Abstract
Background
Alcoholic and non-alcoholic steatohepatitis are leading causes of liver diseases worldwide. While of different etiology, these share common pathophysiological mechanisms and feature abnormal fat metabolism, inflammation and fibrosis. Micro-RNAs (miRNA) are highly conserved non-coding RNAs that control gene expression at the post-transcriptional level either via the degradation of target mRNAs or the inhibition of translation. Each miRNA controls the expression of multiple targets; miRNAs have been linked to regulation of lipid metabolism and inflammation.
Methods
We fed Lieber-DeCarli alcohol or methionine-choline-deficient (MCD) diets to C57Bl6 and analyzed livers for histopathology, cytokines by ELISA, ALT by biochemical assay, and microRNA profile by microarray.
Results
Both Lieber-DeCarli and MCD diets lead to development of liver steatosis, liver injury, indicated by increased ALT, and elevated levels of serum TNFα, suggesting that animal models portray the pathophysiological features of alcoholic and non-alcoholic fatty liver, respectively. We identified that Lieber-deCarli diet up-regulated 1% and down-regulated 1% of known miRNA; MCD diet up-regulated 3% and down-regulated 1% of known miRNA, compared to controls. Of miRNAs that changed expression levels, five miRNAs were common in alcoholic and non-alcoholic fatty livers: the expression of both miR-705 and miR-1224 was increased after Lieber-DeCarli or MCD diet feeding. In contrast, miR-182, miR-183, and miR-199a-3p were downregulated in Lieber-deCarli feeding, while MCD diet lead to their up-regulation, compared to corresponding controls.
Conclusions
Our findings indicate etiology-specific changes in miRNA expression profile during steatohepatitis models, which opens new avenues for research in the pathophysiology of alcoholic and non-alcoholic fatty liver diesase.
Keywords: liver, alcohol, NASH, in vivo
INTRODUCTION
Steatohepatitis (SH) is an emerging liver disease worldwide that often precedes liver fibrosis, failure and hepatocellular cancer (Bugianesi et al., 2002; Day C.P., 2005; Morgan TR et al., 2004). While the steatohepatitis may occur as a result of chronic alcohol abuse (alcoholic fatty liver and steatohepatitis) or accompany the metabolic syndrome without a history of alcohol abuse (non-alcoholic fatty liver and steatohepatitis), these entities have common histopathology features and share components of common mechanisms of liver injury (Tsukamoto H et al., 2008). Both alcoholic and non-alcoholic liver diseases feature steatosis, inflammation and progression to liver fibrosis (Morita et al., 2003; Brunt 2001). Particularities at the levels of transcription, translation and function of proteins were reported in both types of fatty liver diseases, however an unambiguous difference in protein regulation that could set aside alcoholic and non-alcoholic etiologies is yet to be identified.
The groundbreaking discovery of the first microRNA (miRNA) in 1993 followed by intensive research efforts identified over a thousand distinct miRNAs in mammalian cells and indicated that miRNAs negatively regulate protein expression at the point of protein translation; hence, they are fundamental regulators of cellular processes (Ambros V., 2004). More important, the miRNA profile is emerging as an important contributor to many normal homeostatic processes, including metabolic pathways (Wilfred BR et al., 2007; Esau C et al., 2006), cellular stress (S.N. Bhattacharyya et al., 2006), immune defense, inflammation (Chong MM et al., 2008, Tili E et al., 2007, Liston A et al., 2008), and tissue remodeling (Davis BN et al., 2008). More important, miRNA play a critical role in human diseases, including autoimmunity, pathologic inflammation (Schickel R et al., 2008; Mattes J et al., 2008), and tumor control (Wong QW et al., 2008; Varnholt H et al., 2008; Tryndyak VP et al., 2008; Connolly E et al., 2008). More recently Pietrzykowski et al discovered that posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol (Pietrzykowski AZ. et al., 2008), and Tang et al identified an effect of alcohol on miR-212 expression in intestinal epithelial cells (Tang Y. et al., 2008); these reports indicated that miRNA could play a role in alcohol-induced pathology. The microRNA expression pattern and their role in liver diseases are largely unknown.
