Abstract
Aromatic-aromatic and aromatic-hydrophobic interactions have been proposed to play a role in amyloid formation by a range of polypeptides including islet amyloid polypeptide (IAPP, Amylin). IAPP is responsible for amyloid formation during type-2 diabetes. The polypeptide is 37 residues in length and contains three aromatic residues Phe-15, Phe-23, and Tyr-37. The ability of all single aromatic to leucine mutants, all double aromatic to leucine mutants and the triple leucine mutant to form amyloid were examined. Amyloid formation was almost twice as rapid for the F15L mutant relative to wild-type, but was almost three fold slower for the Y37L mutant and almost two fold slower for F23L mutant. Amyloid fibrils formed from each of the single mutants were effective at seeding amyloid formation by wild-type IAPP, implying that the fibril structures are similar. The F15LF23L double mutant has a larger effect than the F15LY37L double mutant on the rate of amyloid formation, even though a Y37L substitution has more drastic consequences in the wild-type background than does the F23L mutation, suggesting non-additive effects between the different sites. The triple leucine mutant and the F23LY37L double mutant are the slowest to form amyloid. F15 has been proposed to make important contacts early in the aggregation pathway, but the F15L mutant data indicates that they are not optimal. A set of variants containing natural and unnatural amino acids at position 15, which were designed to conserve hydrophobicity, but which alter α-helix and β-sheet propensity, were analyzed to determine the properties of this position that control the rate of amyloid formation. There is no correlation between β-sheet propensity at this position and the rate of amyloid formation, but there is a correlation with α-helical propensity.
Keywords: IAPP, Islet amyloid polypeptide, Amylin, Amyloid, Aromatic interactions, Pi-interactions
Amyloid formation plays a role in a range of human diseases and a much larger universe of proteins can be induced to form amyloid in vitro.1–4 Amyloid formation also plays a functional, beneficial role in certain cases and amyloid like structures have been proposed as biomaterials.5 The proteins and peptides which form amyloid can be divided into two broad classes; those which are largely unstructured, “intrinsically disordered,” in their monomeric state and those which form compact globular structures. Important examples of intrinsically disordered sequences which form amyloid include the Aβ peptide of Alzheimer’s disease and islet amyloid polypeptide (IAPP, Amylin), the protein responsible for islet amyloid in type-2 diabetes.6–8 IAPP is an endocrine partner to insulin, is synthesized in the pancreatic β-cells, stored in the insulin secretory granules and released in response to the same stimuli that promote insulin release. 9–11 Amyloid formation by IAPP is believed to contribute to the loss of β-cell mass in type-2 diabetes and to graft failure after islet transplantion.12–14
The details of amyloid formation are still not well understood, especially for those proteins which are intrinsically disordered in their monomeric states. Aromatic-aromatic interactions have been proposed to play an important role in amyloid formation, particularly for IAPP.15–17 IAPP contains three aromatic residues, F15, F23 and Y37 (Figure 1), and a triple aromatic to leucine mutant forms amyloid at a significantly reduced rate compared to wild-type,16 but the effect of single site substitutions or double mutations have not been examined. Phe to Tyr FRET studies suggest that aromatic residues may make contacts during the early stages of amyloid formation18. However, experiments with variants that contain the fluorescent Tyr analog 4-cyanophenylalanine argue that the aromatic side chains remain solvated during the lag phase, and suggest that aromatic-aromatic interactions involving Y37 do not develop during the lag phase.19 Thus, the role of the aromatic residues in amyloid formation by IAPP is not clear. In fact, the details of the early stages of IAPP amyloid formation are not well understood. This is a topic of current interest because intermediates along the pathway amyloid formation may be the most toxic entities, and could be targets for inhibitor design.7,20,21 The role of F15 is particularly interesting since this position is believed to make contacts important for initial oligomerization.22 Here we examine a comprehensive set of aromatic to leucine mutants as well as a set of F15 variants containing natural and unnatural amino acids which preserve hydrophobicity but alter secondary structure propensity to deduce the role aromatic residues play in amyloid formation by IAPP.
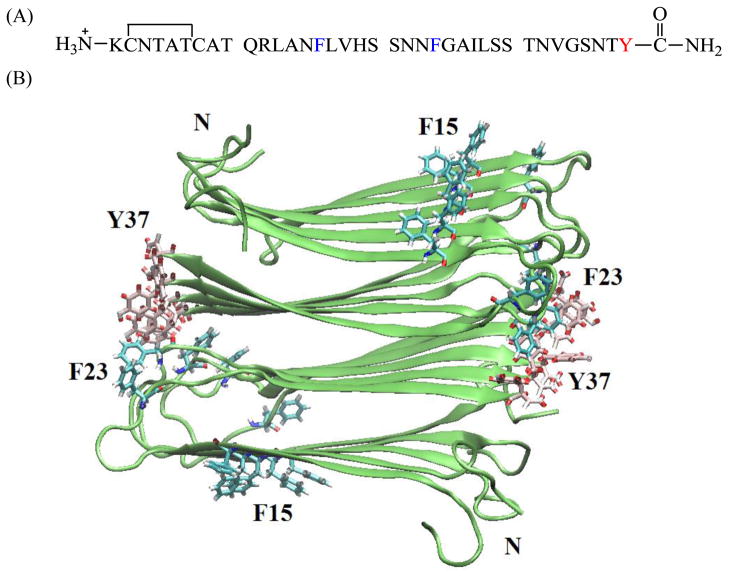
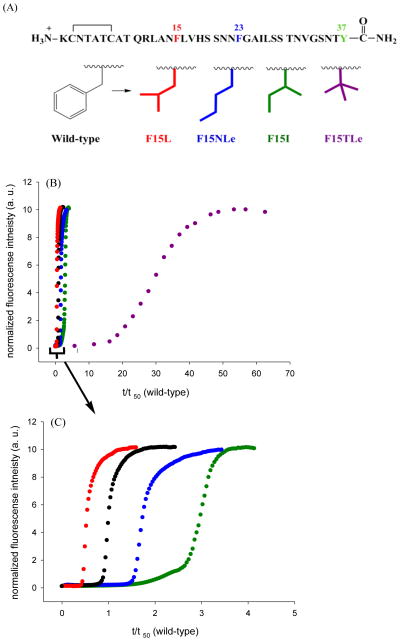
Figure 1.
(A) Primary sequence of human IAPP. IAPP contains a disulfide bridge between residues 2 and 7. The C-terminus is amidated and the N-terminus is free. The aromatic residues are colored in blue (F15 and F23) and red (Y37). (B) Diagrams of the solid-state NMR derived model of IAPP protofibrils.26 The aromatic residues and the N-terminus are labeled. F23 (blue) and Y37 (red) are colored differently as an aid to the eye.
