Abstract
Mutations of Desert hedgehog (DHH) have been associated to 46,XY pure gonadal dysgenesis (PGD) and to mixed gonadal dysgenesis (MGD); however, there have been no functional studies of mutations described in DHH. To determine if mutations p.L162P and Δ1086delG yield functional impairment, we performed in vitro and in silico analysis of both DHH mutants. In complementary DNA of DHH, we performed site-directed mutagenesis, which was confirmed by DNA sequencing. Protein extracts were obtained from HEK293cells transfected with different constructs and analyzed by Western blot; besides, densitometric analysis of chemiluminescent signals was performed. In addition, the structure of the wt-DHH and its two mutant proteins was inferred using in silico protein molecular modeling. In the Western blot analysis, we observed the absence of signal for p.L162P in DHH-N and a diminished signal for Δ1086delG in DHH-C, when compared to wt-DHH. Protein modeling showed notable conformational changes for the side chains of p.L162P, while the secondary structure was drastically modified in Δ1086delG, when compared to wt-DHH. To our knowledge, this is the first study focused to determine by in vitro studies, the effect of two specific mutations in DHH associated with 46,XY PGD and MGD. Our results suggest that both mutations have a deleterious effect on the expression of the DHH mutant proteins.
This study observed decreased expression of mutant DHH proteins and modeled the conformational changes in protein structure associated with those mutations associated with gonadal dysgenesis.
Introduction
Male sex determination in mammals depends on the presence of the SRY gene located on the Y chromosome, as well as on the action of several other genes located on autosomal and X-linked loci, which are involved in the testis-determining pathway. A determinant event in testicular organogenesis is the specification of somatic cell lineages which include Leydig cells, Sertoli cells, and peritubular myoid cells. The specification of these lineages is critical for establishing testis morphology and hormone production (Yao et al., 2002).
One of the genes involved in testis development is Desert hedgehog (DHH), a member of the hedgehog family of signaling proteins, which also includes Sonic hedgehog (SHH) and Indian hedgehog (IHH) (Ingham, 1998). In humans, DHH is constituted by three exons and encodes a protein of 396 amino acids (Tate et al., 2000). Diverse evidences support that Dhh/DHH is involved in testis differentiation, that is, it has been demonstrated that differentiation of peritubular myoid cells and the consequent formation of testis cords is regulated by Dhh (Clark et al., 2000; Pierucci-Alves et al., 2001). Likewise, Dhh/Ptch1 signaling triggers Leydig cell differentiation by upregulating the Steroidogenic Factor 1 and P450 Side Chain Cleavage enzyme expression in Ptch1-expressing precursor cells, which are located outside the testis cords (Yao et al., 2002). Kawai et al. (2011) reported a missense mutation in the Dhh gene that resulted in impaired Leydig cell development in mp/mp rats, suggesting that this mutation was responsible for the presence of pseudohermaphrodite phenotypes of mp/mp rats. Furthermore, in 46,XY subjects with gonadal dysgenesis, mutations in this gene have been described (Umehara et al., 2000; Canto et al., 2004, 2005; Das et al., 2011; Paliwal et al., 2011).
In previous studies, our research group reported mutations in the DHH gene in three individuals with 46,XY complete pure gonadal dysgenesis (PGD), as well as in two individuals with mixed gonadal dysgenesis (MGD) (Canto et al., 2004, 2005). One individual with complete PGD had a mutation located in the amino-terminal domain of the DHH protein (p.L162P), while four individuals (two with complete PGD and two with MGD) had a mutation in the carboxy-terminal domain, consisting of a deletion of one nucleotide in exon 3 (Δ1086delG, p.L362).
To our knowledge, there have been no functional studies of mutations described in the DHH gene. Therefore, to determine if mutations p.L162P and Δ1086delG yield functional impairment, we performed in vitro analysis of both DHH mutants. In addition, we analyzed the structure of both mutated DHH proteins by using molecular modeling.
