Abstract
Introduction
Organic anion transporting polypeptide (OATP) uptake transporters are important for the disposition of many drugs and perturbed OATP activity can contribute to adverse drug reactions (ADRs). It is well documented that both genetic and environmental factors can alter OATP expression and activity. Genetic factors include single nucleotide polymorphisms (SNPs) that change OATP activity and epigenetic regulation that modify OATP expression levels. SNPs in OATPs contribute to ADRs. Environmental factors include the pharmacological context of drug--drug interactions and the physiological context of liver diseases. Liver diseases such as non-alcoholic fatty liver disease, cholestasis and hepatocellular carcinoma change the expression of multiple OATP isoforms. The role of liver diseases in the occurrence of ADRs is unknown.
Areas covered
This article covers the roles OATPs play in ADRs when considered in the context of genetic or environmental factors. The reader will gain a greater appreciation for the current evidence regarding the salience and importance of each factor in OATP-mediated ADRs.
Expert opinion
A SNP in a single OATP transporter can cause changes in drug pharmacokinetics and contribute to ADRs but, because of overlap in substrate specificities, there is potential for compensatory transport by other OATP isoforms. By contrast, the expression of multiple OATP isoforms is decreased in liver diseases, reducing compensatory transport and thereby increasing the probability of ADRs. To date, most research has focused on the genetic factors in OATP-mediated ADRs while the impact of environmental factors has largely been ignored.
Keywords: adverse drug reaction, drug metabolism, non-alcoholic fatty liver disease, organic anion transport polypeptide, pharmacogenetics
1. Introduction
Organic anion transporting polypeptides (OATPs) are important uptake transporters involved in the pharmacokinetics (PK) of many drugs. OATPs transport drugs into cells of epithelial barriers such as enterocytes, hepatocytes and renal tubule cells, while other families of transporters are required for cellular efflux to complete the transport across the epithelial barrier. Perturbations in OATP-mediated drug uptake from any source can dramatically influence drug PK and pharmacodynamics (PD), manifested as either sub-therapeutic or toxic effects due to altered exposure concentrations and/or times [1,2]. There is emerging evidence to implicate changes in OATP transporter activity in the efficacy and safety of drugs as well as the occurrence of adverse drug reactions (ADRs) [3–5]. In this review, we have classified factors that can impact OATP activity, and therefore hepatic drug uptake, into two general categories: genetic (i.e., pharmacogenetics and epigenetic regulation) and environmental (i.e., pharmacological and physiological context; Figure 1). Areas such as transporter genotype (genetic factor) and drug–drug interactions (DDIs; environmental factor) have received considerable attention in the literature whereas there is a dearth of knowledge regarding epigenetic regulation of transporters and the impact of chronic liver diseases on OATP activity. Although both genetics and environmental effects have been studied in isolation, there is a need for comparative studies to better inform healthcare providers regarding the most salient and influential risk factors to consider when prescribing drugs.
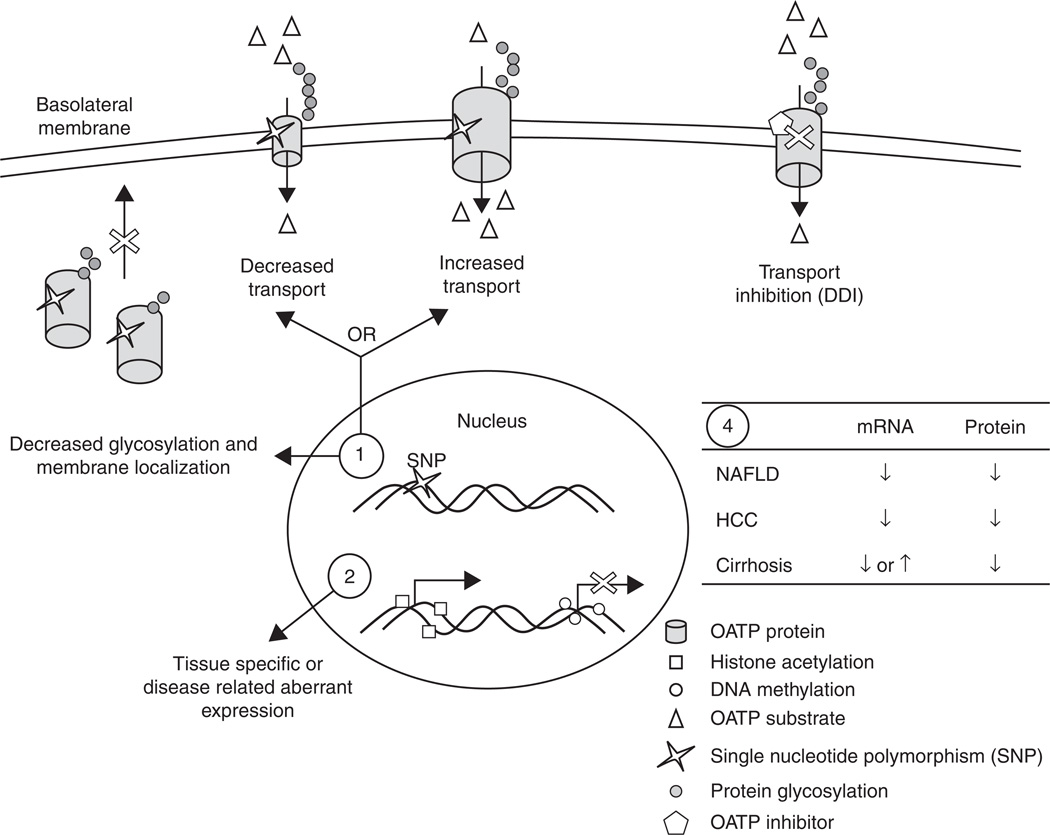
Figure 1. Factors that can affect organic anion transporting polypeptide (OATP) expression or activity and potentially drug uptake.
Genetic factors: 1) single nucleotide polymorphisms (SNPs) in SLCO genes can decrease or increase transport activity of OATP substrates (open triangles). SNPs can also impair protein glycosylation (grey circles) and membrane trafficking. 2) Epigenetic changes such as DNA methylation (open circles) and histone acetylation (open squares) can activate (histone acetylation) or inhibit (DNA methylation) transcription of OATPs. Environmental factors: 3) OATP inhibitors (open pentagon) can interact with drugs that require OATP transport and cause drug--drug interactions (DDIs). 4) Several disease states of the liver such as non-alcoholic fatty liver disease (NAFLD), hepatocellular carcinoma (HCC) or cirrhosis, in most cases, decreases the expression of OATP mRNA and protein, although it has been reported that several OATPs are increased in cholestasis-induced cirrhosis.
2. OATP structure and function
All OATP proteins are membrane proteins predicted to contain 12 transmembrane (TM) helices with the amino and carboxy termini exposed to the cytoplasmic surface of the membrane. These transporters share several common structural characteristics such as a large extracellular loop between TMs 9 and 10, N-glycosylation sites in extracellular loops 2 and 5 and the OATP ‘superfamily signature’ between extracellular loop 3 and TM helice 6 [6]. Due to the rapid and independent identification of OATPs, many OATPs were given ambiguous and inconsistent names which created confusion in the literature. A species-independent nomenclature system was proposed and accepted by the HUGO Gene Nomenclature Committee in which genes are distinguished using the SLCO prefix while proteins are given the OATP prefix [6]. Human genes and proteins are identified by capital letters while rodent genes and proteins are represented with an initial capital letter followed by lower case letters. The OATP superfamily is subdivided into families and subfamilies according to amino acid sequence identity. Each of the six families of OATPs share ≥ 40% sequence identity and are designated by Arabic numerals (i.e., OATP1, OATP2, OAPT3, OAPT4, OAPT5 and OATP6). A subfamily shares ≥ 60% sequence identity and is designated by a letter (e.g., OATP1A, OATP1B, OATP3A, etc.). Within each subfamily Arabic numerals are used to identify individual proteins based on chronology of identification (e.g., OATP1B1, OATP1B3, OATP2B1, etc.).
