Abstract
The penultimate event in the intoxication of mammalian cells by ricin toxin is the reduction, in the endoplasmic reticulum (ER), of the intermolecular disulfide bond that links ricin’s enzymatic (RTA) and binding (RTB) subunits. In this report we adapted an in vitro protein disulfide isomerase (PDI)-mediated reduction assay to test the hypothesis that the RTA-specific neutralizing monoclonal antibody (mAb) IB2 interferes with the liberation of RTA from RTB. IB2 recognizes an epitope located near the interface between RTA and RTB and, like a number of other RTA-specific neutralizing mAbs, is proposed to neutralize ricin intracellularly. In this study, we found that IB2 virtually eliminated the reduction of ricin holotoxin into RTA and RTB in vitro. Surprisingly, three other neutralizing mAbs (GD12, R70 and SyH7) that bind epitopes at considerable distance from ricin’s disulfide bond were as effective (or nearly as effective) as IB2 in interfering with PDI-mediated liberation of RTA from RTB. By contrast, two non-neutralizing RTA-specific mAbs, FGA12 and SB1, did not affect PDI-mediated reduction of ricin. These data reveal a possible mechanism by which RTA-specific antibodies may neutralize ricin intracellularly, provided they are capable of trafficking in association with ricin from the cell surface to the ER.
1. Introduction
Ricin is a member of the superfamily of A–B plant- and bacteria-derived toxins that includes cholera and Shiga toxins (Sandvig et al., 2002; O'Hara et al., 2012) . Ricin’s A subunit (RTA) is an RNA N-glycosidase whose exclusive substrate is a highly conserved adenosine residue within the so-called sarcin-ricin loop (SRL) of eukaryotic 28s ribosomal RNA (Endo et al., 1987). Ricin’s B subunit (RTB) is a galactose (Gal)-/N-acetylgalactosamine (GalNac)-specific lectin that mediates attachment, entry, and intracellular trafficking of ricin in host cells (Rutenber et al., 1987). Ricin exploits multiple endocytic pathways to gain entry into host cells, and then traffics retrograde to the trans-Golgi network (TGN) and, eventually, the endoplasmic reticulum (ER) (van Deurs et al., 1986; Rapak et al., 1997; Lord and Spooner, 2011). In the ER, the disulfide bond that links RTA and RTB is reduced by protein disulfide isomerase (PDI), a step that is critical for ricin cytotoxicity (Spooner et al., 2004). Once liberated from RTB, RTA retrotranslocates across the ER membrane to the cytoplasm where it assumes its catalytically active form (Spooner and Lord, 2012).
Protective immunity to ricin is antibody (Ab)-mediated, although the exact mechanism(s) of toxin neutralization are not well understood. While a number of RTB-specific monoclonal antibodies (mAbs) with potent toxin-neutralizing activity have been identified (Prigent et al., 2011; Yermakova and Mantis, 2011; Yermakova et al., 2012; Yermakova and Mantis, 2013), the majority of ricin-neutralizing mAbs that have been described to date are against RTA (Colombatti et al., 1986; Lemley et al., 1994; Maddaloni et al., 2004; O'Hara et al., 2010; O'Hara et al., 2012). Characterization of these RTA-specific mAbs, including R70, PB10, GD12 and SyH7 has revealed that epitope specificity, and not antibody subclass or affinity per se is the primary determinant of toxin-neutralizing activity (O'Hara et al., 2012). As RTA plays no known role in ricin uptake, internalization or intracellular trafficking, it is not immediately obvious how antibodies against this subunit might influence ricin cytotoxicity. Deciphering this issue is critically important because the primary objective of the two candidate ricin toxin vaccines currently in Phase I clinical trials is to elicit RTA-specific toxin neutralizing antibodies (Meagher et al., 2011; Reisler and Smith, 2012; Vitetta et al., 2012; O'Hara et al., 2013).
In this study, we put forth in vitro evidence to suggest that a recently identified RTA-specific mAb, known as IB2, neutralizes ricin intracellularly, possibly by interfering with the capacity of PDI to reduce the single disulfide bond that links RTA and RTB. We demonstrate that IB2 (i) neutralizes ricin after the toxin has bound to cell surfaces; (ii) is internalized and co-localizes with ricin in Vero cells; (iii) recognizes an epitope that is adjacent to the cysteine residue on RTA that forms a disulfide bridge with RTB; and (iv) virtually eliminated PDI-mediated reduction of ricin holotoxin in a cell free assay. While further studies will be necessary to demonstrate that IB2 can actually localize with ricin in the ER of mammalian cells, these data are intriguing in that they raise the possibility that RTA-specific antibodies may incapacitate ricin at a key step in its intracellular pathway.
2. Materials and methods
2.1 Chemicals and biological reagents
Biotin-labeled, FITC -labeled and unlabeled ricin toxin (Ricinus communis II; RCA-II) and RTA were purchased from Vector Laboratories (Burlingame, CA). ΔRTA was kindly provided by Dr. Ralph Tammariello (USAMRIID, Fort Detrick, MD). Ovalbumin (OVA; Sigma-Aldrich, Inc, St. Louis, MO) was biotinylated using sulfo-NHS-LC-biotin (Pierce, Rockford, IL). Ricin and OVA preparations were dialyzed against phosphate buffered saline (PBS) at 4°C in 10,000 MW cutoff Slide-A-Lyzer dialysis cassettes (Pierce) prior to use in cell-based and PDI-mediated reduction assays. Unless noted otherwise, chemicals were obtained from Sigma-Aldrich. NADPH, tetrasodium salt was obtained from Roche Diagnostics (Indianapolis, IN). 10% SDS-PAGE precast gels, Laemmli sample buffer (devoid of β-mercaptoethanol; BME) and 0.45 µm pore size nitrocellulose membrane were obtained from Bio-Rad Laboratories (Hercules, CA). Avidin-HRP was obtained from Southern Biotech (Birmingham, AL). THP-1 and Vero cells were purchased from the American Type Culture Collection (Manassas, VA) and maintained in a humidified incubator at 37°C with 5% CO2. MAb IB2 was produced by B-cell hybridoma technology using splenocytes from female BALB/c mice immunized four times with ricin toxoid, essentially as described (O'Hara et al., 2010). IB2 was purified using IEX and protein G chromatography under endotoxin free conditions by the Wadsworth Center protein expression core. IB2 was labeled with Alexa-fluor®-633 according to the instructions of the Alexa-fluor® 633 protein labeling kit, cat # A20170 (Molecular Probes, Eugene, OR). 633-Alexa-fluor® -labeled polyclonal goat anti-mouse IgG isotype control Ab for confocal microscopy studies was also obtained from Molecular Probes.