Methods
Site-directed mutagenesis and constructs
Complementary DNA of DHH was obtained from the OriGene Company, TrueClone Human Collection, SC122903, Homo sapiens DHH as transfection-ready DNA (OriGene Technology, Inc.). On this plasmid, site-directed mutagenesis of DHH was carried out using the QuickChange Mutagenesis Kit (Stratagene), according to the manufacturer's instructions. The mutagenic primer used for DHH-c.T485C (p.L162P) was 5′-CAAGTATGGGTTGCCGGCGCGCCTCG -3′ (the bold and underlined base indicate the mutant nucleotide); and for DHH-ΔG1086 was 5′-CCTTGAGACT*CTGCACGCGCTAGG-3′ (the * means that a G nucleotide was deleted). Polymerase chain reaction conditions consisted of an initial denaturing step at 95°C for 1 min, followed by 18 cycles of amplification, divided into a denaturing step at 95°C for 50 s, an annealing step at 60°C for 50 s, an extension step at 68°C for 7 min, and a final extension at 68°C for 7 min. By this procedure, we obtained the constructs pCMV6-XL5+DHH-c.T485C and pCMV6-XL5+DHH-ΔG1086. All constructs were screened in Escherichia coli DH5α strain and transformed into an E. coli BL21 (DE3) strain for expression. The integrity of the construct was confirmed by DNA sequencing, on an automated DNA sequencer ABI 377 (Perkin-Elmer, Applied Biosystems Division) using the DNA Sequencing Kit BigDye™ Terminator Cycle Sequencing Ready Reaction (PE Biosystems), following the protocol supplied by the manufacturer.
Cell culture and transfection
HEK293 cells were kindly provided by Dr. Felipe Vilchis and Dr. Bertha Chávez (Instituto Nacional de Ciencias Médicas y Nutrición “Salvador Zubirán,” México City) and were cultured in growing conditions in the Dulbecco's modified Eagle's medium (DMEM; Gibco Life Technology, Co.), supplemented with 10% fetal bovine serum in a humidified atmosphere at 37°C and 5% CO2. Once the cells were confluent, differentiation was induced substituting the growing medium by the DMEM containing 1% horse serum.
One microgram of each construct was transiently transfected using Lipofectamine 2000 (Life Technologies, Invitrogen) according to the manufacturer's instructions. Control cells were obtained from HEK293 cells transfected with the empty vector.
Western blot and densitometry analysis
Protein extracts were obtained from HEK293cells transfected with the different constructs using the ProteoJet membrane protein extraction kit (Fermentas, Thermo Scientific Molecular Biology, Life Science), according to the manufacturer's instructions, and quantified by Bradford Protein Assay (BioRad Laboratories, Inc.).
An aliquot of each of the protein extracts (30 μg of each one) was mixed with an equal volume of 2×sample buffer (1.5 mM Tris-HCl [pH 6.8], 10% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol, 8% glycerol, 0.05% bromophenol blue) and boiled for 5 min before analysis by 10% SDS-polyacrilamide gel electrophoresis and Coomassie brilliant blue staining. A replica of the gel was electrotransferred onto a polyvinyl difluoride membrane (Amersham Pharmacia Biotech), which was incubated 1 h at 37°C in a blocking buffer (phosphate-buffered saline, 6% low-fat dry milk) and 1 h at 37°C with the polyclonal goat antibody anti-DHH-N-17 (for the c.T485C-DHH) or anti-DHH-C-terminus (for the DHH-ΔG1086) (SC-1197 and SC-33940; Santa Cruz Biotechnology) a 1:100 dilution, followed by 1 h at 37°C with rabbit anti-goat HRP-conjugated IgG (1:5000 dilution). As a negative control, a rabbit anti-goat HRP-conjugated IgG (Santa Cruz Biotechnology) was used. Immunoblots were developed using an ECL Plus detection kit (Amersham Biosciences). Negative controls were HEK293 cells without the constructs; glyceraldehyde 3-phosphate dehydrogenase (GAPDH) detection was used for normalization of densitometric data. Experiments for each kind of DHH proteins were repeated in four independent experiments.
Densitometric analysis of chemiluminescent signals was performed with the Alpha Imager Software from Alpha Imager Gel Documentator (Alpha Innotech Corporation). Significance was assayed using the Mann–Whitney U test. Data were obtained from at least four independent experiments, carried out with different preparations of wt-DHH and with both mutant DHH proteins.
