Abstract
MicroRNA (miRNA) are small, non-coding RNA molecules that negatively regulate gene expression and control a wide range of cellular processes. Extracellular forms of miRNA circulating in the bloodstream (circulating miRNA, c-miRNA) are of increasing interest for their potential as biomarkers and long-range physiological signaling molecules. Precise measurement of intracellular miRNA expression is possible but can be challenging, especially in the context of specialized tissue niches in vivo. The accurate measurement of extracellular miRNA presents other obstacles stemming from their low concentrations and confounding sources of intracellular miRNA that contaminate RNA extraction protocols. Here, we describe multiple methods to isolate extracellular miRNA from cell culture media, serum, and plasma in order to accurately measure their variable expression under different conditions. We additionally describe an in situ staining protocol designed not only to quantify, but also to localize miRNA in formalin-fixed paraffin-embedded (FFPE) tissue, that may prove useful in describing the action of c-miRNA before they leave their tissue of origin and after they potentially arrive at their target destination.
Keywords: circulating miRNA, qRT-PCR, in situ staining, serum biomarkers, RNA extraction
1. Introduction
MicroRNA (miRNA) are small, non-coding RNA that directly regulate gene expression by binding to messenger RNA transcripts. They regulate a wide variety of cellular processes and disease phenotypes and, as such, investigation into their roles in physiology and disease has increase exponentially in recent years. Typical methods used to measure miRNA include extraction from tissue or cells of interest followed by Northern blot or reverse transcriptase polymerase chain reaction (RT-PCR). Importantly, high throughput screens have been developed based on RT-PCR methods to detect differences in miRNA samples, but these, as of yet, are limited in their scope by their inability to easily distinguish among different stages of miRNA maturity (primary, pre-miRNA and mature miRNA). In addition to variance in their maturity, miRNA exist in several different niches, including extracellular and in situ contexts. New methods for the quantification of miRNA have recently been developed to distinguish among these miRNA niches, including extracellular and intracellular miRNA isoforms.
Extracellular forms of microRNA circulating in the bloodstream (c-miRNA) have recently been recognized as biomarkers in health and disease and are potential regulators in pathology and physiology.(1) Importantly, extracellular miRNA are thought to be especially stable.(1) By quantifying expression levels of certain c-miRNA, our previous work demonstrated their specific and dynamic modulation in plasma from athletes undergoing exhaustive and sustained aerobic exercise.(2) In addition, we have used similar methods described below to isolate and measure miRNA levels from cell culture media, allowing the detection and manipulation of release of what likely, in vivo, represent c-miRNA.
Technical challenges exist in measuring extracellular miRNA. The optimized methods described here attempt to address the following issues: 1) the low concentration of extracellular miRNA compared to intracellular levels presents a substantial challenge for detection, and therefore raise concern regarding the accuracy and sensitivity of high-throughput assays in detecting differences between samples (3); 2) because of the relatively low expression of RNA isoforms in serum and plasma, conventional methods of spectrophotometry are not reliable to control for total amount of RNA extracted from samples; 3) the high concentration of miRNA in blood cells can easily confound results if these are not removed carefully and/or cellular lysis is not prevented. (4–6)
Moreover, to facilitate the propensity of such c-miRNA to be released and potentially taken up by recipient tissue and to further characterize their actions therein, quantification of intracellular miRNA is useful not merely in en block tissue, but rather in specific cell types. In situ hybridization offers a unique opportunity to visualize and compare miRNA expression across cell types, and is particularly useful for studying miRNA in rare and/or specialized niches of tissue. While initial attempts have been hampered by the digestion of miRNA binding proteins, leading to release of a large percentage of intracellular miRNA into fixative solutions during the staining process (7), optimized protocols have been developed to preserve and more accurately measure miRNA levels in situ. Here, we describe methods proposed by others but refined in our lab for the in situ localization and quantification of miRNA in formalin-fixed paraffin-embedded tissue. (8,7)
Regardless of the technical challenges associated with these methods, measurement of these molecules intracellularly and extracellularly has broad implications not only in the study of the basic biology of miRNA but also in their quantification and categorization as important biomarkers of human physiology and disease as well as disease risk stratification. Thus, we offer well-tested methods to measure miRNA in plasma and serum; in cell media; and in formalin-fixed paraffin-embedded tissue in situ.
