Abstract
Flowering is a critical event in the life cycle of plants; the WRKY-type transcription factors are reported to be involved in many developmental processes sunch as trichome development and epicuticular wax loading, but whether they are involved in flowering time regulation is still unknown. Within this study, we provide clear evidence that GsWRKY20, a member of WRKY gene family from wild soybean, is involved in controlling plant flowering time. Expression of GsWRKY20 was abundant in the shoot tips and inflorescence meristems of wild soybean. Phenotypic analysis showed that GsWRKY20 over-expression lines flowered earlier than the wild-type plants under all conditions: long-day and short-day photoperiods, vernalization, or exogenous GA3 application, indicating that GsWRKY20 may mainly be involved in an autonomous flowering pathway. Further analyses by qRT-PCR and microarray suggests that GsWRKY20 accelerating plant flowering might primarily be through the regulation of flowering-related genes (i.e., FLC, FT, SOC1 and CO) and floral meristem identity genes (i.e., AP1, SEP3, AP3, PI and AG). Our results provide the evidence demonstrating the effectiveness of manipulating GsWRKY20 for altering plant flowering time.
Introduction
In higher plants, a phase transition from vegetative to reproductive development is one of the most important events in their life history [1,2]. This transition is tightly coordinated through a diverse array of signaling networks that integrate various endogenous and exogenous signals [3]. Flowering time is a key trait in adaptation, as it is vital for reproductive success. Arabidopsis thaliana contains at least four flowering pathways that are responsive to these cues: the photoperiod pathway monitors changes in day length; the gibberellin pathway plays a promotive role in flowering under non-inductive photoperiods; the vernalization pathway senses the prolonged exposure to low temperature; and the autonomous pathway mediates flowering by perceiving plant developmental status [3–5]. Most recently, an endogenous pathway that adds plant age to the control of flowering time has been described [6]. Several genes, such as CONSTANS (CO), FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), and FLOWERING LOCUS C (FLC) have been identified as key components in these flowering signal pathways [3]. CO, which encodes a zinc-finger transcriptional activator, controls the timing of flowering by positively regulating two floral integrators, FT [7] and SOC1 [8]; FLC, a flowering repressor gene, also acts as an upstream regulator gene of FT and SOC1 [9]. Moreover, these flowering integrators have been shown to exhibit both overlapping and independent functions in the determination of flowering time and they integrate signals from multiple flowering pathways and their expression levels eventually determine the exact flowering time [3,10].
During the signaling of flowering regulation, a number of transcription factors (TFs) are included. MADS-domain TF family is one of the most important TF families that function in flowering regulation. Among the floral transition genes, FLC, SOC1, APETALA1 (AP1), APETALA3 (AP3), PISTILLAT (PI), AGAMOUS (AG) and SEPALLATA3 (SEP3) are members of the MADS-box gene family [11]. Furthermore, members of other transcription factor families have been identified for their role in the regulation of floral MADS-domain proteins and /or other flowering time genes directly or indirectly [11], such as NACs [12], MYBs [13], DREBs [14].
WRKY proteins are a class of DNA-binding transcriptional factors which contain one or two highly conserved WRKY domains typically having a conserved WRKYGQK motif at N-termini as well as a C2H2 or C2HC zinc-finger structure which is distinct from other known zinc-finger motifs at C-termini [15]. To date, numerous WRKY proteins have been experimentally identified from more than 10 plant species, and it has become clear that WRKY TFs play key roles in responses to biotic and abiotic stress along with various hormones [16–18]. Some WRKY genes also have been reported to be involved in developmental processes. For example, SUSIBA2 [19] and MINISEED3 [20] playing roles in the regulation of seed development; also, Testa Glabra 2 (TTG2)/AtWRKY44 playing a role in trichome development and mucilage and tannin synthesis in the seed coat [21], and OsWRKY89 increasing epicuticular wax loading [22]. We have recently reported that the GsWRKY20, isolated from wild soybean, played a role in the developmental processes of stomata and cuticle, mediated ABA signaling and improves the drought tolerance [23].
In this paper, we will report a novel physiological function of GsWRKY20 in planta, the ectopic overexpression of GsWRKY20 in Arabidopsis (Col-0) accelerating flowering time. qRT-PCR analysis showed that overexpression of GsWRKY20 altered the transcriptional profiles of the genes which were involved in flowering control, implicating that GsWRKY20 may play an important role, not only in in stress [23] but also in flowering transition. Furthermore, we conclude that GsWRKY20 accelerates Arabidopsis flowering may mainly through an autonomous pathway.
Materials and Methods
Sequence analysis of GsWRKY20
Sequence alignments were performed with ClustalW. Phylogenetic analysis was performed using MEGA 4.1. The amino acid sequence of GsWRKY20 and other homologues were retrieved and compared to decipher their relationship. The accession numbers of the genes were listed in Information S1.
Plant Materials and Growth Conditions
The landrace G07256 of wild soybean (Glycine soja) was obtained from Jilin Academy of Agricultural Sciences (Changchun, China). The seeds were sown in soil in 10-liter pots in a growth chamber and grown at a consistent air temperature of 25°C and 16 h light/8 h dark cycles. The light source SON-T ARGO 400 W generated constant illumination of 30000 lx. At the seedling stage, each pot was thinned to 6 plants. These plants were grown till the cotyledons opened or until the unifoliates fully expanded. The plants were then treated with different photoperiods of SD (8 h light/16 h dark) or LD (16 h light/8 h dark). All other parameters for plant growth and treatments were described by Zhu et al [24].