2.2 Ricin binding to cell surfaces
MAbs were incubated with FITC-ricin (0.6 µg) for 30 min before being added to 96 well Microtest™ U-bottom plates (BD Falcon™, Bedford, MA) containing THP-1 cells (1–2 × 106) that had been washed with ice cold sorting buffer (calcium/magnesium-free PBS, 1mM EDTA, 25 mM HEPES, pH 7.0, 1% FBS). THP-1 cells were incubated with ricin-mAb or ricin-lactose mixtures in the dark for 30 min on ice, and then washed with cold sorting buffer and fixed with 4% paraformaldehyde (PFA) in PHEM, pH 7.2 (60 mM PIPES, 21 mM HEPES, 10 mM EGTA, 2 mM magnesium chloride, 685 mM sodium chloride). Cells were subjected to flow cytometry using a FACS Calibur (BD Biosciences, Franklin Lakes, NJ). A minimum of 10,000 cells were analyzed per sample. One-way ANOVA with Tukey’s posttest was used to compare the percent of ricin binding to cell surfaces in the samples treated with mAb relative to the samples treated with ricin only.
2.3 Ricin-induced apoptosis
Ricin-induced apoptosis was assessed using the Annexin V-FITC Apoptosis Detection Kit II (BD Pharmagin). THP-1 cells (5×105) were incubated on ice with ricin (0.5 µg) for 30 min, washed to remove unbound toxin, and then “chased” for an additional 30 min on ice with ricin-specific mAbs (4 µg). The cells were then incubated at 37°C for 5 hr, washed with PHEM buffer, and then incubated with propidium iodide (PI) to assess necrosis and Annexin V-FITC (5 µl) to assess apoptosis and then subjected to flow cytometry using a FACS Calibur (BD Biosciences), as described previously (Yermakova and Mantis, 2011). Alternatively, THP-1 cells (5×105) were incubated for 30 min on ice with pre-formed ricin-mAb mixtures and then assessed for early apoptosis, as described above. A minimum of 10,000 cells were analyzed per sample. Differences in levels of apoptosis between cells treated with ricin-mAbs complexes (ricin + mAb) and those treated with ricin and then chased with mAbs (ricin -> mAb) was determined using an unpaired t-test.
2.4 Visualization of mAb-ricin complexes by laser scanning confocal microscopy (LSCM)
Vero cells (5 × 104 cells/ml) were seeded onto 22 mm sterile glass coverslips (Corning, Lowell, MA) in 6-well tissue culture plates and incubated at 37˚C for 24 h to achieve 50–70% confluency. Cells were then cooled to 4°C, overlaid with ricin or FITC-ricin (10 µg/ml) for 40 min at 4°C. The coverslips were rinsed twice with ice cold DMEM containing 10% FBS and then inverted onto droplets (100 µl) of 633-Alexa-fluor®-labeled IB2 or a relevant isotype control mAb (60 µg) on parafilm and incubated at 4°C for 30 min. To observe cell surface staining, cells were fixed with 1% glutaraldehyde in PHEM buffer for 10 min at room temperature (RT) before reduction with sodium borohydride in PHEM (1 mg/ml) for 3 min, followed by another 3 min in sodium borohydride in PBS (1 mg/ml). Cells were washed 4×, 5 min each in PHEM. To visualize ricin internalization, cells were transferred to 37°C for 30 min before being fixed as described above. Following fixation, samples were treated with Hoechst (1µg/ml in PHEM) for 2 min. Coverslips were rinsed in PHEM, and mounted onto microscope slides with ProLong® Gold. Cells were visualized using a TCS SP5 AOBS (acousto-optical beam splitter) confocal microscope with multi-photon laser (Leica Microsystems Inc., Buffalo Grove, IL).
2.5 ELISA and SPR
ELISAs were done as described previously (O'Hara et al., 2010). Competitive mAb binding assays using surface plasmon resonance (SPR) were done using a Biacore 3000 (GE Healthcare) with ricin attached to a CM5 chip surface, essentially as described previously (O'Hara et al., 2010). The first mAb was injected until saturation was achieved, (i.e., when no significant additional rise in resonance units (RU) was observed after antibody injection.) The second, mAb was then injected using a 2-min injection time. The amount of the second mAb bound to the chip, in RU, was calculated as the RU value at 15s after the injection minus the RU value at 15s preceding the start of the injection. The chip surface was regenerated by short pulses with 10 mM glycine, pH 1.5, until the RU values had returned to baseline.
2.6 In vitro protein disulfide isomerase (PDI)-mediated ricin reduction assay
The in vitro PDI-mediated ricin reduction assays were performed as described by Bellisola and colleagues (Bellisola et al., 2004) with some minor modifications. PDI (1.2 µM) was activated by thioredoxin reductase (TrxR; 90nM) by incubation in KPE buffer (100 mM potassium phosphate, 2mM EDTA, pH 7.4) containing 200 µM NADPH at 25°C in the dark for 20 min. Reduced glutathione (GSH; 750µM) and oxidized gluthathione (GSSH; 250µM) were then added to the reaction, followed by the anti-ricin mAbs of interest (1–2 µM each), biotin-labeled ricin (20 nM) and biotin-labeled OVA (20nM). Biotin-OVA was added to each sample as a SDS-PAGE loading control. The final reaction volume was 100µl. The reaction mixtures were incubated at 37°C in the dark for 1 hr. The reaction was stopped by the addition of 20µl of 1× Laemmli sample buffer. A total of 20µl of the reaction mixture was subjected to SDS-PAGE. As controls, biotin-ricin (20nM) and biotin-OVA (20nM) were diluted in sample buffer with or without 2% (v/v) BME and subjected to SDS-PAGE in parallel. For Western blot analysis, proteins were transferred to nitrocellulose membrane as previously described (Neal et al., 2010) and then probed using avidin-horseradish peroxidase (HRP; 0.25 µg/ml). The membranes were developed using an enhanced chemiluminescent detection (ECL) kit (Pierce, Rockford, IL), and then exposed to CL-Xposure film (Thermo Scientific, Rockford, IL). Bands on the blot were imaged and quantitated by densitometry using a Bio-Rad Chemidoc XRS imaging system and Quantity One (version 4.6.7.) software and graphed with GraphPad Prism 5 (GraphPad Software, San Diego, CA). The amount of PDI-mediated reduction of ricin holotoxin into RTA/RTB in the absence or presence of mAbs was expressed as a percentage of RTA/RTB present in control samples (i.e., ricin plus PDI). One-way ANOVA with Tukey’s posttest was used to compare the percent of RTA/RTB in the samples treated with mAb relative to the percent of RTA/RTB in the PDI-treated ricin only sample.