2. Materials
2.1 Sample Collection and handling
EDTA treated vacutainers
Centrifuge capable of 16,000 × g
Cell culture media (appropriate for cells of choice)
2.2 Concentrating miRNA from cell culture media
100kDa Ultracel Ultrafiltration discs
MagniRIP RNA Binding Protein Immunoprecipitation Kit (Millipore)
RIPAb+ anti-Ago2 and IgG control kit (Rabbit IgG, Millipore)
Proteinase K solution (can be stored in a 20X solution (containing 10 ml TE buffer, 10 ml glycerol and 8mg proteinase K. This stock should then be diluted 1:20 in TE buffer (pH 7.4))
Control miRNA-mimic that has been found empirically to be poorly expressed in the situation of interst. For example, miR-422b mimic (Ambion/Applied Biosystems) Alternatively, a C. elegans miRNA can be used, such as celmiR-54 (Ambion/Applied Biosystems). (9)
Beckman-Couter ultracentrifuge apparatus (e.g., Optima™ XPN 100K Preparative Ultracentrifuge) accompanied by a swinging bucket rotor (e.g., Allegra® X-12R Benchtop Centrifuge, Refrigerated, 230 V, 50 Hz - Cell Culture)
2.3 miRNA extraction
RNA extract kit (Benevbio) or miRNeasy kit (Qiagen) (containing a protein denaturant, guanidium thiocyanate, and a nonpolar solvent, phenol, e.g., “qiazol”).
RNAse free water
RNase-free DNase
Filtered, RNAse free pipette tips
2.4 qRT PCR
Taqman Reverse transcription kit (Applied Biosystems)
Taqman miRNA Assay of interest and miR-422b (or alternate control) (Applied Biosystems, Assay ID will vary, e.g., for miR-422b. Note, these assays consist of hair-pin loop primers: one for the RT-amplification step and a second for PCR itself. Using specific RT primers instead of random primers increases the specificity of the assay. Their specificity ensures that even sequences with one base pair substitution will not be amplified.)
50 μl thermal cycling tubes (e.g., Applied Biosystems)
RNAse free water
Thermal cycler (e.g., BioRad c1000 96-well thermal cycler)
Taqman PCR mix (Applied Biosystems)
384-well plates (e.g., MicroAmp™ Optical 384-Well Reaction Plate, Applied Biosystems)
Optical adhesive covers (e.g., MicroAmp® Optical Adhesive Film, Applied Biosystems)
PCR instrument (e.g., Applied Biosystems 7900HT, Life Technologies)
SDS software for data analysis (See Life Technologies website for download)
Filtered, RNAse-free pipette tips
2.5 In situ miRNA detection
Buffers and Solutions
-
1
4% PFA in Tris-buffered saline (TBS)
-
3
Dry ice and 100% ethanol
-
4
1-methylimidazole solution: 0.13M 1-methylimidazole, 300 mM NaCl at pH 8.0 (Add HCl to adjust pH).
-
5
EDC solution: 300 mM NaCl, 0.1 M 1-methylimidazole (≥99%), 0.1M EDC- -HCl. Keep under argon gas. Make immediately before use.
-
6
Acetylation buffer: 873 μL of triethanolamine (Sigma) and 375 μL acetic anhydride (Fisher) in 75 ml distilled water
-
7
Hybridization buffer
-
8
20X SSC (Standard Socidum Citrate) (pH. 7.4): 175.3 g NaCL, 88.2 g Sadium Citrate, 800 ml H2O, Adjust pH = 7.0 and then add dH2O up to 1000 mL
-
9
Blocking buffer: 0.5% Blocking Reagent, 10% heat inactivated goat serum, and 0.1% Tween 20 in TBS
-
10
TNT buffer: 0.1M Tris-HCL at pH 7.5, 0.15M NaCl and 0.1% Tween.