Based on the pathogenesis of steatohepatitis, we analyzed the hypothesis that alcoholic and non-alcoholic fatty livers may exhibit signature miRNA profiles. Our data indicate that animals fed Lieber-deCarli alcohol diet or methionine-choline deficient (MCD) diet share common alterations in miRNA profile, however they also have distinct microRNA signatures.
MATERIALS AND METHODS
Animals and Experimental Protocols
Three-month old, female C57BL/6 mice were housed in a specific pathogen-free, climate-controlled animal facility under a 12-hour light-dark cycle, and were cared for in accordance with the IACUC regulations at the University of Massachusetts Medical School. All animals had unrestricted access to water during the entire duration of the experiment. The starting weight in all mice was 22.6±2.3g.
To induce alcoholic liver disease, mice received Lieber-DeCarli diet (Bio-Serv, Frenchtown, NJ) with 4.5% (volume/volume) ethanol (32.4% ethanol-derived calories) for 5 weeks; pair-fed control mice were fed an equal amount of calories as their alcohol-fed counterparts with the alcohol-derived calories substituted with dextran-maltose. During the 5 weeks feeding the mice gained comparable weigh in both alcohol-fed (+15±3.38g) and pair-fed (+15.6±3.54g) groups.
To induce non-alcoholic steatohepatitis, mice were fed with a methionine-choline-deficient (MCD) diet or a methionine-choline-supplemented (MCS) diet for 5 weeks; the latter control diet was identical in composition to the MCD diet but was supplemented with L-methionine (1.7 g/kg) and choline bitartrate (14.48 g/kg) (Dyets Inc., Bethlehem, PA). Methionine and choline deprivation blocks hepatic lipid secretion by disruption of phosphatidylcholine transfer and thus creates steatotic livers (Vance et al., 1985; Yao et al., 1988). During the MCD diet feeding the mice lost weight (−6.7±0.7g), in contrast to MCS diet-fed (+7.2±2.1g) control group.
Biochemical assays
Serum alanine aminotransferase (ALT) was determined using a kinetic method (ALT Liquid, Advanced Diagnostics Inc., South Plainfield, NJ). Liver triglyceride levels were measured as previously described, using commercial kits (Triglyceride TG H R2 994-40491, Triglyceride TG H R1 994-40391, and Lipid Calibrator 996-41791, Wako Chemicals USA Inc., Richmond, VA).
Histopathological Analysis
Sections of formalin-fixed, paraffin-embedded livers were stained with hematoxylin and eosin to assess for histologic features of steatohepatitis. The histopathology analysis was performed in a blinded fashion.
Cytokine Measurements
Serum TNFα level was determined using the Pierce SearchLight Multiplex Cytokine array (Pierce Biotechnology, Inc., Woburn, MA).
microRNA analysis
Total RNA was isolated from snap-frozen livers with organic and solid-phase extraction using mirVana miRNA Isolation Kit, which allows for miRNAs preservation (Applied Biosystems/Ambion, Austin, TX, catalog #1560), and quantified by spectrophotometry (BioPhotometer, Eppendorf, Westbury, NY). Total liver RNA (5 μg) samples were analyzed for miRNA profile at LC Sciences LLC (Houston, TX) using miRNA Microarray Service. The miRNA probe layout in the selected region is shown in Supplemental Table 1. All probe sequences in the array (MRA#1002) were based on Sanger miRBase Release 8.1, which represents the updated validated mouse miRNA sequences. The samples were paired randomly and RNA was labeled with Cy3 dye (in green color) or Cy5 dye (in red color); within the same experiments the colors were rotated to provide a color bias-free analysis. The labeled RNA was hybridized with the probe-containing chip, which detected miRNA transcripts listed in Sanger miRBase Release 10.1. The Cy3 and Cy5 provided information on the level of expression of distinct miRNAs in individual samples, and the ratio sensed the differential expression of miRNAs between the corresponding samples. The plots of representative Cy3, Cy5 and Cy3/Cy5 ratio for Lieber-deCarli and MCD diet fed groups are shown in Supplemental Figure 1. The background was calculated from the median of 5% and 25% of low intensity wells; the blank wells were excluded from the background calculation. The probes where the systematic dye bias was identified were excluded from the calculations. The signals are presented after background subtraction, normalization and detection evaluation.
Statistical Analysis
There were 3–6 mice in each experimental group. Statistical significance was determined using Student t-test for microRNA Microarray analysis and non-parametric Kruskal-Wallis and Mann-Whitney tests for all other parameters; data were considered statistically significant at a p<0.05.