Surface representation of ricin and relevant B cell epitopes
The PyMOL Molecular Graphics System (Version 1.3. Schrödinger, LLC) was used to model B cell epitopes on ricin holotoxin. Ricin structure was based on accession 2AAI (Rutenber et al., 1991) from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB).
3. Results
IB2 is a murine IgG1 mAb that is sufficient to passively protect mice from a 5xLD50 ricin challenge (Table 1; J. O’Hara and N. Mantis, manuscript submitted). We subjected IB2 to both SPR and ELISA analysis and found that it bound to ricin holotoxin with high affinity, and to RTA to a slightly lesser degree (Table 1; Fig. 1). IB2 did not react with purified RTB (data not shown). To assess IB2’s capacity to neutralize ricin in vitro, IB2 was incubated with toxin for 30 min at room temperature and then applied to THP-1 cells, which are known to undergo apoptosis within a matter of hours in response to ricin (Yermakova and Mantis, 2011). Parallel THP-1 apoptosis assays were done with two additional mAbs: PB10 and FGA12 (Table 1). As expected, the non-neutralizing mAb FGA12 failed to prevent ricin-induced apoptosis of THP-1 cells, whereas the potent neutralizing mAb PB10 completely protected THP-1 cells from toxin-mediated death (Table 2). IB2 proved as effective as PB10 in neutralizing ricin in vitro, confirming the activity we observed in vivo.
Table 1.
Characteristics of anti-RTA and RTB mAbs used in this study.
| mAb | Isotype | Epitope | TNA | Protectiond | KD[M] |
|---|---|---|---|---|---|
| FGA12 | IgG1 | D37–R48 | − | ND | 4.12 × 10−8 |
| IB2 | IgG1 | T34–P43b | + | + | 6.7 × 10−10 |
| R70 | IgG1 | N97–F108 | + | + | 3.2 × 10−9 |
| GD12 | IgG1 | T163–S174 | + | + | 2.9 × 10−9 |
| SyH7 | IgG1 | E187–S198 | + | + | 2 × 10−8 |
| SB1 | IgG2a | Q223–F240 | − | − | 1.07 × 10−8 |
| SylH3a | IgG1 | L247–P254c | + | + | 3.38 × 10−9 |
SylH3 is an RTB-specific mAb. Tentative localization of epitope on
RTA or
RTB
as determined in a mouse model of passive protection. ND = not determined.
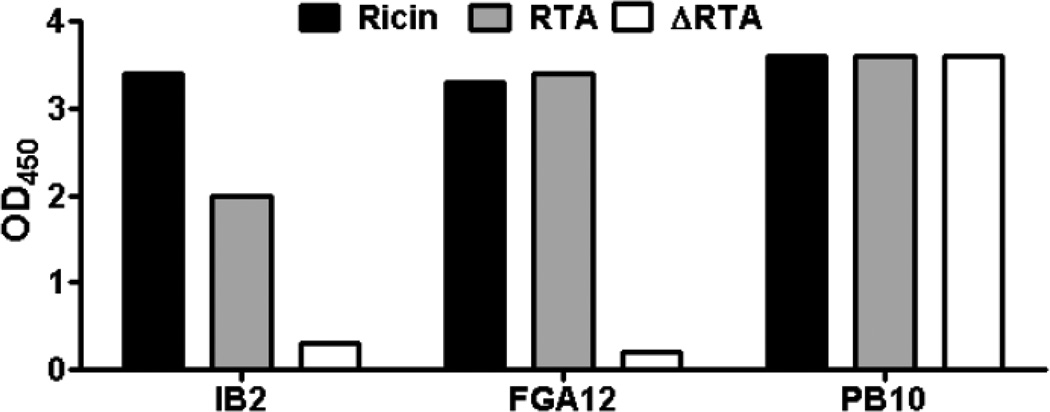
Fig. 1. Reactivity profile of IB2.
Reactivity by ELISA of RTA-specific mAbs IB2, FGA12 and PB10 with ricin holotoxin, RTA and ΔRTA.
Table 2.
Prevention of ricin-induced apoptosis by RTA-specific mAbs
mAbs were either mixed with ricin and applied to THP-1 cells (ricin + mAb) or added to THP-1 cells that had been pre-incubated (on ice) with ricin (ricin->mAb).
Percentage of cells positive for ricin-induced early apoptosis, as described in Materials and Methods.
Ricin-neutralizing mAbs can be grouped into two classes; those that prevent toxin attachment to cell surfaces (“Type I”), and those that interfere with one or more step(s) downstream of attachment (“Type II”) (O'Hara et al., 2012). In the case of IB2, we found that it was largely ineffective at blocking ricin binding to cell surfaces (Figure S1), suggesting that IB2 is a Type II antibody. To test this directly, we repeated the THP-1 apoptosis assays except that ricin was prebound to the cells at 4°C prior to the addition of IB2. As shown in Table 2, IB2 was nearly as effective at neutralizing ricin when the toxin was prebound to cells (ricin -> mAb) as compared to when IB2 and ricin were mixed (ricin + mAb) and then applied to THP-1 cells, indicating that it functions at a step post ricin cell attachment.