-
11
Maleate buffer: 0.09M maleic acid, 0.175 M NaOH, 1 M NaCl, 0.5% Tween 20 at pH 7.5
-
12
TMN buffer: 0.1 M Tris base, pH 9.5, 0.05 M MgCl2, 0.5 M NaCl, 0.5% Tween 20, 2 mM (−)-tetramisole hydrochloride. (Store at 4°C)
Wash solutions
-
13
1X TBST: TBS/0.1% Tween 20
-
14
Post-hybridization wash buffer: 50% formamide, 1x SSC, and 0.1% Tween
-
15
Sodium-Tris-EDTA solution: 10 mM Tris-HCl (pH 8.0), 500 mM NaCl, 5mM EDTA, 0.05% Tween 20
Reagents
-
16
Proteinase K
-
17
Xylene
-
18
l–ethyl–3– (3–dimethylaminopropyl) carbodimide (“EDC”)
-
19
LNA miRCURY probe for miRNA of interest and control probe (Exiqon Life Sciences)
-
20
Blocking Agent: 1:500 solution of anti-DIG-FAB peroxidase (POD) (Roche) in blocking buffer (see 2.5 #8)
-
21
3% hydrogen peroxide
-
22
Anti-FITC horseradish peroxidase-conjugated secondary antibody (DAKO)
-
23
Individual Indirect Tyramide Reagent kit (Perkin Elmer Life Sciences)
-
24
NeutrAvidin–conjugated alkaline phosphatase (Thermo Scientific)
-
25
Nitro blue tetrazolium chloride 5–Bromo–4–chloro–3-indolyl phosphate (NBT/BICP stock solution, Roche)
-
26
Vectashield with DAPI (Vector Laboratories)
Equipment
-
27
Tissue sectioning blade (e.g., Vibratome 3000, Vibratome Company)
-
28
Tissue processor for formalin fixation and paraffin embedding such as Hypercenter XP System and Embedding Center (Thermo Fischer Scientific, Shandon, Pittsburg, PA)
-
29
Siliconized LifterSlips (Thomas Scientific)
3. Methods
3.1 miRNA measurement by RT-PCR
Sample collection and preparation: c-miRNA from serum or plasma
-
1
Collect blood (animal or human) in vacutainers. If plasma is desired, use anticoagulant (EDTA, not heparin, see Note 1) treated vacutainers. If serum is desired, use untreated tubes for collection and allow the whole blood to clot at room temperature for 15–30 minutes before proceeding to step 2.
-
2
Centrifuge samples at 2000 g for 10 minutes at 4 °C to remove cellular contents and debris, which will form a pellet at the bottom of each tube. 3. Immediately remove supernatant plasma or serum, being careful not to disturb the pellet. Color should be straw colored (serum) or clear (plasma). Pink or red color suggests significant contamination with erythrocytes or erythrocyte lysis, which could release intracellular miRNA into the sample and confound results. Store at −80°C in 100 ul aliquots to avoid freeze-thaw cycles (see Note 2).
-
4
When ready to extract c-miRNA from plasma samples, thaw plasma samples on ice. It is important that the temperature of samples remain as close to 0°C as possible.
-
5
Centrifuge samples in cold room (4°C) at 15,700 g for 10 min (see Note 3).
-
6
Carefully remove plasma supernatant and transfer to new polyethylene tube.
-
9
Before extracting RNA, it is necessary to add to each sample a control miRNA mimic that has been previously or empirically validated to be expressed at low or absent levels in the plasma or serum (see Note 4). We suggest hsa-miR-422b, which we have previously validated, but C. elegans miRNA can be used as well. (2,10) We add 2 fmol of miRNA mimic per sample.
Sample collection and preparation: miRNA in cell culture supernatant
Select the desired cell type and media, plate cells and incubate with media for an appropriate amount of time for cells to attach (if non-suspension cells). Expose to desired treatment (see Note 5).
Carefully extract media from over adherent cells. If growing cells in suspension, transfer to 50 ml conical tubes and centrifuge at 2,000 g for 5 minutes before extracting media. Note, it is extremely important to avoid aspirating cellular debris and cross contamination between samples (see Note 6).
Immediately centrifuge samples at 2000 g for 10 minutes at 4°C to remove cellular contents and debris, which will form a pellet at the bottom of each tube (see Note 6).
Immediately remove supernatant media. Centrifuge samples at 4000 g for 10 minutes at 4°C to remove residual cellular contents and debris.
Immediately remove supernatant media and either freeze at −80°C or move on to c- miRNA concentration, keeping samples on ice.
Concentrating miRNA in cell culture media
For poorly expressed microRNA, different techniques may be used to concentrate miRNA.
Centrifugal filtration by mass
By filtering media with a Millipore Amicon 100kDa filter apparatus, extracellular RNA can be concentrated in a sample by up to 100-fold (i.e., beginning with ~ 50 ml of media and concentrating to 0.5 ml (see Note 7).
After adding up to 12 ml of media to centrifugal filters, centrifuge at no faster than 4000 g for a swinging bucket rotor or 5000 g for a fixed angle rotor for approximately 20 minutes at 4°C.
Remove concentrate and store at −80°C. Flow-through can be collected for RNA extraction in order to confirm that extracellular miRNA is retained in complexes larger than 100 kDa and thus retained in the concentrate.