Arabidopsis thaliana (ecotype Col-0 background) seeds were obtained from the Nottingham Arabidopsis Stock Centre (NASC). The GsWRKY20 over-expression lines have been described previously [23]. Arabidopsis thaliana seeds were pretreated at 4°C for 3 days and sown in pot soil or on half-strength Murashige and Skoog (MS)-agar plates (0.6 g L-1 MES pH 5.8 and 0.8% w/v agar, hereafter referred to as 0.5× MS-agar plates) for germination and growth at 22°C air temperature, 100 µmol photons m-2 s-1and 60% relative humidity.
Gene Expression Analyses
GsWRKY20 tissue-specific expression levels in G. soja cv G07256 plants were analyzed by quantitative real-time RT-PCR (qRT-PCR). Total RNA was isolated from root, trifoliate leaf, stem, flower bud, and pod. To analyze the diurnal expression of GsWRKY20 in wild soybean leaves, the fully developed young trifoliate leaves from the plants grown under SD or LD were sampled every 4 h starting at dawn for a total of 20 hours. For analyzing the expression of flowering regulating genes, 10-day-old to three-week-old wild type (WT) and GsWRKY20 overexpression line 28 transgenic Arabidopsis seedlings were harvested from 0.5× MS agar plates at the given indicated time intervals for qRT-PCR and microarray (the Arabidopsis ATH1 Genome Arrays, Affymetrix) assays. All microarray experiments including data analysis were carried out as described previously [25]. For the expression analysis of the FLC, CO, SOC1 and FT at different growing days, GsWRKY20ox line 28 and WT plants were grown in soil, and from the first occurrence of bolting to the last flowering, the leaves were sampled every day. All of the above samples were taken for three biological replicates at the indicated time after treatments.
Quantitative real-time RT-PCR
Total RNA was extracted using RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) and on-column DNA digestion was performed to remove any contamination of genomic DNA using RQ1 RNase-free DNase (Promega, USA). RNA quality was verified by agarose gel electrophoresis, and cDNAs were synthesized by using oligo d(T)18 reverse primer from 2μg of total RNA in a total volume of 20 μL by using the SuperScript™ III Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA).
Prior to the qRT-PCR assays, the quality of the cDNA samples were assessed by PCR using GAPDH specific primers for wild soybean, and ACTIN2 specific primers for Arabidopsis . qRT-PCR reactions were carried out in 96-well (25 μL) format by using the SYBR Green Master Mix (Invitrogen, Carlsbad, CA, USA), and were performed in an Agilent Technologies Stratagene Mx3005p Real-Time PCR system. GAPDH and ACTIN2 were used to normalize all values in the qRT-PCR assays in wild soybean and in Arabidopsis , respectively. All of the reactions were performed in biological triplicates using RNA samples extracted from three independent plant materials and the gene-specific primers designed using Primer5 software were listed in Information S1. Expression levels for all candidate genes were determined using the 2-△△CT method, relative transcript levels were calculated and normalized as described previously [26]. The locus of the candidate genes were listed in Information S1.
Flowering time
After 3-day cold stratification, the WT and GsWRKY20ox seeds were sown and germinated in pot soil. Plants were grown under different conditions till flowering. Flowering time was measured by counting the number of rosette leaves and the number of days to flower (when the floral buds are visible) [27]. For the LD experiment, the plants were grown under 16/8 h light/dark photoperiod. For the SD experiment, the plants were grown under 16/8 h dark/light photoperiod. For gibberellic acid (GA3, Sigma-Aldrich) and paclobutrazol (PAC) treatments, when the two cotyledons fully opened, the plants were sprayed with 100 μM GA3 twice a week until flowering, or watered with 37mg/L PAC solution once a week [28].
For vernalization treatments, the seeds were transferred to 4°C for one month before they were continued to grow at 22°C under the light illumination of 50 μmol photons m-2 s-1. The time spans seed germination (measured as the time until stem elongation [bolting] was observed) and was tabulated starting from the beginning of the first day at the higher temperature (22°C) [29,30].
Results
Sequence analysis of GsWRKY20
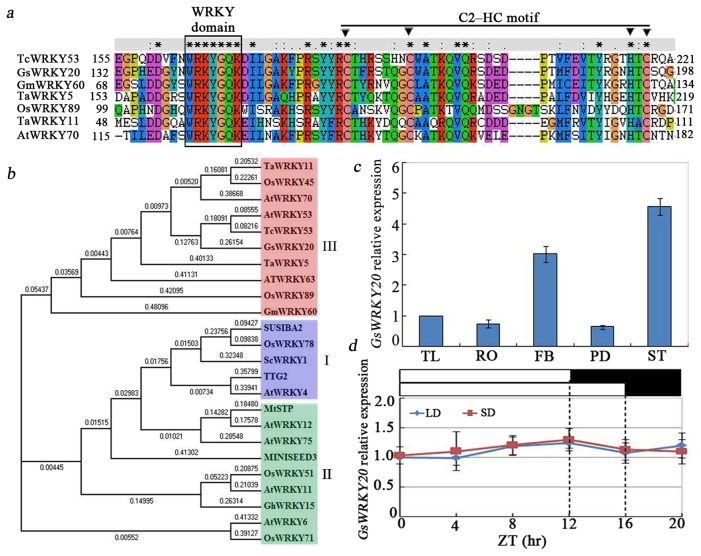
In our previous study, GsWRKY20 was identified as an ABA signaling regulator and drought stress response gene [23]. Based on the sequence analysis, the predicted GsWRKY20 protein contains one conserved WRKY domain and a C2HC-type zinc finger motif (C-X7-C-X23–27-H-X1-C) (Figure 1a). These conserved motifs suggest that GsWRKY20 belongs to the type Ⅲ WRKY subgroup. An alignment analysis by ClustalX revealed that GsWRKY20, together with TcWRKY53, AtWRKY70, OsWRKY89, GmWRKY60, TaWRKY5 and TaWRKY11, belongs to the type Ⅲ WRKY TFs. GsWRKY20 shares 81.8%, 70.9%, 53.8%,76.4%, 74.5, 54.5 identity in WRKY domains and 36.2%, 23.1%, 18.5%, 28.8%, 24.3% and 17% identity in complete sequences with the above six WRKY TFs (Figure 1a), indicating that apart from the WRKY domains the sequences of WRKY proteins are highly divergent.