Molecular modeling of wild/mutant DHH proteins
To analyze the potential effects of mutations on the human DHH structure and to infer the potential consequences on the protein folding and functionality, homology molecular modeling of the proteins was done; we built DHH models submitting to the Swiss-model (Schwede et al., 2003; Arnold et al., 2006) and I-Tasser (Zhang, 2008) servers the original protein sequence of human DHH ([www.ncbi.nlm.nih.gov/sites/entrez], ID: O43323.1, GI: 6166118, 396 amino acids), a manually edited sequence with a change at position 162 (p.L162P), a sequence with a deletion at residue 362, and a sequence with a deletion that spanned from residues 362 to 370. For each protein, seven models were generated and evaluated by submitting them to different assessment tools as previously described (Soriano-Ursúa et al., 2011). The evaluation and selection of the models were carried out by taking into consideration their root mean square deviation, TM-score, C-score (Zhang and Skolnick, 2004), the stereochemistry of polypeptide chain configurations from Ramachandran plots, as well as the score and observations from Swiss-model server tools.
Prediction of structural consequences
To find out possible folding changes in the selected models, they were analyzed by the straightforward Molecular Dynamic minimization process. Briefly, samples were subjected to a refinement process in vaccuo during 10,000 steps at 0 K by using the steepest descendent protocol and the CHARMM27 parameters implemented in the Nanoscale Molecular Dynamics (NAMD 2.6) program (Phillips et al., 2005). The resultant refined models were structurally analyzed as described for their respective unrefined forms.
Additionally, to make a more accurate prediction on the protein stability changes resulting from single amino acid mutations, the structure with the point mutation at position 162 was submitted to three servers: MU-Pro, which uses support vector machines to predict protein stability changes for single amino acid mutations (Cheng et al., 2006), SDM, a server that predicts effects of mutations on protein stability and malfunction (Worth et al., 2011), and the SNAP (screening for nonacceptable polymorphisms) server, which predicts the functional effects of single amino acid substitutions (Bromberg et al., 2008).
Results
Constructs codifying for wt-DHH and both mutant DHH proteins (p.L162P and DHH-ΔG1086) were transiently transfected in HEK293 cells, and their expression was monitored by Western blot analysis using two polyclonal antisera recognizing specifically DHH-N or DHH-C.
The immunoblotting analysis, showed an immunoreactive band of 43.5 kDa in cells transfected with the wt-DHH construct, which corresponds to the full-length uncleaved DHH protein (Fig. 1A, lane 2). Regarding cells transfected with the DHH-ΔG1086 mutant construct, a band corresponding to 39 kDa (Fig. 1A, lanes 3 and 4) was observed, which was in concordance with the predicted molecular mass, after taking into consideration the deletion of 30 amino acids. However, the intensity of the band of the mutant DHH-ΔG1086 was diminished when compared to the full-length wt-DHH band. Protein loading was verified using GADPH, which corresponded to 37 kDa (Fig. 1B).
FIG. 1.
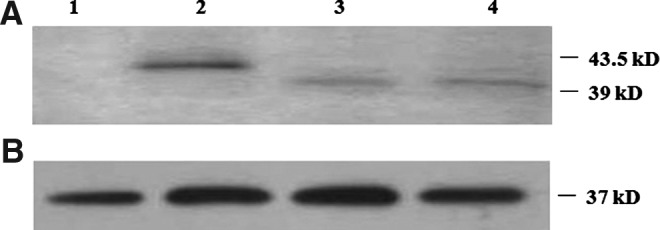
Identification and analysis of full-length Desert hedgehog (DHH) and the Δ1086delG mutant of DHH-C. Analysis was performed by Western blot using antibodies against DHH-N, DHH-C, and GADPH whose signal is presented as loading controls (B). In (A), the expected immunoreactive bands for wt-DHH (43.5 kD) (lane 2) and Δ1086delG mutant DHH-C (39 kD) (lanes 3 and 4) are shown. A rabbit anti-goat HRP-conjugated IgG control was used as negative control (lane 1).