RESULTS and DISCUSSIONS
Experimental models of alcoholic and non-alcoholic fatty liver share features of liver steatosis, impaired lipid metabolism, pro-inflammatory activation and liver damage
Steatohepatitis of alcoholic and non-alcoholic etiology share multiple features, such as steatosis, inflammation and sometimes fibrosis, to result in liver damage and progression to liver failure (Day et al., 2005; Tsukamoto et al, 2008). Our goal in this study was to identify if alcoholic and non-alcoholic fatty livers share the same similarity at the level of miRNA expression, by using the Lieber-DeCarli (alcoholic) and the MCD (non-alcoholic) diet-induced fatty liver models.
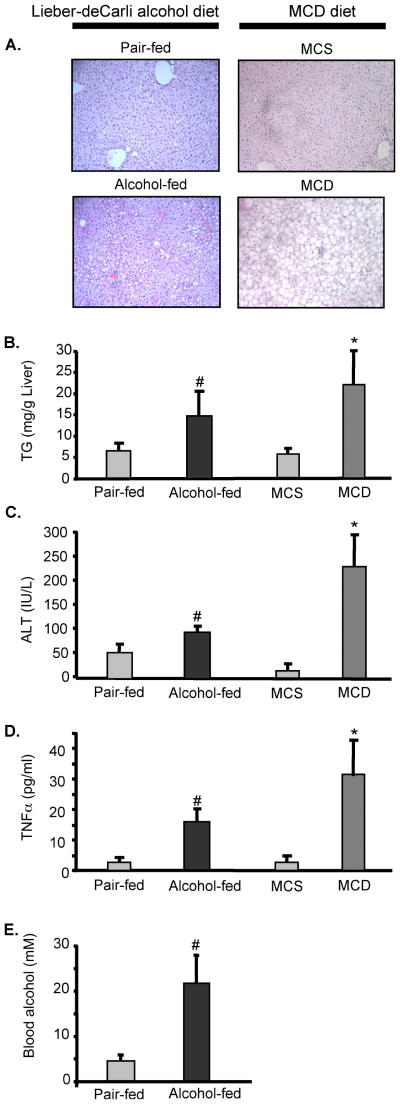
Feeding of alcohol-containing Lieber-deCarli diet (Lieber et al., 1965) to C57Bl6 mice resulted in liver steatosis (Fig 1A) and increased liver triglycerides (Fig 1B), compared to pair-fed animals. Alcohol-enriched diet caused liver damage, as suggested by elevated levels of serum ALT (Fig 1C) and created a pro-inflammatory state, indicated by increased levels of serum TNFα (Fig 1D), compared to alcohol-free counterparts. These characteristics closely resembled the features of alcoholic liver disease in humans (McCullough et al., 1998); we thus concluded that Lieber-deCarli diet induced features of alcoholic liver disease in mice. We next compared the blood alcohol levels (BAL) in Lieber-deCarli alcohol diet fed animals. As shown in Fig 1E, no alcohol was detected in blood of dextrane-maltose-enriched Lieber-deCarli pair-fed animals. In contrast, alcohol was detected in blood of animals fed alcohol-containing Lieber-deCarli diet.
Figure 1.
Lieber-deCarli and methionine-choline deficient (MCD) diet feeding feature liver steatosis, impaired fat metabolism, liver injury and pro-inflammatory activation. C57Bl6 mice were fed with Lieber-deCarli or mehionine-choline deficient diets to establish ASH and NASH models, respectively. Liver sections were stained with H&E and analyzed for morphology (A); serum levels of triglycerides (B), alanine aminotransferase (C), TNFα (D) and alcohol (E) were analyzed. In panel A, representative images out of n=6/group are shown. In panels B–E, data are shown as mean±SD from 6 mice/group; the * indicate p<0.05 compared to pair-fed controls; the # indicate p<0.05 compared to MCS controls.
