Abstract
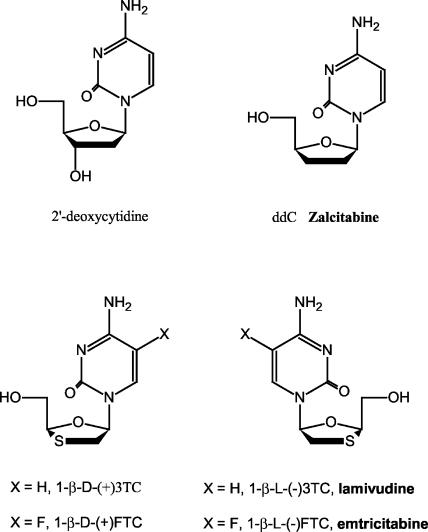
Emtricitabine [(−)FTC; (−)-β-l-2′-3′-dideoxy-5-fluoro-3′-thiacytidine] is an oxathiolane nucleoside analog recently approved by the Food and Drug Administration for the treatment of human immunodeficiency virus (HIV). Structurally, (−)FTC closely resembles lamivudine [(−)3TC] except that the former is 5-fluorinated on the cytosine ring. In HIV-1 reverse transcriptase (RT) enzymatic assays, the triphosphate of (−)FTC [(−)FTC-TP] was incorporated into both DNA-DNA and DNA-RNA primer-templates nearly 3- and 10-fold more efficiently than (−)3TC-TP. Animal studies and clinical trial studies have demonstrated a favorable safety profile for (−)FTC. However, a detailed study of the incorporation of (−)FTC-TP by human mitochondrial DNA polymerase γ, a host enzyme associated with nucleoside toxicity, is required for complete understanding of the molecular mechanisms of inhibition and toxicity. We studied the incorporation of (−)FTC-TP and its enantiomer (+)FTC-TP into a DNA-DNA primer-template by recombinant human mitochondrial DNA polymerase in a pre-steady-state kinetic analysis. (−)FTC-TP was incorporated 2.9 × 105-, 1.1 × 105-, 1.6 × 103-, 7.9 × 103-, and 100-fold less efficiently than dCTP, ddCTP, (+)3TC-TP, (+)FTC-TP, and (−)3TC-TP, respectively. The rate of removal of (−)FTC-MP from the corresponding chain-terminated 24-mer DNA by polymerase γ's 3′→5′ exonuclease activity was equal to the removal of (+)FTC-MP, 2-fold slower than the removal of (−)3TC-MP and (+)3TC-MP, and 4.6-fold slower than the excision of dCMP. These results demonstrate that there are clear differences between HIV-1 RT and polymerase γ in terms of preferences for substrate structure.
Nucleoside analogs have played an essential role in treating human immunodeficiency virus (HIV) infection since the beginning of the AIDS epidemic. Today, 99% of all regimens for HIV treatment contain at least one nucleoside (33). However, long-term use of nucleoside analogs has been associated with adverse effects, many of which are believed to be directly or indirectly associated with mitochondrial dysfunction (18, 23). In many cases, a correlation has been found between mitochondrial toxicity and the inhibition of mitochondrial DNA synthesis, which is carried out by human mitochondrial DNA polymerase γ. Once transformed into their active 5′-triphosphate forms, the nucleoside analogs serve as alternative substrates for HIV-1 reverse transcriptase (RT) and result in chain termination of viral DNA synthesis. However, the same 5′-triphosphate forms of nucleoside analogs can be incorporated into a mitochondrial DNA chain by polymerase γ, leading to termination of DNA synthesis and mitochondrial dysfunction. The 3′→5′ exonuclease activity of polymerase γ has been proposed to play a role in rescuing mitochondrial DNA from this nucleoside-induced chain termination. Our studies and the studies of others have shown that polymerase γ excises 3′-terminal nucleotide monophosphates with drastically different efficiencies depending on the specific nucleotide (9, 15).
Emtricitabine [(−)FTC; (−)-β-l-2′-3′-dideoxy-5-fluoro-3′-thiacytidine], a 5-fluorinated derivative of lamivudine [(−)3TC; (−)-β-l-2′-3′-dideoxy-3′-thiacytidine], is the latest Food and Drug Administration-approved nucleoside reverse transcriptase inhibitor for HIV (Fig. 1). In vitro, (−)FTC is 4 to 10 times more potent than (−)3TC against HIV (30, 31, 32). In detailed enzymatic studies of HIV-1 RT with a pre-steady-state analysis (10), we observed several interesting structure-activity relationships by comparing the incorporation of dCTP, ddCTP, (+)-β-d-2′-3′-dideoxy-3′-thiacytidine 5′-triphosphate [(+)3TC-TP, also commonly referred to as BCH-189], (−)3TC-TP, (+)-β-d-2′-3′-dideoxy-5-fluoro-3′-thiacytidine 5′-triphosphate [(+)FTC-TP], and (−)FTC-TP (Fig. 1). Most interestingly, while (+)FTC-TP has the same incorporation efficiency as its nonfluorinated counterpart (+)3TC-TP, (−)FTC-TP is incorporated ≈10-fold more efficiently than (−)3TC-TP by HIV-1 RT during RNA-dependent DNA synthesis.
FIG. 1.
Structures of dCTP analogs.
Although halogen substitution has long been a practice in searching for higher activity for nucleoside analogs (19, 24, 27), the fluorinated nucleoside analogs have been questioned for their safety, especially after the fatal mitochondrial toxicities which occurred during FIAU [fialuridine; 1-(2′-deoxy-2′-fluoro-β-d-arabinofuranosyl)-5-iodouracil] clinical trials (13). In an earlier effort to understand the mechanism of mitochondrion-related toxicity, we studied the incorporation of dCTP, ddCTP, (+)3TC-TP, and (−)3TC-TP into a DNA-DNA primer-template by recombinant human mitochondrial DNA polymerase and also the excision of these dCMP analogs from the 3′ terminus of a DNA by the exonuclease activity of polymerase γ (9). In that study, we demonstrated that compared to HIV-1 RT, polymerase γ is much more sensitive to the structural modification of ddCTP, including the substitution of a sulfur atom on the sugar ring and the absolute stereochemistry.
