Abstract
We have recently proposed that the shared epitope (SE) may contribute to rheumatoid arthritis (RA) pathogenesis by acting as a ligand that activates pro-arthritogenic signal transduction events. In order to examine this hypothesis, here we characterized a novel small SE-mimetic compound, c(HS4-4), containing the SE primary sequence motif QKRAA, which was synthesized using a backbone cyclization method. The SE-mimetic c(HS4-4) compound interacted strongly with the SE receptor calreticulin, potently activated nitric oxide and reactive oxygen species production and markedly facilitated osteoclast (OC) differentiation and function in vitro. The pro-osteoclastogenic potency of c(HS4-4) was 100,000 to 1,000,000-fold higher than the potency of a recently described linear SE peptidic ligand. When administered in vivo at nanogram doses, c(HS4-4) enhanced Th17 expansion, and in mice with collagen-induced arthritis it facilitated disease onset, increased disease incidence and severity, enhanced OC abundance in synovial tissues and osteoclastogenic propensities of bone marrow-derived cells and augmented bone destruction. In conclusion, c(HS4-4), a highly potent small SE-mimetic compound enhances bone damage and disease severity in inflammatory arthritis. These findings support the hypothesis that the SE acts as signal transduction ligand that activates a CRT-mediated pro-arthritogenic pathway.
Introduction
It has long been observed that individuals carrying HLA-DRB1 alleles that code a particular 5 amino acid sequence motif in position 70-74 of the DRβ chain are at a significantly higher risk of developing severe rheumatoid arthritis (RA) (1). The mechanism by which this motif, commonly called ‘shared epitope’ (SE), affects disease susceptibility and severity has been enigmatic since its identification more than 2 decades ago (2-5). We have recently demonstrated that the SE acts as a signal transduction ligand that interacts with cell surface calreticulin (CRT), an established innate immune system receptor (6). In other studies, we have identified the SE binding site on CRT (7) and determined the spatial requirements for optimal receptor binding and signal transduction potency during SE-CRT interaction (7, 8).
Engagement of cell surface CRT by the SE ligand activates signal transduction events that result in lineage-dependent functional consequences. For example, in CD8+CD11c+ dendritic cells, the SE inhibits the activity of indoleamine 2, 3 deoxygenase, an enzyme known to play an important role in regulatory T (Treg) cell activation. In CD8-CD11c+ dendritic cells the SE triggers production of IL-6 and IL-23, cytokines known to be involved in activation and expansion of IL-17-producing T (Th17) cells. The end result of these two complementing effects is a potent SE-activated Th17 polarization, both in vitro and in vivo(9).
More recently, using transgenic mice expressing SE-positive human HLA-DR molecules, as well as 15-mer linear synthetic peptides expressing the SE sequence motif, we have demonstrated that the SE facilitates osteoclast (OC) differentiation and functional activation. Moreover, when administered in vivo, the 15-mer SE peptidic ligand enhanced erosive bone damage in collagen-induced arthritis (CIA) in mice (10).
Beyond shedding light on RA pathogenesis, the findings with linear peptides expressing allele-specific sequences suggest that in addition to their putative role in antigen presentation, HLA-coded molecules can contribute to the pathogenesis of autoimmune diseases through allele-specific ligands that activate aberrant signaling events with resultant pathology (10). Given the disease specificity and well-defined structure-function properties of such ligands, one cannot avoid wondering whether these properties could be exploited for structure-based rational design of new therapeutic strategies.
The utility of short linear peptides has two important limitations: First, such peptides lack metabolic stability due to rapid degradation in biologic milieus. Second, they are usually in fast equilibrium among many conformations in solution with no single restricted conformation, which results in sub-optimal target engagement. In order to overcome these limitations, we have recently synthesized and screened a library of backbone-cyclyzed 5-mer peptides, all carrying the primary sequence QKRAA (the most common SE motif, coded by the RA-associated allele HLA-DRB1*04:01). We identified one particular compound with high potency that could activate NO signaling in low nM concentrations (approximately 50,000-fold more active on a molar basis than the linear 65-79*0401 synthetic peptide). This low-molecular (969 Dalton) compound, designated c(HS4-4), was remarkably stable towards trypsin/chymotrypsin degradation, while the linear peptidic SE ligand completely degraded within 30 minutes (11).
Given its signaling potency and biologic stability, we have undertaken to study the arthritogenic effect of c(HS4-4). Here we report that compound c(HS4-4) showed high affinity to the SE-binding receptor, CRT, and activated in vitro pro-osteoclastogenesis at low nM-range concentrations. Moreover, when administered in vivo to mice with CIA at low-nanogram doses, the SE mimetic significantly facilitated arthritis onset, increased the incidence and severity of the disease, and enhanced OC-mediated erosive bone damage. These findings substantiate the SE ligand hypothesis. Moreover, given the known structure-function properties and receptor-binding characteristics of the c(HS4-4) agonist, the knowledge acquired here could lay the foundation for future design of SE antagonists as a novel class of therapeutic agents in RA.
Materials and Methods
Reagents, cells and mice
Ficoll-PaqueTM plus was purchased from GE Healthcare (Piscataway, NJ). 2′,7′ dichlorodihydrofluorescein (DCFH-DA) was purchased from Invitrogen (Carlsbad, CA). Murine macrophage colony-stimulating factor (M-CSF) and receptor activator for nuclear factor κ-B ligand (RANKL) were purchased from PeproTech (Rocky Hill, NJ). Denatured chicken collagen type II (CII) was purchased from Chondrex Inc. (Redmond, WA). Complete Freund's Adjuvant (CFA) containing Mycobacterium tuberculosis H37Ra was purchased from BD Difco™ (Franklin Lakes, NJ). AlexaFluor 647 anti-mouse CD4 (clone GK1.5), FITC anti-mouse CD3 (clone 17A2), PE -conjugated anti-mouse IL-17A mAb (clone TC11-18H 10.1) and their corresponding isotype controls were purchased from BioLegend (San Diego, CA). All other commercial reagents were purchased from Sigma (St Louis, MO)
Synthetic peptides corresponding to position 65-79 on the HLA-DRβ chain, coded by the SE-positive allele DRB1*04:01, designated here as 65-79*0401 (sequence: 65-KDLLEQKRAAVDTYC-79), were synthesized and purified (> 90%) as we previously described (12). The 5-mer linear peptide 70-74*0401 (carrying the 70-QKRAA-74 sequence) was synthesized by the University of Michigan peptide synthesis core. The c(HS4-4) compound is a backbone cyclic mimetic peptide, which was synthesized as previously described (11). Recombinant mouse CRT was purified as described (6).
