Abstract
Twenty-eight novel clorobiocin derivatives obtained from mutasynthesis experiments were investigated for their inhibitory activity towards Escherichia coli DNA gyrase and for their antibacterial activities towards clinically relevant gram-positive and gram-negative bacteria in comparison to novobiocin and clorobiocin. Clorobiocin was the most active compound both against E. coli DNA gyrase in vitro and against bacterial growth. All tested modifications of the 3-dimethylallyl-4-hydroxybenzoyl moiety reduced biological activity. The highest activities were shown by compounds containing a hydrophobic alkyl substituent at position 3 of the 4-hydroxybenzoyl moiety. Polar groups in this side chain, especially amide functions, strongly reduced antibacterial activity. Replacement of the alkyl side chain with a halogen atom or a methoxy group at the same position markedly reduced activity. Transfer of the pyrrole carboxylic acid moiety from O-3" to O-2" of l-noviose moderately reduced activity, whereas the complete absence of the pyrrole carboxylic acid moiety led to a loss of activity. Desclorobiocin derivatives lacking the chlorine atom at C-8 of the 3-amino-4,7-dihydroxycoumarin moiety also showed low activity. Lack of a methyl group at O-4" of l-noviose resulted in an inactive compound. From these findings it appears that clorobiocin represents a “highly evolved” structure optimized for bacterial transport and DNA gyrase inhibition.
Antibiotics of the aminocoumarin family exert their therapeutic activity by binding tightly to the B subunit of bacterial DNA gyrase, thereby inhibiting this essential enzyme (19, 24-26). The toxicity of novobiocin towards eukaryotes, as well as its poor activity towards most gram-negative bacterial pathogens and the proclivity of staphylococci to develop endogenous resistance to aminocoumarins during therapy (14, 31), led pharmaceutical companies to direct their antibacterial drug development efforts towards other antibiotic classes, such as β-lactams, fluoroquinolones, or macrolides, rather than aminocoumarins (24), even though the efficacy of novobiocin has been confirmed in several recent clinical trials (35, 36, 40). With the continuing spread of nosocomial pathogens into the community (2, 23) as well as the emergence of new types of resistance in bacteria (1, 9, 22), there has been renewed interest in reevaluating “old” or discontinued antibiotic classes for new uses (32). Given the mode of action of aminocoumarins and the efficacy of novobiocin against many clinically relevant bacteria (12), development of novel aminocoumarins lacking the toxicity and susceptibility range limitations associated with this family of antibiotics would be of great chemotherapeutic benefit (25, 26).
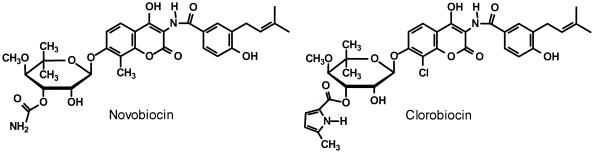
The aminocoumarin antibiotics novobiocin and clorobiocin consist of a 3-amino-4,7-dihydroxycoumarin (ADHC) moiety flanked on one side by l-noviose and on the other side by a 3-dimethylallyl-4-hydroxybenzoyl (DMAHB) moiety (Fig. 1). Early investigations on structure-activity relationships among aminocoumarins (10, 37) demonstrated that both ADHC and l-noviose are essential for antibacterial activity and that the substituents attached to these fragments have a significant impact on their bioactivities. Structurally modified aminocoumarins have been synthesized and investigated for their bioactivities with respect to that of novobiocin (6, 16, 17, 27, 29, 30), and enhancement of the bioactivity of aminocoumarins while simultaneously improving their toxicological and pharmacological properties appears to be a realizable goal.
FIG. 1.
Structures of novobiocin and clorobiocin.
X-ray crystallographic examination of antibiotic-enzyme complexes (15, 18, 39) showed that the ADHC and l-noviose moieties are each involved in antibiotic binding to the B subunit of DNA gyrase. The main difference between the structures of clorobiocin and novobiocin (Fig. 1) complexed with a 24-kDa fragment of the Escherichia coli DNA gyrase B subunit is that the 5-methyl-1H-pyrrole-2-carboxylate group at the O-3" position of l-noviose occupies a hydrophobic pocket and displaces two water molecules that are present when novobiocin (which is carbamoylated at O-3") is complexed with the enzyme. This displacement is regarded as being entropically favorable and is postulated to account for the tighter binding of clorobiocin to DNA gyrase (19). The benzoyl moiety attached to the 3-amino group of the ADHC ring probably contributes weakly to aminocoumarin antibiotic affinity for the B subunit of DNA gyrase through hydrophobic interactions, although it may profoundly influence bacterial aminocoumarin transport (15, 18).
Heide and coworkers recently reported on the preparation of new aminocoumarins by combinatorial biosynthesis (5, 20, 41). The present investigation focused on the antimicrobial and DNA gyrase-inhibitory properties of novel aminocoumarin antibiotics obtained by mutasynthesis with a strain of the clorobiocin producer Streptomyces roseochromogenes (34).
