Abstract
Effective clinical management of prostate cancer (PCA) has been challenged by significant intratumoural heterogeneity on the genomic and pathological levels and limited understanding of the genetic elements governing disease progression1. Here,we exploited the experimental merits of the mouse to test the hypothesis that pathways constraining progression might be activated in indolent Pten-null mouse prostate tumours and that inactivation of such progression barriers in mice would engender a metastasis-prone condition. Comparative transcriptomic and canonical pathway analyses, followed by biochemical confirmation, of normal prostate epithelium versus poorly progressive Pten-null prostate cancers revealed robust activation of the TGFβ/BMP–SMAD4 signalling axis. The functional relevance of SMAD4 was further supported by emergence of invasive, metastatic and lethal prostate cancers with 100% penetrance upon genetic deletion of Smad4 in the Pten-null mouse prostate. Pathological and molecular analysis as well as transcriptomic knowledge-based pathway profiling of emerging tumours identified cell proliferation and invasion as two cardinal tumour biological features in the metastatic Smad4/Pten-null PCA model. Follow-on pathological and functional assessment con-firmed cyclin D1 and SPP1 as key mediators of these biological processes, which together with PTEN and SMAD4, form a four-gene signature that is prognostic of prostate-specific antigen (PSA) biochemical recurrence and lethal metastasis in human PCA. This model-informed progression analysis, together with genetic, functional and translational studies, establishes SMAD4 as a key regulator of PCA progression in mice and humans.
Adenocarcinoma of the prostate (PCA) is the most common form of cancer and the second leading cause of cancer death in American men2. Current methods of stratifying tumours to predict outcome are based on clinical-pathological factors including Gleason grade, PSA and tumour stage3. These parameters are widely considered inadequate, which has motivated the genetic and biological study of PCA progression with the goal of identifying progression risk biomarkers capable of improving patient management4.
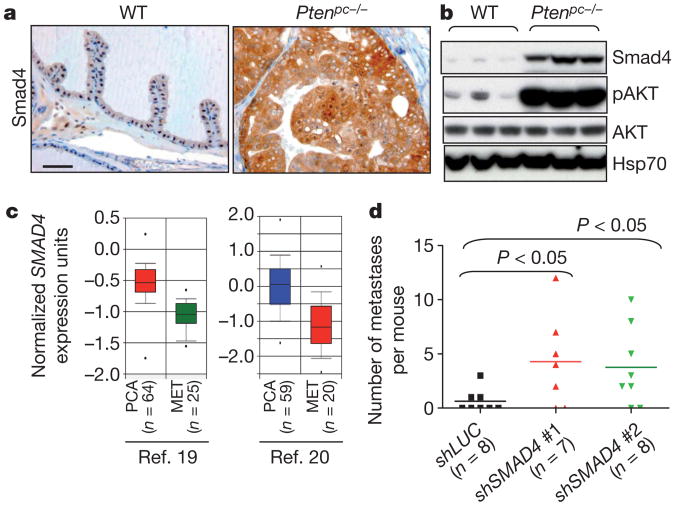
Genetic studies of human PCA has identified signature pathogenetic events5, a number of which have been validated and mechanistically defined in genetically engineered mouse models of PCA6. Prostate-specific Pten deletion (Ptenpc−/−) results in prostate intraepithelial neoplasia (PIN) which, following a long latency, can progress to high-grade adenocarcinoma, albeit with minimally invasive and metastatic features7–10. To understand this feeble progression phenotype, we conducted transcriptome comparison of Ptenpc−/− PIN relative to wild-type prostate epithelium (Supplementary Data 1). In addition to the expected PI3K and p53 (also known as TRP53) pathway representation8, knowledge-based pathway analysis revealed prominent TGFβ/BMP signalling in Ptenpc−/− PIN (Supplementary Fig. 1). Immunohistochemical and western blotting analyses of Smad4 expression confirmed robust increase in Ptenpc−/− PIN compared to wild-type prostate epithelium (Fig. 1a, b). In line with reported down-regulated expression of SMAD4 in a subset of human primary prostate tumours11, Oncomine expression analysis showed consistent SMAD4 downregulation in human PCA metastasis (Fig. 1c and Supplementary Fig. 2).Loss of SMAD4 in advanced PCA is further supported by recent report of frequent epigenetic silencing of the SMAD4 promoter in advanced disease12. On the functional level, SMAD4 knockdown in PC3 showed significantly enhanced frequency of metastases to the lung from renal capsule implantation (Fig. 1d and Supplementary Fig. 3). These observations prompted speculation that a SMAD4-dependent barrier constrains PCA progression.
Figure 1. SMAD4 is a putative suppressor of prostate tumour progression.
a, b, Immunohistochemical (a) and western blot analysis (b) of wild-type (WT) and Ptenpc−/− use prostate tissues. Scale bar, 50 μm. c, Oncomine boxed plot of SMAD4 expression levels between human PCA and metastasis in multiple data sets including those from ref. 19 and ref. 20. d, SMAD4 knockdown enhanced metastatic potential to lung from PC3 cells implanted in renal capsule of immunocompromised nude mice.
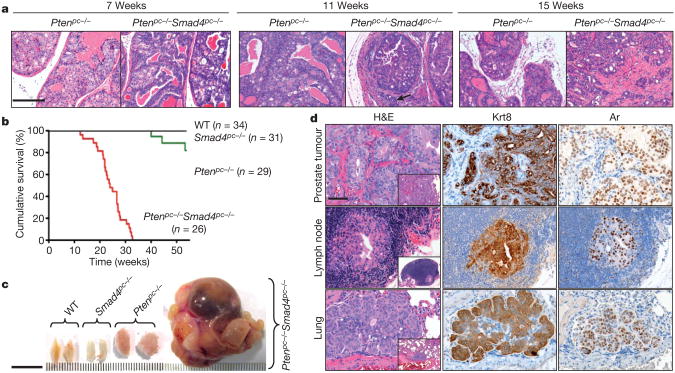
To obtain genetic evidence that Smad4 extinction enables progression, we engineered mice harbouring Pb-Cre4 and conditional knockout alleles of Pten and/or Smad4 (designated Ptenpc−/− and Smad4pc−/−) and confirmed prostate-specific deletion (Supplementary Fig. 4). At 7 weeks of age, both Ptenpc−/− and Ptenpc−/− Smad4pc−/− models develop low-grade PIN (Fig. 2a). Consistent with previous studies7,8, Ptenpc−/− mice acquired invasive features after 19 weeks of age and most survived beyond 1year of age (Fig. 2b). In contrast, Ptenpc−/− Smad4pc−/− mice developed focally invasive PCA by 11 weeks (Fig. 2a, arrow) and highly aggressive invasive PCA with stromal reaction by 15weeks of age (Fig. 2a and Supplementary Fig. 5). All Ptenpc−/− Smad4pc−/− mice died by 32 weeks of age due largely to bladder outlet obstruction which caused hydronephrosis and renal failure (Fig. 2b,c and Supplementary Fig.6),where as Smad4pc−/− mice showed no prostate neoplasia beyond 2years of age (Fig.2b and Supplementary Fig. 7).