In the present study, we asked the following questions. With the 5-fluorine substitute, will (−)FTC-TP and its enantiomer (+)FTC-TP be a more or less efficient substrate for polymerase γ-catalyzed DNA synthesis compared to their corresponding nonfluorinated analogs, (−)3TC-TP and (+)3TC-TP? Will there be a difference in the rate of excision between a terminally incorporated (−)FTC-MP and that of (+)FTC-MP and the two 3TC-MP isomers? Is (−)FTC-TP incorporated more efficiently than (−)3TC-TP by HIV-1 RT during DNA-dependent DNA synthesis? And finally, by comparing the substrate structure-activity relationship for HIV-1 RT with that for polymerase γ, what can we learn about fine tuning between antiviral activity and host toxicity?
MATERIALS AND METHODS
Reagents.
dCTP was purchased from Sigma. (+)FTC-TP and (−)FTC-TP were kindly provided by R. F. Schinazi at Emory University, Atlanta, Ga. The purity of these compounds (>99%) was verified by high-pressure liquid chromatography analysis as well as liquid chromatrography-electrospray ionization mass spectrometry. A synthetic DNA duplex, the D23/D45-mer, 5′-* GCC TCG CAG CCG TCC AAC CAA CT CGG AGC GTC GGC AGG TTG GTT GAG TTG GAG CTA GGT TAC GGC AGG-5′ in which the next correct base for incorporation was dCTP and the asterisk indicates the position of the 32P label, was used. The concentrations of the oligonucleotides were estimated spectrophotometrically at 260 nm with the calculated extinction coefficients 226,750 M−1 cm−1 and 491,960 M−1 cm−1 for the 23-mer and 45-mer, respectively. Before annealing, the primer was 5′-end labeled with T4 polynucleotide kinase and [γ-32P]ATP. The homoduplex 23/45-mer DNA-DNA primer-template was formed by annealing a ≈1:1.3 molar ratio of the 23-mer and 45-mer at 80°C for 5 min and at 50°C for 30 min, followed by cooling on ice.
Purification of HIV-1 RT and human polymerase γ.
The C-terminally His-tagged heterodimer of recombinant HIV-1 RT (p66/p51) was purified from an Escherichia coli expression system as previously reported (17). The recombinant large and small subunits of polymerase γ were purified separately from a baculovirus and an Escherichia coli expression system, as described previously (12, 16). The protein concentration was determined spectrophotometrically at 280 nm, with extinction coefficients of 260,450, 234,420, and 71,894 M−1 cm−1 for HIV-1 RT and the large and the small subunits of polymerase γ, respectively.
Rapid chemical quenching and kinetic analysis.
Rapid chemical quenching experiments were performed with a KinTek Instruments model RQF-3 rapid quench-flow apparatus. Unless mentioned otherwise, all concentrations refer to concentrations during reactions after mixing. All of the reactions were carried out at 37°C. As described earlier (10), the reactions catalyzed by HIV-1 RT were performed under burst conditions, for which a preincubated mixture of 100 nM HIV-1 RT and 300 nM 5′-32P-labeled D23/D45-mer was mixed with a solution containing 10 mM MgCl2 and various concentrations of dCTP analogs, or under single-turnover conditions, for which the reaction mixture contained 250 nM HIV-1 RT and 50 nM primer-template substrate. The reactions catalyzed by polymerase γ used conditions very similar to those described previously (9). All of the incorporation studies were carried out at protein concentrations high enough to maintain the holoenzyme polymerase γ complex and on a time scale short enough to follow a single turnover of enzyme.
A preincubated mixture of large (67 nM) and small (440 nM) subunits of polymerase γ and D23/D45-mer (250 nM) in reaction buffer (50 mM Tris-HCl, 100 mM NaCl, pH 7.8) was mixed with a solution containing MgCl2 (2.5 mM) and various concentrations of dCTP analogs. Some experiments were conducted under single-turnover conditions, for which the enzyme concentrations (250 nM large subunit and 1.25 mM small subunit) were higher than the D23/D45-mer primer-template concentration (50 nM). The reactions were quenched by addition of EDTA (pH 8.0) to a final concentration of 0.3 M. All reaction mixtures were analyzed on 20% denaturing polyacrylamide sequencing gels (8 M urea), imaged on a Bio-Rad GS-525 molecular image system, and quantified with Molecular Analyst (Bio-Rad). Data were fitted by nonlinear regression with KaleidaGraph (Synergy Software version 3.51).
The pre-steady-state burst experiments were fitted to a burst equation, [product] = A[1 − exp(−kobsdt) + ksst], where A represents the amplitude of the burst, which correlates with the concentration of enzyme in the active form, kobsd is the observed first-order rate constant for deoxynucleoside triphosphate (dNTP) incorporation, and kss is the observed steady-state rate constant. For single-turnover experiments, the data were fitted to a single exponential equation, [product] = A[1 − exp(−kobsdt)]. The dissociation constant, Kd, for dNTP binding to the enzyme-DNA complex was calculated by fitting the data into the hyperbolic equation kobsd = (kpol × [dNTP])/(Kd + [dNTP]), where kpol is the maximum rate of incorporation, [dNTP] is the corresponding concentration of dNTP, and Kd is the equilibrium dissociation constant for the interaction of dNTP with the enzyme-DNA complex.
The exonuclease activity of polymerase γ was studied under steady-state conditions as described previously (9). The 24-bp primers listed in Table 2 were prepared by incorporating the corresponding dCTP analogs into the 23-bp DNA (9). A mixture of 5′-32P-labeled 23-mer/45-mer DNA duplex (1,000 pmol), HIV-1 RT (300 pmol), dCTP analogs (5 to 50 μM), and 10 mM MgCl2 in reaction buffer (50 mM Tris-HCl, 50 mM NaCl, pH 7.8) was incubated at 37°C for 10 to 30 min. The 24-mer DNA products were purified by gel electrophoresis and annealed to 45-mer DNA to form the D24/D45-mer duplex at a ratio of 1:1.3 at 80°C for 5 min and 50°C for 30 min, followed by cooling on ice.