The OATP transporters mediate the sodium-independent transport of a wide spectrum of amphipathic compounds including bile acids, bilirubin, eicosanoids, steroid and thyroid hormones, prostaglandins, statin drugs, methotrexate, bromosulfophthalein (BSP) and many others [7]. There is overlap in substrate specificity among the different OATPs. For example, pitavastatin is transported by OATP1B1, OATP1A2 and OATP1B3 [1]. Although the precise transport mechanism by OATPs is still largely unknown, it appears to be independent of ATP hydrolysis as well as sodium, potassium and chloride gradients [8], whereas pH gradients can have a significant impact on transport activity [9–12]. The tissue expression profiles and cellular localization (apical or basolateral) of different OATP isoforms varies both within and between species. For example, OATP1B1, OATP1B3 and Oatp1b2 are primarily expressed in the liver and are localized to the basolateral (sinusoidal) membrane of hepatocytes, highlighting the importance of these transporters in hepatic drug uptake [13]. By contrast, human OATP4A1 is expressed in multiple tissues with the highest expression being observed in both lung and placenta [14], whereas Oatp4a1 is expressed primarily in the mouse placenta [15]. For a thorough review of the expression patterns of OATPs, please refer to previously published review articles [6,13]. The diversity of substrates and expression patterns for OATPs within and between species underscores the necessity for appropriate studies for specific OATP substrates with careful consideration when translating results between mice and humans.
3. Factors affecting OATP-mediated drug uptake
3.1 Genetic
3.1.1 Genotype
Single nucleotide polymorphisms (SNPs) in drug metabolizing enzymes and drug transporters can alter drug PK and PD, a field known as pharmacogenetics [3,5,16,17]. A list of SLCO SNPs and their impact on drug transport is shown in Table 1. SNPs in SLCO1B1 and their interaction with statin drugs have been well studied due to the integral role of OATP1B1 in hepatic uptake of statins. More than 41 non-synonymous variants for SLCO1B1 have been identified, although not all of these have been investigated and demonstrated to impact PK. Two relatively common SNPs (c.388A > G and c.521T > C) form four distinct haplotypes (*1A [c.388A-c.521T], *1B [c.388G-c.521T], *5 [c.388A-c.521C] and *15 [c.388G-c.521C]). In healthy Caucasian subjects who are heterozygous or homozygous for SLCO1B1*5, the plasma AUC (area under curve) of simvastatin acid was 120 and 221% higher, respectively, compared with subjects homozygous for SLCO1B1*1A [18]. Similarly, another study reported a 45% higher AUC for pravastatin in subjects heterozygous for SLCO1B1*15 and 92% higher in subjects homozygous for SLCO1B1*15 compared with subjects homozygous for SLCO1B1*1A [19]. By contrast, in a study in healthy Japanese males, subjects with the SLCO1B1*1B genotype had a lower plasma AUC for pravastatin compared with subjects with the SLCO1B1*1A genotype, indicating greater hepatic uptake in subjects with *1B genotype [20]. These data clearly demonstrate that SLCO1B1 SNPs can alter the PK of statin drugs and that the magnitude and direction of response varies between SNPs.
Table 1.
Common OATP SNPs and haplotypes that alter transport activity.
| Transporter | Polymorphisms/ haplotypes |
Substrate studied | Cell line or patient population |
Change due to SNP | Ref. |
|---|---|---|---|---|---|
| OATP1A2 | c.516A > C | Estrone 3-sulfate, Deltorphin II, DPDPE | Transfected HeLa | Decreased transport | [30] |
| c.404A > T | Estrone 3-sulfate, Deltorphin II, DPDPE | Transfected HeLa | Decreased transport | [30] | |
| OATP1B1 | Rs11045879C > T and Rs4149081A > G | Methotrexate | Pediatric patients with acute lymphoblastic leukemia | Increased clearance and GI toxicity with C/T (het) and T/T (homo) polymorphism | [28] |
| c.388A and c.521C (*5) | Estrone 3-sulfate and estradiol 17β-d-glucuronide | Transfected HeLa | Decrease transport | [29] | |
| Simvastatin hydroxyl acid | Healthy Caucasians | Increased AUC | [18] | ||
| c.388G and c.521T (*1B) | Pravastatin | Healthy Japanese men | Decreased AUC | [20] | |
| c.388G and c.521C (*15) | Pravastatin | Healthy mixed population | Increased AUC and Cmax | [19] | |
| OATP1B3 | c.1564G > T | Cholyltaurine | Transfected MDCKII cells | Abolished | [77] |
| c.334T > G and c.699G > A | Testosterone | Transfected Cos-7 cells | Decreased uptake | [26] | |
| OATP2B1 | c.312A > G | DHEAS | LNCaP cells | Increased uptake | [27] |
| c.1457C > T | Fexofenadine | Healthy men | Decreased AUC | [51] |
AUC: Area under curve; OATP: organic anion transporting polypeptide; Ref: Reference; SNP: Single nucleotide polymorphism.
There is also evidence that altered PK caused by SLCO1B1 SNPs can translate to altered therapeutic and adverse drug response. In a retrospective study, patients heterozygous for the SLCO1B1 c.521C allele had a 22.3% attenuation in the total cholesterol lowering action of pravastatin, atorvastatin and simvastatin compared with subjects homozygous for c.521T [21]. In the Heart Protection Study, a randomized placebo control study of 20,536 UK adults, the reduction of low-density lipoprotein (LDL) cholesterol by 40 mg of simvastatin was 1.25 ± 0.25% smaller per copy of c.521C and 0.62 ± 0.18% larger per copy of c.388G [22]. It is noteworthy that these effects are relatively modest and may not translate to dramatic alterations in patient outcomes. However, it has been hypothesized that the increased plasma concentrations of statins caused by SLCO SNPs leads to the increased risk of statin-induced myopathy and several studies have linked simvastatin-induced myopathy with the c.521T > C SNP [22–24]. In a genome-wide association study (GWAS) performed by the SEARCH collaborative group, CT and CC genotypes had a cumulative risk of 3 and 18%, respectively, during the first year. Interestingly, in patients carrying both the c.521C and c.388G SNPs (SLCO1B1*15) there was a reduced risk of myopathy (p = 0.03) [22]. In a follow-up study, it was confirmed that patients with the c.521C SNP taking simvastatin had a threefold increased risk for myopathy. By contrast, there was no association between the c.521C SNP and myopathy in patients taking atorvastatin, indicating that this association is substrate specific [23]. In another GWAS it was reported that occurrence of a composite adverse event (CAE) was associated with the presence of SLCO1B1*5 only in patients taking simvastatin but not in those taking atorvastatin or pravastatin, although a trend toward increased CAE was evident in SLCO1B1*5 patients taking atorvastatin. Perhaps the most interesting result from this study was a gene dosage effect where the proportion of patients with CAE increased with the number of SLCO1B1*5 risk alleles present (0.19, 0.27 and 0.50 for patients with 0, 1 and 2 alleles, respectively) [24]. In the Heart Protection Study, it was also seen that the c.521C SNP was associated with myopathy in patients taking simvastatin with a relative risk for myopathy of 2.6 per SNP copy [22]. Collectively, these data provide strong evidence that SNPs in SLCO1B1 can alter the PK and PD of statin drugs and contribute to the statin-induced ADR of myopathy, setting the precedence for studies involving other substrates of SLCO1B1.
SNPs causing altered OATP transport of endogenous and exogenous compounds can impact the progression and treatment of diseases such as cancer. OATP2B1 and OATP1B3 are two examples of steroid hormone transporters [18], with OATP2B1 shown to transport estrone-3-sulfate and dehydroepiandrosterone sulfate (DHEAS) [14,25] and OATP1B3 shown to transport testosterone [26]. In prostate cancer, continual androgen signaling is required for cancer progression and thus androgen ablation therapy is still a leading treatment. It has been shown that the SLCO2B1 genotype c.312A > G (rs12422149A > G) results in augmented DHEAS uptake, androgen receptor activation and cell proliferation in cell culture [27]. In prostate cancer patients, three SNPS in SLCO2B1, including c.312G, were associated with resistance to androgen depravation therapy as well as between 7- and 12-month decrease in the time to disease progression [27]. A gene dosage effect was observed in these cancer patients such that increasing the number of risk SLCO2B1 SNPs decreases the time to progression, indicating a worse prognosis for patients with SLCO2B1 SNPs [27]. In contrast to the increased uptake of DHEAS with SLCO2B1 SNPs, the combination of SLCO1B3 SNPs c.334T > G and c.699G > A resulted in impaired testosterone uptake in cell culture. Also, although SLCO2B1 SNPs were detrimental to the prognosis in prostate cancer patients due to the increased uptake of DHEAS, patients with the SLCO1B3 haplotype 334GG/699AA showed longer median survival and improved survival probability at 10 years compared with patients with TT/AA and TG/GA haplotypes [26]. In pediatric patients with acute lymphoblastic leukemia two SNPs in SLCO1B1, rs11045879C > T and rs4149081A > G, were in complete linkage disequilibrium with each other and the presence of these SNPs resulted in increased methotrexate clearance. Most importantly, the gastrointestinal (GI) toxicity common in patients receiving methotrexate was more likely to occur in SLCO1B1 polymorphic patients. These two observations are consistent with methotrexate concentrations being lower in plasma and higher in the GI tract and enterocytes, resulting in increased GI toxicity [28]. These data clearly indicate that understanding the interplay between OATP SNPs and drug or hormone action is vital in the context of disease, and considerations need to be taken for both exogenous as well endogenous OATP substrates.