The mouse OC precursor cell line RAW 264.7 was purchased from ATCC (Manassas, VA) and cultured as described (10). Human peripheral blood mononuclear cells (PBMCs) were isolated from healthy blood donors as previously described (13). Procedures involving human subjects were conducted under an institutional IRB-approved protocol.
DBA/1 mice, 6 to 10 weeks old, were purchased from the Jackson Laboratory (Bar Harbor, Maine). Mice were maintained and housed at the University of Michigan-Unit for Laboratory Animal Medicine facility, and all experiments were performed in accordance with protocols approved by University of Michigan Committee on Use and Care of Animals.
Surface plasmon resonance (SPR)
Receptor-ligand interactions were determined using SPR assays as previously described (6-8). All assays were performed at 25°C in a binding buffer containing 10 mM HEPES, pH 7.4, 50 mM KCl, 0.5 mM CaCl2, 100 μM ZnCl2, and 0.005% surfactant P-20. The immobilized level of mouse CRT was 3000 response units (RU). The analyte was injected at a flow rate of 10μl/min.
Measurement of ROS
Production of reactive oxygen species (ROS) was determined as previously described (6,12), using the ROS probe CM-H2DCFDA. The fluorescence level was recorded every 5 minutes over a period of 500 minutes, using a Fusion αHT system (PerkinElmer Life Sciences) at an excitation wavelength of 488 nm and emission wavelength of 515 nm. ROS levels are shown as fluorescence units per minute (FU/min).
In vitro assay for OC differentiation
Murine OCs were generated from primary bone marrow cells (BMCs) isolated from femurs and tibias as previously described (14, 15). Briefly, bone marrows cells were cultured in 48-well plates (2×105 per well) in α-MEM medium supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin, in the presence of 10 ng/ml of M-CSF alone during the first 2 days, followed by 4 additional days in the presence of 10 ng/ml of M-CSF, plus 20 ng/ml of RANKL. Human OCs were differentiated from PBMCs isolated from healthy blood donors as previously described (13). PBMCs were cultured for 7 days in 100 ng/ml of M-CSF and 100 ng/ml of RANKL supplemented in 10% FBS αMEM. To quantify the number of OCs, cultures were fixed and stained for tartrate-resistant acid phosphatase (TRAP) activity using an acid phosphatase kit (Kamiya Biomedical Company, Seattle, WA) according to the manufacturer's instructions. TRAP-positive multinucleated OCs (>3 nuclei) were counted using a tissue culture inverted microscope.
In vitro bone degradation assays
Degradation of osteoblast-derived bone matrix was quantified as previously described (16) with some modifications. Briefly, 12,000 osteosarcoma cells (SaOS-2) per well were cultured in McCoy's 5A medium supplemented with 15% FBS in 48-well polystyrene culture plates. When cultures reached 80–90% confluence, the medium was changed to osteoblast differentiation medium (Gibco), containing 10% FBS, 2mM glutamine, 300 mM ascorbic acid, 10mM b-glycerol phosphate. After 20–25 days, osteoblasts were removed using 15mM NH4OH. Mouse BMCs (200,000 cells/well in 48-well plates) were plated on the matrix in an OC differentiation medium as above. After 15 days in culture, cells were removed using 15mM NH4OH and matrix was stained with Von Kossa dye. Photographs of individual wells were taken using a transmitted light microscope and matrix abundance was quantified by Image J software.
To determine ex-vivo bone degradation, 5-mm-diameter bovine cortical bone disks were prepared and studied as described with some modifications (17). Disks were washed and sonicated in distilled water, and stored dry at room temperature. Before use, bone disks were sterilized by immersion in ethanol and placed under UV light for 30 min. Single disks were placed in individual wells of 48-well culture plates with 0.5 ml alpha MEM + 10 ng/ml M-CSF + 20 ng/ml RANKL. Mouse BMCs, 400′000 cells per well, were incubated for 10 days with replenishment of fresh media every other day. At the end of incubation, bone disks were removed and stained for TRAP and number of OC per disk was determined as above. Cells and debris were then removed by 2 burst of 15-second sonication in concentrated ammonium hydroxide. Disks were stained with 1% toluidine blue for 30 seconds, and resorption pits were counted by scanning the entire surface of each disk with a reflected light microscope.
CIA induction and in vivo compound administration
DBA/1 mice (7 week old) were immunized with chicken CII in CFA as previously described (10). In brief, 50 μl of an emulsion containing 100 μg of CII in 25 μl of 0.05 M acetic acid and 25 μl of CFA was injected intradermally at the base of the tail. Mice were injected once per week intraperitoneally with c(HS4-4) at one of the following doses: 0.0, 0.8, 8 or 80 ng/gm BW, in 50 μl of PBS. Arthritis severity was determined as previously described (18), using a visual scoring system on a 4-point scale for each paw: 0 = no arthritis, 1 = swelling and redness confined to digits, 2 = minor swelling and redness spreading from the digits to the distal paw, and 3 = major swelling and redness extending proximally from the paw.
To determine the effect of the compound on Th17 cell differentiation in vivo, DBA/1 mice were injected subcutaneously in the footpad with 100 μg of CII emulsified in CFA. The inoculums contained 8 or 80 ng/gm BW of c(HS4-4) in 50 μl of PBS, or an equal volume of PBS alone. Animals were sacrificed 7 days after immunization. Draining lymph nodes (DLNs) were collected and single cell suspensions were prepared. Unfractionated lymph node cells were cultured with PMA, Ionomycin and Brefeldin A for 6 hours. Cells were stained with AlexaFluor 647 anti-mouse CD4, FITC anti-mouse CD3 or isotype controls, followed by fixation and permeabilization using a Cytofix-Cytoperm™ kit. After permeabilization, intracellular staining was performed using PE-conjugated anti-mouse IL17A and cells were analyzed by flow cytometry. The results were analyzed using the FlowJo (TreeStar Inc., Ashland, OR) software.
Joint tissue studies
Limbs were dissected and decalcified in 10% EDTA for 14 days. After decalcification, the specimens were processed for paraffin embedding and serial sectioning. The histological sections were deparaffinized, rehydrated and stained with hematoxylin and eosin (H&E), or stained for TRAP activity using a commercial kit (Kamiya Biomedical Company, Seattle, WA). To determine OC abundance, TRAP-positive multinucleated cells were counted. Data represent mean ± SEM of the total number of OCs in front and rear paws and knees.
Radiological imaging
Bone damage was evaluated by radiography and micro-computed tomography (micro-CT). Front and hind limbs from arthritic mice were dissected, fixed in 10% formalin and stored in 70% ethanol. Limbs were scanned by a micro-CT system (eXplore Locus SP, GE Healthcare Pre-Clinical Imaging, London, ON) in distilled water. The protocol included the source powered at 80 kV and 80 μA. In addition to a 0.508 mm Al filter, an acrylic beam flattener was used to reduce beam hardening artifact (19). Exposure time was defined at 1600 ms per frame with 400 views taken at increments of 0.5°. With 4 frames averaged and binning at 2×2, the images were reconstructed with an 18 μm isotropic voxel size. Regions of interest were defined through a spline-fitting algorithm to create separate masks for the carpals, tarsals, calcaneus, talus, or phalanges. The global threshold for cortical bone was defined at 2000 HU and 1200 HU for trabecular bone.