TABLE 2.
Kinetic parameters for the exonuclease activity of human mitochondrial DNA polymerase γ towards different DNA substratesa
| Primer | Mean holoenzyme kexo (s−1) ± SE |
|---|---|
| D24-dC | 0.022 ± 0.001 |
| D24-ddC | <1 × 10−5b |
| D24-(+)3TCc | 0.012 ± 0.001 |
| D24-(−)3TCc | 0.010 ± 0.001 |
| D24-(+)FTC | 0.0048 ± 0.0001 |
| D24-(−)FTC | 0.0048 ± 0.0001 |
The same 45-mer template was used in all of the duplexes.
Estimated based on lack of cleavage products formed within 12 h.
Values from reference 9.
The exonuclease activity was studied by measuring the rate of formation of the cleavage products in the absence of dCTP or dCTP analogs. The reaction was initiated by adding MgCl2 (2.5 mM) to a preincubated mixture of polymerase γ large subunit (40 nM), small subunit (270 nM), and 1,500 nM D24/D45-mer duplex in reaction buffer (50 mM Tris-HCl, 100 mM NaCl, pH 7.8) and quenched with 0.3 M EDTA at the designated time points. All reaction mixtures were analyzed on 20% denaturing polyacrylamide sequencing gels (8 M urea), imaged on a Bio-Rad GS-525 molecular image system, and quantified with Molecular Analyst (Bio-Rad). Products formed from the early time points were plotted as a function of time. Data were fitted by linear regression with KaleidaGraph (Synergy Software version 3.51). The slope of the line was divided by the active enzyme concentration in the reaction to calculate the kexo for exonuclease activity.
RESULTS
The potential mechanism of toxicity was investigated by examining the kinetics of the two different reactions catalyzed by human mitochondrial DNA polymerase γ, DNA-dependent DNA synthesis and 3′→5′-exonuclease cleavage. This analysis was carried out by examining the single nucleotide incorporation of (+)FTC-TP and (−)FTC-TP opposite a template of 2′-deoxyguanosine for DNA-dependent DNA polymerization by the recombinant polymerase γ holoenzyme complex. Exonuclease cleavage was investigated by evaluating the excision of (+)FTC-MP and (−)FTC-MP from the 3′ terminus of the corresponding DNA 24-mer primer by the holoenzyme complex of polymerase γ.
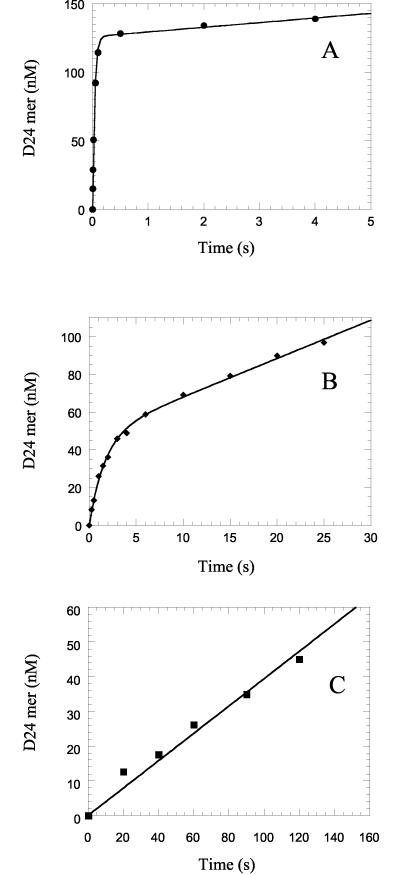
Using a transient kinetic approach, we determined the rate of polymerization (kpol), the dissociation constant for the ground-state nucleotide binding (Kd), and the incorporation efficiency or specificity constant (kpol/Kd) for (+)FTC-TP and (−)FTC-TP. We employed a synthetic 23-mer/45-mer DNA-DNA primer-template (see Materials and Methods) to perform a series of pre-steady-state burst and single-turnover experiments. In the case of dCTP and (+)FTC-TP, a pre-steady-state burst of product formation was observed, indicating that at saturating concentrations of dNTP, a step after the chemical step is rate limiting (Fig. 2). The rate-limiting step after incorporation of a single nucleotide is the release of the DNA from the enzyme (16). In Fig. 2A and B, the solid line represents the best fit of the data to a burst equation, from which the burst amplitude, A, the observed rate of incorporation, kobsd, and the steady-state rate constant, kss, were obtained.
FIG. 2.
Pre-steady-state kinetics of incorporation of dCMP (•) (2.5 μM), (+)FTC-MP (⧫) (2 μM), and (−)FTC-MP (▪) (100 μM) into a DNA-DNA 23-mer/45-mer primer-template by HIV-1 RT. (A) The solid lines represent the best fit of the data (•) to a burst equation with an amplitude A of 126 ± 1 nM, an observed first-order rate constant for the burst phase kobsd of 25.7 ± 0.6 s−1, and an observed rate constant for the linear phase kss of 0.027 ± 0.004 s−1 for dCMP incorporation. (B) The solid lines represent the best fit of the data (⧫) to a burst equation with an A of 48 ± 2 nM, kobsd of 0.60 ± 0.05 s−1, and kss of 0.043 ± 0.004 s−1 for (+)FTC-MP incorporation. (C) The solid line represents the best fit of the data (▪) to a linear equation, y = ax + b, with a slope of 0.39 nM · s−1 and an observed rate of 0.0039 s−1.