MATERIALS AND METHODS
Peptide synthesis and purification
Peptides were synthesized on a 0.25 mmol scale using a CEM microwave peptide synthesizer, and 9-fluornylmethoxycarbonyl (Fmoc) chemistry. Fmoc protected pseudoproline dipeptide derivatives were incorporated at positions 9–10, 19–20, and 27–28 to facilitate the synthesis.23 Solvents used were ACS-grade. All other reagents were purchased from Advanced Chemtech, PE Biosystems, Sigma and Fisher Scientific. 5-(4′-fmoc-aminomethyl-3′, 5-dimethoxyphenol) valeric acid (PAL-PEG) resin was used to form an amidated C terminus. Standard Fmoc reaction cycles were used. The first residue attached to the resin, β-branched residues, Arg, and all pseudoproline dipeptide derivatives were double coupled. The peptide was cleaved from the resin through the use of standard trifluoroacetic acid (TFA) methods. Crude peptides were partially dissolved in 20 % acetic acid (v/v), frozen in liquid nitrogen, and lyophilized to increase their solubility. The dry peptide was dissolved in dimethyl sulfoxide (DMSO) at room temperature to promote the formation of the disulfide bond.24,25 The peptides were purified by reverse-phase HPLC using a Vydac C18 preparative column (10 mm × 250 mm). A two buffer gradient was used: buffer A consists of 100 % H2O and 0.045 % HCl (v/v) and buffer B includes 80 % acetonitrile, 20 % H2O and 0.045 % HCl. HCl was used as the counterion instead of TFA to reduce the residual amount of TFA since TFA can influence amyloid formation. Analytical HPLC were used to check the purity of peptides before each experiment. Peptides were analyzed by mass spectrometry using a Bruker MALDI-TOF MS to confirm the molecular weight. Wild-type IAPP, expected 3903.3, observed 3902.1; F15-IAPP, expected 3871.3, observed 3872.0; F23-IAPP, expected 3871.3, observed 3871.3; Y37-IAPP, expected 3855.4, observed 3853.8; F15LF23L-IAPP, expected 3835.3, observed 3834.0; F15LY37L-IAPP, expected 3821.3, observed 3819.8; F23LY37L-IAPP, expected 3821.3, observed 3820.0; 3XL-IAPP, expected 3787.2, observed 3785.1; F15I-IAPP, expected 3871.3, observed 3871.5; F15NLe-IAPP, expected 3871.3, observed 3872.1; F15TLe-IAPP, expected 3871.3, observed 3872.0
Sample preparation
A 1.6 mM peptide solution was prepared in 100 % hexafluoroisopropanol (HFIP) and stored at −20 °C. For kinetic experiments, 17 μL of the filtered stock solution was diluted into 20 mM Tris-HCl buffer to give a final concentration of 16 μM IAPP in 2% HFIP. This condition was chosen to facilitate comparison with previously published studies. For the seeding assays, preformed wild-type IAPP and single mutant fibrils were produced by diluting a 1.6 mM stock solution of peptide in HFIP into 20 mM Tris–HCl buffer containing thioflavinT at pH 7.4 for a final concentration of 16 μM IAPP in 2% HFIP. The solution was incubated and thioflavin T fluorescence was monitored to ensure fibril formation had occurred. Seeds were used within 12 hours to ensure reproducibility.
Thioflavin-T fluorescence assays
Fluorescence experiments were performed on an Applied Phototechnology fluorescence spectrophotometer using an excitation wavelength of 450 nm and emission wavelength of 485 nm. The excitation and emission slits were set at 6 nm, and a 1.0 cm cuvette was used. Solutions were prepared by diluting filtered stock solution using a 0.45 μm GHP Acrodisc 13 syringe filter into Tris-HCl buffer and Thioflavin-T solution immediately before the measurement. The final concentrations were 16 μM peptide and 25 μM Thioflavin-T in 2% HFIP, 20 mM Tris-HCl at pH 7.4 for all experiments. All solutions were stirred during the fluorescence experiments in order to maintain homogeneity. Stirring is known to accelerate IAPP amyloid formation. For the seeding assays, 1.6 mM peptide stock solutions in HFIP were diluted into the seeded buffer solution to give final conditions of 16 μM peptide, 25 μM thioflavin T, 1.6 μM seed (in monomer units), and 2 % HFIP, 20 mM Tris-HCl at pH 7.4. For kinetic experiments without HFIP, solutions were prepared by adding 20 mM, pH 7.4 Tris–HCl buffer containing thioflavin-T to lyophilized dry peptides for a final peptide concentration of 16 uM. These studies were conducted using a Beckman Coulter DTX880 plate reader without stirring.
Circular Dichroism (CD) experiments
CD experiments were performed at 25 °C using an Applied Photophysics Chirascan circular dichroism spectrometer. The solutions for the CD experiments were prepared by diluting the filtered stock peptide solutions into 20 mM Tris-HCl buffer at pH 7.4. The final concentration of peptide was 16 μM in 2 % HFIP. Spectra were recorded from 190 to 260 nm at 1 nm intervals in a quartz cuvette with 0.1 cm path length at 25 °C. CD experiments used the same stock solutions as the thioflavin-T fluorescence measurements. A background spectrum was subtracted from the collected data.
Transmission electron microscope (TEM)
TEM was performed at the Life Science Microscopy Center at the State University of New York at Stony Brook. Samples were aliquots from the same solutions that were used for the fluorescence measurements. 5 μL of peptide solution was placed on carbon-coated Formvar 300 mesh copper grid for 1 min and then negatively stained with saturated uranyl acetate for 1 min.
RESULTS AND DISCUSSION
Analysis of amyloid formation by aromatic to Leu variants
The sequence of IAPP and the solid-state NMR derived model of IAPP amyloid protofibrils26 are shown in figure 1. The molecule contains a disulfide bond between Cys-2 and Cys-7 and has an amidated C-terminus. F15 and Y37 are conserved among all known IAPP sequences; F23 is not, but it is substituted only by leucine (Supporting information). A set of seven aromatic to leucine variants were prepared: all three single mutants, the complete set of double mutants, and the triple mutant. Leu was used because it is the closest natural non-aromatic substitution for Tyr and Phe in terms of size and hydrophobicity. An additional set of variants was prepared to examine the role of Phe-15 in detail.
The rate of amyloid formation was measured using fluorescence detected Thioflavin- T binding assays. Thioflavin is a small dye whose quantum yield is enhanced when it binds to amyloid fibrils.27 The molecule is believed to bind to groves on the surface of the amyloid fibrils. Thioflavin-T assays reliably report on IAPP amyloid formation kinetics and the compound does not alter the aggregation kinetics under the conditions used here. Amyloid formation by IAPP follows a sigmoidal time course in which no detectable amyloid fibrils are formed during the lag-phase. The lag phase is followed by a growth phase during which fibrils develop and elongate, eventually leading to a steady state where fibrils are in equilibrium with soluble protein. Thioflavin-T curves for the wild-type and single mutants are displayed in Figure 2A, and the data for the double and triple mutants is shown in Figure 2B. Amyloid formation by wild-type IAPP is rapid under the conditions of these experiments (2% HFIP, 20mM Tris-HCl, pH 7.4, constant stirring). IAPP amyloid formation has been widely studied by dilution from HFIP and we choose this protocol to facilitate comparison to earlier studies. Additional studies, described below, were performed without HFIP.