Figure 1. Sequence and expression analysis of GsWRKY20.
(a) Amino acid sequence alignment of WRKY domains among GsWRKY20 and other type Ⅲ WRKY TFs, Sequences were aligned using ClustalW, and gaps were introduced to maximize alignment, filled triangle marks the cystine and histidine in the C2HC-type zinc finger domain.
(b) The phylogenetic tree of the WRKY TFs. The phylogenetic tree was constructed using MEGA 4.1. Total 24 WRKY proteins from Oryza, Arabidopsis, Gossypium, Medicago, Triticum aestivum, Glycine max, Thlaspi caerulescens and Solanum were selected to construct the phylogenetic tree.
(c) Tissue-specific expression analysis of GsWRKY20 by real-time quantitative PCR (qRT-PCR). Tissues included trifoliate leaf (TL), root (RO), stem tip (ST), flower bud (FB), and pod (PD). Expression of GAPDH was used as an internal control. The experiment included three fully independent biological repeats, and three technical repeats and the mean value is shown.
(d) qRT-PCR analysis of GsWRKY20 diurnal expression under SD and LD. Trifoliate leaves were sampled every 4 h at 21 DAE. White and black bars at the top represent light and dark phases, respectively. Relative transcript levels were analyzed by qRT-PCR and normalized by GAPDH. The experiment included three fully independent biological repeats, and three technical repeats and the mean value is shown.
In order to gain insights into the evolutionary pathway of GsWRKY20 among the other WRKY TF orthologs which are involved in development processes, a total of 24 WRKY proteins from Oryza, Arabidopsis , Gossypium , Medicago , Triticum aestivum, Glycine max, Thlaspi caerulescens and Solanum were retrieved, and a phylogenetic tree was constructed using Neighbor–Joining method (Figure 1b). The result revealed that these WRKY proteins were classified into three groups. GsWRKY20 was more closely related to TcWRKY53, AtWRKY53, AtWRKY70 and OsWRKY45 in group Ⅲ. Among these proteins, OsWRKY45 [31] and TcWRKY53 [32] are involved in abiotic stress, while AtWRKY53 [33] and AtWRKY70 [34] are involved in development process. The phylogenetic analysis indicates that GsWRKY20 probably has a distinct role in plant development and should be investigated further.
Expression patterns of GsWRKY20
The Glycine soja cv 07256 seedlings were planted and maintained under LD conditions until the unifoliates were fully expanded. Hereafter, one set of plants was kept growing under LD conditions and another set was kept growing under SD conditions. GsWRKY20 expression profiles were detected by qRT-PCR using GAPDH as a reference. The data showed that GsWRKY20 expression was observed in almost all tissues, including the root, leaf, flower, pod and inflorescence stem (Figure 1c). Notably, the GsWRKY20 expressed significantly higher in flowers and inflorescence stems than in roots and leaves, suggesting that GsWRKY20 may function in reproductive development. To know if GsWRKY20 gene expression has diurnal circadian rhythm, the trifoliate leaves (15 DAE) were sampled every 4 h. Expression of GsWRKY20 under SD and LD conditions did not exhibit diurnal circadian rhythm (Figure 1c), suggesting that GsWRKY20 was not regulated by circadian clock genes. Therefore, GsWRKY20 may have a role in reproductive development, but it is not involved in photoperiodic pathway.
Over-expression of GsWRKY20 in Arabidopsis promotes early flowering
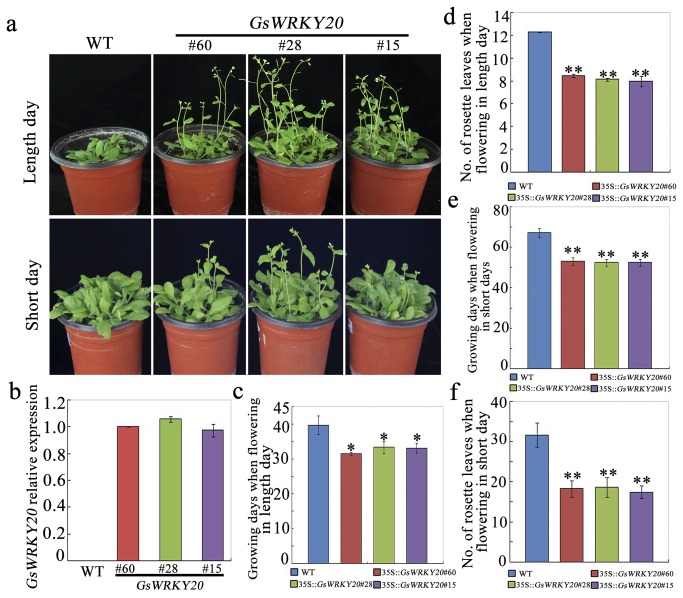
We have recently reported that the GsWRKY20 is intimately related with ABA-mediated drought tolerance [23]. Notably, we also found that all of the three independent homozygous T3 transgenic lines (line 60, line 28, and line 15) with high expression of GsWRKY20 (Figure 2b) also showed early flowering phenotype compared to WT plants grown under the 16/8 h (light/dark) photoperiod (LD) (Figure 2a). The GsWRKY20 over-expression lines, which were referred as GsWRKY20ox plants, flowered with an average of 32.1-day vegetative growth and 8.2 leaves at flowering time under LD, whereas WT plants flowered with an average of 39.7-day vegetative growth and 12.3 leaves at flowering time (Figure 2c, d). The early flowering promotive role of GsWRKY20 suggested that it is an important component plant reproductive development
Figure 2. Over-expression of GsWRKY20 in Arabidopsis accelerates plant flowering.