MATERIALS AND METHODS
Disk diffusion assays.
Antibacterial activities of novobiocin, clorobiocin, and mutasynthesized aminocoumarins were screened by using a disk diffusion protocol with Bacillus subtilis ATCC 14893 as the indicator strain. Different amounts of aminocoumarin dissolved in 1 to 10 μl of methanol were applied to filter paper disks (3 mm in diameter) (MN 440 B blotting paper; Macherey-Nagel, Düren, Germany) placed on the surface of nutrient agar (Difco Laboratories, Detroit, Mich.) plates seeded with ca. 2 × 105 B. subtilis spores per ml of solid medium (13). The plates were cultured overnight at 37°C in ambient air, and the diameters of growth inhibition zones were measured.
MIC determinations.
MICs were determined by using the broth microdilution procedure recommended by the National Committee for Clinical Laboratory Standards (28). Some of the test strains used in this study have been described by the National Committee for Clinical Laboratory Standards (Staphylococcus aureus ATCC 29213 and Enterococcus faecalis ATCC 29212) (28), by Engel et al. (S. aureus 80CR5) (4), and by El Falaha et al. (E. coli UB1005, E. coli DC2, Pseudomonas aeruginosa K799/wt, and P. aeruginosa K799/61) (3). All test strains (see Table 2) were obtained from the bacterial collection of Basilea Pharmaceutica AG (Basel, Switzerland). Microtiter plates (96 wells/plate; final assay volume, 100 μl per well) were inoculated with exponential-phase cells and incubated overnight at 35°C in ambient air. Wells were examined for bacterial growth by using an illuminated microtiter plate reader fitted with a magnifying mirror (MIC-2000; Cooke Laboratory Products, Alexandria, Va.). MICs for Streptococcus pneumoniae were determined in cation-adjusted Mueller-Hinton broth (BBL, Cockeysville, Md.) supplemented with 5% (vol/vol) horse serum (Sigma Chemical Co., St. Louis, Mo.); MICs for all other strains were determined with unsupplemented cation-adjusted Mueller-Hinton broth. Aminocoumarins were dissolved in dimethyl sulfoxide; the final concentration of dimethyl sulfoxide in assay wells never exceeded 2% (vol/vol).
TABLE 2.
Effect of various aminocoumarins on E. coli DNA gyrase in vitro and on growth of selected microorganisms
| Compound | DNA gyrase inhibition ratio (IC50nov/ IC50comp)a | Bioassay with B. subtilis (relative activity [%]) | Growth inhibition (MIC [μg/ml]) of:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus ATCC 29213 | S. aureus 42080 | S. epider- midis CNS184 | S. aureus 80CR5 | S. epider- midis CNS10 | E. faecalis ATCC 29212 | E. faecalis Van B E80-8 | E. faecium ATCC 19434 | E. faecium Van A E25-1 | S. pneumo- niae 1/1 (serotype 6) | E. coli UB1005 | E. coli DC2 | P. aeru- ginosa K799/wt | P. aeru- ginosa K799/61 | |||
| Novobiocin | 1.00 | 300 | ≤0.06 | 0.25 | ≤0.06 | >32 | 4 | 16 | 16 | >32 | 2 | 8 | >32 | 4 | >32 | 2 |
| Clorobiocin | 3.52 | 100 | ≤0.06 | ≤0.06 | ≤0.06 | 4 | 0.125 | 2 | 4 | >32 | 0.25 | 2 | >32 | 2 | 32 | ≤0.06 |
| Clorobiocin derivatives | ||||||||||||||||
| 211 | 0.91 | 50-100 | ≤0.06 | ≤0.06 | ≤0.06 | 8 | 0.25 | 16 | 16 | >32 | 8 | 8 | >32 | 4 | >32 | 0.5 |
| 221 | 1.83 | 25-50 | ≤0.06 | ≤0.06 | ≤0.06 | 32 | 1 | ≥32 | 32 | >32 | 16 | 8 | >32 | 8 | >32 | 0.5 |
| 231 | 1.75 | 25 | ≤0.06 | 0.125 | ≤0.06 | 32 | 2 | >32 | >32 | >32 | 16 | 16 | >32 | 16 | >32 | 1 |
| 241 | 0.86 | 6-12.5 | 4 | 4 | 0.125 | >32 | 16 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 2 |
| 251 | 0.84 | 12.5 | 1 | 0.5 | ≤0.06 | >32 | 8 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 1 |
| 261 | 1.65 | 12.5 | 2 | 1 | ≤0.06 | >32 | 4 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 1 |
| 271 | 1.85 | 12.5-25 | 1 | 1 | ≤0.06 | >32 | 4 | >32 | >32 | >32 | >32 | 32 | >32 | 32 | >32 | 0.5 |