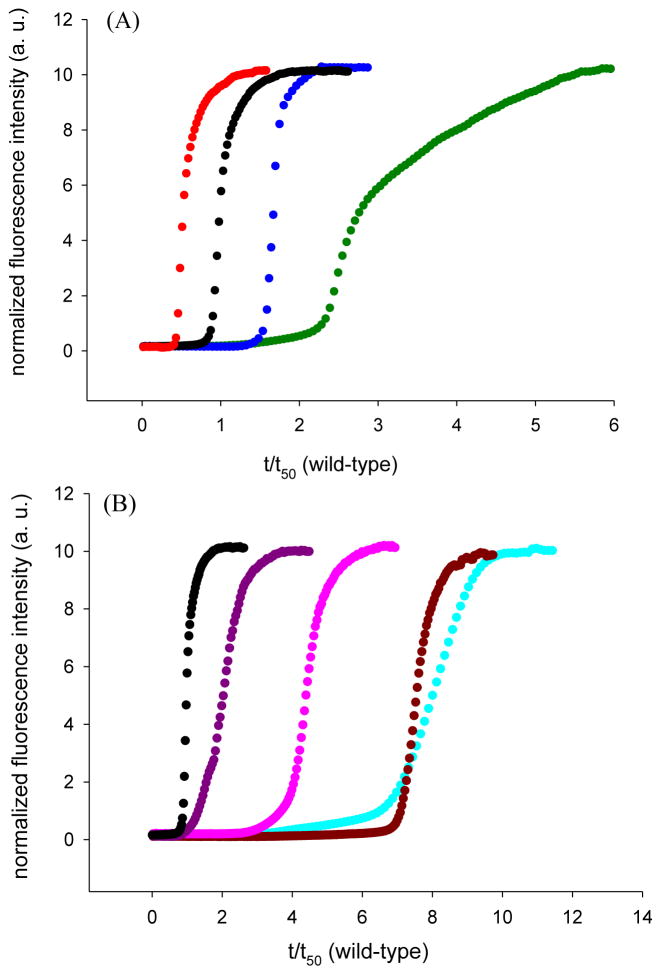
Figure 2.
Substitution of the aromatic residues in IAPP alters the kinetics of amyloid formation. Thioflavin-T fluorescence monitored kinetic experiments. (A) Single mutants compared to wild-type. Black, wild-type IAPP; red, F15L-IAPP; blue, F23L-IAPP; green, Y37L-IAPP (B) Multiple mutants compared to wild-type. Black, wild-type IAPP; pink, F15LF23L-IAPP; purple, F15LY37L-IAPP; brown, F23LY37L-IAPP; cyan, 3XL-IAPP. All experiments were conducted in 20 mM, pH 7.4 Tris buffer, 2% HFIP with constant stirring at 25 °C.
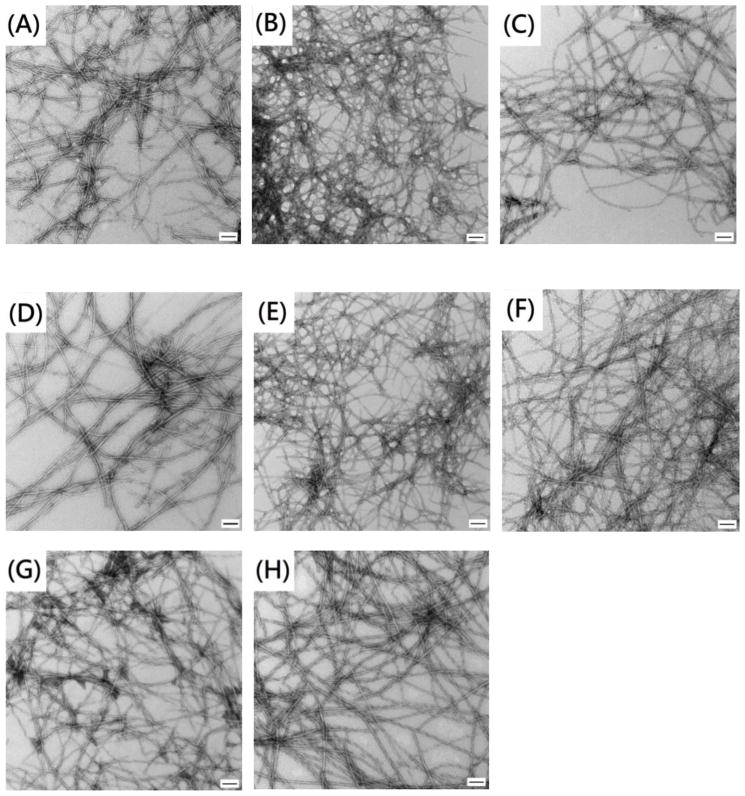
The kinetic analysis reveals a number of interesting features. All of the mutants, with the exception of F15L, show a decreased rate of amyloid formation and an increased lag time. In contrast, replacement of F15 accelerates amyloid formation, decreasing the lag time by almost a factor of two. F15 has been proposed to be involved in the early steps of amyloid formation, but the data shows that the contacts Phe 15 makes are not optimal. The role of F15 is considered in more detail below. The other single mutants form amyloid more slowly with the most pronounced effect observed for the Y37 mutant, which has a lag time that is a factor of three longer than wild-type IAPP. The lag time for F23L is almost two fold longer than wild-type. The F15LY37L double mutant forms amyloid more rapidly than the Y37L single mutant, but more slowly than wild-type, indicating that the acceleration caused by the F15 substitution partially compensates for the slowing caused by the Y37L replacement. The other mutants are much slower to form amyloid than wild-type IAPP. F15L23L-IAPP is four fold slower, while F23LY37L-IAPP and 3XL-IAPP are about 7.5 fold slower. The lag times and t50 values for all of the mutants are listed in table 1. T50 is the time required to reach half the final intensity in a thioflavin-T assay and is commonly used as a quantitative measure of amyloid formation. The differences observed for t50 values and the lag times between different peptides are much larger than the experimental uncertainty. For examples, analysis of a set of 25 independent measurements conducted on wild-type IAPP over the course of two years with more than five separate batches of peptide yield a standard deviation in t50 of less than 20% (Supporting information). The calculated standard deviations for independent measurements conducted on an individual batch of peptide are even smaller, less than 10% in all cases and often significantly smaller. Transmission electron microscopy (TEM) images of aliquots removed at the end of each kinetic run confirm the presence of extensive amyloid fibrils (Figure 3, supporting information). Circular Dichroism (CD) confirms the presence of β-sheet structure in all samples (Supporting information).
Table 1.