Ultracentrifugation
MiRNA released in exosomes or other vesicles can also be isolated by ultracentrifugation.
Using a Beckman-Couter ultracentrifuge apparatus pre-clarified media (from step 2 in section 3.1) or plasma/serum can be ultracentrifuged at 100,000 g × for 1 hour. Resultant pellets will contain the exosomes and vesicles, while freely complexed proteins/RNA remain in the supernatant.
Carefully remove supernatant and store in small aliquots (~250 μl) at −80°C. The remaining pellet can be dissolved in PBS or RNase-free water and stored at −80°C.
When ready to extract RNA, thaw supernatant and/or pelleted samples and add control mimic miRNA as is described in step 9 of the plasma/serum miRNA sample prep above.
Co-immunoprecipitation of extracellular miRNA
Given the documented interaction of some forms of extracellular miRNA with the RNA-binding protein Argonaute2 (Ago2), co-immunoprecipitation of Ago2 complexed with extracellular miRNA is possible. In this assay, such complexes can be isolated from cell culture media or plasma/serum.
First, attach the Ago2 or IgG isotype control antibodies to the Protein A/G labeled magnetic beads. Per the manufacterer’s instructions, a series of washes using Magna RIP wash buffer is followed by 30 min incubation at 25°C with the antibodies of interest. Subsequently, the beads are washed. A magnetic separator is used to draw beads to the side of the tube and allow aspiration of supernatant, which should always be accomplished using RNAse free, filtered pipette tips
Once appropriate antibodies are attached to the magnetic beads, wash and then reconstitute the beads in 900 μl of the kit immunoprecipitation buffer.
Quickly thaw the cell media or concentrate and add 100 μl to each tube. Incubate for 3 hours to overnight on a rotating apparatus at 4°C.
After immunoprecipitation, centrifuge, magnetically separate, and wash the beads as directed with wash buffer.
Per the manufacturer’s instructions, prepare a proteinase K solution. In doing so, ensure that concentrated proteinase K is added to the diluted SDS and not vice versa. Add 150 μl of this final working solution to each sample.
Incubate at 55°C for 30 minutes while shaking to digest proteins and antibodies.
Briefly centrifuge tubes and place on magnetic separator. Remove all supernatant from each sample and store at −80°C or immediately move to step h.
Before moving onto RNA extraction, add 2 fmol control miRNA mimic (as described in Step 9 of the plasma preparation protocol above) to each sample.
RNA extraction
MiRNA extraction may be accomplished using any number of miRNA column-based extraction kits, each with its own manufacterer’s protocol. For extraction, we have previously used miRNA extraction kits from Benevbio or miRNeasy kits from Qiagen. To avoid variations in efficiency of extraction within an experimental set, all samples in a given experiment should be extracted at the same time (see Note 8).
Guanidium thiocyanate-phenol-chloroform extraction of RNA is begun by first mixing samples with Qiazol (or equivalent phenol-containing kit component). For plasma and serum extraction, we use a ratio of 100 μl to 800 μl (~8× volume) of Qiazol; for cell culture media extraction, we use 300 μl of media and 900 μl (~3× volume) of Qiazol.
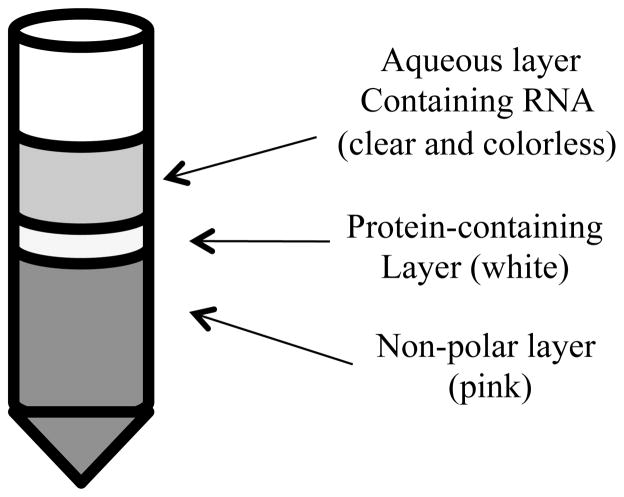
To these samples, chloroform is added (at a v/v ratio of 5:1, sample: chloroform) followed by centrifugation at 12,000 x g for 15 min at 4 °C. This allows for phase separation into a non-polar bottom layer (usually pink), a middle protein-containing layer (white) and an aqueous (clear/colorless) layer that contain nucleic acids such as miRNA (Figure 1).