We next examined by LSCM if IB2 was internalized in complex with ricin into host cells. Vero cells cooled to 4°C (to prevent endocytosis) were treated sequentially with FITC-labeled ricin and Alexa-fluor® −633 labeled IB2. To permit toxin and toxin-antibody internalization, the cells were warmed to 37°C and incubated for 30 min before being fixed and imaged by LSCM. LSCM image analysis revealed that IB2 bound ricin on the surface of Vero cells at 4°C (Figure 2C–E) and was subsequently internalized with the toxin following incubation at 37°C, as evidenced by cytoplasmic vesicles that were positive for both FITC-ricin and Alexa-fluor® −633-IB2 (Figure 2F–H). Neither binding nor uptake of IB2 into Vero cells was observed in the absence of pretreatment with ricin toxin, demonstrating that IB2 is internalized only in complex with the toxin (data not shown). Attempts to determine the fate of ricin-IB2 complexes at later time points were unsuccessful due to the inability to detect by LCSM the small amount of toxin that is known to actually traffic to the TGN and ER (van Deurs et al., 1988; Rapak et al., 1997). Nonetheless, these data strongly suggest that IB2 neutralizes ricin intracellularly.
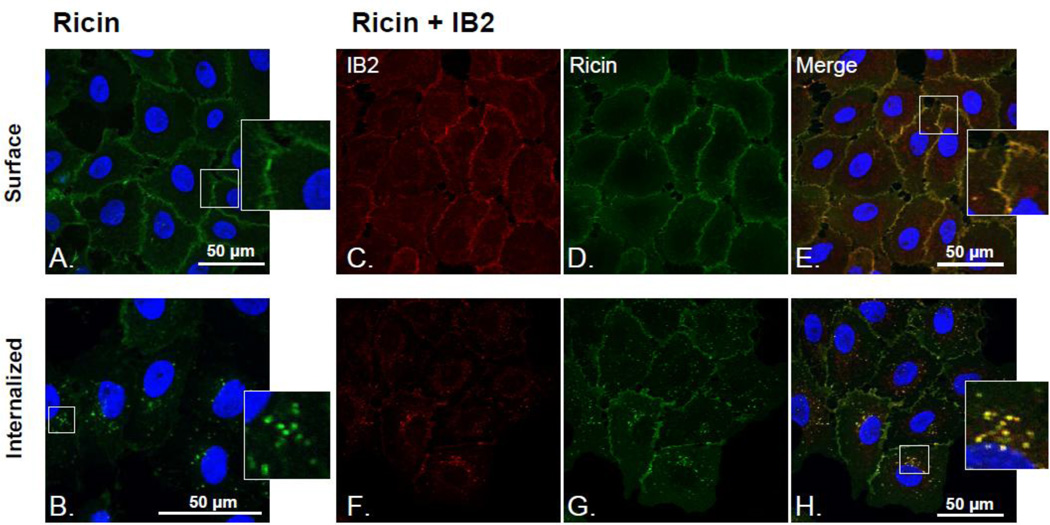
Fig. 2. Uptake of IB2-ricin immune complexes by Vero cells.
Vero cells grown on glass coverslips were chilled to 4°C and then treated sequentially with FITC-ricin and Alexa-fluor® −633-IB2, as described in the Materials and Methods. (A) LSCM image analysis revealed that IB2 bound ricin on the surface of Vero cells at 4°C (Figure 2C–E) and was subsequently internalized with the toxin following incubation at 37°C, as evidenced by cytoplasmic vesicles that were positive for both FITC-ricin and Alexa-fluor® −633-IB2 (Figure 2F–H). Imaging of surface bound and internalized ricin and mAb by LSCM. A) FITC-ricin bound to the cell surface. B) FITC-ricin internalized within vesicular compartments inside the cell after incubating ricin treated cells at 37°C for 30 min. C) 633-Alexa-fluor®-labeled IB2 mAb bound to the cell surface, D) in the presence of FITC-ricin. E) FITC-ricin and 633-Alexa-fluor®-IB2 co-localize at the cell surface. F) 633-Alexa®-IB2 in vesicular compartments within the cell after incubation of ricin and mAb treated cells at 37°C for 30 min, G) FITC-ricin (in the presence of 633-Alexa-fluor®-IB2) H) FITC-ricin and 633-Alexa-fluor®-IB2 co-localize within vesicular compartments inside Vero cells.
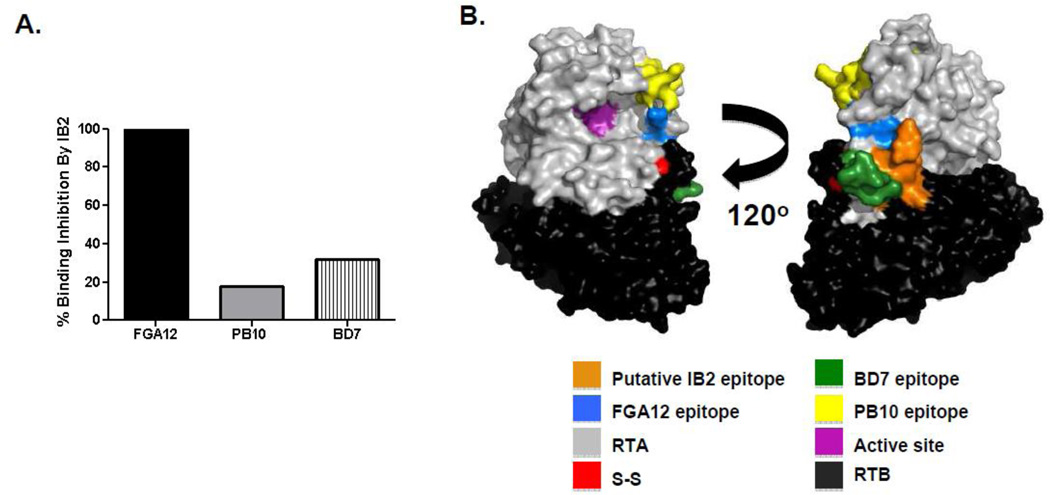
As a strategy to localize IB2’s epitope on RTA and ultimately better understand how the antibody neutralizes ricin intracellularly, we examined IB2’s reactivity with ARTA, a derivative of ricin’ A subunit lacking a small N-terminal loop (residues T34–P43) and virtually the entire C-terminus (residues A199 -F267) (Carra et al., 2007). As controls, we also examined in parallel PB10 and FGA12 reactivity with ARTA. As expected based on their known epitope location, PB10 recognized ΔRTA, while FGA12 did not (Fig. 1A). Like FGA12, IB2 did not react with ΔRTA, suggesting that IB2 binds an epitope situated within residues T34–P43 or A199–F267. To further localize IB2’s epitope to the N- or C-terminus of RTA, we performed competitive binding assays with mAbs FGA12, PB10 and BD7 (Table 1). We observed that IB2 inhibited FGA12, but not PB10 or BD7, from binding to ricin, demonstrating that IB2 likely recognizes an epitope in close proximity to residues T34–P43 in the N-terminus of RTA (Fig. 3).