One of the mechanisms by which certain SLCO SNPs alter OATP function is through impeded cellular trafficking of OATP proteins to the plasma membrane. For example, SLCO1B1*5 resulted in decreased plasma membrane expression and Vmax in HeLa cells transfected with expression vectors, indicating that impaired localization decreased the maximal amount of substrate that can be transported [29]. Similarly, SLCO1A2 polymorphisms c.516A > C, c.404A > T and c.2003C > G have been reported to decrease plasma membrane localization and transport activity in transfected HeLa cells [30]. In these studies, it was shown that the c.404A > T SNP resulted in lower glycosylation of OATP1A2, a factor known to impede protein trafficking to the membrane [30]. Although these data provide insight into one of the mechanisms by which SLCO SNPs can alter drug transport, there remains a great need for a better understanding of the influence of specific OATP SNPs on their structure and function. As a whole these data provide strong evidence to suggest that SNPs in drug transporters impact drug PK, PD, efficacy, toxicity and their associated ADRs.
3.1.2 Epigenetic regulation
Epigenetics, or changes in gene expression without changes in the underlying DNA sequence, can also impact the expression of SLCO transporters [31]. One way this occurs is by variable gene promoter activity caused by changes in DNA methylation and histone modifications. Methylation provides a docking site for methyl binding proteins which in turn recruit co-repressor complexes containing histone deacetylase enzymes facilitating the removal of acetyl groups on histone tails. Unmethylated DNA and acetylated DNA are both associated with actively transcribed DNA. It has been shown that epigenetic regulation is responsible for tissue-specific expression of drug transporters [32]. For example, Slco1b2 was primarily expressed in the liver where its promoter was hypomethylated and hyperacetylated, whereas in the kidney it was not expressed and its promoter was hyper-methylated and hypoacetylated [32]. Epigenetic regulation of SLCO genes also plays an important role in pathologies such as cancer. For example, SLCO2A1 expression was low in colon cancer cell lines but was restored after treatment with 5-aza-2′-deoxycytidine, a demethylating agent, or trichostatin A (TSA), a histone deacetylase inhibitor [33]. It has also been reported that there was an association between the promoter methylation and expression levels of SLCO1B3 in a panel of cancer cell lines. In these cell lines the levels of methylation and expression correlated with each other, but were different across lines. In HepG2 and Caco-2 cell lines, which had low expression of SLCO1B3 and high DNA methylation, treatment with 5-aza-2′-deoxycytidine induced expression of SLCO1B3 by 18- and 14- fold, respectively, further connecting changes in DNA methylation with transporter expression [34]. These data illuminate the potential for cancer-related epigenetic aberrations in transporter expression to impact chemotherapeutic efficacy of drugs that are substrates for SLCO transport. It is noteworthy that the dynamic nature of epigenetic modification makes the elucidation of common changes in gene expression difficult. Perhaps the most promising way epigenetics can be applied in SLCO expression is in relation to specific drugs and/or disease states known to modify the epigenetic status of their promoters. By utilizing high-throughput epigenomic analyses and bioinformatics techniques, epigenetic regulation of OATP transporters will become clearer.
3.2 Environmental
3.2.1 Pharmacologic context
DDIs occur when the presence of one drug affects the PK and/or PD of another drug. Many drugs are known to inhibit OATP-mediated drug uptake and contribute to DDIs [35]. One well-studied example is cyclosporin A, a common immunosuppressant used after organ transplants to prevent organ rejection. Lipid abnormalities are common side effects observed in patients who receive heart or kidney transplants [36,37] and therefore many transplant patients are prescribed statin drugs. As discussed above, many statin drugs are substrates for OATP transporters and an interaction between cyclosporin A and statins has been observed. Cyclosporin A has been shown to inhibit OATP1B1-mediated uptake of cerivastatin into primary human hepatocytes in vitro with Ki values between 0.3 and 0.7 µM. Furthermore, uptake of cerivastatin into OATP1B1-expressing MDCKII cells was inhibited by cyclosporin A with a Ki value of 0.2 µM, indicating that OATP1B1 is at least in part responsible for the uptake of cerivastatin and that cyclosporin A can inhibit its transport [38]. Cyclosporin A can also potently inhibit OATP1B1-mediated transport of pitavastatin in transfected MDCKII cells [39], indicating that it is not a substrate-specific inhibition. In LLC-PK1 cells over-expressing rat Oatp1a1 and Oatp1a4, 30-µM cyclosporin A inhibited > 80% of the transport activity of each isoform [40]. Many other OATP substrates have been reported to be altered by cyclosporin A-mediated OATP inhibition including bosentan, a dual endothelin receptor antagonist for the treatment of pulmonary arterial hypertension. Cyclosporin A caused 4.4- and 17-fold higher Cmax and AUC of bosentan in rats which was attributed to inhibition of Oatp1a1 and Oatp1a4 [41]. These data show that cyclosporin A-mediated inhibition of multiple OATPs is the cause of DDIs between cyclosporin A and OATP substrates.
The DDI between cyclosporin A and statin drugs has also been observed in the clinical setting. Cyclosporin A caused a 20-fold increase in plasma AUC and a sevenfold increase in the Cmax of pravastatin in heart transplant patients [42]. Pediatric heart transplant patients who received triple immunosuppressive therapy including cyclosporin A had approximately a 10-fold higher AUC and Cmax of pravastatin compared with patients who did not receive immunosuppressive therapy. Although a dramatic increase in AUC and Cmax was observed, patients who received cyclosporin A had a similar reduction in total and LDL cholesterol compared with control patients, indicating that the greater exposure did not result in altered PD [43]. In renal transplant patients, Cmax and AUC for simvastatin were increased nearly eightfold when co-administered with cyclosporin A. Again, somewhat surprisingly, liver function and creatinine phosphokinase, both indicators of the common statin ADRs myopathy and rhabdomyolysis, were not changed and no ADRs were reported that were attributable to simvastatin treatment [44]. By contrast, a retrospective analysis of statin-induced rhabdomyolysis, the most severe form of myopathy which, if left untreated, can lead to complications such as acute renal failure or cardiac arrest [45], indicated that interaction with cyclosporin A may have contributed to 8.5% of the cases [46]. Because of these changes in PK and the potential for ADRs it is recommended that transplant patients receive the lowest possible dose of the statin drug, and if rhabdomyolysis occurs, both cyclosporin A and statin should be discontinued [47].
Consumption of several different fruit juices has been reported to influence the uptake and disposition of OATP substrates. For example, naringin is a glycoside found in grapefruit juice that is reported to inhibit OATP drug transport. It has been shown in vitro that 5 µmol/l naringin inhibited rat Oatp1a5-mediated uptake of the antihistamine drug fexofenadine by 64%, and 3.6 µmol/l naringin completely inhibited human OATP1A2-mediated transport of fexofenadine [48,49]. In contrast to cyclosporin A where drug AUC is increased with concomitant administration of OATP substrate and inhibitor, it was demonstrated in healthy human subjects that whole grapefruit juice or aqueous naringin reduced the AUC of fexofenadine by 55 and 75%, respectively [49]. Because the tmax of fexofenadine was not altered in these subjects yet the AUC was significantly decreased, the authors postulated that the naringin-mediated changes in AUC resulted from decreased oral fexofenadine bioavailability. In another study it was eloquently shown that naringin-mediated inhibition of intestinal OATP1A2 was responsible for the decreased oral fexofenadine bioavailability [50]. It has also been reported that concomitant consumption of apple juice with fexofenadine resulted in an 80% reduction in plasma AUC of fexofenadine. In vitro data from the same study showed that OATP2B1 could efficiently transport fexofenadine and that the presence of apple juice decreased its uptake by > 60%. Interestingly, it was shown that subjects carrying the c.1457C > T SNP also had decreased plasma AUC of fexofenadine and when these subjects consumed apple juice with fexofenadine an even greater decrease in AUC was observed [51]. The relative contribution of OATP1A2 and OATP2B1 to the intestinal uptake of various drugs requires further investigation to clarify the importance of each. In the case of naringin and apple juice, inhibition of an OATP isoform in the intestine caused a decrease in systemic drug concentrations, potentially leading to sub-therapeutic doses and altered PD. Collectively, these data indicate that DDIs involving OATPs can dramatically alter the systemic exposure of certain drugs and potentially lead to either altered PD and/or ADRs.