Noteworthy, the signal corresponding to the precursor protein that harbored the p.L162P mutant (located in the N-terminal domain) was not evident in the performed Western blot (data not shown); this result was reproduced in four independent experiments. Besides, we could neither observe the bands corresponding to the amino (19 kDa) or carboxyl fragments (24.3 kDa).
To evaluate the relative protein quantities of wt-DHH and the DHH-ΔG1086 mutant protein, intensity of the band was measured in arbitrary optic density units, which was normalized with GADPH bands intensity; results are presented in percentage. In this regard, the mutant DHH-ΔG1086 protein showed a decreased level of 33.75% when compared to the wt-DHH, this difference was statistically significant (p<0.05).
The DHH models were built using, as templates, data from crystallized fragments of Hedgehog family proteins (PDB codes: 3N1G, 3N1Q, 2WFR, 3K7I, 1AT0, 1VHH); those with the highest score were evaluated. The models obtained were deposited in the Protein Model Data Base (http://mi.caspur.it/PMDB) with the following identification codes: PM0078947 for wt-DHH, PM0078948 for the DHH-p.L162P mutant protein, and PM0078948 for the DHH-ΔG1086 mutant protein.
Moreover, protein segments with an alpha-helix or a beta-sheet secondary structure were conserved in the p.L162P mutant protein when compared with wt-DHH, but differences in coil segments were notable. Furthermore, bioinformatics analysis of the p.L162P mutant protein threw score values associated with defects in the stability or protein folding (Table 1). These findings indicate that the mutation located at position 162 is predicted to be highly destabilizing and could cause protein malfunction.
Table 1.
Results from the Computational Analysis of the p.L162P DHH-N Mutant Form of Desert Hedgehog
| Server | Score | Consequence |
|---|---|---|
| MU-Pro | Pseudo delta delta G: −4.85 | Decrease of the stability of protein structure |
| SDM | Delta delta G: −4.87 | This mutation is predicted to be highly destabilizing and causes protein malfunction and disease |
| Confidence score: −0.18585369 | ||
| SNAP | Reliability: 6 | This is considered a non-neutral mutation, can be related to protein malfunction |
| Expected accuracy: 93% |
DHH, Desert hedgehog.
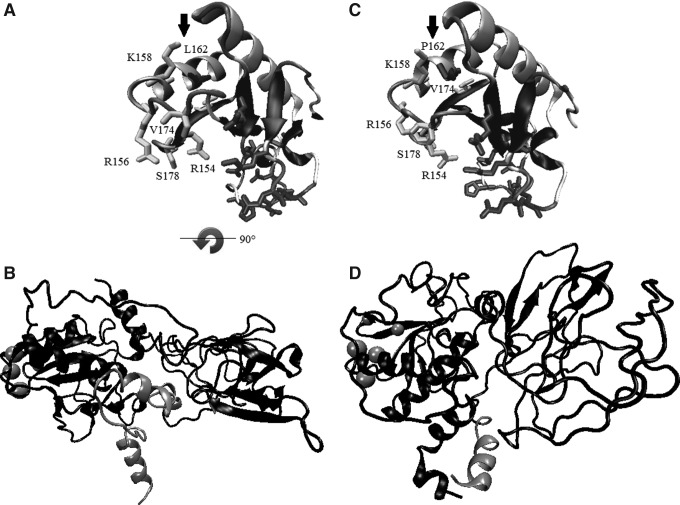
In the refined form of wt-DHH of p.L162P, it was observed that coils are predominant; as well as two clusters with segments of the antiparallel beta-sheet and six alpha-helix segments (Fig. 2). Although this secondary structure was preserved in the p.L162P mutant protein (Fig. 3C), this mutant protein presents remarkable changes in the side chains of residues at position 154, 156, 158, 174, and 178 in comparison to the wt-DHH (Fig. 3A, C).
FIG. 2.