Fig. 3. Localization of IB2’s epitope on RTA.
(Panel A) Competitive mAb binding assays in which preincubation of ricin with IB2 was examined for the ability to block FGA12, PB10 or BD7 from associating with to ricin, as determined using SPR. (B) PyMOL surface depiction of relevant epitopes on the surface of ricin holotoxin. For reference, RTA and RTB are colored grey and black, respectively. IB2’s proposed epitopes (residues T34–P43) is color coated orange. The residues that make up the RTA’s active site are color-coated magenta. The disulfide bond linking RTA to RTB are color-coated red.
Residues T34–P43 are spatially close to the cysteine residue on RTA that forms a disulfide bridge with RTB (Figure 3B). For this reason, we hypothesized that IB2 may interfere with PDI-mediated reduction of ricin holotoxin, a prerequisite for ricin cytotoxicity. To examine the effect of IB2 on the reduction of ricin, IB2 (2 µM) was incubated with biotinylated ricin holotoxin (20 nM) in the presence of TrxR-activated PDI, as described in Materials and Methods. As an internal loading control for these assays, all reactions were “spiked” with a fixed concentration of biotinylated OVA. Following a 1 h incubation, the reactions were subjected to SDS-PAGE and Western blotting in which nitrocellulose membranes were probed with avidin-HRP to simultaneously detect ricin (and its respective subunits) and OVA, which migrates with apparent molecular weights between 45–52 kDa. The amount of biotinylated RTA/RTB in the resulting Western blot was quantitated by densitometry (Figure 4).
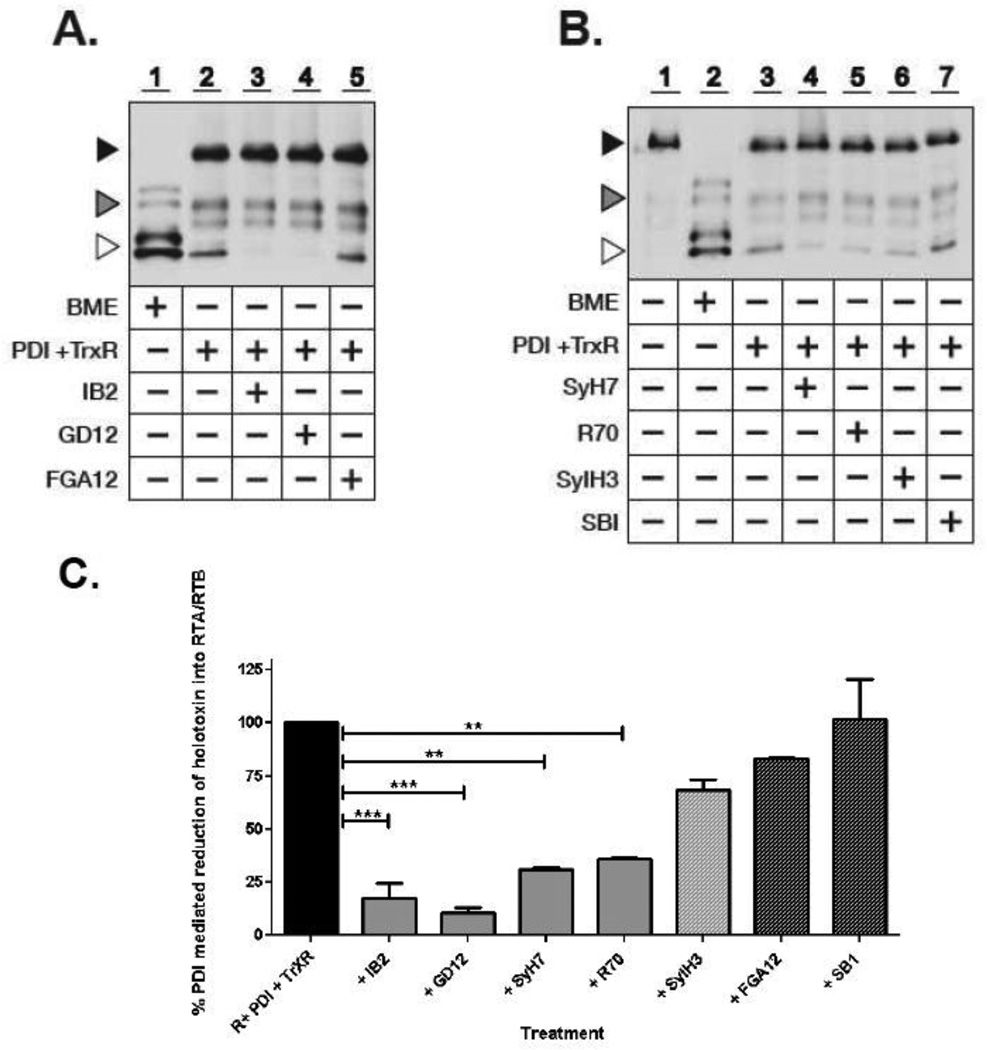
Fig. 4. RTA-specific neutralizing mAbs interfer with PDI-mediated reduction of ricin holotoxin.