| 281 | 0.87 | 25-50 | 1 | 0.5 | ≤0.06 | 32 | 4 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 0.5 |
| 311 | 0.47 | 3-6 | 2 | 2 | 0.125 | >32 | 8 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 4 |
| 381 | 1.91 | 12.5-25 | 0.125 | 0.5 | ≤0.06 | 32 | 4 | 8 | >32 | >32 | >32 | 8 | >32 | 16 | >32 | 1 |
| Isoclorobiocin derivatives | ||||||||||||||||
| 212 | 0.46 | 6-12.5 | 0.25 | ≤0.06 | ≤0.06 | 32 | 1 | >32 | >32 | >32 | >32 | 32 | >32 | 32 | >32 | 1 |
| 222 | 0.23 | 6-12.5 | 0.25 | 0.25 | ≤0.06 | >32 | 4 | >32 | >32 | >32 | >32 | 32 | >32 | >32 | >32 | 1 |
| 232 | 0.05 | 3-6 | 1 | 2 | 0.125 | >32 | 8 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 8 |
| 242 | 0.11 | NDd | 4 | 4 | 0.5 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 4 |
| 252b | 0.21 | ND | 8 | 4 | 0.5 | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 | >16 | 4 |
| 262 | 0.21 | ND | 2 | 2 | 0.25 | >32 | 16 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 4 |
| 272 | 0.23 | 6-12.5 | 2 | 2 | 0.25 | >32 | 8 | >32 | >32 | >32 | >32 | 32 | >32 | >32 | >32 | 2 |
| 282 | 0.11 | 6-12.5 | 2 | 2 | ≤0.06 | >32 | 8 | >32 | >32 | >32 | >32 | 32 | >32 | >32 | >32 | 2 |
| 312 | 0.47 | 3-6 | 2 | 4 | 0.25 | >32 | 8 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 8 |
| 382 | 0.48 | 6-12.5 | 0.5 | 1 | 0.125 | >32 | 16 | 16 | >32 | >32 | >32 | 8 | >32 | >32 | >32 | 4 |
| Desclorobiocin derivatives | ||||||||||||||||
| 214 | 0.48 | 3-6 | 4 | 4 | 0.5 | >32 | 16 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 32 |
| 233 | 0.23 | ND | 4 | 4 | 0.5 | >32 | 16 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 32 |
| 243 | 0.23 | ND | 16 | 32 | 2 | >32 | 16 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 32 |
| 253 | 0.11 | ND | 16 | 16 | 2 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 16 |
| 313 | 0.25 | ND | 8 | 16 | 2 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 32 |
| 383 | 0.50 | 6-12.5 | 8 | 8 | 4 | >32 | >32 | ≥32 | >32 | >32 | >32 | 32 | >32 | >32 | >32 | 8 |
| Other compounds | ||||||||||||||||
| 384c | 0.25 | 3 | 8 | 8 | 2 | >8 | >8 | >8 | >8 | >8 | >8 | >8 | >8 | >8 | >8 | 8 |
| 385 | 0.02 | <1 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| 283 | 0.03 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
IC50 for novobiocin (0.5 μM)/IC50 for compound.
Due to restricted availability of the compound, MICs were measured over the concentration range 0.03 to 16 μg/ml.
Due to restricted availability of the compound, MICs were measured over the concentration range 0.015 to 8 μg/ml.
ND, not determined.
DNA supercoiling assay and determination of inhibitory activities of drugs.
Relaxed pBR322 DNA and E. coli DNA gyrase assay kits were obtained from John Innes Enterprises Ltd. (John Innes Centre and Norwich Research Park, Colney, Norwich, United Kingdom), courtesy of A. Maxwell. Reaction mixtures (20 μl) containing 35 mM Tris-HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 2 mM dithiothreitol, 1.8 mM spermidine, 1 mM ATP, 6.5% (wt/vol) glycerol, 2 μg of bovine serum albumin (Merck, Darmstadt, Germany), 1 U of DNA gyrase, 100 ng of relaxed pBR322 DNA, and various concentrations of aminocoumarins dissolved in methanol-water (0.2 to 20% methanol) were incubated at 37°C for 1 h. Reactions were terminated by cooling to 0°C, and then 3 μl of 30% glycerol containing 0.25% bromophenol blue was added and the DNA was analyzed by electrophoresis in 0.8% agarose.
One unit of DNA gyrase activity was defined as the amount of activity that supercoils 0.5 μg of relaxed pBR322 in 30 min at 37°C. The inhibitory effects of different aminocoumarins were expressed as the aminocoumarin concentration at which the DNA gyrase supercoiling activity was inhibited by 50% (IC50).
RESULTS
Preparation of clorobiocin derivatives by mutasynthesis.