3.2.2 Physiologic context
The liver is the primary site of metabolism within the body and liver diseases such as non-alcoholic fatty liver disease (NAFLD), cirrhosis and hepatocellular carcinoma (HCC) can change the physiology of the liver and reduce its metabolic capacity [52,53]. The primary mechanism by which liver physiology can impact OATP-mediated drug uptake is by altering OATP expression. NAFLD is an increasingly prevalent liver disease reported to affect approximately one-third of the US population and it encompasses a range of disease states from simple steatosis to non-alcoholic steatohepatitis (NASH) [54]. There is growing evidence to implicate NAFLD, and particularly NASH, in altered OATP expression and function. For example, expression of Oatp1a1, Oatp1a4, Oatp1b2 and Oatp2b1 mRNA was decreased in rat models of steatosis and NASH, with the greatest reduction in expression observed in NASH. At the protein level, both Oatp1a1 and Oatp1b2 were reduced in steatosis and NASH. Importantly, the decreases in Oatp1a1 and Oatp1b2 expression in the NASH model were implicated in decreased plasma clearance of BSP, a known substrate for these OATPs [55]. In support of these findings in rodent models, a recent study in human NAFLD progression reported that uptake transporters are significantly down-regulated in the transition from steatosis to NASH [56], potentially contributing to altered drug disposition for OATP substrates in the clinical setting. Because of the significant prevalence of NAFLD and its association with multiple comorbidities such as obesity, cardiovascular disease and type 2 diabetes, these data highlight the importance of accounting for OATP function in the context of NAFLD and drug metabolism.
Bile duct ligation and lipopolysaccharide (LPS) induced models of intra- and extrahepatic cholestasis in rodents leads to substantial liver injury and eventually fibrosis and cirrhosis. Altered OATP expression and drug disposition has been reported in both mice and rat bile duct ligation cholestasis models. In various mouse strains, mRNA for Oatp1a1 was down-regulated 72 h after bile duct ligation but, by contrast, both Oatp1a4 and Oatp3a1 were up-regulated [57]. Another report in a rat bile duct ligation model, Oatp1a1 and Oatp1a4 were down-regulated after 15 days, although no change in uptake of a putative OATP substrate, gadobenate dimeglumine (Gd-BOPTA), was evident. In this study, the authors speculated that due to decreased Mrp2 expression concomitant with decreased OATP expression similar amounts of Gd-BOPTA were present within control and cirrhotic hepatocytes [58]. Also, because the specific transporters involved in Gd-BOPTA uptake have not been characterized, it is possible that other uptake transporters that were not changed in cirrhosis are involved. The expression differences between these two rodent bile duct ligation models of extrahepatic cholestasis are potentially due to differences in animal species (mouse vs rat) or timing of measurement (3 vs 15 days), although further investigation is needed to clarify these data. LPS-induced intrahepatic cholestasis resulted in down-regulation of Oatp1a1, Oatp1a4 and Oatp1b2 in both mice and rats between 6 and 12 h after LPS administration [59–61]. In the case of Oatp1b2, LPS down-regulation was found to occur through toll-like receptor 4 [62] but was independent of TNF-α, IL-1β, IL-6 or inducible nitric oxide synthase [63]. In another study, Oatp1a1, Oatp1a4 and Oatp1b2 were down-regulated in various mouse strains 16 h after LPS administration but, by contrast, Oatp3a1 and Oatp1c1 were up-regulated [57]. Because many of the examples discussed thus far involve decreased OATP expression or function, it is interesting to note that several OATPs were up-regulated in both models of cholestasis potentially leading to decreased plasma drug concentrations, more rapid drug clearance and sub-therapeutic drug exposures. These data show altered OATP expression in the physiological context of bile duct ligation and LPS models of cholestasis as a possible confounding factor in drug PK.
HCC is another liver disease that has been reported to alter the expression and function of OATPs resulting in changes in drug clearance. Multiple reports in the context of HCC have shown reduced mRNA and protein expression of the principle OATP uptake transporters found in the liver, namely OATP1B1, OATP1B3 and OATP2B1. For example, reduced expression of OATP1B1 and OATP1B3 has been reported in various HCC cell lines and tissue samples [64–68]. Furthermore, OATP1B1 and OATP2B1 protein expression have been reported to be lower in liver cancer compared with healthy tissues. In these samples, the decrease in OATP1B1 trended with lower cancer grade while the decrease in OATP2B1 trended with decreased cellular differentiation [69], indicating that as HCC progresses these OATPs are down-regulated. Decreased expression of OATP1B1 and OATP1B3 in HCC has been implicated in the decreased uptake of the gadoxetic acid, a liver-specific contrast agent used in magnetic resonance imaging [70]. Because HCC is a pervasive disease comprising one-third of cancer-related deaths worldwide, these data have potential implications for the transport and disposition of endogenous and exogenous OATP substrates in the context of HCC. Collectively, the environmental factors of DDI and pathophysiology have great potential to influence the role of OATPs in drug metabolism.
4. Conclusion
Transepithelial transport of drugs requires membrane-bound transporters that facilitate entry into and exit out of cells in epithelial barriers. In this review, we have discussed multiple factors that influence OATP-mediated drug uptake, thereby contributing to altered PK and PD of various drugs. These changes in drug concentrations can have far-reaching implications in the clinic due to sub-therapeutic or toxic doses, potentially leading to off-target effects such as ADRs. From the literature reviewed we conclude that both genetic (pharmacogenetic and epigenetic) and environmental (pharmacologic and physiologic) factors influence the role of OATP transporters in drug metabolism as summarized in Figure 1. Genetic factors include SNPs in OATPs that change transport activity and epigenetic modifications that regulate OATP expression in a disease and/or tissue-specific manner. Environmental factors include DDIs, particularly OATP inhibitors, and liver pathophysiology such as NAFLD, HCC and cirrhosis that change transporter expression or activity. Because OATPs play a vital role in the disposition of many endogenous and exogenous compounds, these factors may ultimately have a significant effect on the physiological and pharmacological action of these compounds leading to adverse outcomes. Expanding our understanding of the comparative roles played by genetics and environment on OATP activity is important and will aid clinicians when prescribing drugs or treating patients in cases where one or more of these factors are involved.
5. Expert opinion
Identifying alterations in drug metabolism and disposition, regardless of the source of that variation, is of paramount concern in our attempt to safely and effectively treat each patient. Accounting for the impact of both genetic and environmental factors in this variation is an important step in reducing the number of ADRs. In 2010, the Adverse Events Reporting System maintained by the US Food and Drug Administration reported 82,724 deaths and 471,291 serious outcomes which were defined as hospitalization, life-threatening, disability, congenital anomaly and/or other serious outcomes [71]. A recent study on ADRs performed at the Royal Liverpool University Hospital concluded that one in seven hospital inpatients experience an ADR [72]. With such a high occurrence, there is need for greater understanding of the mechanism and risk factors associated with ADRs.
There is a paucity of direct information regarding whether genetic or environmental factors contribute more to altered PK, PD and ADRs. One of the greatest hindrances to the progress of this work is the lack of research on the role of environmental factors in OATP-mediated drug transport. Recently, the association between altered OATP function and disease state has been suggested to potentially result in increased toxicity and ADRs for various drugs [8], but the experimental data to support this are lacking. The International Transporter Consortium (ITC), a group focused on investigating the role of drug transporters in therapeutic and adverse drug response, has made great strides in forming a consensus regarding the role of transporters in drug development [73,74]. Unfortunately, the ITC has focused primarily on pharmacogenetics and DDIs involving OATPs while the effects of disease status on OATP activity has largely been ignored. The burgeoning interest in how physiological status impacts the expression, regulation and function of OATP transporters will help define the relevance of disease state when prescribing drugs.