(Panels A, B) Avidin-HRP blot analysis of biotinylated-ricin in its non-reduced and reduced forms following treatment with 2% BME or TrxR-activated PDI in the absence or presence of ricin-specific neutralizing and non-neutralizing mAbs. Biotinylated-OVA was added to each PDI reaction as an internal loading control. Symbols: Black triangle, ricin holotoxin (∼64 kDa); white triangle, reduced RTA/RTB (32– 34 kDa); grey triangle, OVA (doublet 45–52 kDa). Treatment with BME resulted in the complete reduction of ricin (Lane 1, Panel A; Lane 2, Panel B), whereas TrxR-activated PDI resulted in only partial reduction of ricin (Lane 2, Panel A; Lane 3, Panel B). PDI-mediated reduction of ricin was inhibited by the RTA-specific neutralizing mAbs IB2, GD12, SyH7, R70, but not the RTA-specific non-neutralizing mAbs FGA12 or SB1. The RTB-specific neutralizing mAb SylH3 did not alter PDI-mediated reduction of ricin. (Panel C) Western blots in panels A and B were subjected to densitometry. Show are the relative amounts of ricin in its reduced form after treatment with TrxR-activated PDI in the absence or presence of indicated mAbs. R, ricin. ** = P<0.001, *** = P<0.0001. It should be noted that the relative proportion of reduced RTA to RTB differs depending on whether ricin is reduced by BME or PDI (e.g., lanes 1 and 2). We postulated that this apparent difference in RTB is due to two factors. First, BME is significantly more effective than PDI in reducing the intermolecular disulfide bond that links RTA and RTB, as well as RTB’s intramolecular disulfide bonds. As a result, ricin holotoxin is reduced in its entirely, whereas PDI results in only partial reduction. Second, we have consistently observed that RTA tends to be more heavily biotinylated that RTB, likely due to more accessible lysine residues. As a result, at low concentrations RTB is actually more difficult to detect by avidin blot than is RTA.
It has been reported that PDI in the presence of TrxR and NADPH are sufficient to partially (∼30%) reduce ricin holotoxin into its respective subunits (Bellisola et al., 2004). Indeed, we confirmed that ∼30% of ricin holotoxin was reduced to RTA and RTB under these conditions (lane 2, Fig. 4A). Complete reduction of ricin to RTA and RTB was not achieved, probably because PDI in its oxidized form can also rapidly catalyze the formation of disulfide bonds in proteins (Holmgren, 1984). The addition of 2 µM IB2 (roughly equivalent to a 1:100 toxin:mAb ratio) markedly reduced the appearance of bands corresponding to free RTA and RTB (lane 3, Fig. 4A). Quantitation of the RTA/RTB bands by densitometry revealed that in IB2-treated samples the ability of PDI-mediated reduction of ricin’s intersubunit disulfide bond was less than 25% of the PDI-ricin control (Fig. 4C). IB2’s effects were dose-dependent, as the addition of two-fold less IB2 (roughly equivalent to a toxin:mAb ratio of 1:50) was approximately two-fold less effective at blocking ricin reduction (data not shown). To determine whether IB2’s effect on PDI-mediated reduction of ricin was specific and not due simply to the addition of an immunoglobulin (which may possibly serve as a competitive substrate for PDI), we performed parallel experiments in the presence of the anti-RTA non-neutralizing mAb FGA12. We observed that FGA12 did not prevent the appearance of ricin in its reduced form (lane 5, Fig. 4A; C), thereby demonstrating that the mere presence of an immunoglobulin like FGA12 is itself not sufficient to interfere with PDI-mediated reduction. Rather, IB2’s effect on PDI’s activity is specific.
Based on these findings, we next wished to investigate whether IB2’s effect on PDI-mediated reduction of ricin was unique or whether it is a property associated with other neutralizing RTA-specific antibodies. Ricin was incubated with neutralizing mAbs R70, GD12 and SyH7, or the non-neutralizing mAb SB1, and then assessed for PDI-mediated reduction of ricin’s interchain disulfide bond (Table 1; Figure S2) (Baillie et al., 2010; Neal et al., 2010; O'Hara et al., 2012). Interestingly R70, GD12 and SyH7 each interfered with PDI-mediated reduction of ricin in vitro, whereas the non-neutralizing mAb SB1 did not (lane 4, Fig 4A; lanes 4, 5, 7, Fig. 4B). When quantitated by densitometry, all three neutralizing mAbs significantly reduced the appearance of reduced holotoxin, although to different degrees. GD12, for example, which is known to bind an epitope distant from the RTA-RTB interface (Figure S2) was slightly more effective than IB2 at preventing PDI-mediated reduction of ricin, while SyH7 and R70 were slightly less effective (Figure 4C). SB1-treated samples had levels of RTA/RTB that were equivalent to PDI-ricin controls (Figure 4C), confirming that non-neutralizing mAbs do not influence PDI’s ability to mediate ricin reduction. As an additional control, we also evaluated RTB-specific neutralizing mAb SylH3 in the PDI-mediated ricin reduction assay. We observed that SylH3 had no demonstrable effect on the appearance of free RTA/RTB, as compared to the ricin only control samples (lane 6, Figure 4B; C). These data are consistent with the fact that SylH3 is proposed to neutralize ricin by blocking toxin attachment to cell surfaces, and not intracellular neutralization.
4. Discussion
The PDI family of redox enzymes and chaperones are central players in regulating protein folding and homeostasis in the ER (Hatahet and Ruddock, 2007). It was only recently, however, that it was discovered that viruses and toxins exploit this family of redox enzymes as a means to gain entry into the cytosol of host cells (Walczak et al., 2012). PDI, for example, serves as an ER-specific unfoldase for cholera toxin (CT) during the process of retrotranslocation (Tsai et al., 2001) and a possible reductase in the case of Pseudomonas exotoxin A (PE) (McKee and FitzGerald, 1999). In the case of ricin, PDI is sufficient to reduce the holotoxin into RTA and RTB, as well as promote the reassociation of the toxin subunits (Spooner et al., 2004). Another PDI family member known as thioredoxin-like transmembrane protein (TMX) has just recently been shown to also play a role in the reductive activation of ricin (Pasetto et al., 2012).