For a model of non-alcoholic stateohepatitis we fed a methionine-choline deficient diet to C57Bl6 mice; this diet is well established and frequently employed in hepatology research to create steatotic livers (Yamaguchi et al, 2007, 2008; Rahman et al., 2008). The MCD feeding resulted in liver steatosis (Fig 1A) and increased liver triglycerides (Fig 1B), compared to methionine-choline supplemented (MCS) diet animals. Further, MCD diet caused elevation in serum levels of ALT (Fig 1C) and TNFα (Fig 1D), indicative of liver damage and inflammation, respectively. These data suggested that both alcohol-containing Lieber-deCarli and MCD diets cause steatohepatitis.
Alcoholic and non-alcoholic fatty livers show different microRNA expression profiles
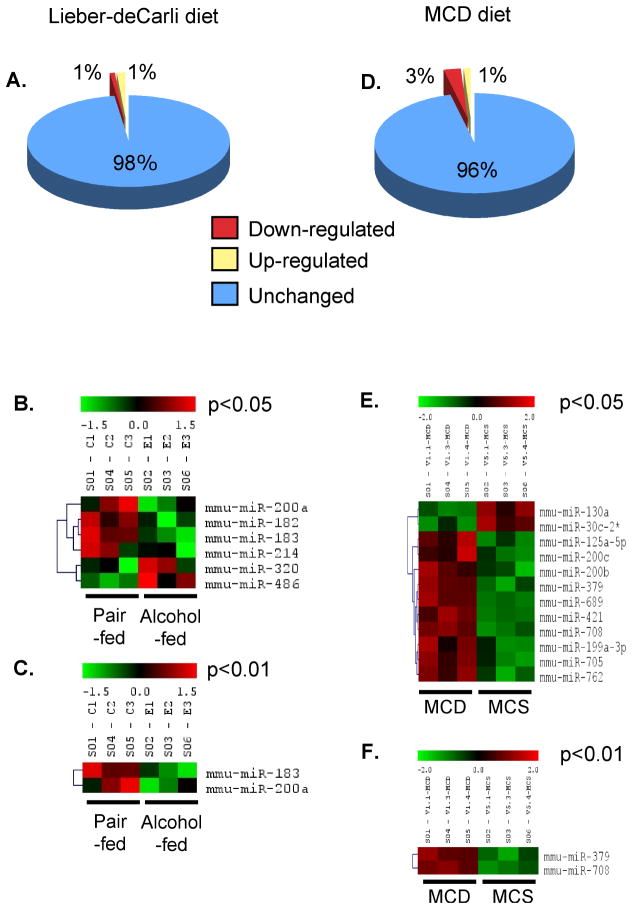
We next quantified the expression of known and predicted microRNAs in the liver of animals fed Lieber-deCarli alcohol diet or MCD diet, and their respective controls. As shown in Fig 2, clustering of microRNA probes revealed that multiple microRNA were expressed in the liver of control, healthy animals. The majority of liver microRNA were expressed at similar levels in animals fed Lieber-DeCarli diet (Fig 2A) or methionine-choline deficient diet (Fig 2E), compared to corresponding diet controls (Supplementary Figure 1). We identified that 2% of the known microRNA showed changes their expression levels upon Lieber-deCarli feeding compared to pair-fed controls (Fig 2B). Likewise, 4% of microRNA were modulated in animals fed MCD diet compared to MCS-diet-fed controls (Fig 2F). Among genes that changed their expression levels as a result of steatohepatitis, 1% of known miRNA were up-regulated and 1% were down-regulated during Lieber-deCarli alcohol diet feeding (Fig 2B), while MCD feeding caused up-regulation of 3% and down-regulation of 1% of known microRNA (Fig 2F). Figure 2C,D shows the patterns of microRNA expression in ASH model which was modulated significantly (featuring p<0.05 (Fig 2C) or p<0.01 (Fig 2D)); similar information from the MCD diet model is shown in Fig 2G,H. We further observed that three microRNAs were differentially expressed depending on the etiology of steatohepatitis: feeding of alcohol-containing diet caused down-regulation of miR-182, miR-183, and miR-199a-3p, while feeding of MCD diet resulted in up-regulation of these three microRNAs (Table 3). In contrast, miR-705 and miR-1224 expression was increased in steatohepatitis of both alcoholic and non-alcoholic etiology (Table 3). These data suggested that steatohepatitis of alcoholic and non-alcoholic etiology modulates expression of microRNA in the liver. Further, similarities and distinct signature of microRNA expression were identified in alcoholic and non-alcoholic fatty liver models.
Figure 2.
Steatohepatitis modulates liver microRNA expression.