A great deal of effort has been focused on identifying SNPs that contribute to ADRs through genome-wide studies. These types of studies have identified many of the SNPs in SLCO genes discussed here. Importantly, there are several shortcomings and considerations that need to be discussed for these types of data. First of all, many SNPs that are associated with ADRs identified in GWAS are at intronic or intergenic locations in the genome which do not code for any known proteins, and therefore identifying the biological significance of these SNPs is difficult [75]. To overcome these shortcomings it is imperative that SNPs identified from GWAS be confirmed molecularly and pharmacologically. It is also important to note that GWAS do not capture environmental factors that can change OATP expression, such as changes in liver physiology, DDIs or transporter localization. As an example, it has been reported that the membrane localization and function of ABCC2, an efflux transporter, is impeded in NAFLD progression due to changes in glycosylation status [76]. This type of change would not be identified in GWAS analyses. In spite of shortcomings in the methods used to identify SNPs that contribute to ADRs, it is clear that SNPs play an important role in OATP-mediated ADRs. However, it is equally imperative that environmental factors that alter OATP function be identified and accounted for in OATP-mediated ADRs.
When considering whether genetics or environment is more influential in OATP-mediated ADRs there are several key points that need to be discussed. Most importantly, OATPs have overlapping substrate specificity and therefore genetic polymorphisms resulting in reduced function of one isoform could be compensated for by other isoforms that transport the same drug. In the case of SLCO SNPs, only individual SLCO genes are affected by each SNP and therefore there is great potential for compensatory transport by other fully functional OATPs (Figure 2B). By contrast, environmental factors such as liver diseases may downregulate multiple OATPs, thereby reducing the capacity for compensatory transport (Figure 2C). The cumulative effect of multiple transporters being down-regulated in liver diseases could potentially have a greater impact on drug PK, PD and OATP-mediated ADRs than reduction in activity of individual OATPs by SNPs.
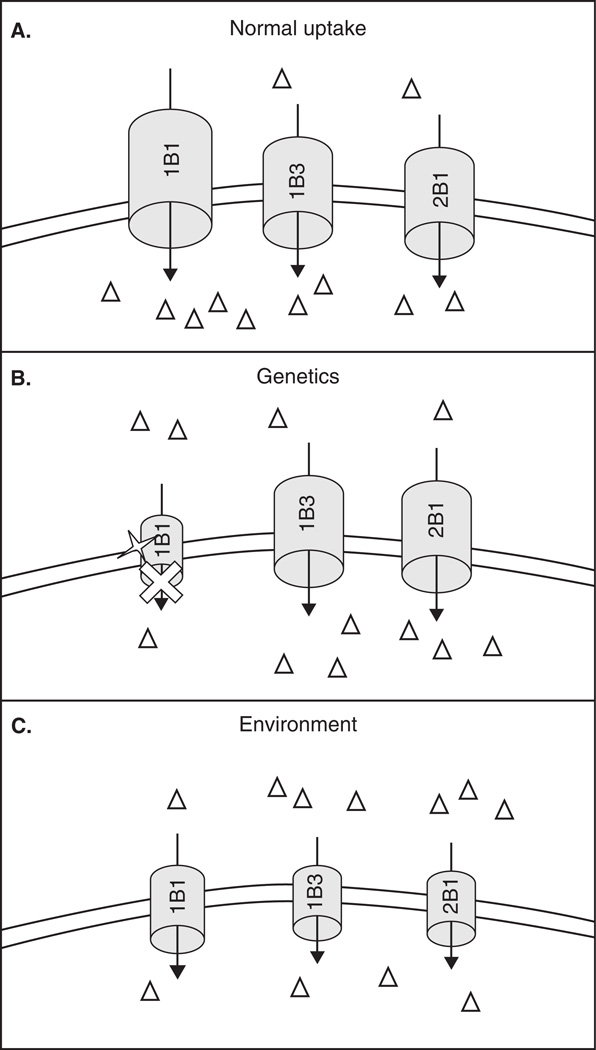
Figure 2. Representation of the influence of genetics vs environment on the disposition of OATP substrates.
A. In healthy liver, an OATP substrate (open triangle) is transported across the sinusoidal membrane into the hepatocyte predominately by one OATP isoform but other isoforms can potentially participate. B. A single nucleotide polymorphism (open star) in a particular OATP isoform (OATP1B1 in this case) results in lower transport capacity of that isoform. Due to overlapping substrate specificity, the OATP substrate will still be transported into the hepatocyte by other OATP isoforms, although the uptake will be less than normal. C. Environmental factors can decrease the expression of multiple OATP isoforms, thereby reducing the capacity for compensatory transport and potentially result in a greater difference in overall transport of an OATP substrate compared with transport in the context of an OATP SNP.
Another point to consider when discussing whether genetic or environmental factors contribute more to OATP-mediated ADRs is the prevalence of each factor in the population. In the case of genetic factors, the occurrence of each SNP varies in different populations (Table 2) and how each SNP affects PK differs in both direction and magnitude. For example, as discussed above, the AUC of simvastatin acid was 120 and 221% higher in subjects heterozygous and homozygous for SLCO1B1*5 but, importantly, the allele frequency of this SNP is only reported to be 14% in European Americans and 2% in African Americans. The SLCO1B1*15 SNP had a smaller effect on plasma AUC of pravastatin (45 and 92% higher in heterozygous and homozygous subjects, respectively) and its allele frequency is reported to be slightly higher at 24% in America, 16% in Europe, 16% in North Africa and 9% in north/central Asia. Arguably among all of the SNPs these two are most pertinent to altered disposition of statins but the prevalence in the population is relatively low and therefore may not account for all of the OATP-mediated ADRs. SNPs c.334T > G and c.699G > A in OATP1B3 are very common in several populations (Table 2) but these SNPs do not change the transport of several substrates in vitro [77] and, therefore, likely do not contribute to OATP-mediated ADRs. In the case of environmental factors, the prevalence of liver diseases such as NAFLD, biliary cirrhosis and HCC also varies and may represent specific populations that are at higher risk for ADRs associated with altered OATP function. Importantly, the prevalence of NAFLD is increasing and has been estimated to be present in 30 – 40% of all adults and up to 90% of obese adults in the USA, potentially impacting drug disposition in these people [78]. Primary biliary cirrhosis, another liver disease discussed herein, is estimated to be present in 1 in 1000 women over the age of 40 [79]. Also, the incidence of HCC in the USA tripled from 1975 to 2005 [80] and the American Cancer Society estimates that 26,190 new cases of liver and intrahepatic bile duct cancers will be diagnosed in 2011. Although the latter two liver diseases are not common, all three diseases may represent populations at a higher risk for ADRs associated with OATP substrates. Together these data on the prevalence of genetic and environmental factors indicate that some populations will be at greater risk but, importantly, no single factor has 100% penetrance and therefore each factor will likely contribute to the occurrence of OATP-associated ADRs. Furthermore, interaction between multiple factors can compound changes in PK and PD. For example, an interaction between pharmacogenomic and pharmacologic context has been reported. Patients who received rifampin, a known inhibitor of OATB1B1, had increased AUC of atorvastatin which was dependent on the SLCO1B1 genotype (nine-, six- and fourfold increase in individuals with c.521TT, c.521TC and c.521CC genotype, respectively) [81]. These types of interactions between genetic and environment are to be expected and need to be considered in order to account for the occurrence of all OATP-mediated ADRs.
Table 2.
OATP SNP allele frequencies (%).
| EA | AA | AM | HA | EU | AF | HC | ME | AS | FI | Ref. | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| OATP1A2 | |||||||||||
| c.516A > C | - | 2.1 | - | 5.7 | 5.3 | - | - | - | - | - | [30] |
| c.404A > T | - | 0 | - | 0 | 0 | - | - | - | - | - | [30] |
| OATP1B1 | |||||||||||
| c.521C (*5) | 14 | 2 | - | - | 2 | 2 | - | - | - | - | [29,82] |
| c.388G and c.521T (*1B) | - | - | 39 | - | 26 | 48 | - | - | 39 | - | [82] |
| c.388G and c.521C (*15) | - | - | 24 | - | 16 | 16 | - | - | 9 | - | [82] |
| OATP1B3 | |||||||||||
| c.1564G > T | - | - | 1.9 | - | - | - | - | - | [77] | ||
| c.334T > G | 88 | 41 | - | - | 81 | - | 80 | 78 | - | [83] | |
| c.699G > A | 87 | 41 | - | - | 83 | - | 77 | 79 | - | [83] | |
| OATP2B1 | |||||||||||
| c.935G > A | - | 13 | - | - | - | - | - | - | - | 13.6 | [84] |
| c.1457C > T | - | - | - | - | - | - | - | - | - | 2.8 | [84] |
AA: African American; AF: North African population; AM: Population in America as defined in Pasanen et al. [82]; AS: South/central Asian population; EA: European American; EU: European Caucasian; FI: Finnish; HA: Hispanic American; HC: Han Chinese; ME: Mexican; OATP: Organic anion transporting polypeptide; (−): Not determined; Ref: References; SNP: Single nucleotide polymorphism.