The fact that PDI-mediated reduction of ricin is essential for cytotoxicity, led us to examine the possibility that certain neutralizing mAbs can interfere with this event. Indeed, we found that four RTA-specific neutralizing mAbs, IB2, R70, GD12 and SyH7, were highly effective in inhibiting PDI-mediated reduction of ricin in an in vitro assay. In contrast, two non-neutralizing mAbs, SB1 and FGA12, did not influence the reduction of ricin, even though they are known to target epitopes near the RTA-RTB interface.
We postulate that the observed interference of PDI-mediated reduction of ricin by IB2, R70, GD12 and SyH7 could be due to one of two mechanisms. First, the mAbs could sterically prevent direct contact between PDI and ricin toxin. Although this is likely for IB2 based on the putative localization of its epitope we think this is unlikely to be the case for R70 and GD12 considering they are known to bind epitopes on RTA that are a considerable distance from the RTA-RTB interface. Alternatively, the mAbs could exert allosteric effects on ricin such that when the holotoxin is complexed with IB2, R70, GD12 or SyH7 it is simply a poor substrate for PDI. While we have no direct evidence to suggest that these mAbs do induce conformational changes in ricin or affect PDI recognition of ricin, there are numerous examples in the literature in which antibodies exert effects “at a distance” (Ganesan et al., 2010; Sela-Culang et al., 2012). Such a model is particularly compelling considering that Reynolds and colleagues demonstrated using dihydrofolate reductase (DHFR) as a model that physically contiguous and co-evolving amino acids (“sectors”) represent evolutionarily conserved “hot spots” for allosteric regulation of protein structure and function (Reynolds et al., 2011). If this were to apply to ricin, then perhaps the association of GD12, R70 or SyH7 with key sectors or hot spots of the holotoxin impacts the ability of the toxin to engage with PDI. In this context, it is interesting to note that we have shown that GD12, R70 and SyH7 interfere with RTA’s capacity to inactivate ribosomes in an in vitro translation assay even though those antibodies do not bind near RTA’s active site (Neal et al., 2010; O'Hara et al., 2010). Clearly further experiments are required to understand how PDI and related enzymes interact with ricin and how the association of mAbs with RTA (or RTB) impact ricin conformation and functionality.
Supplementary Material
Acknowledgments
We would like to thank Dr. Karen Chave of the Wadsworth Center’s Protein Expression Core for assistance with monoclonal antibody purification, Richard Cole of the Wadsworth Center’s Advanced Light Microscopy Core for assistance with imaging, and Drs. Jane Kasten-Jolly and Renjie Song of the Wadsworth Center’s Immunology Core for performing the SPR and flow cytometry, respectively. We thank Dr. Anastasiya Yermakova for providing us with SylH3. We thank Dr. David Vance for critical reading of the manuscript. This work was supported by grant AI097688 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baillie LW, Huwar TB, Moore S, Mellado-Sanchez G, Rodriguez L, Neeson BN, Flick-Smith HC, Jenner DC, Atkins HS, Ingram RJ, Altmann DM, Nataro JP, Pasetti MF. An anthrax subunit vaccine candidate based on protective regions of Bacillus anthracis protective antigen and lethal factor. Vaccine. 2010;28:6740–6748. doi: 10.1016/j.vaccine.2010.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellisola G, Fracasso G, Ippoliti R, Menestrina G, Rosen A, Solda S, Udali S, Tomazzolli R, Tridente G, Colombatti M. Reductive activation of ricin and ricin A-chain immunotoxins by protein disulfide isomerase and thioredoxin reductase. Biochem Pharmacol. 2004;67:1721–1731. doi: 10.1016/j.bcp.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Carra JH, Wannemacher RW, Tammariello RF, Lindsey CY, Dinterman RE, Schokman RD, Smith LA. Improved formulation of a recombinant ricin A-chain vaccine increases its stability and effective antigenicity. Vaccine. 2007;25:4149–4158. doi: 10.1016/j.vaccine.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Colombatti M, Pezzini A, Colombatti A. Monoclonal antibodies against ricin: effects on toxin function. Hybridoma. 1986;5:9–19. doi: 10.1089/hyb.1986.5.9. [DOI] [PubMed] [Google Scholar]
- Endo Y, Mitsui K, Motizuki M, Tsurugi K. The mechanism of action of ricin and related toxins on eukaryotic ribosomes. J. Biol. Chem. 1987;262:5908–5912. [PubMed] [Google Scholar]
- Ganesan R, Eigenbrot C, Kirchhofer D. Structural and mechanistic insight into how antibodies inhibit serine proteases. Biochem J. 2010;430:179–189. doi: 10.1042/BJ20100634. [DOI] [PubMed] [Google Scholar]
- Hatahet F, Ruddock LW. Substrate recognition by the protein disulfide isomerases. FEBS J. 2007;274:5223–5234. doi: 10.1111/j.1742-4658.2007.06058.x. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Enzymatic reduction-oxidation of protein disulfides by thioredoxin. Methods Enzymol. 1984;107:295–300. doi: 10.1016/0076-6879(84)07019-1. [DOI] [PubMed] [Google Scholar]
- Lemley PV, Amanatides P, Wright DC. Identification and characterization of a monoclonal antibody that neutralizes ricin toxicity in vitro and in vivo. Hybridoma. 1994;13:417–421. doi: 10.1089/hyb.1994.13.417. [DOI] [PubMed] [Google Scholar]
- Lord JM, Spooner RA. Ricin trafficking in plant and Mammalian cells. Toxins (Basel) 2011;3:787–801. doi: 10.3390/toxins3070787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddaloni M, Cooke C, Wilkinson R, Stout AV, Eng L, Pincus SH. Immunological characteristics associated with the protective efficacy of antibodies to ricin. J. Immunol. 2004;172:6221–6228. doi: 10.4049/jimmunol.172.10.6221. [DOI] [PubMed] [Google Scholar]
- McKee ML, FitzGerald DJ. Reduction of furin-nicked Pseudomonas exotoxin A: an unfolding story. Biochemistry. 1999;38:16507–16513. doi: 10.1021/bi991308+. [DOI] [PubMed] [Google Scholar]