Figure 2. Smad4 deletion drives progression of Pten-deficient prostate tumour to highly aggressive prostate cancer metastatic to lymph node and lung.
a, Haematoxylin and eosin (H&E) stained sections of representative anterior prostates (AP) at 7, 11 and 15weeks. Scale bar, 200 μm. b, Kaplan– Meier cumulative survival analysis showing significant (P < 0.0001) decrease in
Molecular pathological analysis of PCA-bearing Ptenpc−/− Smad4pc−/− mice showed metastatic spread of Krt8 and androgen receptor-positive (Krt8+, Ar+) tumour nodules to draining lumbar lymph nodes in 25/ 25 cases and lung metastases in 3/25 cases (0.3–3 mm diameter metastatic nodules)(Fig. 2d, Supplementary Fig.8 and Supplementary Table 1). The histological features of these metastases resembled those of the primary prostate tumour (Fig. 2d). These observations are in contrast to the Ptenpc−/− PCA-bearing mice which never developed metastatic lesions when examined at 1year of age (n = 10), and only two mice(2/8)older than 1.5 years of age contained a solitary lumbar lymph node metastasis and one of these mice also possessed a solitary lung micrometastasis (Supplementary Table 1), a constrained progression phenotype that aligns with previous reports7–9. Similarly, 0/20 Ptenpc−/− p53pc−/− PCA-bearing mice developed metastasis during the same observation period (data not shown).
Having demonstrated the distinctly different metastatic potential of the Ptenpc−/−, Ptenpc−/− Smad4pc−/−, and Ptenpc−/− p53pc−/− models, we then compared transcriptomes of primary PCAs from each to gain insight into the molecular determinants of their phenotypic differences. First, primary anterior prostate tumours with comparable sizes were harvested from 15-week-old animals from each model for mRNA profiling. Comparisons of Ptenpc−/− Smad4pc−/− (n = 5) versus Ptenpc−/− (n = 5) or Ptenpc−/− p53pc−/− (n = 3) with Ptenpc−/− (n = 5) prostate tumour transcriptomes defined the Ptenpc−/− Smad4pc−/− or Ptenpc−/− p53pc−/− signatures (Supplementary Data 2, 3). Ingenuity Pathway Analysis (IPA) was used to generate hypotheses on the biological processes that underlie the metastatic phenotype in the Ptenpc−/− Smad4pc−/− PCAs. In contrast to the Ptenpc−/− p53pc−/− signatures, we found that the two most significantly enriched gene-categories in the Ptenpc−/− Smad4pc−/− signature are ‘cellular movement’ and ‘cellular growth and proliferation’ (Supplementary Fig. 9).
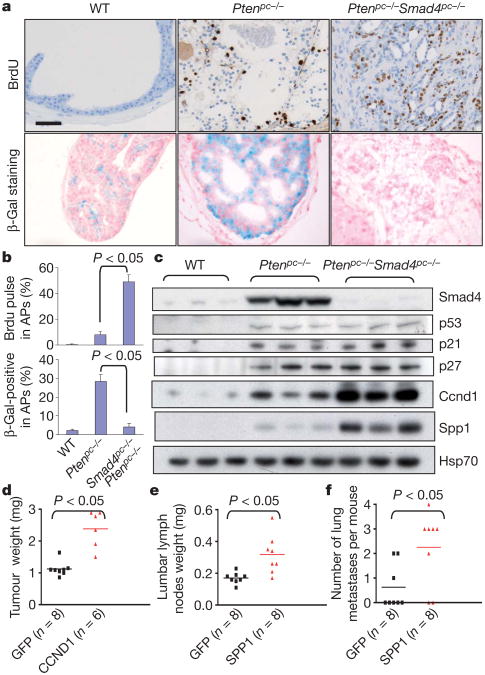
Enrichment of cell growth and proliferation genes in Ptenpc−/− Smad4pc−/− PCA concurs with histopathological observations of markedly increased proliferation index relative to Ptenpc−/− tumours (Fig. 3a, b). Increased proliferation index was not associated with changes in apoptosis (Supplementary Fig. 10), but rather neutralization of oncogene-induced senescence (OIS) as reflected by loss of senescence-associated β-galactosidase staining (Fig. 3a, b). A survey of key regulators of G1/S transition and OIS revealed significant induction of cyclin D1 protein but without significant changes in p53, p21 (also known as Cdkn1a) and p27 (also known as Cdkn1b) in Ptenpc−/− Smad4pc−/− relative to Ptenpc−/− tumours (Fig. 3c and Supplementary Fig. 11). Complementing this hypothesis-driven survey, cyclin D1 was computationally identified as the only cell cycle regulator in the Ptenpc−/− Smad4pc−/− signature that both exhibits human PCA progression-correlated expression in Oncomine and harbours putative SMAD-binding elements (SBEs) in its promoter lifespan in the Ptenpc−/− Smad4pc−/− compared with the Ptenpc−/− cohort. c, Gross anatomy of representative prostates at 22weeks of age. Scale bar, 10 mm. d, H&E-stained sections and immunohistochemical analyses of primary PCA, lumbar lymph nodes and lung of Ptenpc−/− Smad4pc−/−. The tumour context is depicted in low-magnification insets. Scale bar, 50 μm. (Supplementary Data 2). Indeed, chromatin immunoprecipitation (ChIP) assays confirmed that SMAD4 can bind to one of the SBEs in the cyclin D1 gene promoter (Supplementary Figs 12 and 13). Correspondingly, TGFβ1 (also known as TGFB1)-treated SMAD4-transduced Ptenpc−/− Smad4pc−/− prostate tumour cells show down-regulated cyclin D1 expression (Supplementary Fig. 14a). Finally, enforced cyclin D1 expression significantly enhanced xenograft tumour growth in vivo (Fig. 3d). Together, these data support the thesis that cyclin D1 is a key mediator of the cardinal tumour biological feature of increased proliferation in the metastatic Ptenpc−/− Smad4pc−/− model.