Continued research on the role of the various genetic and environmental factors, as well as the interplay between them, will solidify the role of OATPs in drug metabolism and ADRs. Currently, there is a shortage of studies that have directly tested the role environmental factors play in altered OATP-mediated drug PK and PD, and how these changes impact ADRs. This is an essential area of future research because environmental factors that impact OATP function potentially carry more gravity than genetic factors. Although studies designed to investigate each factor in isolation strengthen the foundational understanding of how these factors impact OATP function and thus drug PK and PD, studies designed to directly compare which factors are most influential will provide valuable information regarding potential at risk populations (SNPs vs pathophysiology) for altered drug metabolism and action. Such comparative studies are sorely needed and should be encouraged at every level.
Article highlights.
Genetic (pharmacogenetics and epigenetics) and environmental (pharmacological and physiological context) factors cause changes in organic anion transporting polypeptide (OATP) expression and activity.
OATP single nucleotide polymorphisms (SNPs) are known to cause altered drug pharmacokinetics (PK) and contribute to adverse drug reactions (ADRs).
The role of physiological factors such as liver diseases in OATP-mediated ADRs is unknown.
Liver diseases can potentially cause dramatic changes in PK and contribute to the incidence of ADRs, because multiple OATP isoforms are down-regulated in liver disease reducing the capacity for compensatory transport.
More research is needed to better understand how environmental factors affect OATP-mediated drug metabolism in the occurrence of ADRs.
This box summarizes key points contained in the article.
Footnotes
Declaration of interest
NJ Cherrington is the holder of a patent licensed to Raptor Pharmaceuticals to diagnose non-alcoholic steatohepatitis. JD Clarke declares that he has no conflict of interest and has received no payment in the preparation of this manuscript.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1. Kalliokoski A, Niemi M. Impact of OATP transporters on pharmacokinetics. Br J Pharmacol. 2009;158:693–705. doi: 10.1111/j.1476-5381.2009.00430.x. • A review on pharmacogenetics and drug-drug interactions of multiple OATP isoforms.
- 2.Kalliokoski A, Neuvonen M, Neuvonen PJ, Niemi M. Different effects of SLCO1B1 polymorphism on the pharmacokinetics and pharmacodynamics of repaglinide and nateglinide. J Clin Pharmacol. 2008;48:311–321. doi: 10.1177/0091270007311569. [DOI] [PubMed] [Google Scholar]
- 3.Lee NH. Pharmacogenetics of drug metabolizing enzymes and transporters: effects on pharmacokinetics and pharmacodynamics of anticancer agents. Anticancer Agents Med Chem. 2010;10:583–592. doi: 10.2174/187152010794474019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giorgi MA, Caroli C, Arazi HC, Di GG. Pharmacogenomics and adverse drug reactions: the case of statins. Expert Opin Pharmacother. 2011;12:1499–1509. doi: 10.1517/14656566.2011.563734. [DOI] [PubMed] [Google Scholar]
- 5.Zair ZM, Eloranta JJ, Stieger B, Kullak-Ublick GA. Pharmacogenetics of OATP (SLC21/SLCO), OAT and OCT (SLC22) and PEPT (SLC15) transporters in the intestine, liver and kidney. Pharmacogenomics. 2008;9:597–624. doi: 10.2217/14622416.9.5.597. [DOI] [PubMed] [Google Scholar]
- 6. Hagenbuch B, Meier PJ. Organic anion transporting polypeptides of the OATP/SLC21 family: phylogenetic classification as OATP/SLCO superfamily, new nomenclature and molecular/functional properties. Pflugers Arch. 2004;447:653–665. doi: 10.1007/s00424-003-1168-y. •• A meritorious paper that standardized the previously confusing OATP nomenclature system.
- 7.Hagenbuch B, Gui C. Xenobiotic transporters of the human organic anion transporting polypeptides (OATP) family. Xenobiotica. 2008;38:778–801. doi: 10.1080/00498250801986951. [DOI] [PubMed] [Google Scholar]
- 8.Svoboda M, Riha J, Wlcek K, et al. Organic anion transporting polypeptides (OATPs): regulation of expression and function. Curr Drug Metab. 2011;12:139–153. doi: 10.2174/138920011795016863. [DOI] [PubMed] [Google Scholar]
- 9.Kanai N, Lu R, Bao Y, et al. Transient expression of oatp organic anion transporter in mammalian cells: identification of candidate substrates. Am J Physiol. 1996;270:F319–F325. doi: 10.1152/ajprenal.1996.270.2.F319. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi D, Nozawa T, Imai K, et al. Involvement of human organic anion transporting polypeptide OATP-B (SLC21A9) in pH-dependent transport across intestinal apical membrane. J Pharmacol Exp Ther. 2003;306:703–708. doi: 10.1124/jpet.103.051300. [DOI] [PubMed] [Google Scholar]
- 11.Leuthold S, Hagenbuch B, Mohebbi N, et al. Mechanisms of pH-gradient driven transport mediated by organic anion polypeptide transporters. Am J Physiol Cell Physiol. 2009;296:C570–C582. doi: 10.1152/ajpcell.00436.2008. [DOI] [PubMed] [Google Scholar]
- 12.Nozawa T, Imai K, Nezu J, et al. Functional characterization of pH-sensitive organic anion transporting polypeptide OATP-B in human. J Pharmacol Exp Ther. 2004;308:438–445. doi: 10.1124/jpet.103.060194. [DOI] [PubMed] [Google Scholar]
- 13.Klaassen CD, Aleksunes LM. Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev. 2010;62:1–96. doi: 10.1124/pr.109.002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamai I, Nezu J, Uchino H, et al. Molecular identification and characterization of novel members of the human organic anion transporter (OATP) family. Biochem Biophys Res Commun. 2000;273:251–260. doi: 10.1006/bbrc.2000.2922. [DOI] [PubMed] [Google Scholar]
- 15.Cheng X, Maher J, Chen C, Klaassen CD. Tissue distribution and ontogeny of mouse organic anion transporting polypeptides (Oatps) Drug Metab Dispos. 2005;33:1062–1073. doi: 10.1124/dmd.105.003640. [DOI] [PubMed] [Google Scholar]
- 16.Kerb R. Implications of genetic polymorphisms in drug transporters for pharmacotherapy. Cancer Lett. 2006;234:4–33. doi: 10.1016/j.canlet.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 17. Niemi M, Pasanen MK, Neuvonen PJ. Organic anion transporting polypeptide 1B1: a genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol Rev. 2011;63:157–181. doi: 10.1124/pr.110.002857. •• A comprehensive review of OATP1B1 pharmacogenetics.