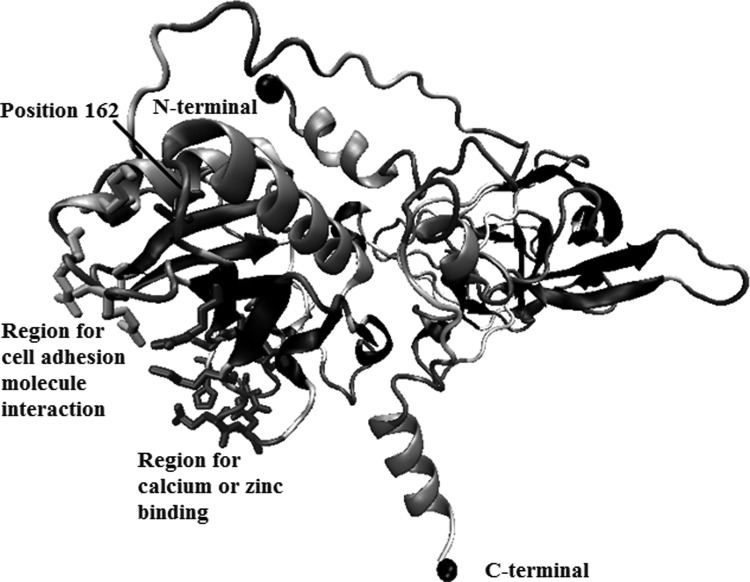
Computational approach of the structure for the wt-DHH. Residues of amino acids involved in interactions with cell adhesion molecules downregulated by oncogenes (CDO) or brother of CDO, or those included in a cluster that interacts with calcium and/or zinc are shown as bonds. Residue in position 162 also is represented as bonds and with a black line. A black sphere was depicted in the amine or carboxyl-terminal region of DHH.
FIG. 3.
Computational approach of the similarities/differences between the wt-DHH (A, B), the p.L162P DHH-N mutant (C), and the Δ1086delG DHH-C mutant (D). In (A, C), the residue in position 162 is represented as bonds and with an arrow. In these panels, only residues in positions 80 to 180 were depicted for clarity. Notable conformational changes were found for the p.L162P mutant protein (C), specifically in the side chains of residues at positions 154, 156, 158, 174, and 178 when compared to the wt-DHH (A). In (B, D) the complete structure of DHH and residues after position 351 are depicted in lighter gray. Residues showed as spheres are those in positions 154, 156, 158, 174, and 178 for spatial reference. The secondary structure for the Δ1086delG DHH-C mutant (D) is modified when compared to the wt-DHH (A).  Indicates a 90° rotation of the (B) figure in relation to the (A) figure.
Indicates a 90° rotation of the (B) figure in relation to the (A) figure.
Regarding the DHH-ΔG1086 mutant protein, it presents secondary structure modifications when compared to wt-DHH (Fig. 3A). This mutant form showed only a cluster of antiparallel beta-sheet and five alpha-helix segments. Besides, there were evident changes in the secondary structure of the regions that interact with cell adhesion molecules or with the calcium or zinc binding region (Fig. 3D).
Discussion
The hedgehog (hh) gene was identified and isolated from Drosophila melanogaster (Nüsslein-Volhard and Wieschaus, 1980). D. melanogaster has a single ortholog hh gene, while mammals have three paralogous genes called SHH (MIM *600725), IHH (MIM *600726), and DHH (*MIM605423), which encode for signaling molecules that play an important role in regulating morphogenesis (Ingham and McMahon, 2001). All known hedgehog proteins are synthesized as pre-pro-proteins, whose signal peptide is removed during their traffic through the endoplasmic reticulum, followed by an autocatalytic cleavage into an N-terminal domain (DHH-N), and a C-terminal domain (DHH-C) implicated in the autoproteolysis reaction (Porter et al., 1996).
Studies in animals show that Dhh is involved in testicular differentiation (Clark et al., 2000; Pierucci-Alves et al., 2001; Yao et al., 2002; Kawai et al., 2011), and that this function is conserved among mammals (O'Hara et al., 2011); besides, mutations have been described in this gene in individuals with 46,XY PGD (Umehara et al., 2000; Canto et al., 2004; Das et al., 2011; Paliwal et al., 2011) and MGD (Canto et al., 2005). However, DHH is the signaling molecule of the HH family least-studied since; so far, there are only five reports of mutations in this gene and there are no functional studies of mutated DHH proteins.