Figure 3. Ccnd1 and Spp1 are mediators of prostate tumour cell proliferation and metastasis.
a, BrdU pulse-labelling and SA-β-galactosidase (β-Gal) staining of 15-week-old APs. b, Quantification of BrdU pulse labelling and β-Gal staining. Error bars represent s.d. for a representative experiment performed in triplicate. c, Western blot analysis demonstrating elevated Ccnd1 and Spp1 levels in Ptenpc−/− Smad4pc−/− compared to Ptenpc−/− prostate tumours. d, Enforced CCND1 expression significantly enhanced prostate xenograft tumour growth of PC3 cells. e, f, Enforced SPP1 expression significantly increases metastatic activity of PC3 cells from prostate xenograft to lumbar lymph nodes (e) and to lung (f).
We next obtained available ORFs corresponding to 21 of the 84 ‘Cellular Movement’ genes (Supplementary Table 2) and assayed their ability to enhance invasion of human prostate cancer cells. Using the modified Boyden chamber assay, 10/21 ORFs enhanced invasion of prostate cancer cells including PC3 (Supplementary Table 2). Among these validated invasion genes, SPP1 was selected for deeper analysis given its PCA progression-correlated expression in Oncomine, its prognostic potential for BCR in univariate COX proportional hazard analysis in a data set comprising of transcriptome and outcome data on 79 PCA patients (Supplementary Tables 3 and 4)13, and its known link to TGFβ signalling under different cellular contexts1–6. Western blotting and immunohistochemical analyses confirmed increased Spp1 expression in Ptenpc−/− Smad4pc−/− compared to Ptenpc−/− tumours (Fig. 3c and Supplementary Fig. 11) and promoter analysis17 identified a conserved SBE in the Spp1 promoter which was confirmed by ChIP assay in cells treated with TGFβ1 (Supplementary Fig. 15). In contrast to previous studies showing Smad4 as an inducer of Spp1 expression through displacement of transcription repressors from Spp1 promoter in a mink lung epithelial cell line and a preosteoblastic cell line14,16, loss of Smad4 in the Ptenpc−/− Smad4pc−/− prostate tumour cells results in markedly increased Spp1 expression(Fig.3c and Supplementary Data 2). TGFβ1 treatment correspondingly suppressed Spp1 expression in SMAD4-dependent manner in Ptenpc−/− Smad4pc−/− prostate tumour cells (Supplementary Fig. 14b). These observations underscore the context-specific actions of TGFβ-SMAD4 signalling on its downstream targets18. Next, to verify that Spp1 functionally contributes to the metastatic phenotype in our model, we showed significant inhibition of invasive activity in vitro upon knockdown of Spp1 in Ptenpc−/− Smad4pc−/− mouse PCA cells (Supplementary Fig. 16). Conversely, enforced SPP1 expression enhanced invasion in vitro of several human lines (Supplementary Fig. 17). Finally, orthotopic implantation of SPP1-transduced PC3 cells in the prostate exhibited increased lumbar lymph node metastasis and enhanced metastasis to lung (Fig. 3e–f and Supplementary Fig. 18). These results strongly indicated that SPP1 is a pro-metastasis invasion gene in human PCA and in the Ptenpc−/− Smad4pc−/− PCA model.
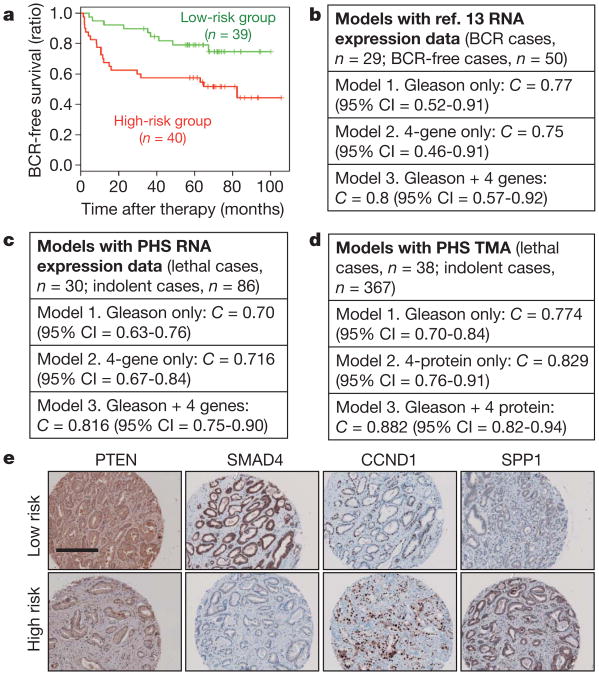
The in vivo genetic modelling studies, the in silico transcriptomic and pathway analyses, along with the tumour biological and functional characterizations collectively point to the inactivation of Pten and Smad4 as well as activation of cyclin D1 (also known as Ccnd1) and Spp1 as drivers of PCA progression. As such, we posited that these four key PCA metastasis progression relevant genes may carry prognostic value for metastasis risk in human PCA (see Supplementary Fig. 19). To this end, we assessed how robustly these four genes can stratify risk of BCR (> 0.2 ng ml−1) in the data set from ref. 13. Although only SPP1 was significantly correlated with BCR in univariate analysis, an overall risk score integrating the four-gene signature by multivariate Cox regression showed significant association with BCR as well (P-value = 0.0025, and overall C-index = 0.66, see Supplementary Tables 4 and 5). Furthermore, the four-gene model robustly stratified the ref. 13 cohort by K-mean clustering into two groups that exhibited significant difference in risk for BCR by Kaplan–Meier analysis (Fig. 4a; hazard ratio = 2.6, log-rank test P = 0.012). Importantly, by C-statistics, this four-gene signature carries independent prognostic information as it can enhance the prognostic accuracy of Gleason score from C-index from 0.77 to 0.8 (Fig. 4b), even though by itself, the four-gene signature (C-index as 0.75) performs only as well as Gleason score alone (Fig. 4b).