Radiographs were taken using a microradiography system (Faxitron X-ray Corporation, Wheeling, IL, USA) with the following operating settings: peak voltage, 27; anode current, 25mA; an exposure time 4.5 seconds. Coded radiographs were evaluated by an experienced rheumatologist who was blinded to the treatment. The presence of bone destruction was assessed separately for front and rear paws and scored on a scale of 0-5, ranging from no damage to complete destruction of the joints as previously described (20).
Virtual ligand docking experiments
c(HS4-4) was docked against an NMR-derived conformation of the CRT P-domain (PDB ID: 1HHN) using the following two steps. First, the binding pocket was identified using the pipeline COACH (21, and J. Yang, A. Roy and Y. Zhang, manuscript in preparation). COACH is a consensus-based ligand-binding site prediction approach that combines four template-based methods [COFACTOR (22), TM-SITE, S-SITE, and FINDSITE (23)] and ab initio method ConCavity (24). In the template-based approaches, sequence profile and structural comparisons are used to recognize the template proteins from the known function library BioLip (21), from which the ligand-binding information of the target is transferred; in the ab initio approach, ligand binding site is derived from the cavity distribution on the surface of the target structures. A consensus binding pocket in the receptor structure was predicted by COACH, which is tethered by residues 216—225 and 255—260. The c(HS4-4) ligand was then docked onto the predicted binding pocket using the ligand-docking software DOCK6 (25).
Statistical analysis
Data are expressed as mean ± SEM. Unless stated otherwise, statistical analyses were performed using a 2-tailed Student's t-test (*, p < 0.05; **, p < 0.01; ***, p < 0.001)
Results
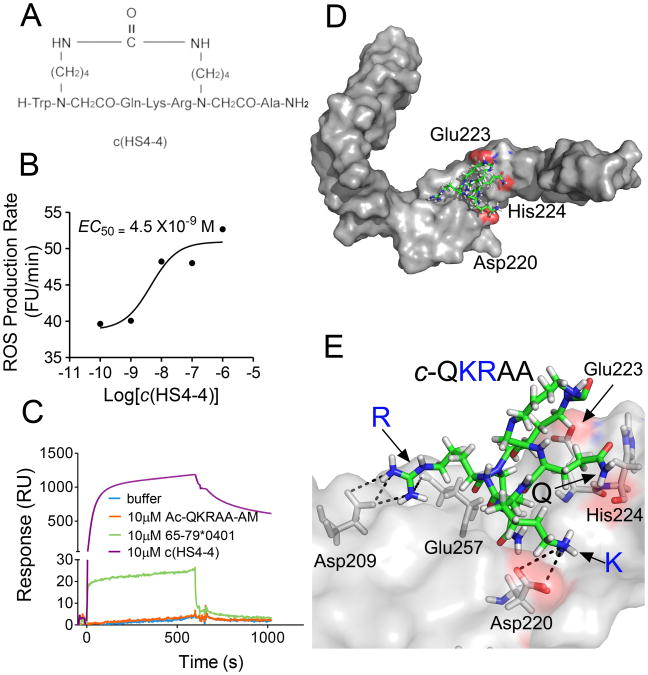
We have previously described 15-mer synthetic peptides carrying the SE sequence motif that acted as signal transduction ligands and activated pro-osteoclastogenic and pro-arthritogenic events (6, 10,12). Because linear peptides are conformationally and biochemically unstable, the efficiency of linear SE peptides as signal transduction ligands was found to be rather low. To address this limitation we have synthesized and screened a library of small (< 1000 Dalton) backbone cyclic analogs, all containing a core SE sequence, QKRAA (11). The structural formula of the compound studied here, designated c(HS4-4), is shown in Figure 1A. This 969-dalton molecule has been previously shown to activate NO signaling in low nM concentrations, and displayed remarkable resistance to trypsin/chymotrypsin degradation (11).
Figure 1.
Chemical structure and ligand properties of c(HS4-4). A. The chemical structure of c(HS4-4), a backbone cyclic peptide analog containing the SE consensus motif QKRAA was designed to induce a stable active conformation. A backbone cyclization methodology was used to keep the functional groups of the side chain residues intact. This feature assures that all the functional groups in a peptide sequence are available for biological activity. B. RAW 264.7 pre-OC cells (3 × 104 /well) were plated in flat-bottom 96-well plates in the presence of various concentrations of c(HS4-4) and ROS production was measured over time. C. SPR data showing binding interactions between recombinant CRT immobilized on a biosensor chip, and c(HS4-4), 15-mer linear SE peptide 65-79*0401, or A 5-mer linear peptide 70-74*0401 (expressing the SE core sequence QKRAA), applied in the analyte. D. A low-power docking image of the c(HS4-4) compound (identified here in its sequence structure cQKRAA, and shown in green) onto the previously identified SE binding site on the CRT P-domain (gray surface). CRT amino acid residues previously found to play critical SE ligand binding roles (7) are highlighted in red. E. A high-power view of c(HS4-4)-CRT molecular interactions. For details see Table 2.
Similar to the 15-mer linear SE peptide, c(HS4-4) activated ROS signaling in the pre-osteoclast cell line RAW 264.7 (Figure 1B), and bound to the SE receptor CRT in surface plasmon resonance (SPR) experiments, albeit at a much higher affinity (Figure 1C). As can be seen, when interacted with a SPR chip-immobilized CRT, c(HS4-4) produced a much higher SPR signal than the linear 15-mer SE ligand 65-79*0401, or the linear 5-mer peptide 70-74*0401, carrying the SE core sequence QKRAA. Kinetic analyses revealed that c(HS4-4) had a 10 to 100-fold higher affinity (lower KD) to CRT compared to the linear peptides (Table I). As could be expected from its higher affinity, c(HS4-4) demonstrated potent signal transduction activities with 100-10,000 –fold lower EC50 values, compared to the linear 15-mer SE ligand 65-79*0401 (Table I).