(a) Flowering time of GsWRKY20ox plants was accelerated under both LD and SD conditions.
(b) qRT-PCR analysis of GsWRKY20 transcript levels in WT and the three homozygous 35S :: GsWRKY20 lines. Expression of ACTIN2 was used as an internal control. The experiment included three fully independent biological repeats, and three technical repeats and the mean value is shown. (c) Average flowering time of WT and GsWRKY20ox plants at the time of flowering under LD conditions.
(d) Average rosette leaf numbers of WT and GsWRKY20ox plants at the time of flowering under LD conditions.
(e) Average flowering time of WT and GsWRKY20ox plants at the time of flowering under SD conditions.
(f) Average rosette leaf numbers of WT and GsWRKY20ox plants at the time of flowering under SD conditions.
All values in (c, d, e, f) are means (±S.E.) from three independent experiments (At least 30 seedlings per experiment). Data were analyzed statistically using the t-test, Asterisk and double asterisks indicate significant differences from the corresponding WT at 0.01 <P< 0.05 and P< 0.01, respectively.
Plant flowering time is regulated by many genes mainly via four genetically distinguishable pathways, i.e., autonomous, photoperiod, vernalization, and GA pathways [3]. In order to further identify in which pathways GsWRKY20 may be involved, WT and GsWRKY20ox plants were given different treatments (photoperiods, vernalization, and GA3).
Under the SD condition, the average rosette leaf numbers of WT and GsWRKY20ox plants at the time of flowering were 32.6 and 18.7 respectively, and GsWRKY20ox plants flowering occurred almost more than two weeks ahead of the WT plants (Figure 2e, f). These results demonstrate that GsWRKY20 overexpression can promote precocious flowering independent of the photoperiod.
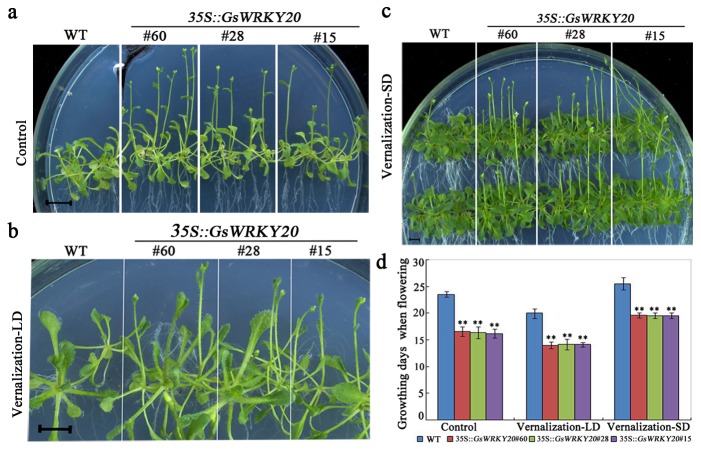
The functional activity of GsWRKY20 in the vernalization pathway was also investigated under the LD and SD conditions. Our results showed that GsWRKY20ox plants and the WT plants responded normally to vernalization, vernalization treatment promoted flowering of GsWRKY20 over-expression lines and the WT plants compared to the normal condition, but GsWRKY20ox plants exhibited earlier flowering after vernalization treatment (Figure 3b, c). After one month of vernalization, GsWRKY20ox plants which were germinated and grown on 0.5 × MS agar plates flowered more than about 6 days ahead of the WT plants both in LD and SD conditions (Figure 3d).
Figure 3. GsWRKY20ox plants exhibited early flowering under vernalization treatment.
(a) Flowering phenotype of GsWRKY20ox plants which were germinated on 0.5 × MS agar plates was accelerated under LD conditions, scale bar: 0.5cm.
(b) Flowering phenotype of GsWRKY20ox plants after one month of vernalization under LD. The seedlings were vernalized for one month at 4°C before transferred to LD photoperiods and grown at 22°C.
(c) Flowering phenotype of GsWRKY20ox plants after one month of vernalization under SD. The seedlings were vernalized for one month at 4°C before transferred to SD photoperiods and grown at 22°C.
(d) Average flowering time of WT and GsWRKY20ox plants which were described in (a, b, c) ; Times to flowering were determined as the time until stem elongation [bolting] was observed. All values are means (±S.E.) from three independent experiments (at least 18 seedlings per experiment). Data were analyzed statistically using the t-test, double asterisks indicate significant differences from the corresponding WT at P < 0.01.