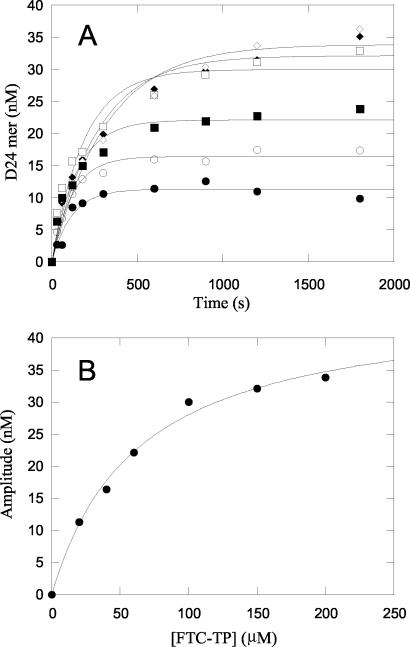
For the incorporation of (−)FTC-MP, a pre-steady-state burst of product formation was not observed (Fig. 2C), and therefore the reaction was studied under single-turnover condition with enzyme in excess of DNA as described previously (8, 16). Incorporation of (−)FTC-MP showed complex kinetics because the amplitude changed as the (−)FTC-TP concentration increased under single-turnover conditions (Fig. 3A) and the rates were independent of (−)FTC-TP concentration under the conditions tested (data not shown). A similar behavior was also seen previously with zidovudine monophosphate incorporation into a DNA-DNA primer-template by polymerase γ (15).
FIG. 3.
Kinetic traces of incorporation of (−)FTC-MP into a DNA-DNA 23-mer/45-mer primer-template by polymerase γ. (A) Time-dependent DNA 24-mer product formation was plotted at 20 (•), 40 (○), 60 (▪), 100 (□), 150 (⧫), and 200 μM (◊) (−)FCT-TP. The data were fitted to a single exponential equation. (B) Concentration dependence of (−)FTC-TP on amplitude. The amplitudes from single exponential fits were plotted against the (−)FTC-TP concentration. A Kd value of 62.9 μM was calculated based on a hyperbolic fit.
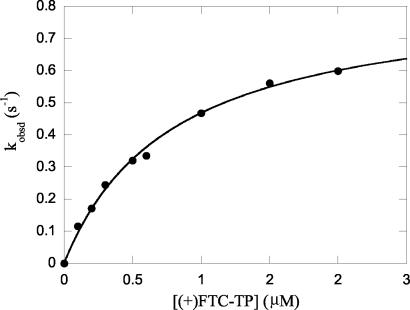
A series of experiments were conducted with various concentrations of (+)FTC-TP and (−)FTC-TP. The interaction of (+)FTC-TP and (−)FTC-TP with the enzyme-DNA complex was assessed by fitting the concentration dependence of the burst rates and the amplitudes, respectively, to the hyperbolic equation. The concentration dependence of the amplitude for (−)FTC-TP incorporation is shown in Fig. 3B. The concentration dependence of the observed polymerization rate for the incorporation of (+)FTC-TP into a DNA-DNA 23-mer/45-mer primer-template by polymerase γ holoenzyme is shown in Fig. 4.
FIG. 4.
Concentration dependence of (+)FTC-TP on the observed rate. The first-order rates were plotted against the (+)FTC-TP concentration, and the data were fitted to hyperbolas to yield a Kd value of 0.79 ± 0.07 μM and a maximum rate of incorporation (kpol) of 0.84 ± 0.03 s−1 for (+)FTC-TP.
A complete summary of the kpol, Kd, and kpol/Kd values for each dCTP analog incorporation by polymerase γ is shown in Table 1. Since the rate of (−)FTC-TP incorporation was independent of the substrate concentration, the kpol for (−)FTC-TP was estimated from the fastest rate observed under the conditions tested. For comparison, the kpol and Kd values of other dCTP analogs which were reported in earlier studies (8, 9), including dCTP, ddCTP, (+)3TC-TP, and (−)3TC-TP, are also shown in Table 1. A comparison of the analogs reveals that the natural substrate, dCTP, served as the best substrate for polymerase γ, as demonstrated by the observed tight binding and fast rate of incorporation. The trend of incorporation efficiency followed the order dCTP > ddCTP > (+)FTC-TP > (+)3TC-TP > (−)3TC-TP > (−)FTC-TP. The (+)FTC-TP and its enantiomer (−)FTC-TP were incorporated 52- to 5,100-fold more slowly than dCTP, respectively.
TABLE 1.
Comparison of kinetic parameters for incorporation of dCTP analogs into D23/D45 by human mitochondrial DNA polymerasea
| Analog | kpol (s−1) | Kd (μM) | kpol/Kd (μM−1 s−1) | Relative substrate specificity |
|---|---|---|---|---|
| dCTP | 44 ± 2b | 1.1 ± 0.1 | 40 | 2.9 × 105 |
| ddCTP | 0.66 ± 0.02b | 0.041 ± 0.007 | 16 | 1.1 × 105 |
| (+)3TC-TP | 0.35 ± 0.01b | 1.5 ± 0.1 | 0.23 | 1.6 × 103 |
| (−)3TC-TP | 0.125 ± 0.005b | 9.2 ± 0.9 | 0.014 | 100 |
| (+)FTC-TP | 0.84 ± 0.03 | 0.79 ± 0.07 | 1.1 | 7.9 × 103 |
| (−)FTC-TP | (8.6 ± 1.5) × 10−3 | 62.9 ± 8.4 | 1.4 × 10−4 | 1 |
Relative substrate specificity = (kpol/Kd)dNTP/(kpol/Kd)(−)FTC-TP, where the relative substrate specificity for (−)FTC-TP is defined as 1. The values reported are means ± standard errors.
From reference 9.
All of the natural d-isomer analogs including dCTP, ddCTP, (+)3TC-TP, and (+)FTC-TP have higher affinity for the enzyme-DNA complex than the l-isomers. In addition, all of the above compounds demonstrated a burst phase followed by a linear steady-state phase under pre-steady-state burst conditions, indicating that the release of the elongated primer-template product is the rate-limiting step for the incorporation of the dCTP analogs with natural configuration. The rates for the steady-state phase were similar between different analogs, ranging from 0.03 to 0.06 s−1 (data not shown). The maximum rate for polymerase γ-catalyzed incorporation of (−)FTC-TP was 98- and 15-fold slower than that of (+)FTC-TP and (−)3TC-TP, while the affinity of (−)FTC-TP for the enzyme-DNA complex was 80- and 7-fold weaker than that of (+)FTC-TP and (−)3TC-TP, respectively.