Table I. Relative potencies of SE ligands.
| SE ligand | Affinity to CRT | EC50 (M) | |||
|---|---|---|---|---|---|
|
|
|
||||
| ka (1/Ms) | kd (1/s) | KD(M) | NO | ROS | |
|
|
|
|
|
|
|
| c(HS4-4) | 2.7 ± 0.5 ×103 | 1.4 ± 0.2 ×10−3 | 5.1 ± 0.2 ×10−7 | 1.0 ×10 −9 | 4.5 ×10 −9 |
| 70-74*0401 | 2.9 ± 2.9 ×102 | 5.6 ± 5.1 ×10−3 | 9.5 ± 7.6 ×10−5 | ND | ND |
| 65-79*0401 | 5.1 ± 4.8 ×104 | 7.4 ± 3.1 ×10−3 | 1.3 ± 0.6 ×10−6 | 6.0 ×10 −5 | 4.5 ×10 −7 |
|
|
|
||||
Using the linear 15-mer SE ligand 65-79*0401 the binding site has been previously mapped to the region 217-224 of the CRT P-domain (7). To determine the likelihood that c(HS4-4) interacts with that binding site as well, we have taken a virtual docking strategy. Using the COACH algorithm (21, and J. Yang, A. Roy and Y. Zhang, manuscript in preparation), a binding pocket that overlaps with our previously mapped SE binding site (7) was identified in residues 216—225 of the CRT P-domain. The c(HS4-4) ligand was then docked onto that binding pocket using the DOCK6 (25) software. As can be seen in Figures 1D and 1E, c(HS4-4) interacts with the same binding site as the linear SE ligand, particularly with 3 CRT residues that have been previously identified as playing a role in SE-CRT interaction (7). The molecular interactions between individual CRT residues and c(HS4-4) are displayed in Table II. Thus, taken together, our findings indicate that compound c(HS4-4) is a bone fide SE mimetic that potently interacts with the SE receptor CRT and activates signal transduction effects at low-nM concentrations.
Table II. Molecular interactions between c(HS4-4) and CRT.
| c(HS4-4) component | Interacting CRT residue | Distance (Å) | Type of Interaction |
|---|---|---|---|
|
|
|
|
|
| HS4-4 Ring | Glu223* | < 4.0 | Proximity |
| Q** | His224 | < 4.0 | Proximity |
| K | Asp220 | 3.3 | Electrostatic |
| 3.8 | Electrostatic | ||
| R | Asp209 | 3.0 | Electrostatic |
| 3.0 | Electrostatic | ||
| 3.9 | Electrostatic | ||
| Glu257 | 3.9 | Electrostatic | |
| Trp258 | 2.1 | H-bond | |
| A | None | N/A | N/A |
| A | Asp220 | 2.1 | H-bond |
CRT residues are shown in a three-letter format.
c(HS4-4) amino acid residues are shown in a single-letter format.
N/A, not applicable.
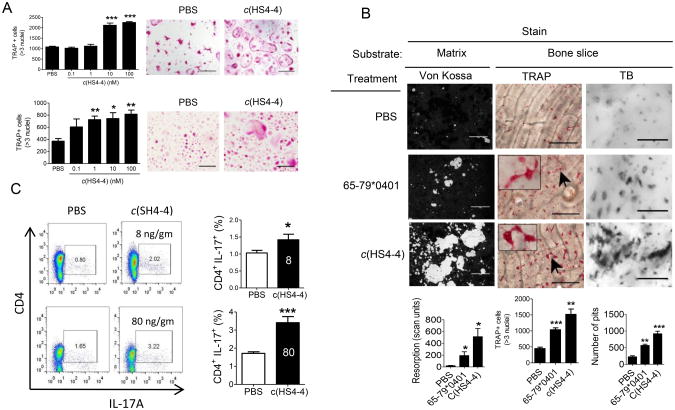
We have recently demonstrated that the 15-mer peptide SE ligand 65-79*0401 can activate osteoclastogenesis (10). Given the strong SE ligand properties of c(HS4-4), we have sought to determine whether it could activate osteoclastogenesis as well. To that end, BMCs derived from DBA/1 mice were cultured in an OC-differentiating medium as previously described (10) in the presence of different concentrations of c(HS4-4), or PBS. After 6 days, cells were stained for TRAP, and OCs were counted. As seen in Figure 2A, c(HS4-4) markedly enhanced OC differentiation in a dose-dependent manner (EC50 = 3.3 ×10−9 M). c(HS4-4) facilitated OC differentiation in healthy human PBMCs as well (EC50 = 4.5 × 10−10 M). These potencies are approximately 100,000 and 1,000,000-fold higher than those of the linear 65-79*0401 15-mer peptidic SE ligand in the two respective cell systems (10). The osteoclastogenic effect of c(HS4-4) was accompanied by higher capacity to degrade artificial bone matrix and bovine bone disks ex vivo (Figure 2B). Additionally, similar to the immune dysregulatory effect of the linear 65-79*0401 15-mer peptidic SE ligand (9,10), compound c(HS4-4) facilitated Th17 polarization (Figure 2C). Thus, c(HS4-4) is a highly potent SE ligand that reproduces the entire spectrum of the previously identified SE immune dysregulatory and OC activation effects.
Figure 2.
Functional activity of c(HS4-4). A. BMCs harvested from DBA/1 mice (upper panel) or human PBMCs (lower panel) were cultured in triplicates with M-CSF and RANKL in the presence of various concentrations of c(HS4-4) or PBS and TRAP-positive multinucleated cells were counted. Representative microscopic fields from treated with PBS or 100 nM c(HS4-4) are shown on the right upper and lower panels. B. c(HS4-4) facilitates bone degradation in vitro. Primary DBA/1 mouse BMCs were cultured in triplicates with M-CSF and RANKL atop bone matrices (left panel), or bone slices (middle and right panels) in the presence of PBS, 58 μM of the SE ligand 65-79*0401, or 100nM of c(HS4-4) in 48-well culture plates. Left panel: Matrix degradation was quantified on day 15 by digital scanning of tissue culture wells. Black areas represent intact matrix; white areas correspond to matrix degradation. Middle panel: BMCs cultures atop bone slices were stained for TRAP and OCs were counted. Representative OCs are identified by arrow and their enlarged image is shown in the box in the left upper corner. Right panel: Bone slice-attached OCs were removed and wells were stained with toluidine blue (TB). Bone pits are shown as black dots. Horizontal bars in all photos correspond to 100 μm. Bar graphs in the bottom represent mean ± SEM events from quintuplicate cultures in each of the three respective panels. C. c(HS4-4) facilitates Th17 polarization. Mice were injected with CII/CFA together with either 8 or 80 ng/gm BW of c(HS4-4), or PBS as detailed in Materials and Methods. Abundance of CD4+ IL-17A+ cells in DLNs was determined on day 7 by flow cytometry. Left-side images display representative dot plots. Bar graphs on the right side display mean ± SEM of data obtained from 6 DLN (3 mice) in each group. All flow cytometry data were determined in duplicate samples from each DLN. White-color numbers within the black bars represent the administered dose (ng/gm BW) of c(HS4-4).