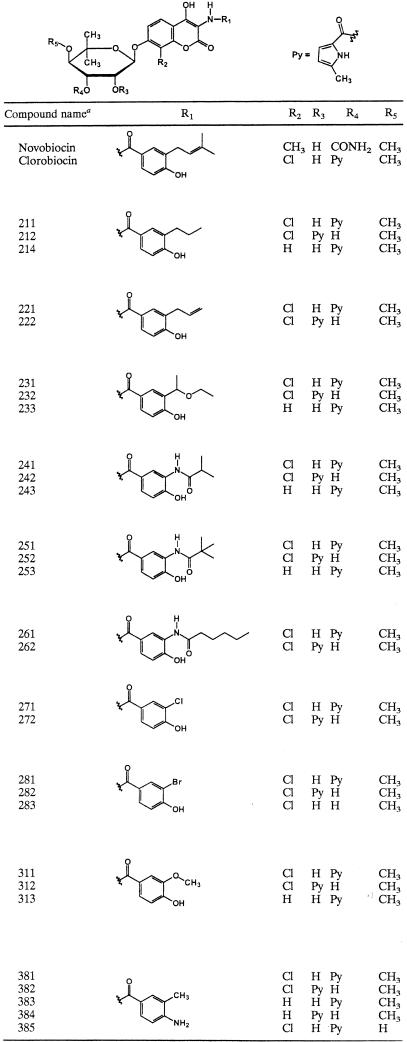
To obtain novel clorobiocin derivatives, the biosynthesis of the DMAHB moiety of clorobiocin was blocked by inactivation of the dimethylallyl transferase gene cloQ (34). Various analogs of DMAHB (Table 1) were prepared by chemical synthesis and added to the cultivation medium of the mutant, which efficiently incorporated these compounds into aminocoumarin antibiotics. The resulting novel clorobiocin derivatives were recovered from the spent media, and their structures were elucidated by 1H nuclear magnetic resonance spectroscopy and mass spectrometry. Details of the chemical synthesis of DMAHB analogues, the feeding and isolation procedures, and the spectroscopic data for the recovered compounds will be reported elsewhere (7).
TABLE 1.
Chemical structures of the test compounds
Compound numbers designate novclobiocin 211, novclobiocin 212, etc.
For each feeding experiment (Table 1), the expected clorobiocin derivative (i.e., the compound in which the DMAHB moiety was replaced by its synthetic analogue) was obtained. One or more of the following additional derivatives could be isolated for each synthetic benzoyl unit supplied: (i) isoclorobiocin derivatives, in which the 5-methyl-1H-pyrrole-2-carboxyl moiety was attached to l-noviose at O-2" instead of O-3"; (ii) desclorobiocin derivatives, which lacked a chlorine atom at C-8 of the ADHC moiety; (iii) derivatives lacking a 5-methyl-1H-pyrrole-2-carboxyl moiety; and (iv) in one case a derivative devoid of a methyl group at the O-4" atom of l-noviose. Similar compounds have been found in previous studies with both wild-type and mutant strains of clorobiocin and novobiocin producers (33, 38).
In accordance with our previous studies on the generation of novobiocin and clorobiocin derivatives (5, 20, 41), the new compounds are called novclobiocins, with different numbers indicating different structures (Tables 1 and 2).
Inhibitory activities of novclobiocins towards E. coli DNA gyrase.
The new aminocoumarin derivatives were tested in vitro for their inhibitory effect on E. coli DNA gyrase, in comparison with the natural compounds novobiocin and clorobiocin. The IC50 of novobiocin was 0.5 μM, and the activities of the other substances were expressed relative to that of novobiocin (Table 2). As observed previously (26), clorobiocin was the most active substance in vitro.
Clorobiocin derivatives with structural modifications in the DMAHB moiety retained marked DNA gyrase-inhibitory activity, ranging from 50 to 200% of the activity of novobiocin. However, even the most active new compounds, i.e., novclobiocins 221, 231, 261, 271, and 381, showed only half of the activity of clorobiocin. Isoclorobiocin derivatives bearing the pyrrole carboxylic acid moiety at O-2" instead of O-3" of l-noviose and desclorobiocin derivatives lacking a chlorine atom at C-8 of the ADHC moiety were less active than clorobiocin. The absence of a pyrrole carboxylic acid moiety (novclobiocin 283) led to a nearly complete loss of activity, as observed earlier (10). Interestingly, the lack of the methyl group at O-4" of l-noviose resulted in a nearly inactive compound (novclobiocin 385), indicating that the 4"-methoxy group is important for the interaction of aminocoumarin antibiotics with DNA gyrase.
Antibacterial activities of novclobiocins.
In a preliminary series of experiments, growth-inhibitory activity against B. subtilis ATCC 14893 was determined in a disk diffusion assay for all new clorobiocin derivatives, as well as for several isoclorobiocin and desclorobiocin derivatives (Table 2). In contrast to results of the DNA gyrase assay, novobiocin was more active than clorobiocin in the disk diffusion assay. Most of the new clorobiocin derivatives showed antibacterial activity, albeit less than that of either novobiocin or clorobiocin. Growth inhibition did not correlate strictly with in vitro DNA gyrase-inhibitory activity.