The aqueous layer should be carefully removed by pipet and transfered to a clean tube. Ethanol is added (at a v/v ratio of 1:1.5, sample: ethanol) followed by vortex mixing.
Samples are then loaded onto spin columns. Depending on the size of the column used for fractionation, these volumes may require multiple centrifugations over the column.
After washing columns with an ethanol-based buffer per the manufacterer’s instructions, on-column treatment with RNase-free DNAse is pursued for 15 min at 25°C to eliminate DNA from the extracted samples. 6. Following two additional column washes, RNA is then eluted from the column using a RNase-free water. (30–50 μl per column) At this point in the protocol, samples can be stored at −80°C.
As is explained above (see Note 4), RNA spectrophotometry is not necessary or helpful at this stage due to the low RNA content of samples.
Figure 1. RNA extraction.
After initial centrifugation for phase separation, each sample will contain three layers. The upper (aqueous) layer should be clear and colorless and the lower layer is dark pink and contains non-polar molecules. The middle layer may appear somewhat granular and is white in color. Both middle and lower layers should be avoided during removal of the aqueous layer containing the RNA. If the tube is jostled, it should be re-centrifuged to again separate the layers appropriately.
3.1.5 Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-QPCR)
Reverse transcription is pursued to produce cDNA from the extracted RNA at quantitatively precise concentrations that reflect endogenous miRNA levels. cDNA is then quantitatively amplified using miRNA Taqman-based primers and fluorescent probes that report a quantitative value Ct, representing “real time” cycle number at which the increase in miRNA probe fluorescence is exponential. Fold-change of RNA species among comparison groups can then be calculated using the formula 2(−ΔΔct).
-
1
Reverse transcription: Prepare a master mix for the reverse transcription reaction, making approximately 20% excess solution (See Table 1). Divide the solution into aliquots for each miRNA to be measured (10μl per reaction plus 15% extra). Use 1.5 ml tubes and keep on ice.
-
3
Add 2 μl per reaction (plus 15%) of miRNA specific primers to each tube: one for the control miRNA selected above and the other for each miRNA of interest.
-
4
Vortex tubes and spin down briefly. Divide into polyethylene tubes, putting 12 μl into each tube.
-
5
Add 3μl of RNA samples to the desired reaction tubes. Be careful to discard micropipette tips after loading sample into each tube to avoid cross-contamination of primers and samples. All tips used should be filtered and RNAse free to prevent contamination with RNAse.
-
6
Vortex polyethylene tubes and spin down.
-
7
Incubate tubes in a thermal cycler with the following parameters: 15 minutes at 30°C, then 30 minutes at 60°C, then 5 minutes at 95°C, then 4°C for at least 5 minutes. Store samples at −20°C unless you plan to move directly to the quantitative PCR step, in which case, cDNA should be kept on ice.
-
8
For PCR, we use the Taqman Applied Biosystems kit as well. It is important to thaw all components on ice and keep them as cold as possible.
-
9
First create a master mix including 8 μl of Nuclease free water and 10 μl of Taqman 20× Universal PCR Master Mix (See Table 2). Divide into aliquots according to the number of miRNA being measured.
-
10
Add 1 μl of Taqman microRNA assay (primer) per desired reaction to each aliquot. Remember to add 10–20% extra volume in order to account for losses. Keep on ice at all times.
-
11
Load 19 μl of the master mix into each desired well of the 384 well plate. Leave two wells blank for subtraction.
-
12
Load 1 μl of cDNA from the RT reactions above into at least triplicate wells on a 384-well plate.
-
13
Seal the plate with optical adhesive cover, making sure to isolate the wells and spin the plate down for 5 minutes at 4°C.
-
14
Configure the plate document per PCR instrument instructions and begin thermal cycling: Run mode should be set at 9600 emulation, volume of 20 μl. Thermal cycles should include a 10 minutes at 95 °C for enzyme activation, then 40 cycles of 15 seconds at 95°C for denaturing followed by 60 seconds at 60°C for annealing and extension.
Table 1. Reverse Transcriptase Reaction Volume Calculation Table (Section 3.1.5 Q-RT PCR, Step 2).