- 18.Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet Genomics. 2006;16:873–879. doi: 10.1097/01.fpc.0000230416.82349.90. [DOI] [PubMed] [Google Scholar]
- 19.Ho RH, Choi L, Lee W, et al. Effect of drug transporter genotypes on pravastatin disposition in European- and African-American participants. Pharmacogenet Genomics. 2007;17:647–656. doi: 10.1097/FPC.0b013e3280ef698f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda K, Ieiri I, Yasuda K, et al. Effects of organic anion transporting polypeptide 1B1 haplotype on pharmacokinetics of pravastatin, valsartan, and temocapril. Clin Pharmacol Ther. 2006;79:427–439. doi: 10.1016/j.clpt.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Tachibana-Iimori R, Tabara Y, Kusuhara H, et al. Effect of genetic polymorphism of OATP-C (SLCO1B1) on lipid-lowering response to HMG-CoA reductase inhibitors. Drug Metab Pharmacokinet. 2004;19:375–380. doi: 10.2133/dmpk.19.375. [DOI] [PubMed] [Google Scholar]
- 22.Link E, Parish S, Armitage J, et al. SLCO1B1 variants and statin-induced myopathy -- a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 23.Brunham LR, Lansberg PJ, Zhang L, et al. Differential effect of the rs4149056 variant in SLCO1B1 on myopathy associated with simvastatin and atorvastatin. Pharmacogenomics J. 2011 doi: 10.1038/tpj.2010.92. published online 18 Jan, 2011. [DOI] [PubMed] [Google Scholar]
- 24.Voora D, Shah SH, Spasojevic I, et al. The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol. 2009;54:1609–1616. doi: 10.1016/j.jacc.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pizzagalli F, Varga Z, Huber RD, et al. Identification of steroid sulfate transport processes in the human mammary gland. J Clin Endocrinol Metab. 2003;88:3902–3912. doi: 10.1210/jc.2003-030174. [DOI] [PubMed] [Google Scholar]
- 26.Hamada A, Sissung T, Price DK, et al. Effect of SLCO1B3 haplotype on testosterone transport and clinical outcome in caucasian patients with androgen-independent prostatic cancer. Clin Cancer Res. 2008;14:3312–3318. doi: 10.1158/1078-0432.CCR-07-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang M, Xie W, Mostaghel E, et al. SLCO2B1 and SLCO1B3 may determine time to progression for patients receiving androgen deprivation therapy for prostate cancer. J Clin Oncol. 2011;29:2565–2573. doi: 10.1200/JCO.2010.31.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trevino LR, Shimasaki N, Yang W, et al. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J Clin Oncol. 2009;27:5972–5978. doi: 10.1200/JCO.2008.20.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tirona RG, Leake BF, Merino G, Kim RB. Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J Biol Chem. 2001;276:35669–35675. doi: 10.1074/jbc.M103792200. [DOI] [PubMed] [Google Scholar]
- 30.Lee W, Glaeser H, Smith LH, et al. Polymorphisms in human organic anion-transporting polypeptide 1A2 (OATP1A2): implications for altered drug disposition and central nervous system drug entry. J Biol Chem. 2005;280:9610–9617. doi: 10.1074/jbc.M411092200. [DOI] [PubMed] [Google Scholar]
- 31.Kacevska M, Ivanov M, Ingelman-Sundberg M. Perspectives on epigenetics and its relevance to adverse drug reactions. Clin Pharmacol Ther. 2011;89:902–907. doi: 10.1038/clpt.2011.21. [DOI] [PubMed] [Google Scholar]
- 32.Imai S, Kikuchi R, Kusuhara H, et al. Analysis of DNA methylation and histone modification profiles of liver-specific transporters. Mol Pharmacol. 2009;75:568–576. doi: 10.1124/mol.108.052589. [DOI] [PubMed] [Google Scholar]
- 33.Holla VR, Backlund MG, Yang P, et al. Regulation of prostaglandin transporters in colorectal neoplasia. Cancer Prev Res (Phila) 2008;1:93–99. doi: 10.1158/1940-6207.CAPR-07-0009. [DOI] [PubMed] [Google Scholar]
- 34.Ichihara S, Kikuchi R, Kusuhara H, et al. DNA methylation profiles of organic anion transporting polypeptide 1B3 in cancer cell lines. Pharm Res. 2010;27:510–516. doi: 10.1007/s11095-010-0064-3. [DOI] [PubMed] [Google Scholar]
- 35.Shitara Y, Sato H, Sugiyama Y. Evaluation of drug-drug interaction in the hepatobiliary and renal transport of drugs. Annu Rev Pharmacol Toxicol. 2005;45:689–723. doi: 10.1146/annurev.pharmtox.44.101802.121444. [DOI] [PubMed] [Google Scholar]
- 36.Lindenfeld J, Page RL, Zolty R, et al. Drug therapy in the heart transplant recipient: part III: common medical problems. Circulation. 2005;111:113–117. doi: 10.1161/01.CIR.0000151609.60618.3C. [DOI] [PubMed] [Google Scholar]
- 37.Kasiske BL, Umen AJ. Persistent hyperlipidemia in renal transplant patients. Medicine (Baltimore) 1987;66:309–316. doi: 10.1097/00005792-198707000-00003. [DOI] [PubMed] [Google Scholar]
- 38.Shitara Y, Itoh T, Sato H, et al. Inhibition of transporter-mediated hepatic uptake as a mechanism for drug-drug interaction between cerivastatin and cyclosporin A. J Pharmacol Exp Ther. 2003;304:610–616. doi: 10.1124/jpet.102.041921. [DOI] [PubMed] [Google Scholar]
- 39.Hirano M, Maeda K, Shitara Y, Sugiyama Y. Drug-drug interaction between pitavastatin and various drugs via OATP1B1. Drug Metab Dispos. 2006;34:1229–1236. doi: 10.1124/dmd.106.009290. [DOI] [PubMed] [Google Scholar]
- 40.Shitara Y, Sugiyama D, Kusuhara H, et al. Comparative inhibitory effects of different compounds on rat oatpl (slc21a1)- and Oatp2 (Slc21a5)-mediated transport. Pharm Res. 2002;19:147–153. doi: 10.1023/a:1014264614637. [DOI] [PubMed] [Google Scholar]
- 41.Treiber A, Schneiter R, Delahaye S, Clozel M. Inhibition of organic anion transporting polypeptide-mediated hepatic uptake is the major determinant in the pharmacokinetic interaction between bosentan and cyclosporin A in the rat. J Pharmacol Exp Ther. 2004;308:1121–1129. doi: 10.1124/jpet.103.061614. [DOI] [PubMed] [Google Scholar]
- 42.Regazzi MB, Iacona I, Campana C, et al. Altered disposition of pravastatin following concomitant drug therapy with cyclosporin A in transplant recipients. Transplant Proc. 1993;25:2732–2734. [PubMed] [Google Scholar]
- 43.Hedman M, Neuvonen PJ, Neuvonen M, et al. Pharmacokinetics and pharmacodynamics of pravastatin in pediatric and adolescent cardiac transplant recipients on a regimen of triple immunosuppression. Clin Pharmacol Ther. 2004;75:101–109. doi: 10.1016/j.clpt.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 44.Ichimaru N, Takahara S, Kokado Y, et al. Changes in lipid metabolism and effect of simvastatin in renal transplant recipients induced by cyclosporine or tacrolimus. Atherosclerosis. 2001;158:417–423. doi: 10.1016/s0021-9150(01)00438-5. [DOI] [PubMed] [Google Scholar]
- 45.Poels PJ, Gabreels FJ. Rhabdomyolysis: a review of the literature. Clin Neurol Neurosurg. 1993;95:175–192. doi: 10.1016/0303-8467(93)90122-w. [DOI] [PubMed] [Google Scholar]
- 46.Omar MA, Wilson JP. FDA adverse event reports on statin-associated rhabdomyolysis. Ann Pharmacother. 2002;36:288–295. doi: 10.1345/aph.1A289. [DOI] [PubMed] [Google Scholar]
- 47.Page RL, Miller GG, Lindenfeld J. Drug therapy in the heart transplant recipient: part IV: drug-drug interactions. Circulation. 2005;111:230–239. doi: 10.1161/01.CIR.0000151805.86933.35. [DOI] [PubMed] [Google Scholar]
- 48.Dresser GK, Bailey DG, Leake BF, et al. Fruit juices inhibit organic anion transporting polypeptide-mediated drug uptake to decrease the oral availability of fexofenadine. Clin Pharmacol Ther. 2002;71:11–20. doi: 10.1067/mcp.2002.121152. [DOI] [PubMed] [Google Scholar]
- 49.Bailey DG, Dresser GK, Leake BF, Kim RB. Naringin is a major and selective clinical inhibitor of organic anion-transporting polypeptide 1A2 (OATP1A2) in grapefruit juice. Clin Pharmacol Ther. 2007;81:495–502. doi: 10.1038/sj.clpt.6100104. [DOI] [PubMed] [Google Scholar]
- 50.Glaeser H, Bailey DG, Dresser GK, et al. Intestinal drug transporter expression and the impact of grapefruit juice in humans. Clin Pharmacol Ther. 2007;81:362–370. doi: 10.1038/sj.clpt.6100056. [DOI] [PubMed] [Google Scholar]
- 51.Imanaga J, Kotegawa T, Imai H, et al. The effects of the SLCO2B1 c.1457C >T polymorphism and apple juice on the pharmacokinetics of fexofenadine and midazolam in humans. Pharmacogenet Genomics. 2011;21:84–93. doi: 10.1097/fpc.0b013e32834300cc. [DOI] [PubMed] [Google Scholar]