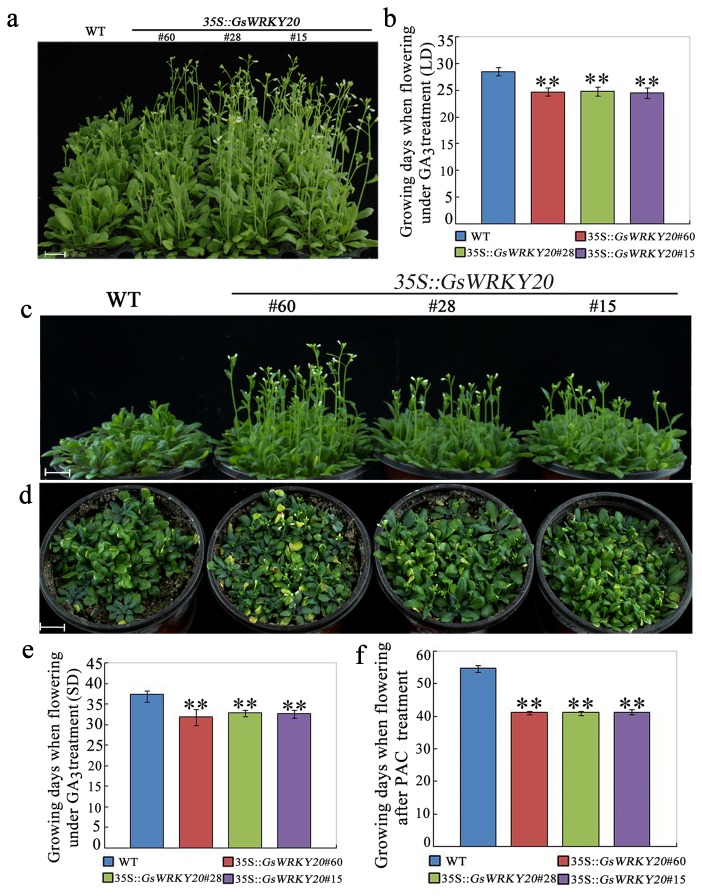
In order to determine the involvement of GsWRKY20 in the GA-regulated flowering pathway, GsWRKY20ox and WT plants, were sprayed with 100 μM GA3 twice a week. GsWRKY20ox and WT plants treated with GA3 flowered significantly earlier than their non-GA3-treated controls under both LD (Figure 4a) and SD conditions (Figure 4c). After exogenous GA3 application, GsWRKY20ox plants flowered more than 4 days ahead of the WT plants in LD conditions (Figure 4b) and 7 days ahead of the WT plants in SD conditions (Figure 4e). In LD conditions, the WT and GsWRKY20ox plants began flowering with an average of 12.3 and 8.2 total rosette leaves, respectively, and the mean total rosette numbers decreased to 10.26 and 6.51 after GA3 application (shown in Information S1). Under SD, the mean rosette leaves numbers also decreased from 32.6 and 18.7 to 18.15 and 13.16 (shown in Information S1), respectively. The experimental results show that GsWRKY20ox plants exhibited earlier flowering after spraying with exogenous GA3. To further ascertain this conclusion, we watered the GsWRKY20ox and WT plants with the GA biosynthesis inhibitor paclobutrazol (PAC) to block endogenous GA biosynthesis under LD condition. We found that the GsWRKY20ox plants still exhibited earlier flowering than the WT plants (Figure 4d). However, after the PAC treatment, the vegetative growth phases of GsWRKY20ox and WT plants were prolonged to 41.7 and 54.3 days, respectively (Figure 4f). These results suggest that GsWRKY20 should not be involved in the GA-induced flowering pathway.
Figure 4. GsWRKY20ox plants exhibited early flowering under GA3 treatment.
(a, c) Flowering phenotype of WT and GsWRKY20ox plants which were treated with GA3 under LD (a) and SD (c) condition. The plants were sprayed with 100 μM GA3 twice a week when the two cotyledons fully opened, scale bar: 1.0 cm.
(d) Flowering phenotype of WT and GsWRKY20ox plants under PAC treatment. The plants were watered with 37mg/L PAC concentrated solution once a week under LD condition, scale bar: 1.0 cm.
(b, e, f) Average flowering time of WT and GsWRKY20ox plants which were described in (a, c, d) at the time of flowering; Times to flowering were determined as the time until stem elongation [bolting] was observed. All values are means (±S.E.) from three independent experiments (at least 30 seedlings per experiment). Data were analyzed statistically using the t-test, double asterisks indicate significant differences from the corresponding WT at P < 0.01.
In all, these results indicated that GsWRKY20 was involved in the regulation of flowering time through regulatory pathways other than the above three pathways.
GsWRKY20 promotes the expression of flowering-related genes
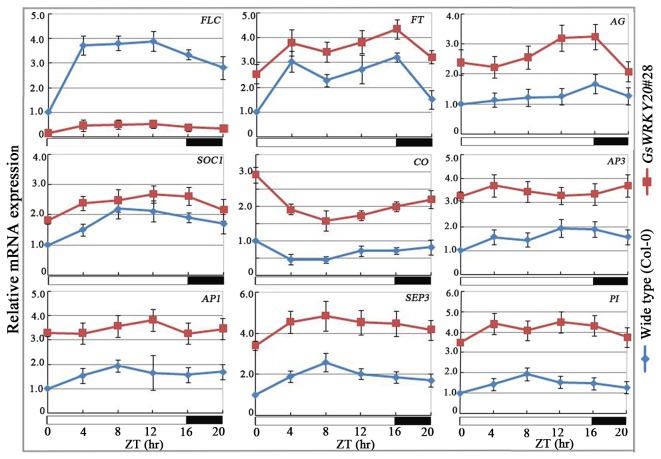
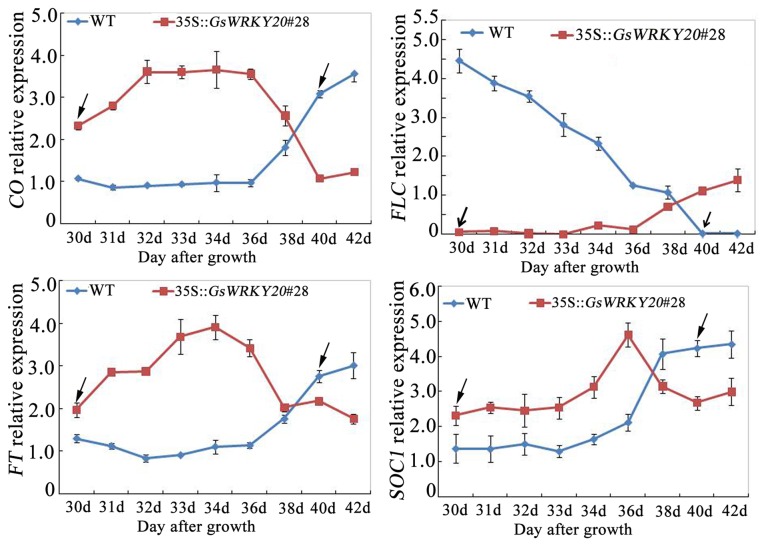
The results mentioned above strongly suggested that GsWRKY20 may be involved in the autonomous flowering pathway. Considering the genetically distinguishable pathways that regulate the flowering time of A. thaliana are integrated by the expression of flowering pathway integrators, to explore the evidence to support the hypothesis that GsWRKY20 may be involved in the autonomous flowering pathway, the expression levels of the genes which are involved in the determination of flowering time were monitored by qRT-PCR. FLC is a key regulator gene of the autonomous pathway [3], so we first analyzed the expression of FLC, and found that GsWRKY20 significantly suppressed FLC expression (Figure 5), and on the other hand we found that GsWRKY20 promoted the expression