Liver microRNA expression was analyzed using miRNA Microarray. Panels A and D show the percentage of microRNA that changed during feeding of Lieber-deCarli (A) and MCD (D) diets, compared to corresponding controls. Panels B and E show the heat-maps of microRNAs expression in Lieber-deCarli diet feeding at p<0.05 and p<0.01, respectively. Panels C and F show the expression of microRNAs in MCD diet feeding at p<0.05 and p<0.01, respectively. In panels B,C,E and F (shown n=3), the black color indicates no change, the red color represents up-regulation, and the green color indicates down-regulation of the gene expression; the intensity of the color correlates with the extent of changes.
Steatohepatitis, either of alcoholic or non-alcoholic etiology, is a major clinical challenge; identification of fundamentals and fine details of the pathogenesis of steatohepatitis is emergent and will likely influence the disease management. To date there is no clear understanding of the sequence of events in development of steatohepatitis and pathogenesis-based approaches to diagnosis, management and disease prediction are emergent. Here we investigated the microRNA profile of steatohepatitis using animal models of ASH and NASH and discovered common features and unique microRNA signatures of these diseases.
We identified that SH changes was associated with changes of a limited number of known microRNA. Based on our results, we predict that the combined changes in the levels of expression of miR-182, miR-183, miR-199a-3p, miR-705 and miR-1224 provide a signature of SH. More importantly, we identified specificity of each microRNA profile with regard to SH pathogenesis. We found that alcohol (Lieber-deCarli diet)-induced steatohepatitis leads to down-regulation of several microRNA, including miR-27b, miR-214, miR-199a-3p, miR-182, miR-183, miR-200a, and miR-322. microRNAs are thought to control expression of more then one, and sometimes thousands of target mRNAs (Ambros 2004). Mammalian microRNAs generally negatively regulate gene expression by repressing translation, possibly through effects on mRNA stability and/or the translation process itself (Wong et al., 2008; Varnholt et al., 2008; Tryndyak et al., 2008). Since the microRNAs inhibit protein expression, we predict that impaired expression of some microRNAs would allow over-expression of target proteins. For example, miR-27 levels are rapidly altered by histone deacetylase inhibition (Scott et al., 2006) and impaired miR-27 expression in ASH could result from emerging role of epigenetic events in the actions of alcohol (Choudhury M et al., 2008; Shukla et al, 2008). miR-214 is a marker of aging in the liver (Maes et al., 2008). TGFβ inhibits miR-200a expression (Gregory et al., 2008), and relevant to this, an increased TGFβ signaling was reported in ASH (Tsukamoto et al 1990). Further, impaired expression of miR-200 family, including miR-200a, results in upregulation of ZEB1 and ZEB2 proteins (Gregory et al., 2008). Consistent with down-regulation of miR-200a, we identified increase in ZEB1 expression in Lieber-deCarli alcohol diet-fed animals; there was no change in miR-200a in MCD diet-fed mice and their ZEB1 expression was comparable to pair-fed controls (data not shown). miR-200a also regulates Hedgehog signaling (Katoh et al, 2008), which has a role in alcoholic liver disease severity in mice and humans (Jung et al., 2008). miR-322 tightly regulates the function of epidermal growth factor receptor (Ramasamy et al., 2007), which affects hepatocyte proliferation (Zhang et al., 1999) and is key for liver protection against alcohol-induced injury and sensitization to bacterial lipopolysaccharide in alcoholic liver diseae models (Deaciuc et al., 2002). It is intriguing that miR-182, miR-183, and miR-199a-3p were up-regulated in NASH model, in contrast to their down-regulation in ASH. miR-182 is reportedly up-regulated in polycythemia vera granulocytes (Bruchova et al., 2008) and miR-183 is overexpressed in colorectal cancer (Bandrés et al., 2006), suggesting that their yet unknown target genes are most likely tumor suppressors.