Figure 4. Prognostic potential of a four-gene signature in human PCA.
a, The four-gene set of PTEN/SMAD4/CCND1/SPP1 can dichotomize PCA cases for BCR in the ref. 13 data set. b, c, C-statistic analysis revealed that this four-gene set can enhance the prognostic accuracy of Gleason score in the ref. 13 data set (b) and in an independent PHS cohort (c). d, TMA-based four-protein model also significantly improve the prognostic ability of Gleason (P = 0.015) from the PHS cohort. e, Representative immunohistochemical staining with specific antibody against PTEN, SMAD4, CCND1 and SPP1 in the Directors Challenge TMA. Scale bar, 200 μm.
We repeated this analysis in an independent extreme-case-control cohort derived from the Physicians' Health Study (PHS) (Supplementary Table 6; see Methods for study design), where we showed that the four-gene model was also capable of enhancing the prognostic accuracy of Gleason score in predicting metastatic lethal outcome (Fig. 4c; C = 0.716 by four-gene signature). Although exclusion of non-informative cases may have biased towards a positive association, the prognostic performance by this four-gene signature is unlikely a chance occurrence because, by gene-set-enrichment testing, it outperforms 243 other bidirectional signatures curated in the Molecular Signature Databases of the Broad Institute (MSigDB, version 2.5) in predicting metastatic lethal outcome in this PHS extreme-case-control cohort (Supplementary Fig. 20).
Encouraged by the prognostic value in two independent cohorts using RNA expression yet mindful of the inherent intra-tumoural heterogeneity of PCA which may obscure expression differences in whole-tumour transcriptome profiles, we next performed immunohistochemical staining with validated antibodies against PTEN, SMAD4, cyclin D1 and SPP1 on a tumour tissue microarrays (TMA) comprising a cohort of 405 tumour specimens randomly selected from men diagnosed with prostate cancer who underwent radical prostatectomy in the PHS cohort. Staining results were quantified by expert pathologists (R.L. and M.L.) blinded to the outcome of the cases. Indeed, not only does the four-protein model improve the prognostic accuracy of Gleason score in combination, it performs significantly better than Gleason score alone (Fig. 4d; C = 0.774 for Gleason only, C = 0.829 for four-protein model alone, and C = 0.882 for Gleason + four-protein model; P = 0.015 for improvement). Moreover, the addition of the four-protein model to the clinical parameters (Gleason, age at diagnosis, TNM stage; C = 0.842) leads to a significant seven point increase in the C-statistic (C = 0.913), P-value for difference between full clinical model versus clinical model + four-protein signature = 0.047 (Supplementary Table 7). The enhanced prognostic value of ‘Gleason + four-protein model’ was similarly validated in yet another independent cohort, the Directors Challenge TMA containing 40 prostate cancer patients with recurrence as outcome (Supplementary Table 8) (Fig. 4e and Supplementary Fig. 19c; C = 0.704 for Gleason alone versus C = 0.740 for Gleason + four-protein model).
In summary, concomitant Pten and Smad4 inactivation in the prostate epithelium can bypass OIS, enhance tumour cell proliferation and drive invasion to produce a fully-penetrant invasive and metastatic PCA phenotype in the mouse (Supplementary Fig. 21). The human relevance of this Ptenpc−/− Smad4pc−/− model of metastatic PCA is credentialed by the prognostic significance of a four-marker signature derived from this mouse model in predicting biochemical recurrence or lethal metastasis in human PCAs. Thus this study will facilitate the development of a molecularly-based prognostic assay that may complement the current standard of care to improve evidence-based management of PCA patients, a current major unmet need.
Methods
Pten and Smad4 conditional alleles, genotyping and expression analysis
The PtenloxP and Smad4loxP conditional knockout alleles have been described elsewhere21,22. p53loxP strain was generously provided by A. Berns23. Prostate epithelium-specific deletion was effected by the PB-Cre424 and was obtained from MMHCC (http://mouse.ncifcrf.gov/search_results.asp). All cohorts were in a FVB/n, C57BL/6 and 129/Sv mixed genetic background.
Tissue analysis
Normal and tumour tissues were fixed in 10% neutral-buffered formalin overnight then processed, paraffin-embedded, sectioned and stained with haematoxylin and eosin according to standard protocol. For immunohisto-chemistry, 5 μm sections were incubated with primary antibodies overnightat4 °C in a humidified chamber. Primary antibodies: rabbit polyclonal anti-androgen receptor (06-680, Millipore), Smad4 (1676-1, Epitomics), Ck8 (also known as Krt8) (GTX15465, GeneTex); p53 (VP-P956, Vector Laboratories), p21 (C-19, sc-397, Santa Cruz), p27 (2747-1, Epitomics) and Cyclin D1 (RM-9104-R7, Thermo Scientific); and mouse monoclonal Spp1 (sc-21742, Santa Cruz). For rabbit antibodies, sections were subsequently developed using Dako Envision. Mouse monoclonal staining was developed using MOM kit (Vector). To assay senescence in prostate tissue of the various genotypes, frozen sections were stained for SA-β-Gal as described elsewhere7. Representative sections from at least three mice were counted for each genotype.