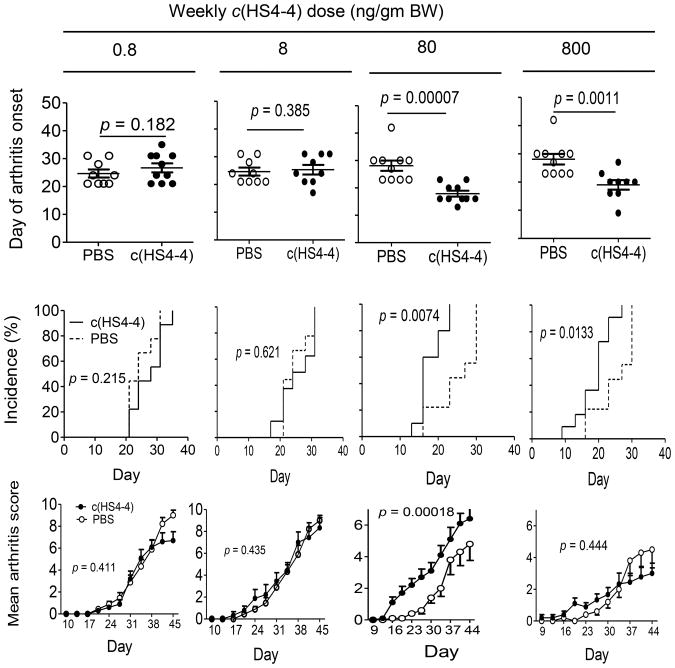
We next addressed the impact of c(HS4-4) on arthritis, using the CIA mouse model. CII-immunized DBA/1 mice were injected intra-peritoneally once a week with different doses of c(HS4-4) or PBS. As can be seen in Figure 3, at doses of 80 and 800 ng/gm BW per week, c(HS4-4) significantly facilitated disease onset and increased disease incidence. Additionally, at a 80 ng/gm BW dose, but not in the higher dose, c(HS4-4) significantly increased mean arthritis scores, as determined by joint swelling. The swelling effect was more prominent in the early stage of the disease, yet was clearly significant throughout the duration of arthritis (p = 1.8 × 10−4, in a paired Student t-test). The 0.8 and 8.0 ng/gm BW doses had no effect on any of the three CIA soft tissue disease parameters.
Figure 3.
c(HS4-4) enhances arthritis. CIA mice were injected intraperitoneally weekly with various doses of c(HS4-4), or PBS and monitored for CIA development. Upper panel: Mean day of arthritis onset. p values were calculated using a 2-tailed Student's t-test. Middle panel: Incidence curves. p values were calculated using log-rank (Mantel-Cox) test. Lower panel: Arthritis scores, shown as mean ± SEM. p values were calculated using a paired Student's t-test for the entire disease course. (N=10 per group).
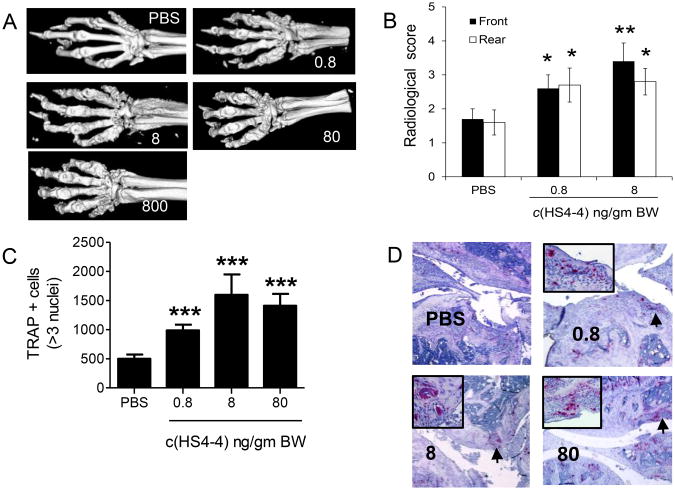
Radiologic analysis showed that irrespective of its effect on tissue swelling, all mice treated with c(HS4-4) had more severe erosive bone damage (Figures 4A). To better quantify the bone effect of c(HS4-4), joints of mice treated with the two lowest concentrations were evaluated radiologically using standard X-ray imaging. As can be seen in Figure 4B, significantly higher bone damage could be seen in mice treated with doses of c(HS4-4) that did not produce measurable effects on clinical scores of CIA.
Figure 4.
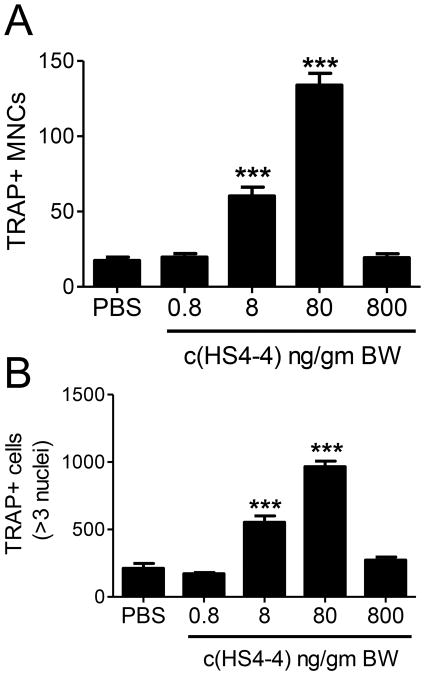
Bone destruction effects of c(HS4-4) A. Representative micro-CT images of CIA mice treated with various doses of c(HS4-4). B. Radiological scores of rear and front paws (N = 10) of CIA mice treated with either 0.8 or 8.0 ng/gm BW c(HS4-4) or PBS. C. Quantification of TRAP-positive cells in CIA joint tissues of mice treated with various concentrations of c(HS4-4), or PBS (N=10 in the PBS group and 5 per treatment group). D. Representative views of TRAP-stained knee joint tissue sections of CIA mice treated with various doses of c(HS4-4), or PBS). Magnification: × 10. An area of high OC abundance is marked by asterisk, and shown in a × 40 magnification in the boxed image in the left upper corner.
Histologic examination of arthritic joints revealed significantly higher numbers of OCs in c(HS4-4)-treated mice, even at ‘subclinical’ doses (Figures 4C). The abundance of TRAP-positive mononuclear (pre-OC) cells was determined in the bone marrow of all c(HS4-4)-treated mice (Figure 5A) and showed a dose-dependent effect. Although the peak effect was seen in the 80 ng/gm BW treatment group, a statistically significant effect was seen in the group treated with a ‘subclinical’ dose of 8 ng/gm BW as well. Upon ex vivo differentiation for 6 days, BMCs harvested from c(HS4-4)-treated CIA mice formed significantly higher numbers of OCs in a similar dose-dependent pattern (Figure 5B).