Subsequently, MICs of the new aminocoumarins, as well as novobiocin and clorobiocin, against representatives of clinically relevant gram-positive and gram-negative bacteria were determined (Table 2). Novclobiocin 283, which was inactive in the DNA gyrase assay, was not available in sufficient amounts for antibacterial testing.
Aminocoumarin antibiotics tend to be highly active towards gram-positive strains, particularly staphylococci. This was confirmed by the very high activities of novobiocin and clorobiocin observed against three Staphylococcus strains, including the methicillin-resistant strains S. aureus 42080 and S. epidermidis CNS184. Novobiocin resistance is uncommon among staphylococci (12). Two staphylococci resistant to novobiocin according to Upjohn's recommended microbiological susceptibility breakpoint of ≤4 μg/ml (G. E. Zurenko, personal communication), S. aureus 80CR5 and S. epidermidis CNS10, were included in the test panel; both of these strains were much more sensitive to clorobiocin than to novobiocin. Three of the four enterococci surveyed (including the vancomycin-resistant strain E. faecalis Van B E80-8 but not the vancomycin-resistant strain Enterococcus faecium Van A E25-1), as well as the penicillin-resistant S. pneumoniae strain 1/1 (serotype 6), were resistant to novobiocin (11) according to Upjohn's microbiological breakpoint, although the pneumococcus and three of four enterococci (including both vancomycin-resistant strains) were ≥2 log2 dilution steps more sensitive to clorobiocin than to novobiocin. Among the four gram-negative microorganisms surveyed, the hyperpermeable strains E. coli DC2 and P. aeruginosa K799/61 were much more sensitive to novobiocin and clorobiocin than their parental strains (E. coli UB1005 and P. aeruginosa K799/wt, respectively), indicating that uptake through the membrane may limit the effectiveness of aminocoumarin antibiotics against gram-negative organisms (though the absence of an active efflux pump in P. aeruginosa K799/61 [21] may contribute to the enhanced antibiotic susceptibility of this strain).
Most of the new clorobiocin derivatives showed marked activity against staphylococci, but clorobiocin was the most active compound among all of those tested, and even conservative changes to its structure led to a loss of activity. Novclobiocins 211 and 221, which contain alkyl side chains at position 3 of the 4-hydroxybenzoyl moiety, were the more active of the new derivatives. Polar groups in this side chain, especially amide functions, strongly reduced antistaphylococcal activity, even though these compounds displayed good inhibitory activity in vitro towards E. coli DNA gyrase. Likewise, replacement of the alkyl side chain with a halogen atom (novclobiocins 271 and 281) markedly reduced antibacterial activity, and a methoxy group at the same position led to an even less active compound (novclobiocin 311). However, the 4-aminobenzoyl compound novclobiocin 381 showed high activity despite lacking a large alkyl group at position 3.
Although the isoclorobiocin derivatives were less active than the corresponding clorobiocin derivatives, the staphylococcal strains were still sensitive to most of these compounds. In contrast, desclorobiocin derivatives showed only weak antibacterial activity, and only S. epidermidis CNS184 was sensitive to these compounds.
Novclobiocin 385, lacking a methyl group at O-4" of l-noviose, was inactive against all test strains, as expected from its lack of activity against DNA gyrase in vitro.
None of the new aminocoumarin derivatives showed any remarkable activity against the novobiocin-resistant S. aureus strain 80CR5, although several compounds were active against the novobiocin-resistant strain S. epidermidis CNS10. With the exception of P. aeruginosa K799/61, all other strains in our panel were insensitive to the new aminocoumarins.
DISCUSSION
The present work aimed at elucidating the contributions of different structural elements to the antimicrobial and DNA gyrase-inhibitory activities of clorobiocin and related compounds. For 28 novel clorobiocin derivatives obtained by mutasynthesis, in vitro inhibitory activities towards E. coli DNA gyrase and MICs against a test panel of representatives of clinically relevant bacterial species were determined.
Clorobiocin was the most active compound in both the E. coli gyrase and bacterial growth experiments. All of the modifications of the 3-dimethylallyl-4-hydroxybenzoate moiety reduced biological activity. The clorobiocin derivatives that showed the highest antistaphylococcal activity were those that contained minimal DMAHB alterations, e.g., novclobiocins 211, 221, 231, and 212. Even conservative structural changes resulted in a marked loss of antibacterial activity against staphylococci, enterococci, pneumococci, and P. aeruginosa. Although the introduction of polar groups, e.g., amide functions, into the side chain only modestly affected DNA gyrase inhibition in vitro, it strongly reduced antibacterial activity, possibly due to reduced uptake.