Comparison of kinetic parameters for wild-type IAPP and aromatic to leucine mutants. Experiments are performed at 25 °C, pH 7.4 20 mM Tris-HCl, 2% (v/v) HFIP with constant stirring.
| peptide | Lag timea (sec) | t50b (sec) | Lag time
|
t50
|
|---|---|---|---|---|
| wild-type Lag time | t50 of wild-type | |||
| Wild-type | 1310±280c | 1470±280 | ||
| F15L-IAPP | 490±50 | 640±40 | 0.4±0.1d | 0.4±0.1 |
| F23L-IAPP | 2230±220 | 2470±220 | 1.7±0.4 | 1.7±0.3 |
| Y37L-IAPP | 2790±90 | 4160±100 | 2.1±0.5 | 2.8±0.5 |
| F15LF23L-IAPP | 5100±60 | 5850±30 | 3.9±0.8 | 4.0±0.8 |
| F15LY37L-IAPP | 1790±140 | 2650±100 | 1.4±0.3 | 1.8±0.4 |
| F23LY37L-IAPP | 9670±150 | 10600±500 | 7.4±1.6 | 7.2±1.4 |
| 3XL | 9210±260 | 10950±250 | 7.0±1.5 | 7.4±1.4 |
The lag time is defined here as the time required to reach 10% of the final fluorescence change in the thioflavin-T assays.
t50 is the time required to reach 50% of the final fluorescence change in the thioflavin-T assays.
The quoted uncertainty is the standard deviation.
The uncertainty was determined by standard propagation of error.
The differences in the rates of amyloid formation are unlikely to be due to small variations in concentration from sample to sample, since the lag time for amyloid formation by IAPP has been shown to be largely independent of peptide concentration for the conditions (2% HFIP with constant stirring) and concentrations used here.28 Small changes in pH can affect the rate of amyloid formation by IAPP since the lag time depends on the protonated state of His-18 and the N-terminus,29 however all samples were at the same pH to within 0.1 pH unit. Control experiments with wild-type IAPP show that the lag time varies by only 10% between pH 7.2 and pH 7.6 in the buffer used here (Supporting information). This is a much wider variation in pH than observed in our experiments and the effect on the lag time in the control studies is much less than the differences in the lag times observed for the different mutants. Variations in the final HFIP concentration can also alter the rate of amyloid formation, but the HFIP concentration was fixed at 2% v/v.
We also plotted the kinetic data as normalized fluorescence intensity versus normalized time, t/t50 (wild-type) to facilitate a comparison of the shape of the kinetic progress curves of the mutants with wild-type (Supporting information). The normalized curves for wild-type, F15L-IAPP, F23L-IAPP, and F15LF23L-IAPP are very similar in shape. This is consistent with the mutants and wild-type IAPP having a similar mode of assembly, but does not prove that they do. The normalized curve for F15LY37L-IAPP shows some differences relative to wild-type in the early stage of fibril formation, while the normalized plot for Y37L-IAPP has a slight difference in the growth phase. The F23LY37L-IAPP and 3XL-IAPP mutants have the slowest rates of amyloid formation, and display the largest differences in normalized progress curves.
The single mutants form amyloid fibrils which are similar to wild-type fibrils
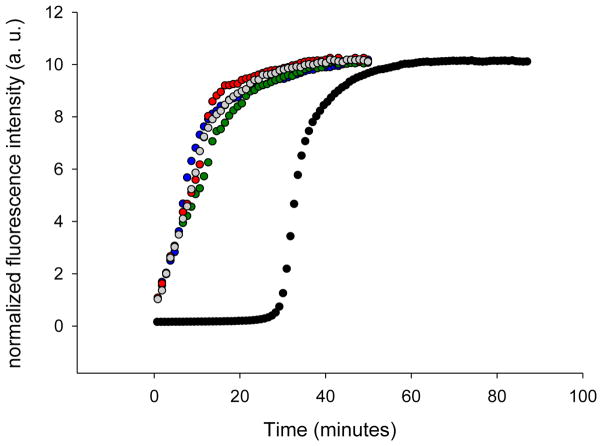
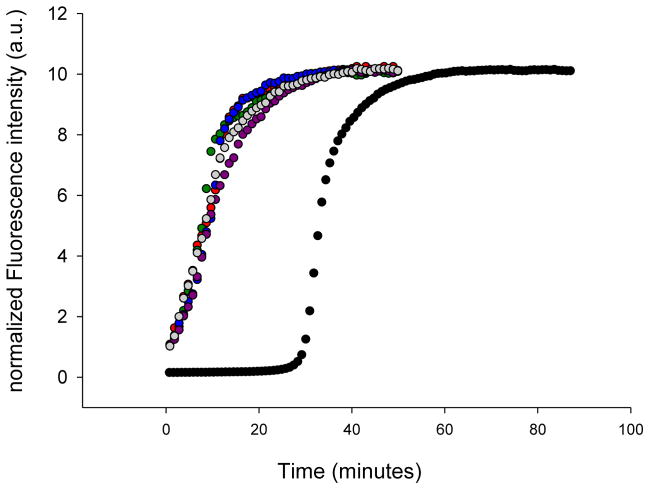
The TEM and CD studies outlined above suggest that the morphology of the fibrils formed by wild-type IAPP and the mutants are similar. We investigated the structural similarity between wild-type and mutants fibrils in more detail by conducting seeding experiments. Preformed amyloid fibrils can serve as a template for the addition of monomers, resulting in a bypassing of the lag phase. Seeding is usually much more effective when fibrils are used as seeds for their own monomers.30,31 We first examined seeding of wild-type IAPP by wild-type fibrils (Figure 4, grey curve). A bypass of the lag phase was observed as expected. We then tested the ability of preformed F15L-IAPP, F23L-IAPP, and Y37L-IAPP amyloid fibrils to seed wild-type IAPP (Figure 4, red, blue, and green curves). The curves perfectly overlap with the curve from the wild-type seeding study, showing that the mutant fibrils seed wild-type IAPP efficiently. This argues that the single mutant fibrils share significant structural similarity with wild-type fibrils. We also used wild-type fibrils to seed the single mutants (Supporting information). The mutants can be seeded by wild-type fibrils, but with different seeding efficiencies. The apparent seeding efficiency correlates with the rate of unseeded reaction in this case.
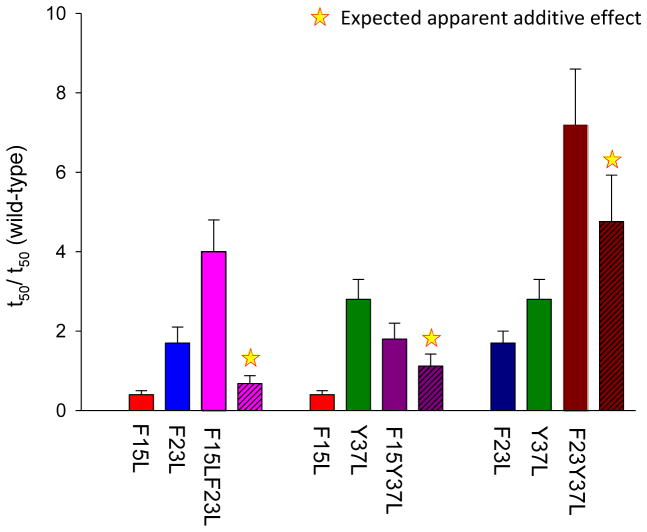
Figure 4.