Figure 5.
In vivo administered c(HS4-4) enhances bone marrow osteoclastogenesis. A. Fresh bone marrow cells harvested from CIA mice treated with various doses of c(HS4-4) or PBS were analyzed by TRAP staining for the abundance of pre-OCs. B. Bone marrow cells harvested from the same CIA mice were cultured for 6 days under OC-differentiating conditions and OCs were quantified by counting multinucleated TRAP-positive cells.
Taken together, these results indicate that c(HS4-4), a potent peptidomimetic SE ligand, is highly arthritogenic. While c(HS4-4) has a clear effect on joint swelling, it is a particularly potent facilitator of OC-dependent bone damage in inflammatory arthritis.
Discussion
We recently proposed a ‘Shared Epitope Ligand Hypothesis’, which postulates that much of the impact of the SE on susceptibility to - and severity of - RA is attributable to signal transduction effects (26-28). In this study we demonstrate that a small, biostable molecule mimicking the 70-74 core sequence motif of the SE is a highly potent ligand that binds to the SE receptor CRT, activates signal transduction, enhances OC functional activity in vitro, and facilitates arthritis and erosive bone damage in CIA. This small SE-mimetic compound reproduced the previously-demonstrated (10) immune dysregulation, OC activation and bone-destroying effects of a 15-mer linear peptide carrying the amino acid sequence of the region 65-79 on the DRβ chain, albeit at a much greater efficiency. These findings have two main implications: First, they shed important light on the biologic role of the SE and strongly support the SE Ligand Hypothesis and; second, they suggest that the new compound may be used in the future as a structural template for RA drug discovery.
The SE is the single most significant genetic risk factor for RA, but the mechanistic basis of its effect is unknown. Interestingly, the SE not only confers a higher risk for RA, but also increases the likelihood of developing a more severe disease. For example, SE-coding HLA-DRB1 alleles are associated with earlier disease onset and more severe bone erosions (29-32). Furthermore, there is evidence supporting gene-dose effect, where the severity of bone destruction in RA correlates positively with the number of SE-coding HLA-DRB1 alleles (31,32). The prevailing hypothesis over the past 20 years has postulated that the basis for SE-RA association is presentation of putative self or foreign arthritogenic antigens by SE-positive HLA-DR molecules (5). However, the identities of such target antigens remain unknown.
We have recently gained new mechanistic insights into the role of the SE in RA, by identifying it as a signal transduction ligand that binds to cell surface CRT and activates NO-mediated pro-oxidative signaling in a strictly allele-specific manner (6-8). In more recent studies, we have provided evidence directly implicating the SE ligand effects in the pathogenesis of autoimmune arthritis, by demonstrating that a cell-free peptidic SE ligand facilitated in vitro and in vivo the differentiation of Th17 cells (9, 10) and OCs (10). Importantly, when administered to mice with CIA, the SE peptidic ligand increased joint swelling, synovial tissue OC abundance and erosive bone damage (10).
In our ongoing efforts to examine the Shared Epitope Ligand Hypothesis, here we applied here a new level of ascertainment, by studying a small non-physiologic synthetic compound. In its natural conformation within the HLA-DR molecule, the SE occupies ∼ 1.5 α helical loops in the third allelic hypervariable region of the HLA-DRβ chain (discussed in 28). Based on structural, functional and evolutionary considerations we have previously proposed that conformationally-equivalent domains in many other MHC molecules are enriched in allele-specific signal transduction ligands that interact with non-MHC cell surface receptors and trigger signaling events. We further proposed that the SE ligand function is just a special case attesting to the broader theory (28). Our previous studies, which demonstrated that a synthetic peptide carrying the SE linear sequence was pro-arthritogenic, were an important advancement toward substantiating the Shared Epitope Ligand Hypothesis. However, due to the random conformation of linear peptides in solution and their exquisite susceptibility to degradation, those studies required relatively high peptide concentrations. In order to address these limitations and examine the SE ligand in a format that more closely resembles its natural tridimensional conformation, we studied a backboned cyclic SE peptide.
As could be expected based on its biostability and predicted conformation (11), the SE-mimetic compound was found to be highly potent. It interacted with the SE receptor CRT at an affinity 10 times higher than the linear 15-mer SE peptide 65-79*0401 and 100 times higher than the linear 5-mer peptide 70-74*0401. Its signaling potency and pro-osteoclastogenic effect in vitro were found to be, respectively, 100,000 and 1,000,000-fold higher than those of 65-79*0401. Importantly, when administered in vivo to CIA mice, compound c(HS4-4) showed potent bone destructive effects at low-nanogram doses, a 250,000-fold higher potency than the linear 65-79*0401 SE peptide (10).
Experiments involving in vivo administration of c(HS4-4) revealed an interesting differential effect in CIA. While joint swelling was affected only by the higher c(HS4-4) dose-range, bone destruction was observed at low concentrations as well, even in the absence of a visible impact on joint swelling. These findings suggest a mechanistic dichotomy between the effect of c(HS4-4) in these two aspects of arthritis. While the molecular basis of this dichotomy requires additional study, it is worth mentioning that discrepancies between joint swelling and bone destruction are not uncommon in RA. In some patients with a substantial DAS28 there is minimal radiological evidence of bone damage, despite RA chronicity; in other cases bone damage continues to progress despite quiescent synovitis. Mechanistic dissociation between inflammation and bone-destruction pathways has been reported in experimental arthritis models in mice as well (20, 33). Evidently there is much to be learned about the interrelationship between synovitis and bone damage. The SE-mimetic described here might be a useful experimental tool to dissect the molecular mechanisms underlying this discrepancy.
The rationale of using a backbone cyclization approach was based on our attempt to accomplish three goals: 1. Maintain the primary amino acid sequence intact. The advantage of the backbone cyclization methodology is its ability to prepare cyclic peptides without utilizing the functional groups of the side chain residues. This feature is very important when functional groups in a peptide sequence are essential for biological activity; 2. Preserve the native α helical conformation of the SE; 3. Increase resistance to biodegradation. The high in vitro and in vivo potencies of c(HS4-4) are likely the end results of all the three design features. However, while biostability is likely accountable to the improved effects of c(HS4-4) in experiments involving in vitro or in vivo biologic milieus, resistance to enzymatic degradation probably had no impact on the higher receptor binding affinity of this compound, since receptor binding studies were carried out in a cell- and serum-free buffer space. Primary and secondary conformational factors, on the other hand, were more likely responsible for the improved receptor affinity of compound c(HS4-4).