The p.L162P mutation, located in the mature amino-terminal domain of the DHH protein (which presents all the known biological activity of the molecule), leads to a nonconservative amino-acid substitution, changing a highly conserved amino-acid among Hh of the D. melanogaster, SHH and IHH proteins (Roessler et al., 2009). Traiffort et al. (2004) characterized for the first time the biochemical and biological properties of several missense mutations located within the SHH-N domain, demonstrating that some of these mutations affect the stability of the SHH precursor protein or the SHH-N fragment and its biological activity. In this sense, we did not observe, by the Western blot analysis, neither the full-length DHH precursor nor the DHH-N peptide harboring the p.L162P mutant; this may be due to the synthesis of an unstable mutant protein, or that the primary antibody may not be able to recognize the structure of the mutant protein. The 3D structural analyses of the DHH p.L162P mutant protein, predicted that the secondary structure is conserved (Fig. 3A, C), but this mutation is highly destabilizing, which may cause protein malfunction and/or susceptibility to proteolytic degradation.
Like it has been proposed by Traiffort et al. (2004), for mutations in SHH-N, our model of DHH-N indicates that the p.L162P mutated residue belongs to a cluster of amino acids that lie on the surface of the protein. Therefore, it is plausible that this region may interact directly with an as yet unidentified protein stabilizing or preventing further degradation of DHH-N or alternatively, of DHH protein before autoproteolysis. On the other hand, although the cluster of residues for interaction with calcium or zinc are conserved in the p.L162P mutant protein with respect to wt-DHH, notable changes were found for the side chains of residues in positions 154, 156, 158, 174, and 178, which are also involved in the interaction with cell adhesion molecules downregulated by oncogenes (CDO) or brother of CDO (BOC) proteins, which have an important role among mammalian Hh proteins, since all of them (SHH, IHH, and DHH) interact with CDO and BOC in a conserved manner (Kavran et al., 2010). Likewise, Das et al. (2011) found a p.D90del in DHH-N in 46,XY subjects with complete gonadal dysgenesis, and their docking studies showed that the mutant protein presents an altered binding model with BOC, thus leading to a less stable complex as compared to the wild DHH. In this regard, the p.L162P mutant protein model obtained showed proximity with the region related to the interaction with cell adhesion molecules; likewise, there were also notable conformational changes in the lateral chains of this region (Fig. 3C) that may reduce the exposed surface and the interaction with other molecules.
Regarding the Δ1086delG mutant DHH protein, this deletion is located in the C-terminal tail of the DHH-C protein and leads to a nonconservative amino acid substitution, changing a highly conserved amino acid among the Hh of the D. melanogaster, SHH and IHH proteins (Roessler et al., 2009). Besides, this mutation induces a decreased expression of the mutant protein, as compared with the wild type. Lee et al. (1994) examined the effects of several distinct types of mutations in the carboxy-terminus domain of hedgehog of D. melanogaster, demonstrating that deletion or modification of residues, within this domain, are associated with the absence or reduced efficiency of autoproteolysis of the protein. Like it has been observed by Lee et al. (1994), the Δ1086delG mutant DHH protein presented a reduced expression (33.75%), when compared to the full length, which may be the effect of an inefficient autoproteolysis. On the other hand, Traiffort et al. (2004) also characterized the biochemical and biological properties of mutations located within the SHH-C domain; one of these mutations was located in the 383 position of the SHH-C protein, which corresponds to the homologous position of our Δ1086delG mutant DHH protein. These authors compared the crystal structure and molecular studies of the autoprocessing domain of hedgehog of D. melanogaster, of a construct lacking the last 63 amino acids at the C-terminal and observed that this mutation was able to form the intermediate thioester, but failed to transfer cholesterol, suggesting a direct role in cholesterol binding for this region. Thus, they proposed that structural modifications introduced by the A383T of SHH-C (which is located in the same region of the deletion in Hh-C described above), could prevent further binding and/or the transfer of the cholesterol molecule.