On the basis of these findings, it may be argued that the DMAHB moiety of clorobiocin and novobiocin is a “highly evolved” structure that has been optimized for bacterial transport and DNA gyrase inhibition. Even minimal alterations in the DMAHB moiety decrease the DNA gyrase-inhibiting and/or antibacterial activity of the corresponding compounds. X-ray crystallographic investigations showed that the dimethylallyl side chain wraps around Pro-79 of the 24-kDa N-terminal fragment of DNA gyrase B, making weak interactions with the hydrophobic surface containing Ile-78, Ala-90, and Ala-94 (15, 18). This may result in an important role of the DMAHB moiety in the binding affinity of aminocoumarin antibiotics to the 43-kDa fragment of DNA gyrase B (15, 26). Therefore, further structural modifications of aminocoumarin antibiotics may be directed towards compounds that contain a sufficiently large, hydrophobic alkyl side chain at position 3 of the 4-hydroxybenzoyl moiety. Interestingly, the 4-aminobenzoyl compound novclobiocin 381 showed high activity despite the lack of a large alkyl group at position 3, suggesting that an investigation of further 4-aminobenzoyl compounds may be worthwhile.
The acyl moieties attached to O-3" of l-noviose (a carbamoyl group in novobiocin and a 5-methyl-1H-pyrrole-2-carboxyl moiety in clorobiocin) are especially important for the hydrogen-bonding network between aminocoumarin antibiotics and the GyrB subunit (18, 39). Comparison of the crystal structures of the complexes of clorobiocin and of novobiocin with a 24-kDa fragment of the DNA gyrase B subunit from E. coli showed that they are very similar. The 5-methyl-1H-pyrrole-2-carboxyl moiety of clorobiocin occupies a hydrophobic pocket and displaces two ordered water molecules which are present in the complex of novobiocin with DNA gyrase. This displacement is entropically favorable and likely to contribute to the higher affinity of DNA gyrase for clorobiocin (18).
In the case of novobiocin, the position of the acyl moiety is of crucial importance: isonovobiocin, resulting from migration of the carbamoyl unit from O-3" to O-2", is reportedly devoid of antibacterial activity (8). In contrast, the isoclorobiocin derivatives investigated in this study still showed marked DNA gyrase-inhibitory activities as well as antibacterial activities. Therefore, for clorobiocin either an O-3" or an O-2" position of the acyl group may allow efficient binding to DNA gyrase, suggesting the possibility of introducing some structural diversity in this region of the molecule. In accordance with the results of Hooper et al. (10), novclobiocin 283, which lacks the 5-methyl-1H-pyrrole-2-carboxyl moiety, had very little DNA gyrase-inhibitory activity.
Novclobiocin 385, which lacks a methyl group at O-4" of l-noviose, was completely inactive, indicating that hydrophobic contacts between this methyl group and a hydrophobic patch of DNA gyrase (18) play a prominent role in the binding of aminocoumarins to the enzyme. Furthermore, aminocoumarins lacking chlorine at position 8′ of the ADHC ring were poor inhibitors of DNA gyrase activity and of bacterial growth.
A better understanding of structure-activity relationships within the aminocoumarin family of anti-infectives may lead to the identification of new antibacterial drugs. However, more radical structural changes than those tested in the present study will be needed to engineer such antibiotics with improved properties for clinical applications.
Acknowledgments
We thank Aventis for the generous gift of authentic clorobiocin and Anthony Maxwell and John Innes Enterprises Ltd. (John Innes Centre and Norwich Research Park, Colney, Norwich, United Kingdom) for providing the DNA gyrase assay kit.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (to L. Heide and S.-M. Li).
REFERENCES
- 1.Cui, L., X. Ma, K. Sato, K. Okuma, F. C. Tenover, E. M. Mamizuka, C. G. Gemmell, M. N. Kim, M. C. Ploy, N. El Solh, V. Ferraz, and K. Hiramatsu. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eady, E. A., and J. H. Cove. 2003. Staphylococcal resistance revisited: community-acquired methicillin resistant Staphylococcus aureus—an emerging problem for the management of skin and soft tissue infections. Curr. Opin. Infect. Dis. 16:103-124. [DOI] [PubMed] [Google Scholar]
- 3.El Falaha, B. M., A. D. Russell, and J. R. Furr. 1983. Sensitivities of wild-type and envelope-defective strains of Escherichia coli and Pseudomonas aeruginosa to antibacterial agents. Microbios 38:99-105. [PubMed] [Google Scholar]