For western blot analysis, tissues and cells were lysed in RIPA buffer (20 mM Tris pH 7.5, 150 mM sodium chloride, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM EDTA, 0.1% SDS)containing complete mini protease inhibitors (Roche) and phosphatase inhibitors. Western blots were obtained using 20–50 μg of lysate protein, and were incubated with antibodies against Smad4 (sc-7966, Santa Cruz), phospho-AktSer473 (4060, Cell Signaling Technology), Akt (3272, Cell Signaling Technology), V5 (R960-25, Invitrogen), Hsp70 (610607, BD Transduction Laboratories), and Spp1 (sc-21742, Santa Cruz), p53 (sc-6243, Santa Cruz), p27 (2747-1, Epitomics), p21 (65961A, BD Biosciences), Cyclin D1 (2926, Cell Signaling), pSmad1/5/8 (9511, Cell Signaling), Smad1 (9743, Cell Signaling), pSmad2 (Ser465/467) (3101S, Cell Signaling), Smad2 (3103, Cell Signaling), pSmad3 (ab52903, Abcam), Smad3 (06-920, Millipore).
Establishment of mouse prostate tumour cell lines
Tumours were dissected from prostates of Ptenloxp/loxp Smad4loxp/loxp PB-Cre4+ (Ptenpc−/− Smad4pc−/−) mice, minced, and digested with 0.5% type I collagenase (Invitrogen) as described previously. After filtering through a 40-μm mesh, the trapped fragments were plated in tissue culture dishes coated with type I collagen (BD Pharmingen). Cells with typical epithelial morphology were collected, and single cells were seeded into each well of a 96-well plate. Three independent cell lines (Ptenpc−/− Smad4pc−/−-1, -2 and -3,) were established and maintained in DMEM plus 10% fetal bovine serum (FBS, Omega Scientific), 25 μg ml−1 bovine pituitary extract, 5 μg ml−1 bovine insulin, and 6 ng ml−1 recombinant human epidermal growth factor (Sigma-Aldrich). The prostate tumour epithelial cells express epithelial marker CK8 detected by immunofluorescence analyses using CK8 (GTX15465, GeneTex) antibody.
Establishment of inducible Ptenpc−/− Smad4pc−/− SMAD4-TetOn cell lines
Ptenpc−/− Smad4pc−/− prostate tumour cells (see above) were used as parental cells for establishment of inducible SMAD4 TetOn cells using TetOn Advanced Inducible Gene Expression System (Clontech). Human SMAD4 coding region inserted into the pTRE-Tight vector, and a TetOn SMAD4 expression system was generated according to the manufacturer's protocol. Stable clones were induced to express SMAD4 using 1 μg ml−1 doxycycline (dox), and expression was verified to be comparable to the SMAD4 level in Ptenpc−/− prostate tumours by western blot analysis of whole-cell extracts, using anti-SMAD4 antibody (sc-21742, Santa Cruz) (Supplementary Fig. 12).
RNA isolation and real-time PCR
Total RNA was extracted using TRIzol followed by RNeasy Mini kit (Qiagen) cleanup and RQ1 RNase-free DNase Set treatment (Promega) according to the manufacturer's instructions. First strand cDNA was synthesized using 1 μg of total RNA and Superscript II (Invitrogen). Real-time quantitative PCR was performed in triplicates with a MxPro3000 and SYBR GreenER qPCR mix (Invitrogen). The relative amount of specific mRNA was normalized to Gapdh. Primer sequences are available upon request.
Transcriptomic and pathway analyses
For transcriptomic analyses, anterior prostate from mice at 15weeks of age were isolated and total mRNA extracted, labelled and hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 Arrays by the Dana-Farber Cancer Institute Microarray Core Facility according to the manufacturer's protocol. Affymetrix mouse MOE430 raw data (CEL files) were pre-processed using robust multi-array analysis (RMA) of the Affy package of Bioconductor. The background-corrected, normalized and summarized probe set intensity data were then analysed using significance analysis of microarrays (SAM) to identify differentially expressed genes. Using a twofold, FDR 5% cut-off, we generated a 3,532 probe set that distinguishes differentially expressed genes in anterior prostate samples from Ptenpc−/− (five mice) versus WT (PB-Cre4) (three mice), 397 probe sets that distinguishes differentially expressed genes in anterior prostate samples from Ptenpc−/− Smad4pc−/− (five mice) versus Ptenpc−/− (five mice), and 370 probe sets that distinguishes differentially expressed genes in Ptenpc−/− p53pc−/− (three mice) versus Ptenpc−/− (five mice). Gene information for all probes was annotated based on ‘Mouse430_2.na28.annot.csv’ downloaded from the Affymetrix website. Probes with multiple genes in the Affymetrix annotation file were mapped against latest mouse genome build (UCSC mm9) for the single matching gene. Probes mapped to more than one position on mm9 were ignored. Human orthologues of mouse genes were extracted from HomoloGene build 64(ftp://ftp.ncbi.nih.gov/pub/HomoloGene/). Intersection of the murine list with the human orthologous genes produced an orthologous set of genes.
All differentially expressed gene lists generated as described above were further analysed with the Ingenuity Pathways Analysis program (http://www.ingenuity.com/index.html) to identify canonical pathways, and molecular and cellular functions enriched in the related gene lists.
cDNA and shRNA constructs
Human cDNAs presented in Supplementary Table 1 were obtained from the Human ORFeome collection, Japan National Institute of Technology and Evaluation (NITE), Japan, and transferred into a modified pMSCV-V5 vector via Gateway recombination. Knockdown of human SMAD4 and mouse Spp1 were performed by infecting the indicated cells with lentivirus containing either shSMAD4 or shSpp1 (provided by W. Hahn). The shRNA constructs for shSMAD4 #1, #2 correspond to clone ID#s TRCN0000040028 (hairpin sequence: CCGGGCAGACAGAAACTGGATTAAACTCGAGTTTAATCCAGT TTCTGTCTGCTTTTTG), and TRCN0000040029 (hairpin sequence: CCGGCC TGAGTATTGGTGTTCCATTCTCGAGAATGGAACACCAATACTCAGGTT TTTG), respectively. The shRNA constructs for shSpp1#1, #2 correspond to clone ID#s TRCN0000054698 (Hairpin sequence: CCGGCTCTTAGCTTA GTCTGTTGTTCTCGAGAACAACAGACTAAGCTAAGAGTTTTTG), and TRCN0000054700 (Hairpin sequence: CCGGCACAAGGACAAGCTAGTCC TACTCGAGTAGGACTAGCTTGTCCTTGTGTTTTTG), respectively, in the RNAi Consortium (TRC).