The apparent dependency of compound c(HS4-4) activity on its tridimensional structure is consistent with our previous observations, which suggested that the SE ligand is much more potent in its native α helical conformation. In previous studies the SE ligand was shown to be effective in several different structural formats: i) in its native conformation as part of cell surface SE-positive HLA-DR molecules, or as SE-expressing HLA-DR tetramers; ii) as cell-free non-HLA recombinant proteins genetically engineered to express the SE motif in its native α helical conformation; or iii) as SE-positive short synthetic peptides. The SE ligand activated signaling in all formats; however, considerably lower concentrations of conformationally-conserved reagents (e.g. tetramers or viral capsids expressing the SE sequence at the tips of α helical spikes) were required to achieve signaling effects comparable to those achieved with linear, conformationally unstable SE peptidic ligands. Although formal structural analysis of c(HS4-4) has not been yet completed, its cyclical structure predicts that it may possess a helical conformation, thereby more closely resembling the native SE ligand (11).
The data presented here, along with the data we recently reported on a library of SE cyclic compounds with a range of biologic activities (11) provide for fairly detailed structure-function-based information that will help decipher the tri-dimensional characteristics of an ‘ideal’ SE ligand. Additionally, in other studies we have mapped the SE binding site on CRT (7) and determined the enhancing and inhibitory influences exerted on SE-CRT interaction by binding site-neighboring amino acid residues (8). As the data shown here (Figures 1D and 1E, and Table II) demonstrate, high-resolution modeling of the tri-dimensional interactive relationships between the SE and its binding site is now achievable.
This study provides for the first time a structural framework to investigate disease-relevant tridimensional conformational parameters in SE-CRT interaction, and suggests that therapeutic targeting of this pathway deserves consideration. In this context, it should be pointed out that the use of global CRT antagonists might not be practical because CRT is a multifunctional protein. CRT has been shown to have many distinct binding sites that interact with other ligands, which, in turn, activate diverse functional activities (34). Consistent with this, crt gene knockout in mice leads to embryonic lethality (35), and knocking-down crt gene expression in vivo by a shRNA approach did not produce a measurable impact on CIA in our hands, despite effectively suppressing crt expression levels (X Pi and J Holoshitz, unpublished data). We therefore propose that selective targeting of SE-CRT interaction by site-specific inhibitors may be more effective than global anti-CRT approaches. The study presented here could lay the foundation for examining the utility of this strategy in the future.
Despite the advent of biologic agents, treating RA remains a challenging endeavor. The crux of the problem relates to a lack of sufficient understanding of the specific mechanisms that trigger disease onset and affect disease severity. As a result, drug ineffectiveness and/or side effects are often encountered. The significance of the data presented here stem from the fact that they provide, for the first time, well-defined structural parameters based on which small molecules with SE pathway inhibitory effects could be rationally designed in the future. Another potential future approach for drug discovery could involve high throughput screening of libraries of small chemical compounds. Hits could be selected for further study based on their ability to compete with c(HS4-4) for CRT binding and to inhibit SE-activated signaling. Evidently, the design, identification and testing of such drugs are all major endeavors. Time will tell if the structure-function properties of the SE-mimicking compound described here could be utilized in the design of future therapies.
Acknowledgments
We thank Ms. Gail Quaderer for administrative assistance.
Footnotes
This study was supported by grants to JH from the US National Institutes of Health (GM088560, AR059085, AR048310)
Abbreviations used in this manuscript: BMCs, bone marrow cells; BW, body weight; CIA, collagen-induced arthritis; CII, collagen type II; CRT, calreticulin; CT, computerized tomography; DCFH-DA, 2′,7′-dichlorodihydrofluorescein; DLN, draining lymph node; FU, fluorescence units; NO, nitric oxide; OC, osteoclast; PBMCs, peripheral blood mononuclear cells; RA, rheumatoid arthritis; RANK, receptor activator for nuclear factor-κB ligand; RANKL, RANK ligand; ROS, reactive oxygen species; SE, shared epitope; SPR, surface plasmon resonance; TRAP, tartrate-resistant acid phosphatase.
References
- 1.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. an approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 2.Wucherpfennig KW, Strominger JL. Selective binding of self peptides to disease-associated major histocompatibility complex (MHC) molecules: a mechanism for MHC-linked susceptibility to human autoimmune diseases. J Exp Med. 1995;181:1597–1601. doi: 10.1084/jem.181.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.La Cava A, Nelson JL, Ollier WE, MacGregor A, Keystone EC, Thorne JC, Scavulli JF, Berry CC, Carson DA, Albani S. Genetic bias in immune responses to a cassette shared by different microorganisms in patients with rheumatoid arthritis. J Clin Invest. 1997;100:658–663. doi: 10.1172/JCI119577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhayani HR, Hedrick SM. The role of polymorphic amino acids of the MHC molecule in the selection of the T cell repertoire. J Immunol. 1991;146:1093–1098. [PubMed] [Google Scholar]
- 5.Klein J, Sato A. The HLA system. Second of two parts. N Engl J Med. 2000;343:782–786. doi: 10.1056/NEJM200009143431106. [DOI] [PubMed] [Google Scholar]
- 6.Ling S, Pi X, Holoshitz J. The Rheumatoid Arthritis Shared Epitope Triggers Innate Immune Signaling via Cell Surface Calreticulin. The Journal of Immunology. 2007;179:6359–6367. doi: 10.4049/jimmunol.179.9.6359. [DOI] [PubMed] [Google Scholar]