- 52.Verbeeck RK. Pharmacokinetics and dosage adjustment in patients with hepatic dysfunction. Eur J Clin Pharmacol. 2008;64:1147–1161. doi: 10.1007/s00228-008-0553-z. [DOI] [PubMed] [Google Scholar]
- 53.Morgan DJ, McLean AJ. Clinical pharmacokinetic and pharmacodynamic considerations in patients with liver disease. An update. Clin Pharmacokinet. 1995;29:370–391. doi: 10.2165/00003088-199529050-00005. [DOI] [PubMed] [Google Scholar]
- 54.Angulo P. GI epidemiology: nonalcoholic fatty liver disease. Aliment Pharmacol Ther. 2007;25:883–889. doi: 10.1111/j.1365-2036.2007.03246.x. [DOI] [PubMed] [Google Scholar]
- 55.Fisher CD, Lickteig AJ, Augustine LM, et al. Experimental non-alcoholic fatty liver disease results in decreased hepatic uptake transporter expression and function in rats. Eur J Pharmacol. 2009;613:119–127. doi: 10.1016/j.ejphar.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lake AD, Novak P, Fisher CD, et al. Analysis of global and absorption, distribution, metabolism, and elimination gene expression in the progressive stages of human nonalcoholic Fatty liver disease. Drug Metab Dispos. 2011;39:1954–1960. doi: 10.1124/dmd.111.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lickteig AJ, Slitt AL, Arkan MC, et al. Differential regulation of hepatic transporters in the absence of tumor necrosis factor-alpha, interleukin-1beta, interleukin-6, and nuclear factor-kappaB in two models of cholestasis. Drug Metab Dispos. 2007;35:402–409. doi: 10.1124/dmd.106.012138. [DOI] [PubMed] [Google Scholar]
- 58.Planchamp C, Montet X, Frossard JL, et al. Magnetic resonance imaging with hepatospecific contrast agents in cirrhotic rat livers. Invest Radiol. 2005;40:187–194. doi: 10.1097/01.rli.0000154587.00638.77. [DOI] [PubMed] [Google Scholar]
- 59.Donner MG, Schumacher S, Warskulat U, et al. Obstructive cholestasis induces TNF-alpha- and IL-1-mediated periportal downregulation of Bsep and zonal regulation of Ntcp, Oatp1a4, and Oatp1b2. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1134–G1146. doi: 10.1152/ajpgi.00079.2007. [DOI] [PubMed] [Google Scholar]
- 60.Cherrington NJ, Slitt AL, Li N, Klaassen CD. Lipopolysaccharide-mediated regulation of hepatic transporter mRNA levels in rats. Drug Metab Dispos. 2004;32:734–741. doi: 10.1124/dmd.32.7.734. [DOI] [PubMed] [Google Scholar]
- 61.Li N, Klaassen CD. Role of liver-enriched transcription factors in the down-regulation of organic anion transporting polypeptide 4 (oatp4; oatplb2; slc21a10) by lipopolysaccharide. Mol Pharmacol. 2004;66:694–701. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 62.Li N, Choudhuri S, Cherrington NJ, Klaassen CD. Down-regulation of mouse organic anion-transporting polypeptide 4 (Oatp4; Oatp1b2; Slc21a10) mRNA by lipopolysaccharide through the toll-like receptor 4 (TLR4) Drug Metab Dispos. 2004;32:1265–1271. doi: 10.1124/dmd.32.11.. [DOI] [PubMed] [Google Scholar]
- 63.Li N, Klaassen CD. Lipopolysaccharide-induced down-regulation of organic anion transporting polypeptide 4 (Oatp4; Slc21a10) is independent of tumor necrosis factor-alpha, Interleukin-1beta, interleukin-6, or inducible nitric oxide synthase. Toxicol Sci. 2005;83:197–203. doi: 10.1093/toxsci/kfi003. [DOI] [PubMed] [Google Scholar]
- 64.Cui Y, Konig J, Nies AT, et al. Detection of the human organic anion transporters SLC21A6 (OATP2) and SLC21A8 (OATP8) in liver and hepatocellular carcinoma. Lab Invest. 2003;83:527–538. doi: 10.1097/01.lab.0000065015.02412.48. [DOI] [PubMed] [Google Scholar]
- 65.Libra A, Fernetti C, Lorusso V, et al. Molecular determinants in the transport of a bile acid-derived diagnostic agent in tumoral and nontumoral cell lines of human liver. J Pharmacol Exp Ther. 2006;319:809–817. doi: 10.1124/jpet.106.106591. [DOI] [PubMed] [Google Scholar]
- 66.Monks NR, Liu S, Xu Y, et al. Potent cytotoxicity of the phosphatase inhibitor microcystin LR and microcystin analogues in OAT. Mol Cancer Ther. 2007;6:587–598. doi: 10.1158/1535-7163.MCT-06-0500. [DOI] [PubMed] [Google Scholar]
- 67.Vavricka SR, Jung D, Fried M, et al. The human organic anion transporting polypeptide 8 (SLCO1B3) gene is transcriptionally repressed by hepatocyte nuclear factor 3beta in hepatocellular carcinoma. J Hepatol. 2004;40:212–218. doi: 10.1016/j.jhep.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 68.Zollner G, Wagner M, Fickert P, et al. Hepatobiliary transporter expression in human hepatocellular carcinoma. Liver Int. 2005;25:367–379. doi: 10.1111/j.1478-3231.2005.01033.x. [DOI] [PubMed] [Google Scholar]
- 69.Pressler H, Sissung TM, Venzon D, et al. Expression of OATP family members in hormone-related cancers: potential markers of progression. PLoS ONE. 2011;6:e20372. doi: 10.1371/journal.pone.0020372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsuboyama T, Onishi H, Kim T, et al. Hepatocellular carcinoma: hepatocyte-selective enhancement at gadoxetic acid-enhanced MR imaging--correlation with expression of sinusoidal and canalicular transporters and bile accumulation. Radiology. 2010;255:824–833. doi: 10.1148/radiol.10091557. [DOI] [PubMed] [Google Scholar]
- 71.Adverse Events Reporting System. [Last accessed 25 August 2011];U S Food and Drug Administration. 2011 Available from: http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/ucm070461.htm.
- 72.Davies EC, Green CF, Taylor S, et al. Adverse drug reactions in hospital in-patients: a prospective analysis of 3695 patient-episodes. PLoS ONE. 2009;4:e4439. doi: 10.1371/journal.pone.0004439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang SM, Zhang L, Giacomini KM. The International Transporter Consortium: a collaborative group of scientists from academia, industry, and the FDA. Clin Pharmacol Ther. 2010;87:32–36. doi: 10.1038/clpt.2009.236. [DOI] [PubMed] [Google Scholar]
- 74. Giacomini KM, Huang SM, Tweedie DJ, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. †† The International Transporter Consortium’s ‘white paper’ regarding the roles of membrane transporters in drug safety and efficacy.
- 75.Gamazon ER, Huang RS, Cox NJ, Dolan ME. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc Natl Acad Sci USA. 2010;107:9287–9292. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hardwick RN, Fisher CD, Canet MJ, et al. Variations in ABC-transporter regulation during the progression of human nonalcoholic fatty liver disease. Drug Metab Dispos. 2011;39:2395–2402. doi: 10.1124/dmd.111.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Letschert K, Keppler D, Konig J. Mutations in the SLCO1B3 gene affecting the substrate specificity of the hepatocellular uptake transporter OATP1B3 (OATP8) Pharmacogenetics. 2004;14:441–452. doi: 10.1097/01.fpc.0000114744.08559.92. [DOI] [PubMed] [Google Scholar]
- 78.Starley BQ, Calcagno CJ, Harrison SA. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51:1820–1832. doi: 10.1002/hep.23594. [DOI] [PubMed] [Google Scholar]
- 79.Hirschfield GM, Invernizzi P. Progress in the genetics of primary biliary cirrhosis. Semin Liver Dis. 2011;31:147–156. doi: 10.1055/s-0031-1276644. [DOI] [PubMed] [Google Scholar]
- 80.Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27:1485–1491. doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He YJ, Zhang W, Chen Y, et al. Rifampicin alters atorvastatin plasma concentration on the basis of SLCO1B1521T > C polymorphism. Clin Chim Acta. 2009;405:49–52. doi: 10.1016/j.cca.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 82. Pasanen MK, Neuvonen PJ, Niemi M. Global analysis of genetic variation in SLCO1B1. Pharmacogenomics. 2008;9:19–33. doi: 10.2217/14622416.9.1.19. • Study showing distribution of SLCO1B1 variants in the global population.
- 83.Smith NF, Marsh S, Scott-Horton TJ, et al. Variants in the SLCO1B3 gene: interethnic distribution and association with paclitaxel pharmacokinetics. Clin Pharmacol Ther. 2007;81:76–82. doi: 10.1038/sj.clpt.6100011. [DOI] [PubMed] [Google Scholar]
- 84.Laitinen A, Niemi M. Frequencies of single-nucleotide polymorphisms of SLCO1A2, SLCO1B3 and SLCO2B1 genes in a Finnish population. Basic Clin Pharmacol Toxicol. 2011;108:9–13. doi: 10.1111/j.1742-7843.2010.00605.x. [DOI] [PubMed] [Google Scholar]