Viral production and transduction
Approximately 2 × 106 293T cells were seeded in 100 mm plates 15h before transfection (∼30% confluent) in 10% FBS/DMEM with antibiotics. For MSCV viral production, 3 μg viral backbone, 2.7 μg gag/pol expression vectors, and 0.3 μg VSV-G expression vector were diluted to 20 μl using Opti-MEM (Invitrogen) and combined with 180ml Opti-MEM containing 12 μl FuGENE-6 (Roche). This mixture was incubated at room temperature (RT) for 20 min and added to the 10 ml media covering the 293T cells. For pLKO shRNA lentivirus production, 10 μg of viral backbone and 10 μg of lentiviral packaging vectors were diluted to 1,000 μl using Opti-MEM (Invitrogen). The resulting mix was combined with 1,000 μl Opti-MEM containing 30 μl Liptofectamine2000 (Invitrogen), incubated at room temperature for 20 min and added to 8 ml media covering the 293T cells. The media was replaced with 10% FBS/DMEM approximately 10 h post-transfection and viral supernatants were collected at 36 h and 60 h after transfection and combined. Viral supernatants (5 ml) containing 8 μg ml−1 polybrene were added to target cells that were seeded 24 h before infection at 70–80% confluence. Cells were infected twice and allowed to recover in 10% FBS/RPMI 1640 with antibiotics for 12 h following the second infection, after which cells were selected with 2 μg ml−1 puromycin for 4 days and allowed to recover in normal medium for 24 h before further experiments.
Transwell invasion assay
Standard 24-well Boyden invasion chambers (BD Biosciences) were used to assess cell invasiveness following the manufacturer's suggestions. Briefly, cells were trypsinized, rinsed twice with PBS, resuspended in serum-free media, and seeded at 2 × 105 cells per well for PC3 cells and Ptenpc−/− Smad4pc−/− cells, 4 × 105 cells per well for BPH1 cells. Chambers in triplicate were placed in 10% serum-containing media as a chemo-attractant and an equal number of cells were seeded in cell culture plates in triplicate as input controls. Following 22 h incubation, chambers were fixedin 10% formalin, stained with crystal violet for manual counting or by pixel quantification with Adobe Photoshop. Data was normalized to input cells to control for differences in cell number (loading control).
Orthotopic and renal capsule implantation
Male SCID mice (6 weeks old) were obtained from Taconic. Orthotopic and renal capsule implantations were performed as described previously25,26. Briefly, a suspension of 1 × 106 cells in 50 μl of a 1:1 mixture of PBS and Matrigel (BD Biosciences) was injected into the anterior prostate lobe. For renal capsule implantation 5 × 105 cells were suspended in 50 μl of neutralized type I rat tail collagen (BD Biosciences), allowed to gel at 37 °C for 15min, covered with growth medium, followed by grafting beneath the renal capsule of mice.
Identification of putative SMAD binding sites (SBEs)
The Smad binding elements (SBEs) in the promoters of the Ptenpc−/− Smad4pc−/− signature of 267 genes were identified computationally by established methods16. Briefly, the conserved nucleotides in the 4kb promoter regions of the promoters were isolated and scanned for enrichment of the SMAD binding motifs in TRANSFAC. Enrichment was assessed by comparing the target regions to matched control regions at the same distance from the transcription start sites of random genes. Promoter analysis on these gene sets for SBEs used the CisGenome software (http://www.biostat.jhsph.edu/∼hji/cisgenome/).
Chromatin immunoprecipitation (ChIP) assay
ChIP assays with 1 μg of normal mouse IgG (Upstate), normal rabbit IgG (Upstate), anti-RNA polymerase II (PoII) (Upstate), anti-acetyl-Histone H3 (Upstate) or anti-SMAD4 IgG (mouse monoclonal, clone B8, sc-7966, Santa Cruz) overnight at 4 °C were conducted by established methods16.
Immunohistochemical evaluation of outcome tissue microarrays (TMAs)
Immunohistochemical staining was performed on 5-μm sections of the TMAs to assess cytoplasmic PTEN (PN37, rabbit polyclonal, 18-0256, Zymed), cytoplasmic SMAD4 (mouse monoclonal, clone B8, sc-7966, Santa Cruz), nuclear cyclin D1 (Rabbit monoclonal, SP4, RM-9104-R7, Thermo Scientific), and cytoplasmic SPP1 expression (Rabbit polyclonal, O17, 18625, IBL) after citrate-based antigen retrieval.
TMA slides were scanned using the CRi Nuance v2.8 (Woburn) slide scanner following the standard bright field TMA protocol. The system acquires images at 20nm intervals and combines them into a stack file which represents one image. This was done automatically to create one image for each core on the TMA. The maximum likelihood method was used to extract the spectra of DAB and haematoxylin, which represent the different elements of IHC. inForm v0.4.2 software (CRi) was used to analyse the spectral images of each core. Initially, a training set comprising two classes of tissue was created: ‘tumor’ and ‘other’. Representative areas for each class were marked on 12–16 images from each TMA. The software was trained on these areas using the spectra of both the counterstain (haematoxylin) and the immunostain (DAB) and tested to determine how accurate it could differentiate between the two classes. This process was repeated until further iterations no longer improved accuracy.