of another major flowering-related gene, CO (Figure 5). Since FLC negatively but CO positively regulate FT and SOC1 [9], we measured the expression levels of FT and SOC1 and the results showed that both of them indeed exhibited higher levels in GsWRKY20ox plants than the WT plants (Figure 5), and the increased expression of FT and SOC1 was independent of sampling time during the diurnal cycle, suggesting that both of them may be implicated in GsWRKY20 signaling. The flower identity gene SEPALLATA3 (SEP3) is known to interact with AP1, and its over-expression can hasten flowering [35]. In GsWRKY20ox plants, we found the expression levels of SEP3 and AP1 were also increased (Figure 5). AP3 and PI are closely related MADS domain proteins that are thought to act as obligate heterodimers [36]. SEP3, AP1 and AG were identified as interaction partners of AP3 and PI [11], so AP3, PI and AG were further determined and we found the expression levels of them were also elevated in GsWRKY20ox plants (Figure 5). Some studies have shown that the expression levels of FLC, CO, SOC1 and FT in Arabidopsis exhibit different during the flowering transition stage [2,3], so we further analyzed the expression of these four flowering integrator genes in the WT plants and GsWRKY20ox plants under different growing days by qRT-PCR, and the results showed that the expression levels of FLC, CO, SOC1 and FT in WT and GsWRKY20ox plants indeed exhibited different during the flowering transition stage, the flowering repressor FLC was down-regulated and the flowering activators CO, SOC1 and FT were all up-regulated much more earlier in GsWRKY20 over-expression lines than in WT (Figure 6).
Figure 5. The effect of GsWRKY20 over-expression on the transcription of FLC, FT, SOC1, CO, AP1, SEP3, AG, PI and AP3.
Ten-day-old WT and GsWRKY20 ox seedlings were harvested every 4 h during LD condition, and mRNA expression level was determined by qRT-PCR. Each value is the mean ±SE of three independent measurements, error bars represent the standard deviation (n=3). White and black bars at the bottom represent light and dark phases, respectively. ZT, Zeitgeber.
Figure 6. Expression analysis of the CO, FLC, FT and SOC1 in GsWRKY20ox and WT plants during the flowering transition stage.
The experiment included three fully independent biological repeats. Arrows indicate the number of growth days from seed germination to flowering (bolting 2 cm), respectively.
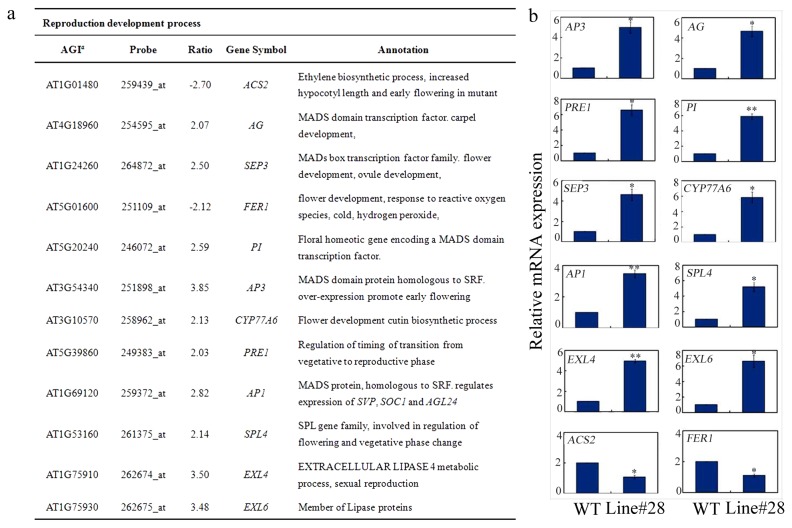
To explore more data about the role of GsWRKY20 in plant flowering, we performed microarray assays using the Affymetrix ATH1 Gene Chip. Differentially expressed genes were identified after statistical analysis, approximately 301 genes were up or down-regulated (≥ 2-fold change) in the GsWRKY20ox lines (shown in Information S2). A major functional category of the differentially expressed genes showed that some of them were involved in ABA signalling, stress regulation, and we also found that there were 12 flowering-related genes were up or down-regulated (≥ 2-fold change) in the GsWRKY20ox plants. The expression levels of these 12 genes were further confirmed by qRT-PCR. ACS2 and FERI negative regulators of flowering time, were down-regulated, whereas AP3, AG, AP1, SPL4, PRE1, PI, EXL4, EXL6, SEP3 and CYP77A6 positive regulators of flowering development, were up-regulated in GsWRKY20 over-expression plants (Figure 7), suggesting the transcription factor GsWRKY20 may act as an upstream regulator to orchestrate the expression of the above flowering-related genes to control plant flowering pattern.