Insert number of desired samples and multiply by the modifiers indicated. Then multiply each reaction component by this number to calculate the total volume necessary to add to the RT reaction master mix. This should then be vortexed, divided evenly into the number of aliquots needed to measure the desired number of miRNAs. Primers should then be added per the protocol (Steps 3–5).
| Component | Volume/sample (μl) | X | Number of Samples x number of miRNA to be measured + 1(control) x 1.2 | = | Total |
|---|---|---|---|---|---|
| 10× RT Buffer | 1.36 | x | = | ||
| dNTP Mix (100 mM) | 0.14 | x | = | ||
| MultiScribe™Reverse | 0.91 | x | = | ||
| Transcriptase | |||||
| RNase Inhibitor | 0.17 | x | = | ||
| Nuclease-free H2O | 7.42 | x | = |
Table 2. Quantitative PCR Reaction Volume Calculation Table (Section 3.1.5 Q-RT PCR, Step 9).
Insert number of desired samples and multiply by the modifiers indicated. Then multiply each reaction component by this number to calculate the total volume necessary to add to the reaction master mix. This should then be vortexed and divided evenly into the number of aliquots needed to measure the desired number of miRNAs. Primers should then be added per the protocol, and the solute divided into a 360 well plate (Steps 10–11).
| Component | Volume/sample (μl) | x | Number of Samples x number of miRNA to be measured + 1 (control) x 3 (replicates) x 1.2 | = | Total |
|---|---|---|---|---|---|
| Nuclease Free Water | 8 μl | x | = | ||
| PCR Master Mix | 10 μl | x | = |
3.2 In situ staining of miRNA in formalin-fixed paraffin-embedded tissue
In situ staining of miRNA in formalin-fixed paraffin-embedded tissue presents challenges related to the need to detect nucleic acids rather than protein, necessitating adaptation of standard immunological staining protocols. Furthermore, these protocols can be hampered by release of miRNA out of tissue when using formaldehyde fixation followed by proteinase K digestion. Thus, we have used previously developed methods employing both formaldehyde and the water-soluble 1-ethyl-3-(3- dimethylaminopropyl) carbodiimide (EDC) to fix miRNA in tissue prior to hybridization. (8,7) We have also utilized miRNA probes that carry complementary Watson-Crick sequences for specific miRNA recognition and subsequent immunological detection.
Tissue harvest and fixation
Enough tissue and treatment groups should be planned to account for the appropriate controls: One that should not have any probe applied to it, and one to which a scrambled probe should be applied. Additionally, if possible, obtain a positive and negative control, preferably from tissue that is known to carry significant differences in miRNA expression by a separate quantitative method, such as RT-PCR after the tissue has been flash-frozen and homogenized.
Perfuse tissue with saline.
Fix in 10% phosphate buffered formalin for 20 h at 4°C.
Process and paraffin-embed tissue using a Hypercenter XP System and Embedding Center or similar equipment.
Create 5-micrometer thin sections of formalin-fixed, paraffin–embedded (FFPE) tissue and mount on glass microscope slides. (see Note 9).
Deparafinization
Deparafinize slides in three consecutive xylene baths for 10 min each, followed by 5 min each in serial dilutions of ethanol (100%, 100%, 95%, 95%) and three changes of RNAse and DNAse-free water.
Proteinase K treatment
Digest with 400 μg/ml of proteinase K at 25 C for 15 min. Wash the slides in 0.1 M glycine diluted in 1× TBS for 5 min at 25°C and air-dry completely.
Wash the slides in 0.1 M glycine diluted in 1× TBS for 5 min at 25°C.
Formaldehyde tissue fixation
Rinse the slides in 1× TBS at 25°C.
Fix tissue sections in 40 mL of 4% formaldehyde diluted in 1× TBS for 10 min at 25°C (see Note 10).
Wash the slides in 0.1M glycine diluted in 1× TBS for 5 min at 25°C.
Rinse the slides in 1× TBS at 25°C.
Fixation of tissues with EDC (see Note 11)
Prepare 300 mL of EDC buffer immediately before use. Keep under Argon gas, and prepare a humidified chamber.
Incubate slides in 120 mL of freshly prepared EDC buffer for 10 min at 25°C. Check pH of EDC buffer and adjust to pH 8.0 with HCl before use.
Wash in 0.1M glycine diluted in 1× TBS for 5 min at 25°C. Prepare EDC solution for step 4 during second incubation.
Add 500 μL of EDC solution to each slide and incubate samples for at least 1 h (up to 2 h) at 25°C in a sealed humidified chamber.
Wash steps after fixation
Wash the slides with 120 mL of 0.1 M glycine diluted in 1× TBS for 5 min at 25°C.