Histological images were then analysed using the ‘nuclear or cytoplasmic’ algorithm. The multispectral imaging capabilities of the Nuanc slide scanner allows the software to isolate or segment the nuclei using the unmixed spectra of the nuclear counterstain and the DAB immunohistochemical stain used in addition for a nuclear biomarker. In turn, cytoplasm is found based on the non-nuclear tumour area. Threshold settings approximated: scale1,offset subtraction0,minimum blob size 30, maximum blob size 10,000, circularity threshold 0, edge sharpness 0, fill hole enabled (nuclear parameters); algorithm 4, area 200, compactness 0.5, Wht threshold 225 (cytoplasmic parameters). The final score was based on the percentage of the cytoplasmic or nuclear tumour area that was positively stained and this was represented as a ten bin histogram. This involved each pixel being placed into one of ten bins based on the intensity of the DAB spectra, with an adjustment of the threshold for the 9th bin by the user in order to create a desirable distribution. By reviewing images and their scores, a threshold level of these bins was determined that represented real staining, and the values from the bins above this threshold were added together to create a final score which represented the per-centage of cytoplasmic or nuclear area that waspositively stained. All samples were also reviewed by pathologists (R.L. and M.L.) to ensure that assigned scores were appropriate. TMA cores that were difficult to classify (due to technical artefacts such as folds in the tissue, air bubbles, cores overlapping or due to difficulty in morphological classification) were either eliminated from the analysis in order to categorize the tissue appropriately. The Directors Challenge TMA originally contained 52 patient samples27. However, as is typical of most heavily used TMAs, some of the samples become exhausted over time from extensive use by the M.L. lab and the community. After careful quality control of each core on the TMA by R.L. in M.L. lab, only 40 high quality core samples were considered usable (Supplementary Table 7). Careful quality control of each core on the PHS TMA by R.L. in the M.L. lab, 405 high quality core samples were considered usable (Supplementary Table 5).
Clinical outcome analysis
The raw Affymetrix HG-U133A expression profiles and clinical information of 79 prostate cancer patients from the ref. 13 cohort (Supplementary Table 2)12 were generously provided by W. Gerald. The raw data set was analysed by MAS5 algorithm. Low-expression probesets with less than 20% present calls across the 79 samples were excluded from the data. The remaining 13,027 probesets map to 8,763 genes with unique symbols, and the mean log-transformed probeset levels were used as the gene expression profiles.
A univariate Cox proportional hazard analysis was conducted using the R ‘survival’ package for invasion assay positive genes to identify those expression in PCA tumours was positively associated with biochemical recurrence (BCR, defined by post-op PSA > 0.2 ng ml−1) in the ref. 13 data set12.
K-means clustering algorithm was used with the PTEN/SMAD4/CCND1/SPP1 four-gene model to identify two cancer sample clusters. The initial centres for the K-means clustering were set at the two cases with the longest Euclidean distance. Kaplan–Meier analysis for the survival difference of the two cancer patient clusters was conducted using the R ‘survival’ package. C-statistics analysis was conducted using the R ‘survcomp’ package. The statistical procedures used in the analyses include a bootstrapping step that estimates the distribution of C-statistics of all models across 10,000 random bootstrapping instances, and a comparative step that uses the paired t-test to compare the C-statistics of models and evaluate the statistical significance28. Multivariate Cox proportional hazards model analysis with the four-gene signature was used to estimate the coefficients of individual genes, which combined the four-gene expression levels into an integrated risk score model defined.
To validate further the prognostic significance of this four-gene model, we repeated this analysis in an independent cohort derived from the Directors Challenge cohort27 (Supplementary Table 7) and the Physicians' Health Study (PHS) cohort. PHS cohort (Supplementary Table 5): the men with prostate cancer included in this study were participants in the Physicians' Health Study (PHS), an ongoing randomized trial among US male physicians29,30. The men were diagnosed with histologically-confirmed prostate cancer after randomization, between January 1983 and December 2004. We obtained archival formalin-fixed, paraffin-embedded tissue specimens, either radical prostatectomy (95%) or TURP (5%) and constructed tumour tissue microarrays for immunohistochemical analyses; 405 had sufficient tumour tissue available for this project. All men in the trial were followed for mortality, and cause of death was confirmed by a study endpoints committee. In addition, we retrieved medical records and questionnaire data on the men with prostate cancer to collect information on treatments, clinical characteristics, as well progression of the cancer. Through March 2010, 38 men of 405 had developed a lethal metastatic phenotype, defined by bony metastases or cancer-specific death.
We undertook gene expression profiling as part of a previous project to define molecular signatures in prostate cancer31 on a subset of the PHS included on the TMAs. As part of the sampling, we sought to maximize efficiency for studies of lethal prostate cancer by devising a study design that included men who either died from prostate cancer or developed metastases during follow up (‘lethal prostate cancer’ cases) or who survived at least 10 years after their diagnosis without any evidence of metastases (men with ‘indolent prostate cancer’).We sought to include all lethal cancers, based on follow-up through March 2007, and took a random sample of indolent cancers for a total sample size of 116 cases. In this design, we exclude men with non-informative outcomes, namely those who died from other causes within 10 years of their prostate cancer diagnosis or had been followed for less than 10 years with no disease progression. The natural history of prostate cancer is quite long, with men dying of prostate cancer even 15 or more years after cancer diagnosis32. Thus, we excluded prostate cancer cases with less than 10 years follow-up to increase confidence on the outcome annotation since we are not seeking to estimate survival time. By focusing on long-follow-up cases, an extreme-case-control study design allows us to maximally identify lethal versus indolent prostate cancer. In addition, to minimize the potential that C-statistics estimation might be biased towards a higher lethal composition by such extreme-case-study-design, we have chosen a logistic regression analysis rather estimating survival analysis.
The tissue based studies were approved by the Institutional Review Boards of Harvard School of Public Health and Partners Healthcare.
We assessed the enrichment of the four-gene signature to that of 244 bidirectional signatures curated in the Molecular Signature Databases of the Broad Institute (MSigDB, version 2.5) by computing an enrichment statistic33.