Figure 7. The differentially expressed flowering-related genes in GsWRKY20ox plants revealed by microarray.
(a) The differentially expressed flowering-related genes in GsWRKY20ox plants. Genes which were up or down-regulated (>2-fold) in the GsWRKY20ox plants compared with the WT are listed. The P values were corrected for multiple testing using FDR methodology. The group of genes was classified based on their biochemical functions. AGI, Arabidopsis Genome Initiative number; FC, fold changes.
(b) Expression validation of the differentially expressed flowering-related genes. Total RNAs were extracted from three-week-old whole plants grown on 0.5 × MS-agar. Transcript levels relative to ACTIN2 are presented for each treatment. Each value is the mean ±SE of three independent measurements. Significant differences from WT are denoted by one, or two stars corresponding to P < 0.05, and P < 0.01, respectively by Student’s t test.
Discussion
WRKY TFs have been reported to regulate plant various developments, but no data is available about whether WRKY TFs are involved in flowering time regulation. In the present study, our data provide clear genetic evidence for the function of GsWRKY20 in controlling floral initiation. We found that GsWRKY20 acts as a positive regulator of flowering, since the transgenic plants over-expressing GsWRKY20 demonstrated early flowering compared to WT. Flowering time is known to be coordinated by at least four pathways, namely autonomous, photoperiod, vernalization, and GA pathways [3]. GsWRKY20ox plants flowered earlier than the WT under both LD and SD conditions. Flower development of Arabidopsis is promoted by LD condition and delayed by SD condition [3]. In our case, GsWRKY20ox plants grown under SD condition flowered significantly earlier than WT plants; however, flower production of both WT and transgenic plants was delayed compared to the plants grown under LD condition. Thus, over-expression of GsWRKY20 can accelerate flower formation, but cannot overcome the photoperiodic effect, suggesting that GsWRKY20ox plants were still sensitive to photoperiod and GsWRKY20 should be independent of photoperiod flowering pathway. Vernalization flowering pathway indicates that low temperature treatment of germinating seed can induce early flowering [30], to judge whether GsWRKY20 was involved in vernalization flowering pathway mainly based on whether vernalization will suppress the early flowering phenotype of the GsWRKY20ox plants, although the transgenic acceptor Col-0 is capable to flower without vernalization, we demonstrated that GsWRKY20ox plants and the WT plants responded normally to vernalization, vernalization treatment promoted flowering of GsWRKY20ox plants and the WT plants compared to the normal condition, but GsWRKY20ox plants still flowered much more earlier than WT plants both in LD and SD conditions, so vernalization cannot suppress the early flowering phenotype of GsWRKY20 transgenic lines. Since the GsWRKY20ox plants showed a normal response to vernalization and exhibited earlier flowering, a role of GsWRKY20 in vernalization flowering pathway was excluded. With this approach, many flowering-related Arabidopsis lines of Columbia (Col-0) background were examined whether they were involved in vernalization flowering pathway. For example, with this approach, the Col-0 background mutants sr45-1 [37] and syp22-1 [38] were demonstrated that they were not involved in vernalization flowering pathway. On the other hand, all GsWRKY20ox plants flowered earlier than WT plant after sprayed with exogenous GA3 or watered with the GA biosynthesis inhibitor PAC, suggesting that GsWRKY20 is excluded from GA3 pathway. From these data, we can conclude that GsWRKY20 was involved in the regulation of flowering time through regulatory pathways other than the above three pathways.
Most recently, an endogenous pathway that adds plant age to the control of flowering time has been described [6], and it is independent on the FT expression and does not affect the expression levels of FLC, so, the likely involvement of GsWRKY20 in the autonomous pathway was further verified by the down-regulation of FLC in GsWRKY20ox plants in this study (Figure 7a), Mockler et al (2004) reported that the FLC transcript level was up-regulated in mutants deficient in the autonomous pathway of Arabidopsis [39]. We also found that the expression of CO was significantly higher in GsWRKY20ox plants, and the previous study has shown that over-expression of CO gene promoted early flowering under any day length [3]. CO is a transcriptional activator and FLC is a suppressor of FT and SOC1. Opposite to the role of FLC, we found the expression of FT and SOC1 were up-regulated in GsWRKY20ox plants.
The family of MADS domain transcription factors play important roles in floral transition, specification of floral organ identity and floral determinacy [40,41]. Flowers develop from floral meristems (FM) that arise in the peripheral zone of the reproductive SAM [41], which is also called the inflorescence meristem (IM). The four different floral organs that are developed from the FM are specified by combinations of different types of MADS transcription factors. Five major floral homeotic MADS-domain proteins (AP1, AP3, PI, AG and SEP3) have been proved playing the positive roles in floral initiation and development. These MADS-domain proteins interact with each other, such as SEP3, AP1, and AG were identified as interaction partners of AP3 and PI [42]. SEP3 was also known to interact with AP1 and AG, and its over-expression hastens flowering, genetic analysis revealed that AP1 and SEP3 could regulate the expression of AP3, PI and AG [43]. As a kind of florigen, the elevated FT protein moves from the leaf to the apex, where it promotes the expression of AP1 and SEP3 [3]. Hence, the different integrators directly or indirectly activate floral meristem identity genes AP1 and SEP3, and then launch the expression of a series of genes and mark the beginning of floral organ formation [3]. In our experiments, the expression levels of AP1, AP3, PI, AG and SEP3 revealed by qRT-PCR and microarray were obvious up-regulated in the GsWRKY20ox plants, indicating that GsWRKY20 may be a critical regulator of these five genes or the function of GsWRKY20 may require the participation of these five proteins.