Wash the samples twice with 120 mL of 1× TBS for 5 min at 25°C.
Acetylation for inactivation of enzymes in tissues
Prepare fresh acetylation buffer immediately before use and mix vigorously by shaking for 30 sec at 25°C.
Incubate slides in 120 mL of acetylation buffer for 30 min at 25°C. Rinse slides twice in 120 mL of 1× TBS for 5 min at 25°C.
Pre–hybridization and hybridization
Add 500 μL of freshly prepared pre-hybridization solution to each slide within the hydrophobic barrier. Incubate slides in a sealed humidified chamber for 1 h at 25°C.
Select the desired probes and pre-heat humidified chambers to the appropriate hybridization temperature, approximately 20°C below the TM.
Add 100 μL of hybridization solution (50nM) to each slide and gently place LifterSlips over the tissue sections
Incubate the slides in a sealed humidified chamber for 16 hours at the hybridization temperature (typically 20°C below the TM of the experimentally determined miRNA–LNA probe duplex).
Post-hybridization washes (see Note 12)
After hybridization, slides are rinsed in 2× SSC, 1× SSC and 0.5×SSC for 2 min each.
Wash for 30 min at 50°C in 0.5× SSC/0.1% Brij35 (Sigma), and rinse twice in TBS.
Inactivation of endogenous peroxidase activity
Incubate the slides in 3% hydrogen peroxide solution for 30 min at 25°C. (see Note 13)
Wash the slides three times 1× TBS–0.1% Tween 20 for 5 min at 25°C.
Anti-FITC horseradish peroxidase-conjugation
Cover slides with anti-FITC horseradish peroxidase-conjugated antibody (1:100 dilution in TBS/1% bovine serum albumin). Incubate for 60 min at room temperature
Wash three times in TBS/0.1% Tween 20 (TBS-T).
Tyramide amplification
Place the slides horizontally, face up in a humidified slide rack and add 250 μL of the Individual Indirect Tyramide Reagent kit working solution 1:50 to each slide and incubate for 30 min at 25°C.
Wash the slides three times in maleate wash buffer for 5 min at 25 °C.
Alkaline phosphatase detection system
Place the slides horizontally, face up in a humidified slide rack and apply a 1:200 dilution of NeutrAvidin-conjugated alkaline phosphatase in maleate wash buffer supplemented with 10 mg/mL blocking reagent
Apply 500 μL of the above NeutrAvidin solution to each slide and incubate for 40 min at 25°C.
Wash the slides two times in maleate buffer for 5 min at 25 °C.
Wash the slides four times in TMN buffer for 5 min at 25 °C.
Dilute 200 μL of NBT/BCIP stock solution in 10 mL of TMN buffer and add 250 μL of the solution to each slide for 30 min at 25°C. While incubating, protect the samples from light. (see Note 14)
Immerse the slides two times in water upon completion of the reaction for 20 sec at 25°C.
Wash the slides three times in Sodium-Tris-EDTA solution for 5 min at 25 °C.
Mounting slides for microscopy
Mount the slides using 2 drops of Vectashield mounting medium with DAPI.
Carefully place a glass coverslip over the tissue sections.
Seal glass coverslip by covering the edges the coverslip with clear nail polish and allow to air dry.
Store samples in a dark slide rack or protect from light using aluminum foil.
Expression (denoted by blue/dark purple stain) can be quantified and compared among experimental groups by assigning each tissue of interest an intermediate grey-scale pixel intensity value relative to a standardized white-black scale (Metamorph 6.1, Molecular Devices).
Figure 2. Appearance of in situ miRNA-21 stain with LNA probes in human pulmonary vasculature.
Arrows indicate areas of particularly high intensity staining in the artery wall, particularly in the media and endothelium as compared to control tissue (Used, with permission, from Parikh et al., Circulation 2012).
Acknowledgments
We thank Ms. Stephanie Tribuna for expert administrative assistance.
Sources of Funding
This work was supported in part by the National Institutes of Health (USA), the Pulmonary Hypertension Association, Gilead Sciences, and the Lerner, Harris, and Watkins funds (S.Y.C.).
Footnotes
It is important to use EDTA and not heparin for anticoagulation, since heparin inhibits the RT reaction. Heparinase has been reported to improve subsequent RT-PCR for mRNA (11), however this has not been shown, to our knowledge, for miRNA.