Non-additive effects of double mutants. The bar graphs show the different effects caused by single mutants and the corresponding double mutants on amyloid formation. The expected additive result is indicated by a star.
Non-additive effects on amyloid formation are observed between different sites
The effect of some of the double mutations is not equal to the additive effect of the two individual mutations (Figure 5). For a simple protein folding reaction non-additive effects can be detected by comparing the effect of mutations on folding rate constants. Independent effects will lead to additive effects on the log of the folding rate, or multiplicative effects on the rate constants. Amyloid formation is more complex, involving multiple steps and possible parallel pathways with heterogeneous intermediates, thus it is not possible to readily relate a simple thioflavin-T progress curve to microscopic rate constants. None the less, a semi-quantitative comparison can be obtained using valves of t50 or the length of the lag phase. We first analyzed the effect caused by a single mutation relative to wild-type by calculating the ratio (t50 of single mutant)/(t50 of wild-type). The expected additive effects (stars in figure 5) are estimated by multiplying the parameters calculated for two single mutations. Assuming independent additive effects, the F15F23L double mutant is expected to form amyloid on the same time scale or even slightly faster than wild-type. However, the value of t50 for the F15F23L double mutant is four fold larger than the wild-type t50. The F15LY37L double mutant forms amyloid close to the rate expected if the effect of the F15 mutation and the Y37 mutation are additive. The F23LY37L double mutant has the most significant effect on the rate of amyloid formation (7.2 fold larger relative to wild-type), but the effect is even greater than expected from simple additivity (4.8 fold). To summarize, both the F15LF23L and F23LY37L double mutants form amyloid more slowly than predicted from simple additive effects, with the largest deviation observed for the F15LF23L double mutant.
Figure 5.
Aromatic mutants effectively seed amyloid formation by wild-type IAPP. Thioflavin-T fluorescence monitored seeding experiments are shown. Black, unseeded wild-type IAPP amyloid formation; grey, wild-type IAPP seeded by wild-type IAPP amyloid fibrils; red, wild-type IAPP seeded by F15L-IAPP amyloid fibrils; blue, wild-type IAPP seeded by F23L-IAPP amyloid fibrils; green, wild-type seeded by Y37L-IAPP amyloid fibrils.
Sensitivity of position 15 to amyloid formation
F15L-IAPP is a rare example of a mutation that speeds up amyloid formation. This region of IAPP has been proposed to be involved in the early stages of oligomerization,22 and it is of interest to determine the properties of this site that impact the rate of amyloid formation. Secondary structure propensity is likely a key feature. Work from several groups suggests that helical intermediates are important during the pathway of fibrillation.22,32–37. For example, crystallographic studies of IAPP fused to maltose binding protein showed that the segments corresponding to residues 8–18 and 22–27 are helical in the construct. NMR studies have shown that human and rat IAPP have a tendency to preferentially sample helical φ,ψ angles between residues 8 to 22 in the monomeric state.32 These observations have led to models which postulate that initial oligomerization is driven by the thermodynamic linkage between helix formation and self- association.22,32,34–37 F15 is within the putative helical region and has been proposed to make important contacts during the early stages of amyloid formation.22,38
We prepared a series of variants with isomeric four-carbon side chains at position 15 in order to probe the factors which control amyloid formation. We compared the effects of Leu, norleucine (four carbon unbranched side chain), Ile, and tert-leucine (t-butyl side chain) on amyloid formation. These amino acids have side chains with similar hydrophobicity39, but different α-helix and β-sheet propensities (Figure 6, supporting information). The substitutions have significantly different effects upon the kinetics of amyloid formation (Figure 6B and supporting information). The F15 to tert-leucine variant, (F15TLe-IAPP), which has three methyl groups on the β-carbon takes much longer to form amyloid than wild-type (Figure 6B, purple), even though TLe has the highest β-sheet propensity.40–42 The F15I-IAPP mutant (Figure 6C, green curve), with its β–branched side chain, also forms amyloid more slowly than wild-type IAPP, or F15L-IAPP, or the F15 to norleucine mutant (F15NLe -IAPP), even though Ile has a higher β-sheet propensity. TEM images confirm that all of the F15 variants form amyloid fibrils and have a similar morphology to wild-type fibrils (Figure 7). CD shows that all of these variant fibrils are rich in β-sheet structure (Supporting information). Seeding experiments were used to further probe the structure of the amyloid fibrils formed by the variants. Each variant was as efficient at seeding amyloid formation by wild-type IAPP as were wild-type seeds (Figure 8), arguing that the structures of the mutant fibrils are similar to wild-type fibrils.
Figure 6.
The rate of amyloid formation is affected by substitution at position 15, but there is no correlation with β-sheet propensity. (A) A series of varients with isomeric four-carbon side chains were analyzed. These “mutations” change α-helix propensity and β-sheet propensity, but maintain hydrophobicity. (B) Thioflavin-T fluorescence monitored kinetic experiments are shown. Black, wild-type IAPP; red, F15L-IAPP; blue, F15NLe-IAPP; green, F15I-IAPP; purple, F15TLe-IAPP. (C) An enlarged plot covering the range from t/t50 (wild-type) = 0 to 5.
Figure 7.

All position-15 variants form amyloid. TEM images recorded at the end of the kinetic runs depicted in figure 6. (A) F15NLe- IAPP, (B) F15I-IAPP, (C) F15TLe-IAPP. The scale bar represents 100 nm.
Figure 8.
The position- 15 variants effectively seed amyloid formation by wild-type IAPP. The results of thioflavin-T fluorescence monitored seeding experiments are shown. Black, unseeded wild-type IAPP amyloid formation; grey, wild-type IAPP seeded by wild-type IAPP amyloid fibrils; red, wild-type IAPP seeded by F15L-IAPP amyloid fibrils; blue, wild-type IAPP seeded by F15NLe-IAPP amyloid fibrils; green, wild-type seeded by F15I-IAPP amyloid fibrils; purple, wild-type seeded by F15TLe-IAPP amyloid fibrils.
The data show that the rate of amyloid formation does not correlate with β-sheet propensity at this position even though residue 15 is located in a β-strand in all known models of IAPP amyloid fibrils.26,43 There is a stronger correlation with α-helical propensity. TLe has by far the lowest α-helical propensity, 40,44 and this variant is by far the slowest to form amyloid. Ile has the second lowest helical propensity and the Ile variant is the second slowest to form amyloid. Phe, the wild-type residue, Leu and NLe have relatively high α-helical propensities and these variants form amyloid more rapidly. Earlier work, using truncated variants of IAPP, has shown that replacement of F15 by Ser or Ala increases that the rate of amyloid formation.22 Both of these residues have higher helical propensity than Phe.