Using the method described in an earlier report (9), we also studied the polymerase γ 3′→5′-exonuclease removal of (+) and (−)FTC 5′-monophosphates (MP) from duplex DNA with highly purified polymerase γ reassociated holoenzyme complex. Our analysis was carried out with DNA substrates that were 3′ terminated with (+)FTC-MP and (−)FTC-MP. We enzymatically synthesized two DNA 24-mer primers that were 3′ terminated with the didexoynucleoside analogs (+)FTC-MP and (−)FTC-MP. Although the presence of dNTPs in the reaction may affect the rate of excision, the experiments were performed in the absence of dNTPs in order to specifically investigate the mechanism of the reaction and not to introduce any complications by adding dNTPs. Furthermore, previous studies have shown that in the case of natural nucleotides, the rate of excision increased significantly when a mismatched base pair was buried in the primer but when the mismatch was at the 3′ end, the presence of the correct dNTP did not affect the rate (14). Therefore, one would not anticipate that the removal rate of a nucleotide analog at the 3′ end of the primer is influenced by dNTPs.
A summary of the rates of exonuclease activity (kexo) for the holoenzyme complex with D24-(+)FTC and D24-(−)FTC is shown in Table 2. For comparison, we also listed other D24-mer substrates that had been tested and reported earlier (9). As previously observed, among all of the D24-mer substrates tested, the unmodified correctly base-paired primer D24-dC exhibited the fastest excision rate. Excision of the (+) and (−) isomers of FTC-MP was 4.6-fold slower than the removal of dCMP and 2.5-fold slower than the removal of the (+) and (−) isomers of 3TC-MP.
In this study, we also examined the incorporation of (+)FTC-MP and (−)FTC-MP into both DNA-DNA and DNA-RNA primer-templates by HIV-1 RT. A complete summary of the kpol, Kd, and kpol/Kd values for each dCTP analog incorporation by HIV-1 RT during both DNA- and RNA-dependent DNA synthesis is shown in Table 3. In general, the incorporation efficiency for the dCTP analogs during DNA-dependent DNA synthesis was 5- to 37-fold lower than for the corresponding RNA-dependent DNA synthesis. For DNA-dependent DNA synthesis, (−)FTC-TP was incorporated twofold faster than (−)3TC-TP, while they had similar binding affinities for the enzyme-DNA complex. When the incorporation efficiency was compared between the two FTC-TP enantiomers for both DNA- and RNA-dependent DNA synthesis, the activity of the unnatural (−) isomer was slightly higher than that of the (+) isomer.
TABLE 3.
Comparison of kinetic parameters for incorporation of dCTP analogs into D23/D45 and DNA-RNA by HIV-1 RTa
| Analog | DNA-DNA
|
DNA-RNA
|
||||||
|---|---|---|---|---|---|---|---|---|
| kpol (s−1) | Kd (μM−1) | kpol/Kd (μM−1 s−1) | Relative substrate specificity | kpol (s−1) | Kd (μM−1) | kpol/Kd (μM−1 s−1) | Relative substrate specificity | |
| dCTP | 2.9 ± 0.2b | 56 ± 10 | 0.052 | 16 | 22.9 ± 0.7c | 30 ± 4 | 0.76 | 13 |
| ddCTP | 0.35 ± 0.02c | 69 ± 11 | 0.0051 | 1.5 | 0.91 ± 0.04c | 5.3 ± 1.1 | 0.17 | 2.8 |
| (+)3TC-TP | 0.036 ± 0.003c | 44 ± 9 | 8.2 × 10−4 | 0.2 | 0.10 ± 0.01c | 3.5 ± 0.4 | 0.030 | 0.5 |
| (−)3TC-TP | 0.019 ± 0.001c | 15 ± 3 | 0.0013 | 0.4 | 0.033 ± 0.002c | 5.0 ± 0.8 | 0.0067 | 0.11 |
| (+)FTC-TP | 0.031 ± 0.003 | 14 ± 2 | 0.0022 | 0.7 | 0.092 ± 0.003d | 3.0 ± 0.4 | 0.031 | 0.5 |
| (−)FTC-TP | 0.039 ± 0.003 | 12 ± 3 | 0.0033 | 1 | 0.082 ± 0.005d | 1.4 ± 0.4 | 0.060 | 1 |
DISCUSSION
Our studies have shown that (−)FTC-TP is much less efficiently incorporated than (+)FTC-TP into DNA by polymerase γ. This observation is similar to that of a previous study showing that (−)3TC-TP is incorporated less efficiently than (+)3TC-TP (9). Between the two (+)-configured oxathiolane nucleoside analogs, (+)FTC-TP is incorporated 4.8-fold more efficiently than its nonfluorinated analog (+)3TC-TP. In contrast, for the two (−)-configured oxathiolane nucleoside analogs (−)FTC-TP and (−)3TC-TP, the 5-fluorine substitution on the cytosine base has an opposite effect, with (−)FTC-TP being incorporated 100-fold less efficiently than (−)3TC-TP.