In general, samples containing RNA in solution should be stored at −80°C and, though c-miRNA are thought to be particularly stable RNAs freeze-thaw cycles should be avoided to protect the RNA content of the sample.(1)
It is extremely important to avoid hemolysis during collection of samples and centrifugation as this can significant alter the miRNA make up measured in plasma.(4–6)
Due to the low concentration of c-miRNA in plasma and cell culture media and the elimination of cell-bound RNA (mRNA, tRNA, etc) from samples, traditional methods of spectrophotometry cannot be relied upon to measure the small amount of total RNA content of these samples. Therefore, a small amount of a control miRNA mimic should be added to each sample PRIOR to RNA extraction in order to control for losses during extraction. We have used hsa-miR-422b for human and mouse plasma, but other miRNA expressed poorly in plasma have been identified previously as well.(1) Regardless of the specific choice of miRNA mimic, levels of its endogenous expression in the plasma or serum samples of interest should be confirmed to be negligible before use as a control. We have also used C. elegans miRNA as well as poorly expressed human miRNA as normalizing standards. Notably, there is no “housekeeping” RNA that has been identified as a reliable endogenous normalization control in cell culture media, plasma, or serum.
We have used conditioned media from human pulmonary artery endothelial cells, pulmonary artery smooth muscle cells, primary human lung fibroblasts, and transformed cell types (293 cells, HT-29 cells). We have also detected endogenously released miRNA as well as miRNA via forced expression by transfection of expression plasmids into the cells of interest.
There can be contamination in conditioned media with cellular debris. Thus we spin down the media 2000 RPM × 5 min sometimes followed by syringe filter of 0.2 μm (Nalgene syringe filter, Thermo Scientific, 190–2520) in order to avoid sampling cell debris prior to extraction. Such syringe filtration does not seem to affect the absolute expression levels of miRNA or their endogenous packaging (i.e., in exosomes, protein complexes, etc.) (data not shown, SY Chan).
Centrifugal filtration-based concentration of miRNA from media is based on the concept that the miRNA are released either complexed with proteins or in exosomes.(12,13) All cell-bound RNA should have been removed by previous centrifugation and clarification steps.
For a given study, inclusion of all samples for extraction at the same time is preferred to avoid technical variability of extraction efficiency (if processed at different times).
We have also successfully detected miRNA expression in situ on zinc-fixed tissue (rather than 10% formalin) prior to paraffin embedding. We have not performed this staining on flash-frozen tissue. (10)
Prepare fresh formaldehyde for each experiment to ensure adequate fixation and to prevent cross-contamination.
It is important for storage of commercial EDC to handle the reagent under anhydrous conditions and protect it with a layer of argon. Aqueous solutions containing EDC and/or 1–methylimidazole cannot be stored.(7)
When working with more than one probe, assign one Coplin jar for each probe. This will help prevent cross-contamination during the wash steps.
Hydrogen peroxide is used to block endogenous peroxidases before the antibody is added.
The time to form deposits (blue color) may vary between samples. Typically the reaction finishes after 30 min.
Disclosures: none
References
- 1.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baggish AL, Hale A, Weiner RB, et al. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J Physiol. 2011;589:3983–3994. doi: 10.1113/jphysiol.2011.213363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Etheridge A, Lee I, Hood L, et al. Extracellular microRNAL a new source of biomarkers. Mutat Res. 2011;717:85–90. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirschner MB, Kao SC, Edelman JJ, et al. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One. 2011;6:e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald JS, Milosevic D, Reddi HV, et al. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57:833–840. doi: 10.1373/clinchem.2010.157198. [DOI] [PubMed] [Google Scholar]
- 6.Pritchard CC, Kroh E, Wood B, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2011;5:492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pena JT, Sohn-Lee C, Rouhanifard SH, et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chau BN, Xin C, Hartner J, et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med. 2012;4:121ra118. doi: 10.1126/scitranslmed.3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho AS, Huang X, Cao H, et al. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol. 2010;3:109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parikh VN, Jin RC, Rabello S, et al. MicroRNA-21 integrates pathogenic signaling to control pulmonary hypertension: results of a network bioinformatics approach. Circulation. 2012;125:1520–1532. doi: 10.1161/CIRCULATIONAHA.111.060269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson ML, Navanukraw C, Grazul-Bilska AT, et al. Heparinase treatment of RNA before quantitative real-time RT-PCR. Biotechniques. 2003;35:1140–142. 1144. doi: 10.2144/03356bm03. [DOI] [PubMed] [Google Scholar]
- 12.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kosaka N, Iguchi H, Yoshioka Y, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]