The effect of F15 substitutions upon amyloid formation is independent of the presence of HFIP
HFIP and other fluoridated alcohols accelerate amyloid formation by IAPP and even low levels of fluoridated alcohol co-solvent can promote helix formation.28,29 We examined the set of F15 variants in the absence of HFIP to ensure that the results were not biased by the presence of the cosolvent. The lag times for wild-type IAPP and the F15 variants range from hours to days without HFIP and without stirring. The order of fibril formation by wild-type IAPP and the F15 variants is the same in the presence or absence of HFIP (Figure 9, table 2). TLe, with the lowest α-helical propensity and highest β-sheet propensity takes the longest time to form amyloid fibrils, while the Ile variant has a longer lag phase and longer growth phase than wild-type. Variants with unbranched side chains at position 15 form amyloid faster more rapidly. We also compared the three single aromatic to leucine mutants in the absence of HFIP and found the same rank order as observed in 2% HFIP; F15L formed amyloid more rapidly than wild-type while both F23L and Y37L did so more slowly than wild-type with the Y37L mutant being the slowest (table 2). These experiments confirm that the results are not an artifact of solvent composition.
Figure 9.
The rank order of amyloid formation by the F15 variants is independent of HFIP. Thioflavin-T fluorescence monitored kinetic experiments in the absence of HFIP without stirring. The final concentration is 16 uM peptide in pH 7.4, 20 mM Tris buffer (A) Black, wild-type IAPP; red, F15L-IAPP; blue, F15NLe-IAPP; green, F15I-IAPP; purple, F15TLe-IAPP. (B) An enlarged plot covering the range from t/t50 (wild-type) = 0 to 6.
Table 2.
(A) Kinetic parameters for wild-type IAPP, single aromatic to Leu mutants, and F15 variants amyloid formation in the absence of HFIP. (B) Comparison of kinetic parameters normalized with respect to wild-type for single aromatic to Leu mutants, and F15 variants in the presence and absence of HFIP. Experiments were conducted at 25 °C, pH 7.4 20 mM Tris-HCl in the presence and absence of HFIP.
| (A) peptide | wild-type | F15L | F23L | Y37L | F15NLe | F15I | F15TLe |
|---|---|---|---|---|---|---|---|
| Lag time (hr) | 16±1.3a | 9.8±0.1 | 38±0.6 | 42±2.3 | 20±1.7 | 41±2.3 | 267±4 |
| t50 (hr) | 23±0.9 | 11±0.1 | 42±1.7 | 63±2.8 | 31±1.3 | 61±5.8 | 276±6 |
| (B) mutant | HFIP | F15L | F23L | Y37L | F15NLe | F15I | F15TLe |
|---|---|---|---|---|---|---|---|
| Lag time/wild-type Lag time | present | 0.4 ±0.08b | 1.7±0.4 | 2.1±0.5 | 1.9±0.4 | 2.3±0.5 | 22±5 |
| t50/t50 of wild-type | present | 0.4±0.08 | 1.7±0.3 | 2.8±0.5 | 1.8±0.4 | 2.9±0.6 | 28±6 |
| Lag time/wild-type Lag time | absent | 0.6±0.08 | 2.4±0.2 | 2.6±0.3 | 1.3±0.1 | 2.6±0.3 | 17±1.4 |
| t50/t50 of wild-type | absent | 0.5±0.02 | 1.8±0.1 | 2.7±0.2 | 1.3±0.1 | 2.7±0.3 | 12±0.5 |
The quoted uncertainty is the standard deviation.
The uncertainty was calculated by standard propagation of error.
CONCLUSIONS
The data presented here reveals that the three aromatic residues make different contributions to IAPP amyloid assembly kinetics with the Y37 substitution having the largest effect. Apparent non-additive effects were observed between F15 and F23, and between F23 and Y37. The effects should be interpreted with caution, because, as noted, amyloid formation is a complex process involving multiple steps and the analysis of non-additive interactions is less straight forward than for the simple folding of globular proteins. Non-the-less, the results are significant with the strongest effect observed between F15 and F23. F23 is located in a region proposed to be important for the early stages of the development of β-sheet structure,45 while F15 has been suggested to be involved in early stages of oligomerization. The observation of non-additivity might reflect synergy between these processes. p-Cyanophenylalanine Tyr FRET measurements argue that F23 and Y37 do not make persistent contacts in the lag phase beyond any formed in the deadtime of kinetic measurements.19 Thus, any non-additive interactions between F23 and Y37 are unlikely to result from long lived direct contacts formed in the lag phase. It is worth noticing that the relative deviation from simple additive behavior is smaller for this pair than for the F15 F23 duo. The two residues are distant from each other in an individual chain, but the supermolecular structure of the protofibrils brings F23 in one chain close to Y37 in the adjacent stack of monomers (Figure 1) and this might account for non-additivity. The F23Cβ to Y37Cβ distances are 12.2± 3.5 Å in the NMR based model of IAPP amyloid.
F15 is within the region proposed to be important for early contact formation during IAPP oligomerization and several groups have proposed that the initial events involve helix formation with the region of residues 8 to 20 or 22.22,34,35,37 In this model, initial association is driven by the thermodynamic linkage between helix formation and oligomerization. Our analysis of the effect of substitutions at position 15 is broadly consistent with this model since the substitutions with high β-sheet propensity, but low α-helical propensity, have longer lag phases than wild-type. Substitutions with high α-helical propensity and lower β-sheet propensity form amyloid more rapidly with shorter lag phases.
It is interesting to compare these results with a recent study of aromatic residues in the Alzheimer’s Aβ peptide46. Aβ contains a pair of adjacent Phe residues at positions 19 and 20. The peptide still forms amyloid if both are replaced with Leu or Ile, indicating that aromatic side chains are not required at these sites. These substitutions have the opposite effect in Aβ compared to IAPP, faster amyloid formation was observed for the substitutions which increased β-sheet propensity (Ile) compared to the one which lowered β-sheet propensity (Leu). In a separate study, the consequences of a series of substitutions at position -19 were examined in a Aβ–GFP constrct.47 Amyloid formation was found to be dependent on the β-sheet propensity with a higher β-sheet propensity correlating with enhanced aggregation although no kinetic data was obtained. Aβ, like IAPP, has been proposed to populate a helical intermediate during amyloid formation in vitro48. Within this context one can still rationalize the effects of F19 and F19, F20 double mutations; perhaps the helical structure does not include this segment of Aβ or perhaps helical structure in this portion of the chain introduces new interactions which slow aggregation.
Supplementary Material
Figure 3.
All aromatic mutants form amyloid. TEM images recorded at the end of the kinetic runs depicted in figure 2. (A) wild-type IAPP, (B) F15L-IAPP, (C) F23L-IAPP, (D) Y37L-IAPP, (E) F15LF23L-IAPP, (F) F15LY37L-IAPP, (G) F23LY37L-IAPP, (H) 3XL-IAPP. The scale bar represents 100 nm.
Acknowledgments
Funding Sources
Grant Sponsor NIH GM078114 to D.P.R.
