belkaldi A, Darnall L, Hu T, Drescher C, Cotleur AC, Padovani-Claudio D, He T, Choi K, Lane TE, Miller RH, Ransohoff RM. CXCR2-positive neutrophils are essential for cuprizone-induced demyelination: relevance to multiple sclerosis. Nat Neurosci. 2010a;13:319–326. doi: 10.1038/nn.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Darnall L, Hu T, Choi K, Lane TE, Ransohoff RM. Myelin repair is accelerated by inactivating CXCR2 on nonhematopoietic cells. J Neurosci. 2010b;30:9074–9083. doi: 10.1523/JNEUROSCI.1238-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara K, Smit MJ, Krajnc-Franken M, Gossen J, Rooseboom M, Dokter W. Human CXCR2 (hCXCR2) takes over functionalities of its murine homolog in hCXCR2 knockin mice. Eur J Immunol. 2005;35:2573–2582. doi: 10.1002/eji.200526021. [DOI] [PubMed] [Google Scholar]
- Murphy PM, Tiffany HL. Cloning of complementary DNA encoding a functional human interleukin-8 receptor. Science. 1991;253:1280–1283. doi: 10.1126/science.1891716. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:116–117. [PubMed] [Google Scholar]
- Tiedt R, Hao-Shen H, Sobas MA, Looser R, Dirnhofer S, Schwaller J, Skoda RC. Ratio of mutant JAK2-V617F to wild-type Jak2 determines the MPD phenotypes in transgenic mice. Blood. 2008;111:3931–3940. doi: 10.1182/blood-2007-08-107748. [DOI] [PubMed] [Google Scholar]
- Tsai HH, Frost E, To V, Robinson S, ffrench-Constant C, Geertman R, Ransohoff RM, Miller RH. The chemokine receptor CXCR2 controls positioning of oligodendrocyte precursors in developing spinal cord by arresting their migration. Cell. 2002;110:373–383. doi: 10.1016/s0092-8674(02)00838-3. [DOI] [PubMed] [Google Scholar]
- Ulyanova T, Priestley GV, Nakamoto B, Jiang Y, Papayannopoulou T. VCAM-1 ablation in nonhematopoietic cells in MxCre+ VCAM-1f/f mice is variable and dictates their phenotype. Exp Hematol. 2007;35:565–571. doi: 10.1016/j.exphem.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Chen S, Zhang Y, Li X, Li Y, Wu X, Yuan J, Robling AG, Kapur R, Chan RJ, Yang FC. Rac1 mediates the osteoclast gains-in-function induced by haploinsufficiency of Nf1. Hum Mol Genet. 2008;17:936–948. doi: 10.1093/hmg/ddm366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Strategy for designing PCR primers to detect the Cre expression under the promoter of the Mx1 gene. (B) PCR amplification of the genomic DNA using specific Mx-Cre primers to detect the Cre gene under the promoter of Mx1 in Mx-Cre mice but not in PLP-Cre mice or C57BL/6 (B6) mice. GAPDH primers were used as a quality control for DNA used for the Mx-Cre detection. These data represent at least three independent experiments.
Peripheral blood cells, bone marrow and spleen cells from Cxcr2fl/fl, Cxcr2fl/+::Mx-Cre+ and Cxcr2fl/fl::Mx-Cre+ mice as indicated stained with Ly6G and CXCR2 antibodies were analyzed by flow cytometry. Cxcr2fl/+::Mx-Cre+ and Cxcr2fl/fl::Mx-Cre+ mice were 8 wks after polyI:C injection. The percentage of CXCR2 negative neutrophils is indicated. Data are showed as gating on Ly6G+ cell population. These data represent at least three independent experiments.