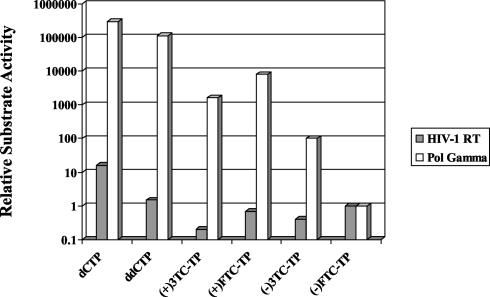
The series of dCTP analogs listed in Table 1 have also been studied for their incorporation into DNA by HIV-1 RT in this report and earlier studies (8, 10). The incorporation efficiencies and the calculated relative substrate specificities of dCTP analogs during DNA-dependent DNA synthesis by HIV-1 RT and polymerase γ, as shown in Tables 1 and 3, are illustrated in Fig. 5. An “ideal” anti-HIV nucleoside analog is expected to have high relative substrate specificity for HIV-1 RT, which corresponds to a high value for the shaded bar in Fig. 5, and at the same time have a low relative substrate specificity for polymerase γ, which corresponds to a low value for the white bar. For both HIV-1 RT- and polymerase γ-catalyzed DNA synthesis, the natural substrate dCTP has the highest incorporation efficiency, followed by ddCTP. ddCTP is incorporated efficiently by both enzymes, in accordance with the high anti-HIV activity and profound mitochondrial toxicity observed with ddC (3-5, 21, 23). (−)FTC-TP is incorporated by HIV-1 RT almost as efficiently as ddCTP, but the latter is a much more potent inhibitor of polymerase γ.
FIG. 5.
Relative substrate specificity of dCTP analogs for HIV-1 RT- and polymerase γ-catalyzed DNA-dependent DNA synthesis. Note that the y axis is plotted on a log scale.
Structural modification of ddCTP through introduction of a sulfur atom at the 3′ position of the sugar moiety led to the analog (+)3TC-TP. For HIV-1 RT, this modification decreases the rate of incorporation but enhances the binding affinity of ddNTP to the HIV-1 RT-DNA complex. In contrast, for polymerase γ, the same structural modification decreases both the rate of incorporation and the binding affinity. The absolute stereochemistry of the 3TC-TP and FTC-TP analogs plays a role in their incorporation by both enzymes; however, the impact is far more profound for polymerase γ. For HIV-1 RT with a DNA-RNA primer-template, (+)3TC-TP was incorporated 4.5-fold more efficiently than its enantiomer (−)3TC-TP, while (+)FTC-TP was incorporated 2-fold less efficiently than (−)FTC-TP. For a DNA-DNA primer-template, the efficiency between the two sets of enantiomers was similar within a twofold range. In contrast, during polymerase γ-catalyzed DNA-dependent DNA synthesis, (+)3TC-TP was incorporated 16-fold more efficiently than (−)3TC-TP. Furthermore, (+)FTC-TP was incorporated an astonishingly 7,900-fold more efficiently than (−)FTC-TP.
The rate of polymerase γ-catalyzed excision offers an indication of the likelihood that the mitochondrial DNA can escape chain termination by dideoxynucleotides. In this study, the rate of excision of incorporated (−)FTC-MP was twofold slower than the that of the removal of (−)3TC-MP, and this difference is considerably less significant than the difference during incorporation. The cellular toxicity studies have shown that (−)3TC, (+)FTC, and (−)FTC are essentially not toxic in a variety of cell lines (32). Cell-based mitochondrial toxicity studies in HepG2 cells have shown that (+)3TC was toxic to mitochondria, whereas (−)3TC, (+)FTC, and (−)FTC were not (7). Weak cellular toxicity of 3TC and FTC was found in human bone marrow progenitor cells, in which (+)3TC was 25- to 45-fold more toxic than (−)3TC (11) and (+)FTC was 3- to 7-fold more toxic than (−)FTC (11, 32). These cell-based results indicate that the cytotoxicity of (+)3TC may be associated with mitochondrial toxicity; however, the weak toxicity of FTC in bone marrow cells may not be due to mitochondria. The apparent discrepancy could be due to other factors, including drug uptake, metabolism, and catabolism, which can affect toxicity.
The introduction of a fluorine atom in the 5-position of the cytosine ring of (+)3TC-TP and (−)3TC-TP had very different effects on the incorporation efficiency for the HIV-1 RT and polymerase γ in this system. For HIV-1 RT, while (+)3TC-TP and (+)FTC-TP had similar incorporation efficiencies, (−)FTC-TP was incorporated three- to ninefold more efficiently than its nonfluorinated counterpart (−)3TC-TP whether DNA or RNA was used as the template. For polymerase γ, the 5-fluorine substitution increased the incorporation efficiency 4.8-fold for (+)FTC-TP compared to (+)3TC-TP, but a surprising 100-fold decrease in the incorporation efficiency was observed when comparing (−)FTC-TP to (−)3TC-TP.
In conclusion, there are clear differences between HIV-1 RT and polymerase γ in terms of preferences for substrate structure changes, even when the changes are subtle. Compared to HIV-1 RT, human mitochondrial DNA polymerase is much more discriminating against the oxathiolane cytosine nucleotide analogs tested versus the natural substrate dCTP. Both 3TC and FTC have high antiviral activity and are well tolerated, with favorable safety profiles (2, 28). The unnatural l-(−) configuration of (−)3TC and (−)FTC seems to play a critical role in preserving the antiviral activity while decreasing the inhibitory effect on host polymerase γ. Furthermore, the introduction of a fluorine group to the 5-position of the cytosine ring apparently makes (−)FTC-TP a better substrate for HIV-1 RT, and this is supported by the in vitro cell culture study and clinical trial data (29, 32).
In this study, we have shown that (−)FTC-TP is a poorer substrate inhibitor for polymerase γ than (−)3TC-TP. The slower rate of incorporation and weaker binding affinity of (−)FTC-TP make its incorporation 100-fold less efficient than that of (−)3TC-TP. To our knowledge, however, there are no substantial differences in toxicity between 3TC and FTC, based on reported in vitro and clinical studies. The discrepancy between this enzymatic study and the other data could be simply that even though 3TC inhibits polymerase γ, it occurs at such a low level that it cannot be manifested at a physiologically appreciable level. Recently, an extensive study of the commercial anti-HIV nucleoside analogs by polymerase γ enzymatic kinetic analysis shed light on the detailed mechanism of mitochondrion-related toxicity (15).