- 4.Engel, H. W., N. Soedirman, J. A. Rost, W. J. van Leeuwen, and J. D. van Embden. 1980. Transferability of macrolide, lincomycin, and streptogramin resistances between group A, B, and D streptococci, Streptococcus pneumoniae, and Staphylococcus aureus. J. Bacteriol. 142:407-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eustáquio, A. S., B. Gust, T. Luft, S.-M. Li, K. F. Chater, and L. Heide. 2003. Clorobiocin biosynthesis in Streptomyces: identification of the halogenase and generation of structural analogs. Chem. Biol. 10:279-288. [DOI] [PubMed] [Google Scholar]
- 6.Ferroud, D., J. Collard, M. Klich, C. Dupuis-Hamelin, P. Mauvais, P. Lassaigne, A. Bonnefoy, and B. Musicki. 1999. Synthesis and biological evaluation of coumarincarboxylic acids as inhibitors of gyrase B. l-Rhamnose as an effective substitute for l-noviose. Bioorg. Med. Chem. Lett. 9:2881-2886. [DOI] [PubMed] [Google Scholar]
- 7.Galm, U., M. A. Dessoy, J. Schmidt, L. A. Wessjohann, and L. Heide. In vitro and in vivo production of new aminocoumarins by a combined biochemical, genetic and synthetic approach. Chem. Biol., in press. [DOI] [PubMed]
- 8.Hinman, J. W., E. L. Caron, and H. Hoeksema. 1957. Novobiocin. V. Carbamoyl migration and isonovobiocin. J. Am. Chem. Soc. 79:5321-5322. [Google Scholar]
- 9.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147-155. [DOI] [PubMed] [Google Scholar]
- 10.Hooper, D. C., J. S. Wolfson, G. L. McHugh, M. B. Winters, and M. N. Swartz. 1982. Effects of novobiocin, coumermycin A1, clorobiocin, and their analogs on Escherichia coli DNA gyrase and bacterial growth. Antimicrob. Agents Chemother. 22:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs, M. R., H. J. Koornhof, R. M. Robins-Browne, C. M. Stevenson, Z. A. Vermaak, I. Freiman, G. B. Miller, M. A. Witcomb, M. Isaacson, J. I. Ward, and R. Austrian. 1978. Emergence of multiply resistant pneumococci. N. Engl. J. Med. 299:735-740. [DOI] [PubMed] [Google Scholar]
- 12.Jones, R. N. 1989. Should novobiocin be clinically re-evaluated? Diagn. Microbiol. Infect. Dis. 12:363-365. [DOI] [PubMed] [Google Scholar]
- 13.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 14.Kirby, W. M., D. G. Hudson, and W. D. Noyers. 1956. Clinical and laboratory studies of novobiocin, a new antibiotic. Arch. Intern. Med. 98:1-7. [DOI] [PubMed] [Google Scholar]
- 15.Lafitte, D., V. Lamour, P. O. Tsvetkov, A. A. Makarov, M. Klich, P. Deprez, D. Moras, C. Briand, and R. Gilli. 2002. DNA gyrase interaction with coumarin-based inhibitors: the role of the hydroxybenzoate isopentenyl moiety and the 5′-methyl group of the noviose. Biochemistry 41:7217-7223. [DOI] [PubMed] [Google Scholar]
- 16.Laurin, P., D. Ferroud, M. Klich, C. Dupuis-Hamelin, P. Mauvais, P. Lassaigne, A. Bonnefoy, and B. Musicki. 1999. Synthesis and in vitro evaluation of novel highly potent coumarin inhibitors of gyrase B. Bioorg. Med. Chem. Lett. 9:2079-2084. [DOI] [PubMed] [Google Scholar]
- 17.Laurin, P., D. Ferroud, L. Schio, M. Klich, C. Dupuis-Hamelin, P. Mauvais, P. Lassaigne, A. Bonnefoy, and B. Musicki. 1999. Structure-activity relationship in two series of aminoalkyl substituted coumarin inhibitors of gyrase B. Bioorg. Med. Chem. Lett. 9:2875-2880. [DOI] [PubMed] [Google Scholar]
- 18.Lewis, R. J., O. M. Singh, C. V. Smith, T. Skarzynski, A. Maxwell, A. J. Wonacott, and D. B. Wigley. 1996. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 15:1412-1420. [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis, R. J., F. T. F. Tsai, and D. B. Wigley. 1996. Molecular mechanisms of drug inhibition of DNA gyrase. Bioessays 18:661-671. [DOI] [PubMed] [Google Scholar]
- 20.Li, S.-M., L. Westrich, J. Schmidt, C. Kuhnt, and L. Heide. 2002. Methyltransferase genes in Streptomyces rishiriensis: new coumermycin derivatives from gene-inactivation experiments. Microbiology 148:3317-3326. [DOI] [PubMed] [Google Scholar]
- 21.Li, X. Z., D. M. Livermore, and H. Nikaido. 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob. Agents Chemother. 38:1732-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchese, A., G. C. Schito, and E. A. Debbia. 2000. Evolution of antibiotic resistance in gram-positive pathogens. J. Chemother. 12:459-462. [DOI] [PubMed] [Google Scholar]
- 23.Marcinak, J. F., and A. L. Frank. 2003. Treatment of community-acquired methicillin-resistant Staphylococcus aureus in children. Curr. Opin. Infect. Dis. 16:265-269. [DOI] [PubMed] [Google Scholar]