Additionally, Kawai et al. (2011) described a mutation in the C-terminal region of Dhh (p.G332R), in a rat with a pseudohermaphrodite phenotype, finding that the DHH protein was not detected by Western blotting in the testicular extracts of the mp/mp rats, suggesting that this mutation has a significant effect on the Dhh protein in the testis.
Moreover, the molecular modeling of the DHH-ΔG1086 mutant protein showed that several changes may be induced by this mutation. The secondary structure was drastically modified since we only found a cluster with segments in antiparallel beta-sheet and five alpha-helix segments. Besides, the deletion in the carboxyl-terminal region impacts the second cluster, with segments in antiparallel beta-sheet (Fig. 3D) and the spatial disposition of nearby segments. All these structural modifications may affect the spatial disposition of residues involved in the interaction with the cell adhesion molecules according with the description by Kavran et al. (2010) (see spheres in Fig. 3B, D).
In conclusion, to our knowledge, this is the first study focused to determine, by in vitro studies, the effect of two specific mutations in the DHH gene associated with the presence of 46,XY gonadal dysgenesis and MGD. Our results suggest that both mutations may induce the synthesis of mutant proteins prone to degradation, and they modify the C-terminal secondary structure as well as the interaction with cell-adhesion molecules. Likewise, these data expand the knowledge about the mammalian family hedgehog proteins.
Acknowledgments
This work was supported by the Fondo de Investigación Básica del Consejo Nacional de Ciencia y Tecnología (CONACYT) to J.P.M., México; Grant: 46683. J.J. Castro was supported by Consejo Nacional de Ciencia y Tecnología (CONACYT). We thank M. Reyes Velázquez social work intern from Universidad Autónoma Metropolitana, Unidad Iztapalapa (UAM-I), México, D.F., México, for technical assistance.
Disclosure Statement
All authors declare that there are no conflicts of interest.
References
- Arnold K. Bordoli L. Kopp J. Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Bromberg Y. Yachdav G. Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;24:2397–2398. doi: 10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto P. Soderlund D. Reyes E. Méndez J.P. Mutations in the desert hedgehog (DHH) gene in patients with 46,XY complete pure gonadal dysgenesis. J Clin Endocrinol Metab. 2004;89:4480–4483. doi: 10.1210/jc.2004-0863. [DOI] [PubMed] [Google Scholar]
- Canto P. Vilchis F. Söderlund D. Reyes E. Méndez J.P. A heterozygous mutation in the desert hedgehog gene in patients with mixed gonadal dysgenesis. Mol Hum Reprod. 2005;11:833–836. doi: 10.1093/molehr/gah216. [DOI] [PubMed] [Google Scholar]
- Cheng J. Randall A. Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62:1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- Clark A.M. Garland K.K. Russell L.D. Desert hedgehog (Dhh) gene is required in the mouse testis for formation of adult-type Leydig cells and normal development of peritubular cells and seminiferous tubules. Biol Reprod. 2000;63:1825–1838. doi: 10.1095/biolreprod63.6.1825. [DOI] [PubMed] [Google Scholar]
- Das D.K. Sanghavi D. Gawde H. Idicula-Thomas S. Vasudevan L. Novel homozygous mutations in Desert hedgehog gene in patients with 46,XY complete gonadal dysgenesis and prediction of its structural and functional implications by computational methods. Eur J Med Genet. 2011;54:e529–e534. doi: 10.1016/j.ejmg.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Ingham P.W. Transducing hedgehog: the history so far. EMBO J. 1998;17:3505–3511. doi: 10.1093/emboj/17.13.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P.W. McMahon A.P. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Kavran J.M. Ward M.D. Oladosu O.O. Mulepati S. Leahy D.J. All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner. J Biol Chem. 2010;285:24584–24590. doi: 10.1074/jbc.M110.131680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y. Noguchi J. Akiyama K. Takeno Y. Fujiwara Y. Kajita S. Tsuji T. Kikuchi K. Kaneko H. Kunieda T. A missense mutation of the Dhh gene is associated with male pseudohermaphroditic rats showing impaired Leydig cell development. Reproduction. 2011;141:217–225. doi: 10.1530/REP-10-0006. [DOI] [PubMed] [Google Scholar]