The efficacy and toxicity of nucleoside analogs used to treat HIV are affected by many factors, including drug uptake, transport, catabolism, and metabolism. HIV-1 RT is a well-studied target for activated nucleoside analogs. However, the removal of the nucleoside monophosphate from the 3′ terminus of the viral DNA by cellular enzymes or other mechanisms may play a role in decreasing drug activity (1, 6, 22, 34). The HIV-1 RT-facilitated phosphorolytic excision is certainly a key issue in the case of zidovudine monophosphate (25) and plays an important role in drug resistance. In the case of 3TC-MP, HIV-1 RT was either not observed or so slow it was insignificant (20; K. L. White [Gilead Sciences], personal communication). A very slow rate for 3TC-MP and FTC-MP has also been confirmed (our unpublished work). Nucleoside analog-related toxicity is a complicated issue, and the disruption of mitochondrial metabolism may account for only part of the toxicity (23). Our intent is not to oversimplify the nucleoside analog potency and toxicity issues but to demonstrate that by fine tuning the structure of nucleoside analogs, we can maximize their inhibitory effect on HIV-1 RT and at the same time minimize the adverse effect on host DNA polymerases. This study will aid us in our effort to find additional anti-HIV agents with high antiviral activity and low host toxicity.
Acknowledgments
This work was supported in part by National Institutes of Health grants GM49551 (to K.S.A.), R37AI-41980 (to R.F.S.), and GM044613 (to K.A.J.) and the Department of Veterans Affairs (to R.F.S.).
R.F.S. receives royalties for the sale of 3TC and is entitled to royalties from sales and marketing of FTC as recognition for his contribution to the discovery and development of these drugs.
REFERENCES
- 1.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 2.Beach, J. W. 1998. Chemotherapeutic agents for human immunodeficiency virus infection: mechanism of action, pharmacokinetics, metabolism, and adverse reactions. Clin. Ther. 20:2-25. [DOI] [PubMed] [Google Scholar]
- 3.Benbrik, E., P. Chariot, S. Bonavaud, M. Ammi-Said, E. Frisdal, C. Rey, R. Gherardi, and G. Barlovatz-Meimon. 1997. Cellular and mitochondrial toxicity of zidovudine (AZT), didanosine (ddI) and zalcitabine (ddC) on cultured human muscle cells. J. Neurol. Sci. 149:19-25. [DOI] [PubMed] [Google Scholar]
- 4.Brinkman, K., and T. N. Kakuda. 2000. Mitochondrial toxicity of nucleoside analogue reverse transcriptase inhibitors: a looming obstacle for long-term antiretroviral therapy? Curr. Opin. Infect. Dis. 13:5-11. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C.-H., M. Vazquez-Padua, and Y.-C. Cheng. 1991. Effects of anti-human immunodeficiency virus nucleoside analogs on mitochondrial DNA and its implication for delayed toxicity. Mol. Pharmacol. 39:625-628. [PubMed] [Google Scholar]
- 6.Chou, K. M., M. Kukhanova, and Y. C. Cheng. 2000. A novel action of human apurinic/apyrimidinic endonuclease: excision of L-configuration deoxyribonucleoside analogs from the 3′ termini of DNA. J. Biol. Chem. 275:31009-31015. [DOI] [PubMed] [Google Scholar]
- 7.Cui, L., R. F. Schinazi, G. Gosselin, J. L. Imbach, C. K. Chu, R. F. Rando, G. R. Revankar, and J. P. Sommadossi. 1996. Effect of beta-enantiomeric and racemic nucleoside analogues on mitochondrial functions in HepG2 cells. Implications for predicting drug hepatotoxicity. Biochem. Pharmacol. 52:1577-1584. [DOI] [PubMed] [Google Scholar]
- 8.Feng, J. Y., and K. S. Anderson. 1999. Mechanistic studies comparing the incorporation of (+) and (−) isomers of 3TCTP by HIV-1 reverse transcriptase. Biochemistry. 38:55-63. [DOI] [PubMed] [Google Scholar]
- 9.Feng, J. Y., A. A. Johnson, K. A. Johnson, and K. S. Anderson. 2001. Insights into the molecular mechanism of mitochondrial toxicity by AIDS drugs. J. Biol. Chem. 276:23832-23837. [DOI] [PubMed] [Google Scholar]
- 10.Feng, J. Y., J. Shi, R. F. Schinazi, and K. S. Anderson. 1999. Mechanistic studies show that (−)-FTC-TP is a better inhibitor of HIV-1 reverse transcriptase than 3TC-TP. FASEB J. 13:1511-1517. [DOI] [PubMed] [Google Scholar]
- 11.Furman, P. A., M. Davis, D. C. Liotta, M. Paff, L. W. Frick, D. J. Nelson, R. E. Dornsife, J. A. Wurster, L. J. Wilson, J. A. Fyfe, J. V. Tuttle, W. H. Miller, L. D. Condreay, D. R. Averett, R. F. Schinazi, and G. R. Painter. 1992. The anti-hepatitis B virus activities, cytotoxicities, and anabolic profiles of the (−) and (+) enantiomers of cis-5-fluoro-1-[2-(hydroxymethyl)-1,3-oxathiolan-5-yl]cytosine. Antimicrob. Agents Chemother. 36:2686-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graves, S. W., A. A. Johnson, and K. A. Johnson. 1998. Expression, purification, and initial kinetic characterization of the large subunit of the human mitochondrial DNA polymerase. Biochemistry 37:6050-6058. [DOI] [PubMed] [Google Scholar]