- Meagher MM, Seravalli JG, Swanson ST, Ladd RG, Khasa YP, Inan M, Harner JC, Johnson SK, Van Cott K, Lindsey C, Wannemacher R, Smith LA. Process development and cGMP manufacturing of a recombinant ricin vaccine: An effective and stable recombinant ricin a-chain vaccine-RVEc. Biotechnol Prog. 2011 doi: 10.1002/btpr.631. [DOI] [PubMed] [Google Scholar]
- Neal LM, O’Hara J, Brey RN, 3rd, Mantis NJ. A monoclonal immunoglobulin G antibody directed against an immunodominant linear epitope on the ricin A chain confers systemic and mucosal immunity to ricin. Infect Immun. 2010;78:552–561. doi: 10.1128/IAI.00796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara JM, Brey RN, Mantis NJ. Comparative Efficacy in Mice of Two Lead Candidate Ricin Toxin A Subunit (RTA) Vaccine. Clinical and Vaccine Immunology. 2013 doi: 10.1128/CVI.00098-13. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara JM, Neal LM, McCarthy EA, Kasten-Jolly JA, Brey RN, 3rd, Mantis NJ. Folding domains within the ricin toxin A subunit as targets of protective antibodies. Vaccine. 2010;28:7035–7046. doi: 10.1016/j.vaccine.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara JM, Yermakova A, Mantis NJ. Immunity to ricin: fundamental insights into toxin-antibody interactions. Curr Top Microbiol Immunol. 2012;357:209–241. doi: 10.1007/82_2011_193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasetto M, Barison E, Castagna M, Della Cristina P, Anselmi C, Colombatti M. Reductive activation of type 2 ribosome inactivating proteins (RIPs 2) is promoted by transmembrane thioredoxin-related protein (TMX) J Biol Chem. 2012 doi: 10.1074/jbc.M111.316828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent J, Panigai L, Lamourette P, Sauvaire D, Devilliers K, Plaisance M, Volland H, Creminon C, Simon S. Neutralising antibodies against ricin toxin. PLoS One. 2011;6:e20166. doi: 10.1371/journal.pone.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapak A, Falnes PO, Olsnes S. Retrograde transport of mutant ricin to the endoplasmic reticulum with subsequent translocation to cytosol. Proc Natl Acad Sci U S A. 1997;94:3783–3788. doi: 10.1073/pnas.94.8.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisler RB, Smith LA. The need for continued development of ricin countermeasures. Adv Prev Med. 2012:149737. doi: 10.1155/2012/149737. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds KA, McLaughlin RN, Ranganathan R. Hot spots for allosteric regulation on protein surfaces. Cell. 2011;147:1564–1575. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutenber E, Katzin BJ, Ernst S, Collins EJ, Mlsna D, Ready MP, Robertus JD. Crystallographic refinement of ricin to 2.5 A. Proteins. 1991;10:240–250. doi: 10.1002/prot.340100308. [DOI] [PubMed] [Google Scholar]
- Rutenber E, Ready M, Robertus JD. Structure and evolution of ricin B chain. Nature. 1987;326:624–626. doi: 10.1038/326624a0. [DOI] [PubMed] [Google Scholar]
- Sandvig K, Grimmer S, Lauvrak SU, Torgersen ML, Skretting G, van Deurs B, Iversen TG. Pathways followed by ricin and Shiga toxin into cells. Histochem.Cell Biol. 2002;117:131–141. doi: 10.1007/s00418-001-0346-2. [DOI] [PubMed] [Google Scholar]
- Sela-Culang I, Alon S, Ofran Y. A Systematic Comparison of Free and Bound Antibodies Reveals Binding-Related Conformational Changes. J Immunol. 2012 doi: 10.4049/jimmunol.1201493. [DOI] [PubMed] [Google Scholar]
- Spooner RA, Lord JM. How Ricin and Shiga Toxin Reach the Cytosol of Target Cells: Retrotranslocation from the Endoplasmic Reticulum. Curr Top Microbiol Immunol. 2012;357:19–40. doi: 10.1007/82_2011_154. [DOI] [PubMed] [Google Scholar]
- Spooner RA, Watson PD, Marsden CJ, Smith DC, Moore KA, Cook JP, Lord JM, Roberts LM. Protein disulphide-isomerase reduces ricin to its A and B chains in the endoplasmic reticulum. Biochem J. 2004;383:285–293. doi: 10.1042/BJ20040742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B, Rodighiero C, Lencer WI, Rapoport TA. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 2001;104:937–948. doi: 10.1016/s0092-8674(01)00289-6. [DOI] [PubMed] [Google Scholar]
- van Deurs B, Sandvig K, Petersen OW, Olsnes S, Simons K, Griffiths G. Estimation of the amount of internalized ricin that reaches the trans-Golgi network. J Cell Biol. 1988;106:253–267. doi: 10.1083/jcb.106.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deurs B, Tonnessen TI, Petersen OW, Sandvig K, Olsnes S. Routing of internalized ricin and ricin conjugates to the Golgi complex. J Cell Biol. 1986;102:37–47. doi: 10.1083/jcb.102.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitetta ES, Smallshaw JE, Schindler J. A Small Phase IB Clinical Trial of an Alhydrogel-Adsorbed Recombinant Ricin Vaccine (RiVax) Clin Vaccine Immunol. 2012 doi: 10.1128/CVI.00381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CP, Bernardi KM, Tsai B. Endoplasmic reticulum-dependent redox reactions control endoplasmic reticulum-associated degradation and pathogen entry. Antioxid Redox Signal. 2012;16:809–818. doi: 10.1089/ars.2011.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yermakova A, Mantis NJ. Protective immunity to ricin toxin conferred by antibodies against the toxin's binding subunit (RTB) Vaccine. 2011;29:7925–7935. doi: 10.1016/j.vaccine.2011.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yermakova A, Mantis NJ. Neutralizing activity and protective immunity to ricin toxin conferred by B subunit (RTB)-specific Fab fragments. Toxicon. 2013 doi: 10.1016/j.toxicon.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yermakova A, Vance DJ, Mantis NJ. Sub-Domains of Ricin's B Subunit as Targets of Toxin Neutralizing and Non-Neutralizing Monoclonal Antibodies. PLoS One. 2012;7:e44317. doi: 10.1371/journal.pone.0044317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.