- Lee J.J. Ekker S.C. von Kessler D.P. Porter J.A. Sun B.I. Beachy P.A. Autoproteolysis in hedgehog protein biogenesis. Science. 1994;266:1528–1537. doi: 10.1126/science.7985023. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C. Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- O'Hara W.A. Azar W.J. Behringer R.R. Renfree M.B. Pask A.J. Desert hedgehog is a mammal-specific gene expressed during testicular and ovarian development in a marsupial. BMC Dev Biol. 2011;11:72. doi: 10.1186/1471-213X-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliwal P. Sharma A. Birla S. Kriplani A. Khadgawat R. Sharma A. Identification of novel SRY mutations and SF1 (NR5A1) changes in patients with pure gonadal dysgenesis and 46,XY karyotype. Mol Hum Reprod. 2011;17:372–378. doi: 10.1093/molehr/gar002. [DOI] [PubMed] [Google Scholar]
- Phillips J.C. Braun R. Wang W. Gumbart J. Tajkhorshid E. Villa E. Chipot C. Skeel R.D. Kale L. Schulten K. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierucci-Alves F. Clark A.M. Russell L.D. A developmental study of the Desert hedgehog-null mouse testis. Biol Reprod. 2001;65:1392–1402. doi: 10.1095/biolreprod65.5.1392. [DOI] [PubMed] [Google Scholar]
- Porter J.A. Ekker S.C. Park W.J. von Kessler D.P. Young K.E. Chen C.H. Ma Y. Woods A.S. Cotter R.J. Koonin E.V. Beachi P.A. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell. 1996;86:21–34. doi: 10.1016/s0092-8674(00)80074-4. [DOI] [PubMed] [Google Scholar]
- Roessler E. El-Jaick K.B. Dubourg C. Vélez J.I. Solomon B.D. Pineda-Alvarez D.E. Lacbawan F. Zhou N. Ouspenskaia M. Paulussen A. Smeets H.J. Hehr U. Bendavid C. Bale S. Odent S. David V. Muenke M. The mutational spectrum of holoprosencephaly-associated changes within the SHH gene in humans predicts loss-of-function through either key structural alterations of the ligand or its altered synthesis. Hum Mutat. 2009;30:E921–E935. doi: 10.1002/humu.21090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T. Kopp J. Guex N. Peitsch M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano-Ursúa M.A. Ocampo-López J.O. Ocampo-Mendoza K. Trujillo-Ferrara J.G. Correa-Basurto J. Theoretical study of 3-D molecular similarity and ligand binding modes of orthologous human and rat D2 dopamine receptors. Comput Biol Med. 2011;41:537–545. doi: 10.1016/j.compbiomed.2011.04.018. [DOI] [PubMed] [Google Scholar]
- Tate G. Satoh H. Endo Y. Mitsuya T. Assignment of desert hedgehog (DHH) to human chromosome bands 12q12→q13.1 by in situ hybridization. Cytogenet Cell Genet. 2000;88:93–94. doi: 10.1159/000015495. [DOI] [PubMed] [Google Scholar]
- Traiffort E. Dubourg C. Faure H. Rognan D. Odent S. Durou M.R. David V. Ruat M. Functional characterization of sonic hedgehog mutations associated with holoprosencephaly. J Biol Chem. 2004;279:42889–42897. doi: 10.1074/jbc.M405161200. [DOI] [PubMed] [Google Scholar]
- Umehara F. Tate G. Itoh K. Yamaguchi N. Douchi T. Mitsuya T. Osame M. A novel mutation of desert hedgehog in a patient with 46,XY partial gonadal dysgenesis accompanied by minifascicular neurophaty. Am J Hum Genet. 2000;67:1302–1305. doi: 10.1016/s0002-9297(07)62958-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth C.L. Preissner R. Blundell T.L. SDM—a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 2011;39:W215–W222. doi: 10.1093/nar/gkr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H.H. Whoriskey W. Capel B. Desert hedgehog/Patched 1 signaling specifies fetal Leydig cell fate in testis organogenesis. Genes Dev. 2002;16:1433–1440. doi: 10.1101/gad.981202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]