Supplementary Material
Acknowledgments
The authors are grateful to the late W. Gerald for providing the primary gene expression data and clinical outcome files13. We thank S. Zhou for excellent mouse husbandry and care, B. Xiong and G. Tonon for bioinformatic assistance, and S. Jia, J. M. Stommel, J. Paik, M. Kim and A. C. Kimmelman for helpful discussion. We thank M. Vidal, the Ellison Foundation and DFCI ISR for support of ORF cloning efforts, R. Maser for MSCV-puro-v5 gateway vector, W. Hahn for shRNA constructs. We thank the DF/HCC Specialized Histopathology Core and the DF/HCC Tissue Microarray and Imaging core for the TMA IHC staining;the DFCI/BWH Center for Molecular Oncologic Pathology (CMOP) for the quantification of the IHC. Z.D was supported by the Damon Runyon Cancer Research Foundation. D.H. was supported by a graduate fellowship from the National Science Foundation. H.Z. was supported by the Helen Hay Whitney Foundation. Y.A.W. was supported by the Multiple Myeloma Research Foundation. This work is supported by the Belfer Institute for Applied Cancer Science, NCI U01-CA84313 (L.C. and R.A.D.), DF/HCC SPORE in Prostate Cancer P50 CA090381-08 (Z.D.), the National Cancer Institute (M.L. RO1CA131945 and P50 CA90381, L.M. RO1 5R01CA136578, M.S. R01CA141298), and the Linda and Arthur Gelb Center for Translational Research (M.L.). R.A.D. was supported by an American Cancer Society Research Professorship and L.M. was supported by the Prostate Cancer Foundation.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Z.D. designed and performed the experiments. L.C. and R.A.D. supervised experiments and computational analysis and contributed as senior authors. C.J.W., Y.X., Y.H., D.H., T.R.G., M.J.S., W.H.W. and L.M. performed the computational analysis. G.C.C. provided pathology analyses. X.W., R.L., S.S. and M.L. performed TMA staining and quantification. N.B. generated Smad4L mouse allele. D.E.H. provided the human ORFeome clones. D.H., J.Z.,S.R.P., E.S.L., B.H., S.J., H.Z., A.H.S. and K.L.S. performed the experiments. Y.A.W. contributed to the writing of the manuscript.
References
- 1.Andreoiu M, Cheng L. Multifocal prostate cancer: biologic, prognostic, and therapeutic implications. Hum Pathol. 2010;41:781–793. doi: 10.1016/j.humpath.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010 doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Walsh PC, DeWeese TL, Eisenberger MA. Localized prostate cancer. N Engl J Med. 2007;357:2696–2705. doi: 10.1056/NEJMcp0706784. [DOI] [PubMed] [Google Scholar]
- 4.Rubin MA. Targeted therapy of cancer: new roles for pathologists—prostate cancer. Mod Pathol. 2008;21(Suppl 2):S44–S55. doi: 10.1038/modpathol.2008.11. [DOI] [PubMed] [Google Scholar]
- 5.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeet V, Russell PJ, Khatri A. Modeling prostate cancer: a perspective on transgenic mouse models. Cancer Metastasis Rev. 2010;29:123–142. doi: 10.1007/s10555-010-9212-9. [DOI] [PubMed] [Google Scholar]
- 7.Wang S, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trotman LC, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma X, et al. Targeted biallelic inactivation of Pten in the mouse prostate leads to prostate cancer accompanied by increased epithelial cell proliferation but not by reduced apoptosis. Cancer Res. 2005;65:5730–5739. doi: 10.1158/0008-5472.CAN-04-4519. [DOI] [PubMed] [Google Scholar]
- 11.Zeng L, Rowland RG, Lele SM, Kyprianou N. Apoptosis incidence and protein expression of p53, TGF-beta receptor II, p27Kip1, and Smad4 in benign, premalignant, and malignant human prostate. Hum Pathol. 2004;35:290–297. doi: 10.1016/j.humpath.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Aitchison AA, et al. Promoter methylation correlates with reduced Smad4 expression in advanced prostate cancer. Prostate. 2008;68:661–674. doi: 10.1002/pros.20730. [DOI] [PubMed] [Google Scholar]
- 13.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hullinger TG, Pan Q, Viswanathan HL, Somerman MJ. TGFβ and BMP-2 activation of the OPN promoter: roles of Smad- and Hox-binding elements. Exp Cell Res. 2001;262:69–74. doi: 10.1006/excr.2000.5074. [DOI] [PubMed] [Google Scholar]
- 15.Packer L, et al. Osteopontin is a downstream effector of the PI3-kinase pathway in melanomas that is inversely correlated with functional PTEN. Carcinogenesis. 2006;27:1778–1786. doi: 10.1093/carcin/bgl016. [DOI] [PubMed] [Google Scholar]
- 16.Shi X, Bai S, Li L, Cao X. Hoxa-9 represses transforming growth factor-β-induced osteopontin gene transcription. J Biol Chem. 2001;276:850–855. doi: 10.1074/jbc.M005955200. [DOI] [PubMed] [Google Scholar]
- 17.Paik JH, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5:540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 19.Yu YP, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 20.Dhanasekaran SM, et al. Delineation of prognostic biomarkersin prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 21.Bardeesy N, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marino S, Vooijs M, van der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, et al. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech Dev. 2001;101:61–69. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 25.Berger R, et al. Androgen-induced differentiation and tumorigenicity of human prostate epithelial cells. Cancer Res. 2004;64:8867–8875. doi: 10.1158/0008-5472.CAN-04-2938. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, et al. A human prostatic epithelial model of hormonal carcinogenesis. Cancer Res. 2001;61:6064–6072. [PubMed] [Google Scholar]
- 27.Singh D, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 28.Haibe-Kains B, Desmedt C, Sotiriou C, Bontempi G. A comparative study of survival models for breast cancer prognostication based on microarray data: does a single gene beat them all? Bioinformatics. 2008;24:2200–2208. doi: 10.1093/bioinformatics/btn374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steering Committee of the Physicians' Health Study Research Group. Final report on the aspirin component of the ongoing Physicians' Health Study. N Engl J Med. 1989;321:129–135. doi: 10.1056/NEJM198907203210301. [DOI] [PubMed] [Google Scholar]
- 30.Christen WG, Gaziano JM, Hennekens CH. Design of Physicians' Health Study II—a randomized trial of beta-carotene, vitamins E and C, and multivitamins, in prevention of cancer, cardiovascular disease, and eye disease, and review of results of completed trials. Ann Epidemiol. 2000;10:125–134. doi: 10.1016/s1047-2797(99)00042-3. [DOI] [PubMed] [Google Scholar]
- 31.Sboner A, et al. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC Med Genomics. 2010;3:8. doi: 10.1186/1755-8794-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johansson JE, et al. Natural history of early, localized prostate cancer. J Am Med Assoc. 2004;291:2713–2719. doi: 10.1001/jama.291.22.2713. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.