- 13.Honkoop, P., H. R. Scholte, R. A. de Man, and S. W. Schalm. 1997. Mitochondrial injury: lessons from the fialuridine trial. Drug Safety 17:1-7. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, A. A., and K. A. Johnson. 2001. Exonuclease proofreading by human mitochondrial DNA polymerase. J. Biol. Chem. 276:38097-38107. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, A. A., A. S. Ray, J. Hanes, Z. Suo, J. M. Colacino, K. S. Anderson, and K. A. Johnson. 2001. Toxicity of antiviral nucleoside analogs and the human mitochondrial DNA polymerase. J. Biol. Chem. 276:40487-40497. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, A. A., Y. Tsai, S. W. Graves, and K. A. Johnson. 2000. Human mitochondrial DNA polymerase holoenzyme: reconstitution and characterization. Biochemistry 39:1702-1708. [DOI] [PubMed] [Google Scholar]
- 17.Kerr, S., and K. S. Anderson. 1997. Pre-steady-state kinetic characterization of wild type and 3′-azido-3′-deoxythymidine (AZT) resistant human immunodeficiency virus type 1 reverse transcriptase: implication of RNA directed DNA polymerization in the mechanism of AZT resistance. Biochemistry 36:14064-14070. [DOI] [PubMed] [Google Scholar]
- 18.Lewis, W., B. J. Day, and W. C. Copeland. 2003. Mitochondrial toxicity of NRTI antiviral drugs: an integrated cellular perspective. Nat. Rev. Drug Discov. 2:812-822. [DOI] [PubMed] [Google Scholar]
- 19.Lin, T. S., J. Y. Guo, R. F. Schinazi, C. K. Chu, J. N. Xiang, and W. H. Prusoff. 1988. Synthesis and antiviral activity of various 3′-azido analogues of pyrimidine deoxyribonucleosides against human immunodeficiency virus (HIV-1, HTLV-III/LAV). J. Med. Chem. 31:336-340. [DOI] [PubMed] [Google Scholar]
- 20.Mas, A., B. M. Vazquez-Alvarez, E. Domingo, and L. Menendez-Arias. 2002. Multidrug-resistant HIV reverse transcriptase: involvement of ribonucleotide-dependent phosphorolysis in cross-resistance to nucleoside analogue inhibitors. J. Mol. Biol. 323:181-197. [DOI] [PubMed] [Google Scholar]
- 21.Medina, D. J., C. H. Tsai, G. D. Hsiung, and Y. C. Cheng. 1994. Comparison of mitochondrial morphology, mitochondrial DNA content, and cell viability in cultured cells treated with three anti-human immunodeficiency virus dideoxynucleosides. Antimicrob. Agents Chemother. 38:1824-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moyle, G. 2000. Toxicity of antiretroviral nucleoside and nucleotide analogues. Drug Safety 23:467-481. [DOI] [PubMed] [Google Scholar]
- 24.Pankiewicz, K. W. 2000. Fluorinated nucleosides. Carbohydr. Res. 327:87-105. [DOI] [PubMed] [Google Scholar]
- 25.Ray, A. S., E. Murakami, A. Basavapathruni, J. A. Vaccaro, D. Ulrich, C. K. Chu, R. F. Schinazi, and K. S. Anderson. 2003. Probing the molecular mechanisms of AZT drug resistance mediated by HIV-1 reverse transcriptase using a transient kinetic analysis. Biochemistry 42:8831-8841. [DOI] [PubMed] [Google Scholar]
- 26.Ray, A. S., E. Murakami, C. N. Peterson, J. Shi, R. F. Schinazi, and K. S. Anderson. 2002. Interactions of enantiomers of 2′,3′-didehydro-2′,3′-dideoxy-fluorocytidine with wild type and M184V mutant HIV-1 reverse transcriptase. Antivir. Res. 56:189-205. [DOI] [PubMed] [Google Scholar]
- 27.Ray, A. S., R. F. Schinazi, E. Murakami, A. Basavapathruni, J.-X. Shi, S. M. Zorca, C. K. Chu, and K. S. Anderson. 2003. Probing the mechanistic consequences of 5-fluorine substitution on pyridimidine nucleotide analog incorporation by HIV-1 reverse transcriptase. Antivir. Chem. Chemother. 14:115-125. [DOI] [PubMed] [Google Scholar]
- 28.Richman, D. D. 2001. Antiretroviral activity of emtricitabine, a potent nucleoside reverse transcriptase inhibitor. Antivir. Ther. 6:83-88. [PubMed] [Google Scholar]
- 29.Rousseau, F. S., C. Wakeford, H. Mommeja-Marin, I. Sanne, C. Moxham, J. Harris, L. Hulett, L. H. Wang, J. B. Quinn, and D. W. Barry. 2003. Prospective randomized trial of emtricitabine versus lamivudine short-term monotherapy in human immunodeficiency virus-infected patients. J. Infect. Dis. 188:1652-1658. [DOI] [PubMed]
- 30.Schinazi, R. F. 2003. Assessment of the relative potency of emtricitabine and lamivudine. J. Acquir. Immune Defic. Syndr. 34:243-245. [DOI] [PubMed] [Google Scholar]
- 31.Schinazi, R. F., C. K. Chu, A. Peck, A. McMillan, R. Mathis, D. Cannon, L. S. Jeong, J. W. Beach, W. B. Choi, S. Yeola, and D. C. Liotta. 1992. Activities of the four optical isomers of 2′,3′-dideoxy-3′-thiacytidine (BCH-189) against human immunodeficiency virus type 1 in human lymphocytes. Antimicrob. Agents Chemother. 36:672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schinazi, R. F., A. McMillan, D. Cannon, R. Mathis, R. M. Lloyd, A. Peck, J. P. Sommadossi, M. St. Clair, J. Wilson, P. A. Furman, G. R. Painter, W.-B. Choi, and C. Liotta. 1992. Selective inhibition of human immunodeficiency viruses by racemates and enantiomers of cis-5-fluoro-1-[2-(hydroxymethyl)-1,3-oxathiolan-5-yl]cytosine. Antimicrob. Agents Chemother. 36:2423-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott-Levin Database. 2003. HIV therapy audit, 2Q03. Verispan, Yardley, Pa.
- 34.Skalski, V., S. H. Liu, and Y. C. Cheng. 1995. Removal of anti-human immunodeficiency virus 2′,3′-dideoxynucleoside monophosphates from DNA by a novel human cytosolic 3′→5′ exonuclease. Biochem. Pharmacol. 50:815-821. [DOI] [PubMed] [Google Scholar]