We observed that Lieber-deCarli alcohol diet feeding induced up-regulation of five microRNAs, while feeding of MCD diet in the non-alcoholic fatty liver disease model lead to overexpression of seventeen microRNAs. Of all microRNAs upregulated during Lieber-deCarli diet feeding, two of them, including miR-705 and miR-1224, were also up-regulated upon feeding of MCD diet. To date there is no information about the function and/or targets of miR-705 and miR-1224; as with many other microRNA included in Sanger Institute microRNA database (website), these two microRNA have a defined sequence but await confirmation of their in vivo functions and physiological role. It is possible that differences in microRNA expression between alcoholic and non-alcoholic fatty livers are due to different models of inducing injury, yet they may also be related to the greater extent of injury in the MCD diet compared to alcohol-enriched diet. It is also conceivable that weight loss, observed only in MCD-fed but not in alcohol-fed animals, may contribute to the differences in microRNA between the two models of liver injury. In these conditions, the specific mechanisms of the effect of alcohol on liver microRNA profile remain to be determined.
To date there are clear indications that miRNA play a role in every step of pathological processes known to lead to development of steatohepatitis, including lipid metabolism (Wilfred et al., 2007; Esau et al., 2006), liver cellular stress response (Bhattacharyya et al., 2006), and control of inflammation via regulation of both innate and adaptive immunity (Chong et al., 2008; Tili et al., 2007; Liston et al., 2008). Further, TGFβ, which is a potent fibrotic factor (Seki et al, 2008), governs a specific set of miRNAs at the pri-miRNA to pre-miRNA step to regulate cell plasticity and possibly oncogenesis (Davis et al., 2008). A number of miRNA reportedly have role in establishment of liver tumorigenesis (Wong QW et al., 2008; Varnholt H et al., 2008; Tryndyak VP et al., 2008; Connolly E et al., 2008); since many liver tumors are preceded by advanced stages of steatohepatitis (Bugianesi et al., 2002; Morgan et al., 2004), the miRNA profile may have predictive value at pre-cancerous stages of liver diseases. Recent reports indicated that alcohol-induced pathology in brain and intestine could be controlled at the levels of miRNAs (Pietrzykowski et al., 2008; Tang et al 2008); our data provide novel insights into the liver-specific microRNA profile during liver damage due to alcohol intake (Lieber-deCarli diet model) or MCD deficiency-induced fatty liver. More excitingly, miRNAs are emerging as therapeutic targets in cancer, diseases of hart and virally induced diseases (Barbarotto et al., 2008; van Rooij et al., 2008; Sall et al., 2008). Recently reported employment of microRNA antagomers for delivery to liver set-up high expectations for miRNA-based therapy strategies of liver diseases (Krutzfeldt et al., 2005); our new data about the baseline status of the microRNA profile in normal liver and upon development of steatohepatitis facilitate better understanding of this future therapeutic tool.
In conclusion, our results provide novel information about the microRNA signatures of statohepatitis of alcoholic and non-alcoholic origin, and suggest new avenues for research in hepatology.
Supplementary Material
Liver microRNA (5μg/sample) was labeled with Cy3 (in green color) or Cy5 dyes (in red color) and hybridized with the probe-containing chip (sequences according to Sanger miRBase Release 10.1). The representative plots of Cy3, Cy5 and Cy3/Cy5 ratio for Lieber-deCarli and MCD diet feeding groups are shown. The Cy3 and Cy5 provided information on the level of expression of distinct miRNAs in individual samples, and the ratio sensed the differential expression of miRNAs between the corresponding samples.
Table 1.