We thank Dr. Peter Marek and Dr. Ping Cao for assistance with experiments and members of the Raleigh group for helpful discussions.
ABBREVIATIONS
- CD
circular dichroism
- F15L-IAPP
a Phe-15 to Leu variant of human islet amyloid polypeptide
- F15NLe-IAPP
a Phe-15 to norleucine variant of human islet amyloid polypeptide
- F15I-IAPP
a Phe-15 to Ile variant of human islet amyloid polypeptide
- F15Tle-IAPP
a Phe-15 to tert-leucine variant of human islet amyloid polypeptide
- F23L-IAPP
a Phe-23 to Leu variant of human islet amyloid polypeptide
- Y37L-IAPP
a Tyr-37 to Leu variant of human islet amyloid polypeptide
- F15LF23L-IAPP
a Phe-15 to Leu, Phe-23 to Leu variant of human islet amyloid polypeptide
- F15LY37L-IAPP
a Phe-15 to Leu, Tyr-37 to Leu variant of human islet amyloid polypeptide
- F23LY37L-IAPP
a Phe-23 to Leu, Tyr-37 to Leu variant of human islet amyloid polypeptide
- 3XL-IAPP
a Phe-15 to Leu, Tyr-37 to Leu, Phe-23 to Leu variant of human islet amyloid polypeptide
- IAPP
human islet amyloid polypeptide
- NLe
norleucine
- TLe
tert-leucine, 2-amino-3,3-dimethylbutyric acid
- t50
the time required for 50% of the total signal change in a kinetic experiment
- TEM
transmission electron microscopy
- tlag
the lag time of a kinetic experiment
Footnotes
A figure showing the results of 25 independently kinetic experiments on wild-type IAPP; figures showing more TEM fibril images collected at the end of the kinetic experiments; a figure showing CD spectra collected at the end of the kinetic experiments for the aromatic mutants; a figure showing the effect of varying the pH on the rate of amyloid formation by wild-type human IAPP; figures comparing the kinetic curves for each aromatic mutant to wild-type IAPP; a figure showing singles aromatic to leucine mutants which were seeded by wild-type fibrils; another CD spectra collected at the end of the kinetic experiments for the F15 variants; a table giving amino acid side chain parameters; another table giving kinetic parameters for F15NLe, F15I, and F15Tle in the presence of HFIP. This material is available free of charge via the Internet at http://pubs.acs.org.”
References
- 1.Sipe JD. Amyloidosis. Crit Rev Clin Lab Sci. 1994;31:325–354. doi: 10.3109/10408369409084679. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe DJ. Cell biology of protein misfolding: The examples of Alzheimer’s and Parkinson’s diseases. Nat Cell Biol. 2004;6:1054–1061. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- 3.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 4.Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443:774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- 5.Mitraki A. Protein aggregation: From inclusion bodies to amyloid and biomaterials. Advances in Protein Chemistry and Structural Biology. 2010;79:89–125. doi: 10.1016/S1876-1623(10)79003-9. [DOI] [PubMed] [Google Scholar]
- 6.Westermark P, Wernstedt C, Wilander E, Hayden DW, Obrien TD, Johnson KH. Amyloid fibrils in human insulinoma and islets of langerhans of the diabetic cat are derived from a neuropeptide-like protein also present in normal islet cells. Proc Natl Acad Sci USA. 1987;84:3881–3885. doi: 10.1073/pnas.84.11.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark A, Lewis CE, Willis AC, Cooper GJS, Morris JF, Reid KBM, Turner RC. Islet amyloid formed from diabetes-associated prptide may be pathogenic in type-2 diabetes. Lancet. 1987;2:231–234. doi: 10.1016/s0140-6736(87)90825-7. [DOI] [PubMed] [Google Scholar]
- 8.Westermark P, Andersson A, Westermark GT. Islet amyloid polypeptide, Islet amyloid, and diabetes mellitus. Physiol Rev. 2011;91:795–826. doi: 10.1152/physrev.00042.2009. [DOI] [PubMed] [Google Scholar]
- 9.Hutton JC. The insulin secretory granule. Diabetologia. 1989;32:271–281. doi: 10.1007/BF00265542. [DOI] [PubMed] [Google Scholar]
- 10.Kahn SE, Dalessio DA, Schwartz MW, Fujimoto WY, Ensinck JW, Taborsky GJ, Porte D. Evidence of cosecretion of islet amyloid polypeptide and Insulin by beta-cells. Diabetes. 1990;39:634–638. doi: 10.2337/diab.39.5.634. [DOI] [PubMed] [Google Scholar]
- 11.Nishi M, Sanke T, Nagamatsu S, Bell GI, Steiner DF. Islet amyloid polypeptide-a new beta-cell secretory product related islet amyloid deposits. J Biol Chem. 1990;265:4173–4176. [PubMed] [Google Scholar]
- 12.Westermark GT, Westermark P, Berne C, Korsgren O Nordic Network Clin Islet T. Widespread amyloid deposition in transplanted human pancreatic islets. N Engl J Med. 2008;359:977–979. doi: 10.1056/NEJMc0802893. [DOI] [PubMed] [Google Scholar]
- 13.Westermark GT, Westermark P, Nordin A, Tornelius E, Andersson A. Formation of amyloid in human pancreatic islets transplanted to the liver and spleen of nude mice. Upsala J Med Sci. 2003;108:193–203. doi: 10.3109/2000-1967-113. [DOI] [PubMed] [Google Scholar]
- 14.Potter KJ, Abedini A, Marek P, Klimek AM, Butterworth S, Driscoll M, Baker R, Nilsson MR, Warnock GL, Oberholzer J, Bertera S, Trucco M, Korbutt GS, Fraser PE, Raleigh DP, Verchere CB. Islet amyloid deposition limits the viability of human islet grafts but not porcine islet grafts. Proc Natl Acad Sci USA. 2010;107:4305–4310. doi: 10.1073/pnas.0909024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gazit E. A possible role for pi-stacking in the self-assembly of amyloid fibrils. FEBS J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- 16.Marek P, Abedini A, Song BB, Kanungo M, Johnson ME, Gupta R, Zaman W, Wong SS, Raleigh DP. Aromatic interactions are not required for amyloid fibril formation by islet amyloid polypeptide but do influence the rate of fibril formation and fibril morphology. Biochemistry. 2007;46:3255–3261. doi: 10.1021/bi0621967. [DOI] [PubMed] [Google Scholar]
- 17.Tracz SM, Abedini A, Driscoll M, Raleigh DP. Role of aromatic interactions in amyloid formation by peptides derived from human amylin. Biochemistry. 2004;43:15901–15908. doi: 10.1021/bi048812l. [DOI] [PubMed] [Google Scholar]
- 18.Padrick SB, Miranker AD. Islet amyloid polypeptide: Identification of long-range contacts and local order on the fibrillogenesis pathway. J Mol Biol. 2001;308:783–794. doi: 10.1006/jmbi.2001.4608. [DOI] [PubMed] [Google Scholar]
- 19.Marek P, Mukherjee S, Zanni MT, Raleigh DP. Residue-specific, real-time characterization of lag-phase species and fibril growth during amyloid formation: A combined fluorescence and IR study of p-cyanophenylalanine analogs of islet amyloid polypeptide. J Mol Biol. 2010;400:878–888. doi: 10.1016/j.jmb.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenzo A, Razzaboni B, Weir GC, Yankner BA. Pancreatic-islet cell toxicity of amylin associated with type-2 diabetes-mellitus. Nature. 1994;368:756–760. doi: 10.1038/368756a0. [DOI] [PubMed] [Google Scholar]