- 24.Maxwell, A. 1993. The interaction between coumarin drugs and DNA gyrase. Mol. Microbiol. 9:681-686. [DOI] [PubMed] [Google Scholar]
- 25.Maxwell, A. 1997. DNA gyrase as a drug target. Trends Microbiol. 5:102-109. [DOI] [PubMed] [Google Scholar]
- 26.Maxwell, A., and D. M. Lawson. 2003. The ATP-binding site of type II topoisomerases as a target for antibacterial drugs. Curr. Top. Med. Chem. 3:283-303. [DOI] [PubMed] [Google Scholar]
- 27.Musicki, B., A. M. Periers, P. Laurin, D. Ferroud, Y. Benedetti, S. Lachaud, F. Chatreaux, J. L. Haesslein, A. Iltis, C. Pierre, J. Khider, N. Tessot, M. Airault, J. Demassey, C. Dupuis-Hamelin, P. Lassaigne, A. Bonnefoy, P. Vicat, and M. Klich. 2000. Improved antibacterial activities of coumarin antibiotics bearing 5′,5′-dialkylnoviose: biological activity of RU79115. Bioorg. Med. Chem. Lett. 10:1695-1699. [DOI] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6, 6th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 29.Peixoto, C., P. Laurin, M. Klich, C. Dupuis-Hamelin, P. Mauvais, P. Lassaigne, A. Bonnefoy, and B. Musicki. 2000. Synthesis of isothiochroman 2,2-dioxide and 1,2-benzooxathiin 2,2-dioxide gyrase B inhibitors. Tetrahedron Lett. 41:1741-1745. [Google Scholar]
- 30.Periers, A. M., P. Laurin, D. Ferroud, J. L. Haesslein, M. Klich, C. Dupuis-Hamelin, P. Mauvais, P. Lassaigne, A. Bonnefoy, and B. Musicki. 2000. Coumarin inhibitors of gyrase B with N-propargyloxy-carbamate as an effective pyrrole bioisostere. Bioorg. Med. Chem. Lett. 10:161-165. [DOI] [PubMed] [Google Scholar]
- 31.Perronne, C. M., R. Malinverni, and M. P. Glauser. 1987. Treatment of Staphylococcus aureus endocarditis in rats with coumermycin A1 and ciprofloxacin, alone or in combination. Antimicrob. Agents Chemother. 31:539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pitlik, S. 2003. Old drugs for new bugs. Br. Med. J. 326:235-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pojer, F., S.-M. Li, and L. Heide. 2002. Molecular cloning and sequence analysis of the clorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. Microbiology 148:3901-3911. [DOI] [PubMed] [Google Scholar]
- 34.Pojer, F., E. Wemakor, B. Kammerer, H. Chen, C. T. Walsh, S.-M. Li, and L. Heide. 2003. CloQ, a prenyltransferase involved in clorobiocin biosynthesis. Proc. Natl. Acad. Sci. USA 100:2316-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raad, I., R. Darouiche, R. Hachem, M. Sacilowski, and G. P. Bodey. 1995. Antibiotics and prevention of microbial colonization of catheters. Antimicrob. Agents Chemother. 39:2397-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raad, I. I., R. Y. Hachem, D. Abi-Said, K. V. Rolston, E. Whimbey, A. C. Buzaid, and S. Legha. 1998. A prospective crossover randomized trial of novobiocin and rifampin prophylaxis for the prevention of intravascular catheter infections in cancer patients treated with interleukin-2. Cancer 82:403-411. [DOI] [PubMed] [Google Scholar]
- 37.Reusser, F., and L. A. Dolak. 1986. Novenamine is the active moiety in novobiocin. J. Antibiot. 39:272-274. [PubMed] [Google Scholar]
- 38.Sasaki, T., Y. Igarashi, N. Saito, and T. Furumai. 2001. TPU-0031-A and B, new antibiotics of the novobiocin group produced by Streptomyces sp. TP-A0556. J. Antibiot. 54:441-447. [DOI] [PubMed] [Google Scholar]
- 39.Tsai, F. T., O. M. Singh, T. Skarzynski, A. J. Wonacott, S. Weston, A. Tucker, R. A. Pauptit, A. L. Breeze, J. P. Poyser, R. O'Brien, J. E. Ladbury, and D. B. Wigley. 1997. The high-resolution crystal structure of a 24-kDa gyrase B fragment from E. coli complexed with one of the most potent coumarin inhibitors, clorobiocin. Proteins 28:41-52. [PubMed] [Google Scholar]
- 40.Walsh, T. J., H. C. Standiford, A. C. Reboli, J. F. John, M. E. Mulligan, B. S. Ribner, J. Z. Montgomerie, M. B. Goetz, C. G. Mayhall, D. Rimland, D. A. Stevens, S. L. Hansen, G. C. Gerard, and R. J. Ragaul. 1993. Randomized double-blinded trial of rifampin with either novobiocin or trimethoprim-sulfamethoxazole against methicillin-resistant Staphylococcus aureus colonization: prevention of antimicrobial resistance and effect of host factors on outcome. Antimicrob. Agents Chemother. 37:1334-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westrich, L., L. Heide, and S.-M. Li. 2003. CloN6, a novel methyltransferase catalysing the methylation of the pyrrole-2-carboxyl moiety of clorobiocin. Chembiochem 4:768-773. [DOI] [PubMed] [Google Scholar]