Liver microRNA changes upon Lieber-deCarli and MCD diet feeding
| MicroRNA | Lieber-deCarli diet | MCD diet |
|---|---|---|
| 182 | Down-regulated | Up-regulated |
| 183 | Down-regulated | Up-regulated |
| 199a-3p | Down-regulated | Up-regulated |
| 705 | Up-regulated | Up-regulated |
| 1224 | Up-regulated | Up-regulated |
Acknowledgments
This work was supported with funds from NIH (1R01DK075635 and 5R01AA008577)
Abbreviations
- ALT
alanine aminotransferase
- MCD
methionine-choline deficient
- MCS
methionine-choline supplemented
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bandrés E, Cubedo E, Agirre X, Malumbres R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M, García-Foncillas J. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29–39. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbarotto E, Calin GA. Potential therapeutic applications of miRNA-based technology in hematological malignancies. Curr Pharm Des. 2008;14(21):2040–2050. doi: 10.2174/138161208785294627. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Bruchova H, Merkerova M, Prchal JT. Aberrant expression of microRNA in polycythemia vera. Haematologica. 2008;93(7):1009–1016. doi: 10.3324/haematol.12706. [DOI] [PubMed] [Google Scholar]
- Brunt EM. Nonalcoholic steatohepatitis: definition and pathology. Semin Liver Dis. 2001;21(1):3–16. doi: 10.1055/s-2001-12925. [DOI] [PubMed] [Google Scholar]
- Bugianesi E, Leone N, Vanni E, Marchesini G, Brunello F, Carucci P, Musso A, De Paolis P, Capussotti L, Salizzoni M, Rizzetto M. Expanding the natural history of nonalcoholic steatohepatitis: From cryptogenic cirrhosis to hepatocellular carcinoma. Gastroenterology. 2002;123:134–140. doi: 10.1053/gast.2002.34168. [DOI] [PubMed] [Google Scholar]
- Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205(9):2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury M, Shukla SD. Surrogate alcohols and their metabolites modify histone H3 acetylation: involvement of histone acetyl transferase and histone deacetylase. Alcohol Clin Exp Res. 2008;32(5):829–839. doi: 10.1111/j.1530-0277.2008.00630.x. [DOI] [PubMed] [Google Scholar]
- Connolly E, Melegari M, Landgraf P, Tchaikovskaya T, Tennant BC, Slagle BL, Rogler LE, Zavolan M, Tuschl T, Rogler CE. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. 2008;173(3):856–864. doi: 10.2353/ajpath.2008.080096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CP. Natural History of NAFLD: Remarkably Benign in the Absence of Cirrhosis. Gastroenterology. 2005;129:375–378. doi: 10.1053/j.gastro.2005.05.041. [DOI] [PubMed] [Google Scholar]
- Deaciuc IV, D’Souza NB, Burikhanov R, Lee EY, Tarba CN, McClain CJ, de Villiers WJ. Epidermal growth factor protects the liver against alcohol-induced injury and sensitization to bacterial lipopolysaccharide. Alcohol Clin Exp Res. 2002;26(6):864–874. [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Jung Y, Brown KD, Witek RP, Omenetti A, Yang L, Vandongen M, Milton RJ, Hines IN, Rippe RA, Spahr L, Rubbia-Brandt L, Diehl AM. Accumulation of hedgehog-responsive progenitors parallels alcoholic liver disease severity in mice and humans. Gastroenterology. 2008;134(5):1532–1543. doi: 10.1053/j.gastro.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Katoh M. Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA. Int J Mol Med. 2008;22(3):271–275. [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lieber CS, Jones DP, deCarli LM. Effects of prolonged ethanol intake: production of fatty liver despite adequate diets. J Clin Invest. 1965;44:1009–1021. doi: 10.1172/JCI105200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A, Lu LF, O’Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205(9):1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes OC, An J, Sarojini H, Wang E. Murine microRNAs implicated in liver functions and aging process. Mech Ageing Dev. 2008;129(9):534–541. doi: 10.1016/j.mad.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Mattes J, Collison A, Foster PS. Emerging role of microRNAs in disease pathogenesis and strategies for therapeutic modulation. Curr Opin Mol Ther. 2008;10(2):150–157. [PubMed] [Google Scholar]
- McCullough AJ, O’Connor JF. Alcoholic liver disease: proposed recommendations for the American College of Gastroenterology. Am J Gastroenterol. 1998;93:2022–2036. doi: 10.1111/j.1572-0241.1998.00587.x. [DOI] [PubMed] [Google Scholar]
- Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology. 2004;127:S87–S96. doi: 10.1053/j.gastro.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Morita Y, Ueno T, Sasaki N, Kuhara K, Yoshioka S, Tateishi Y, Nagata E, Kage M, Sata M. Comparison of liver histology between patients with non-alcoholic steatohepatitis and patients with alcoholic steatohepatitis in Japan. Alcohol Clin Exp Res. 2005 Dec;29(12 Suppl):277S–81S. doi: 10.1097/01.alc.0000191777.36629.33. [DOI] [PubMed] [Google Scholar]
- Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, Siegelmann HT, Treistman SN. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59(2):274–287. doi: 10.1016/j.neuron.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman SM, Schroeder-Gloeckler JM, Janssen RC, Jiang H, Qadri I, Maclean KN, Friedman JE. CCAAT/enhancing binding protein beta deletion in mice attenuates inflammation, endoplasmic reticulum stress, and lipid accumulation in diet-induced nonalcoholic steatohepatitis. Hepatology. 2007;45(5):1108–1117. doi: 10.1002/hep.21614. [DOI] [PubMed] [Google Scholar]
- Ramasamy S, Duraisamy S, Barbashov S, Kawano T, Kharbanda S, Kufe D. The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol Cell. 2007;27(6):992–1004. doi: 10.1016/j.molcel.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sall A, Liu Z, Zhang HM, Yuan J, Lim T, Su Y, Yang D. MicroRNAs-based therapeutic strategy for virally induced diseases. Curr Drug Discov Technol. 2008;5(1):49–58. doi: 10.2174/157016308783769478. [DOI] [PubMed] [Google Scholar]
- Sanger Institute microRNA database, found on-line. at http://microrna.sanger.ac.uk.
- Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27(45):5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66(3):1277–81. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]; Semin Liver Dis. 2001;21(1):3–16. doi: 10.1055/s-2001-12925. [DOI] [PubMed] [Google Scholar]
- Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging role of epigenetics in the actions of alcohol. Alcohol Clin Exp Res. 2008;32(9):1525–34. doi: 10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- Tang Y, Banan A, Forsyth CB, Fields JZ, Lau CK, Zhang LJ, Keshavarzian A. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res. 2008;32(2):355–364. doi: 10.1111/j.1530-0277.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179(8):5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol Carcinog. 2008 doi: 10.1002/mc.20484. [DOI] [PubMed] [Google Scholar]
- Tsukamoto H, Gaal K, French SW. Insights into the pathogenesis of alcoholic liver necrosis and fibrosis: status report. Hepatology. 1990;12(3 Pt 1):599–608. doi: 10.1002/hep.1840120325. [DOI] [PubMed] [Google Scholar]
- Tsukamoto H, She H, Hazra S, Cheng J, Wang J. Fat paradox of steatohepatitis. J Gastroenterol Hepatol. 2008;23 (Suppl 1):S104–107. doi: 10.1111/j.1440-1746.2007.05294.x. [DOI] [PubMed] [Google Scholar]
- Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79(4):581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- Vance JE, Vance DE. The role of phosphatidylcholine biosynthesis in the secretion of lipoproteins from hepatocytes. Can J Biochem Cell Biol. 1985;63:870–881. doi: 10.1139/o85-108. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103(9):919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47(4):1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of metabolic pathways. Mol Genet Metab. 2007;91(3):209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135(1):257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Yang L, McCall S, Huang J, Yu XX, Pandey SK, Bhanot S, Monia BP, Li YX, Diehl AM. Inhibiting triglyceride synthesis improves hepatic steatosis but exacerbates liver damage and fibrosis in obese mice with nonalcoholic steatohepatitis. Hepatology. 2007;45(6):1366–1374. doi: 10.1002/hep.21655. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Yang L, McCall S, Huang J, Yu XX, Pandey SK, Bhanot S, Monia BP, Li YX, Diehl AM. Diacylglycerol acyltranferase 1 anti-sense oligonucleotides reduce hepatic fibrosis in mice with nonalcoholic steatohepatitis. Hepatology. 2008;47(2):625–635. doi: 10.1002/hep.21988. [DOI] [PubMed] [Google Scholar]
- Yao ZM, Vance DE. The active synthesis of phosphatidylcholine is required for very low density lipoprotein secretion from rat hepatocytes. J Biol Chem. 1988;263:2998–3004. [PubMed] [Google Scholar]
- Zhang BH, Farrell GC. Chronic ethanol consumption disrupts complexation between EGF receptor and phospholipase C-gamma1: relevance to impaired hepatocyte proliferation. Biochem Biophys Res Commun. 1999;257(1):89–94. doi: 10.1006/bbrc.1999.0403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Liver microRNA (5μg/sample) was labeled with Cy3 (in green color) or Cy5 dyes (in red color) and hybridized with the probe-containing chip (sequences according to Sanger miRBase Release 10.1). The representative plots of Cy3, Cy5 and Cy3/Cy5 ratio for Lieber-deCarli and MCD diet feeding groups are shown. The Cy3 and Cy5 provided information on the level of expression of distinct miRNAs in individual samples, and the ratio sensed the differential expression of miRNAs between the corresponding samples.