- 21.Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 22.Wiltzius JJW, Sievers SA, Sawaya MR, Eisenberg D. Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process. Protein Sci. 2009;18:1521–1530. doi: 10.1002/pro.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abedini A, Raleigh DP. Incorporation of pseudoproline derivatives allows the facile synthesis of human IAPP, a highly amyloidogenic and aggregation-prone polypeptide. Org Lett. 2005;7:693–696. doi: 10.1021/ol047480+. [DOI] [PubMed] [Google Scholar]
- 24.Tam JP, Wu CR, Liu W, Zhang JW. Disulfide bond formation in peptides by dimethyl-sulfoxide - Scope and applications. J Ame Chem Soc. 1991;113:6657–6662. [Google Scholar]
- 25.Abedini A, Singh G, Raleigh DP. Recovery and purification of highly aggregation-prone disulfide-containing peptides: Application to islet amyloid polypeptide. Anal Biochem. 2006;351:181–186. doi: 10.1016/j.ab.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 26.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine H. Thioflavine-T interaction with synthetic Alzheimers-disease beta-amyloid peptides - Detection of amyloid aggregation in solution. Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Padrick SB, Miranker AD. Islet amyloid: Phase partitioning and secondary nucleation are central to the mechanism of fibrillogenesis. Biochemistry. 2002;41:4694–4703. doi: 10.1021/bi0160462. [DOI] [PubMed] [Google Scholar]
- 29.Abedini A, Raleigh DP. The role of His-18 in amyloid formation by human islet amyloid polypeptide. Biochemistry. 2005;44:16284–16291. doi: 10.1021/bi051432v. [DOI] [PubMed] [Google Scholar]
- 30.O’Nuallain B, Williams AD, Westermark P, Wetzel R. Seeding specificity in amyloid growth induced by heterologous fibrils. J Biol Chem. 2004;279:17490–17499. doi: 10.1074/jbc.M311300200. [DOI] [PubMed] [Google Scholar]
- 31.Krebs MRH, Morozova-Roche LA, Daniel K, Robinson CV, Dobson CM. Observation of sequence specificity in the seeding of protein amyloid fibrils. Protein Sci. 2004;13:1933–1938. doi: 10.1110/ps.04707004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williamson JA, Miranker AD. Direct detection of transient alpha-helical states in islet amyloid polypeptide. Protein Sci. 2007;16:110–117. doi: 10.1110/ps.062486907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nanga RPR, Brender JR, Xu JD, Hartman K, Subramanian V, Ramamoorthy A. Three-dimensional structure and orientation of rat islet amyloid polypeptide protein in a membrane environment by solution NMR spectroscopy. J Am Chem Soc. 2009;131:8252–8261. doi: 10.1021/ja9010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abedini A, Raleigh DP. A critical assessment of the role of helical intermediates in amyloid formation by natively unfolded proteins and polypeptides. Protein Eng Des Sel. 2009;22:453–459. doi: 10.1093/protein/gzp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abedini A, Raleigh DP. A role for helical intermediates in amyloid formation by natively unfolded polypeptides? Phys Biol. 2009;6:15005. doi: 10.1088/1478-3975/6/1/015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao P, Meng F, Abedini A, Raleigh DP. The ability of rodent islet amyloid polypeptide to inhibit amyloid formation by human islet amyloid polypeptide has important implications for the mechanism of amyloid formation and the design of inhibitors. Biochemistry. 2010;49:872–881. doi: 10.1021/bi901751b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williamson JA, Loria JP, Miranker AD. Helix stabilization precedes aqueous and bilayer-catalyzed fiber formation in islet amyloid polypeptide. J Mol Biol. 2009;393:383–396. doi: 10.1016/j.jmb.2009.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilead S, Wolfenson H, Gazit E. Molecular mapping of the recognition interface between the islet amyloid polypeptide and insulin. Angew Chem Int Ed. 2006;45:6476–6480. doi: 10.1002/anie.200602034. [DOI] [PubMed] [Google Scholar]
- 39.Fauchere JL, Charton M, Kier LB, Verloop A, Pliska V. Amino acid side chain parameters for correlation studies in biology and pharmacology. Int J Pept Protein Res. 1988;32:269–278. doi: 10.1111/j.1399-3011.1988.tb01261.x. [DOI] [PubMed] [Google Scholar]
- 40.Paterson Y, Leach SJ. Effect of side-chain branching on theoretically predicted conformational space available to amino-acid-residues. Macromolecules. 1978;11:409–415. [Google Scholar]
- 41.Minor DL, Kim PS. Measurement of the beta-sheet-forming propensities of amino-acids. Nature. 1994;367:660–663. doi: 10.1038/367660a0. [DOI] [PubMed] [Google Scholar]
- 42.Street AG, Mayo SL. Intrinsic beta-sheet propensities result from van der Waals interactions between side chains and the local backbone. Proc Natl Acad Sci USA. 1999;96:9074–9076. doi: 10.1073/pnas.96.16.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiltzius JJ, Sievers SA, Sawaya MR, Cascio D, Popov D, Riekel C, Eisenberg D. Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin) Protein Sci. 2008;17:1467–1474. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyu PC, Sherman JC, Chen A, Kallenbach NR. Alpha-helix stabilization by natural and unnatural amino-acids with alkyl side-chains. Proc Natl Acad Sci USA. 1991;88:5317–5320. doi: 10.1073/pnas.88.12.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shim SH, Gupta R, Ling YL, Strasfeld DB, Raleigh DP, Zanni MT. Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. Proc Natl Acad Sci USA. 2009;106:6614–6619. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Armstrong AH, Chen J, McKoy AF, Hecht MH. Mutations that replace aromatic side chains promote aggregation of the Alzheimer’s A beta peptide. Biochemistry. 2011;50:4058–4067. doi: 10.1021/bi200268w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Groot NS, Aviles FX, Vendrell J, Ventura S. Mutagenesis of the central hydrophobic cluster in A beta 42 Alzheimer’s pepticle - Side-chain properties correlate with aggregation propensities. Febs Journal. 2006;273:658–668. doi: 10.1111/j.1742-4658.2005.05102.x. [DOI] [PubMed] [Google Scholar]
- 48.Kirkitadze MD, Condron MM, Teplow DB. Identification and characterization of key kinetic intermediates in amyloid beta-protein fibrillogenesis. J Mol Biol. 2001;312:1103–1119. doi: 10.1006/jmbi.2001.4970. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.