- 7.Ling S, Cheng A, Pumpens P, Michalak M, Holoshitz J. Identification of the Rheumatoid Arthritis Shared Epitope Binding Site on Calreticulin. PLoS ONE. 2010;5:e11703. doi: 10.1371/journal.pone.0011703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling S, al E. Citrullinated calreticulin potentiates rheumatoid arthritis shared epitope signaling. Arthritis Rheum. 2012 doi: 10.1002/art.37814. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Almeida DE, Ling S, Pi X, Hartmann-Scruggs AM, Pumpens P, Holoshitz J. Immune Dysregulation by the Rheumatoid Arthritis Shared Epitope. J Immunol. 2010;185:1927–1934. doi: 10.4049/jimmunol.0904002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holoshitz J, Liu Y, Fu J, Joseph J, Ling S, Colletta A, Sharma P, Begun D, Goldstein S, Taichman R. An HLA-DRB1-coded signal transduction ligand facilitates inflammatory arthritis. A new mechanism of autoimmunity. J Immunol. 2013;190:48–57. doi: 10.4049/jimmunol.1202150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naveh S, Tal-Gan Y, Ling S, Hoffman A, Holoshitz J, Gilon C. Developing potent backbone cyclic peptides bearing the shared epitope sequence as rheumatoid arthritis drug-leads. Bioorg Med Chem Lett. 2012;22:493–496. doi: 10.1016/j.bmcl.2011.10.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling S, Lai A, Borschukova O, Pumpens P, Holoshitz J. Activation of nitric oxide signaling by the rheumatoid arthritis shared epitope. Arthritis Rheum. 2006;54:3423–3432. doi: 10.1002/art.22178. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Jung Y, Sun H, Joseph J, Mishra A, Shiozawa Y, Wang J, Krebsbach PH, Taichman RS. Erythropoietin mediated bone formation is regulated by mTOR signaling. J Cell Biochem. 2012;113:220–228. doi: 10.1002/jcb.23347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi K, Takahashi N, Jimi E, Udagawa N, Takami M, Kotake S, Nakagawa N, Kinosaki M, Yamaguchi K, Shima N, Yasuda H, Morinaga T, Higashio K, Martin TJ, Suda T. Tumor necrosis factor alpha stimulates osteoclast differentiation by a mechanism independent of the ODF/RANKL-RANK interaction. J Exp Med. 2000;191:275–286. doi: 10.1084/jem.191.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koga T, Inui M, Inoue K, Kim S, Suematsu A, Kobayashi E, Iwata T, Ohnishi H, Matozaki T, Kodama T, Taniguchi T, Takayanagi H, Takai T. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature. 2004;428:758–763. doi: 10.1038/nature02444. [DOI] [PubMed] [Google Scholar]
- 16.Lutter AH, Hempel U, Wolf-Brandstetter C, Garbe AI, Goettsch C, Hofbauer LC, Jessberger R, Dieter P. A novel resorption assay for osteoclast functionality based on an osteoblast-derived native extracellular matrix. J Cell Biochem. 2010;109:1025–1032. doi: 10.1002/jcb.22485. [DOI] [PubMed] [Google Scholar]
- 17.Yang CR, Wang JH, Hsieh SL, Wang SM, Hsu TL, Lin WW. Decoy receptor 3 (DcR3) induces osteoclast formation from monocyte/macrophage lineage precursor cells. Cell Death Differ. 2004;11(1):S97–107. doi: 10.1038/sj.cdd.4401403. [DOI] [PubMed] [Google Scholar]
- 18.Wooley PH, Luthra HS, Stuart JM, David CS. Type II collagen-induced arthritis in mice. I. Major histocompatibility complex (I region) linkage and antibody correlates. J Exp Med. 1981;154:688–700. doi: 10.1084/jem.154.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meganck JA, Kozloff KM, Thornton MM, Broski SM, Goldstein SA. Beam hardening artifacts in micro-computed tomography scanning can be reduced by X-ray beam filtration and the resulting images can be used to accurately measure BMD. Bone. 2009;45:1104–1116. doi: 10.1016/j.bone.2009.07.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joosten LA, Helsen MM, Saxne T, van De Loo FA, Heinegard D, van Den Berg WB. IL-1 alpha beta blockade prevents cartilage and bone destruction in murine type II collagen-induced arthritis, whereas TNF-alpha blockade only ameliorates joint inflammation. J Immunol. 1999;163:5049–5055. [PubMed] [Google Scholar]
- 21.Yang J, Roy A, Zhang Y. BioLiP: a semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic Acids Res. 2013;41:D1096–1103. doi: 10.1093/nar/gks966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy A, Yang J, Zhang Y. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 2012;40:W471–477. doi: 10.1093/nar/gks372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci U S A. 2008;105:129–134. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capra JA, Laskowski RA, Thornton JM, Singh M, Funkhouser TA. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS Comput Biol. 2009;5:e1000585. doi: 10.1371/journal.pcbi.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang PT, Brozell SR, Mukherjee S, Pettersen EF, Meng EC, Thomas V, Rizzo RC, Case DA, James TL, Kuntz ID. DOCK 6: combining techniques to model RNA-small molecule complexes. RNA 15. 2009:1219–1230. doi: 10.1261/rna.1563609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Almeida DE, Ling S, Holoshitz J. New insights into the functional role of the rheumatoid arthritis shared epitope. FEBS Lett. 2011;585:3619–3626. doi: 10.1016/j.febslet.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holoshitz J, Ling S. Nitric oxide signaling triggered by the rheumatoid arthritis shared epitope: a new paradigm for MHC disease association? Ann N Y Acad Sci. 2007;1110:73–83. doi: 10.1196/annals.1423.009. [DOI] [PubMed] [Google Scholar]
- 28.de Almeida DE, Holoshitz J. MHC molecules in health and disease: At the cusp of a paradigm shift. Self Nonself. 2011;2:43–48. doi: 10.4161/self.2.1.15757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez-Gay MA, Garcia-Porrua C, Hajeer AH. Influence of human leukocyte antigen-DRB1 on the susceptibility and severity of rheumatoid arthritis. Semin Arthritis Rheum. 2002;31:355–360. doi: 10.1053/sarh.2002.32552. [DOI] [PubMed] [Google Scholar]
- 30.Mattey DL, Hassell AB, Dawes PT, Cheung NT, Poulton KV, Thomson W, Hajeer AH, Ollier WE. Independent association of rheumatoid factor and the HLA-DRB1 shared epitope with radiographic outcome in rheumatoid arthritis. Arthritis Rheum. 2001;44:1529–1533. doi: 10.1002/1529-0131(200107)44:7<1529::AID-ART275>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 31.Plant MJ, Jones PW, Saklatvala J, Ollier WE, Dawes PT. Patterns of radiological progression in early rheumatoid arthritis: results of an 8 year prospective study. J Rheumatol. 1998;25:417–426. [PubMed] [Google Scholar]
- 32.Weyand CM, Goronzy JJ. Disease mechanisms in rheumatoid arthritis: gene dosage effect of HLA-DR haplotypes. J Lab Clin Med. 1994;124:335–338. [PubMed] [Google Scholar]
- 33.Binder NB, Puchner A, Niederriter B, Hayer S, Leiss H, Blumel S, Kreindl R, Smolen JS, Redlich K. Tumor necrosis factor-inhibiting therapy preferentially targets bone destruction but not synovial inflammation in a tumor necrosis factor-driven model of rheumatoid arthritis. Arthritis Rheum. 2013;65:608–17. doi: 10.1002/art.37797. [DOI] [PubMed] [Google Scholar]
- 34.Gold LI, Eggleton P, Sweetwyne MT, Van Duyn LB, Greives MR, Naylor SM, Michalak M, Murphy-Ullrich JE. Calreticulin: non-endoplasmic reticulum functions in physiology and disease. FASEB J. 2010;24:665–683. doi: 10.1096/fj.09-145482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mesaeli N, Nakamura K, Zvaritch E, Dickie P, Dziak E, Krause KH, Opas M, MacLennan DH, Michalak M. Calreticulin is essential for cardiac development. J Cell Biol. 1999;144:857–868. doi: 10.1083/jcb.144.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]