The early flowering phenotype (the average number of growing days and rosette leaves at flowering time) of the three homozygous T3 transgenic lines were essentially the same, so, to be sure, ectopic expression of GsWRKY20 attributed to the earlier flowering phenotype of these over-expression plants. And the GsWRKY20 levels in these three transgenic lines did not show any significant difference each other (Figure 2b), so only one transgenic line was used to detect the expression levels of the flowering regulation genes which might be regulated by GsWRKY20. Our previous study also suggest that although expression levels of the downstream genes which regulated by the overexpressed gene in different transgenic lines may be not exactly the same, but they usually did not exhibit significant differences each other [44,45], and a lot of researchers also just used only one transgenic line to detect the expression levels of the downstream genes which might be regulated by the overexpressed gene [46–49].
As the over-expression of GsWRKY20 resulted in altered expression patterns of flowering, we postulated that GsWRKY20 functioned as a transcriptional regulator, acting as a master regulator of downstream flowering-related genes. Our current findings appear to rule out a role in transcriptional repression, insofar as a fusion of GsWRKY20 and the GAL4 DNA binding domain could induce LacZ expression in yeast [23], implicating that GsWRKY20 may act as an activator of gene transcription, which is consistent with most current reported WRKY TFs [50,51]. GsWRKY20-over-expressing Arabidopsis characterizes this protein as a putative negative regulator of FLC. In this regard, GsWRKY20 could be indirectly involved in the regulation of FLC possibly via transcriptional activation of a number of negative regulators. However, transcriptional activation assay using yeast system only provides an indirect evidence that GsWRKY20 possesses transcription activation function, so we cannot figure out the possibility that GsWRKY20 has the other regulation mechanisms in plant.
WRKY TFs can regulate many types of genes directly through binding to W-box (TTGACC/T) [51] or non-W box sequences [19,52–55], and they are also found to regulate various genes indirectly. For examples, AtWRKY63 could bind to the W-box so as to directly regulate the expression of ABF2, and indirectly control the expression of RD29A and COR47 [50]. There are also some reports of WRKY proteins binding to non-W box sequences. OsWRKY13 can bind to the PRE4 element (TGCGCTT) as well as to W-box [54], and barley HvWRKY46 [53] can bind to both W boxes and a sugar-responsive (SURE) element (TAAAGATTACTAATAGGAA) whereas tobacco (Nicotiana tabacum) NtWRKY12 appears to bind a SURE-like element but not the W box [19]. On the other hand, although MINI3 (WRKY10) can bind to W-boxes in the MINI3 and IKU2 promoters, but MINI3 cannot activate the transcription of MINI3 and IKU2 [56]. So whether WRKY proteins can bind to W-box is not the necessary requirement for their ability to regulate the expression of downstream genes.
Thus, we established a model to reveal a missing link in the GsWRKY20-mediated flowering signaling pathway between the primary signaling events to downstream gene expression (Figure 8). In this model, GsWRKY20 protein acts as a positive e regulator of floral development in Arabidopsis . GsWRKY20 promotes flowering may mainly via the autonomous pathway by indirectly inhibiting FLC which is a suppressor of flowering-promoting factors, FT and SOC1; and on the other hand, GsWRKY20 enhances the CO expression directly or indirectly, which subsequently promotes the expression of FT and SOC1. The different integrators will then directly or indirectly strengthen the expression of floral meristem identity genes AP1, SEP3, AP3, PI and AG mark the beginning of floral organ formation. In this model, however, some other unknown factors or signaling cascades involved in the repression of FLC expression may exist and are directly or indirectly regulated by GsWRKY20.
Figure 8. Proposed model for the role of GsWRKY20 in the regulation of Arabidopsis flowering time.
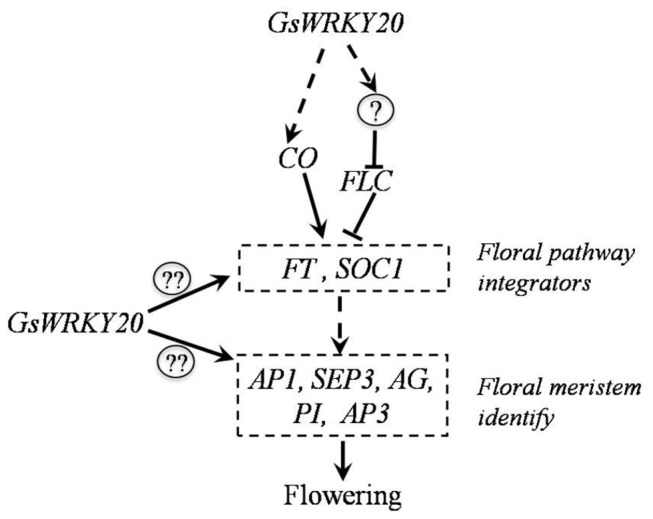
The symbol <?> indicates an unknown factor or signaling cascade that may repress the FLC gene expression, and the symbol <??> indicates the possibility that GsWRKY20 directly regulate the expression of the floral pathway integrators FT and SOC1 and the floral meristem identity genes AP1, AP3, PI, AG and SEP3. The straight dotted lines represent directly or indirectly regulation; the solid straight lines represents directly regulation. Arrows denote the positive effects; and lines terminated with a bar denote inhibitory effect.
Finally, it is noteworthy that, in this model, the underlying mechanisms of several important steps remain open questions. Whether GsWRKY20 directly enhance the expression of CO? And whether GsWRKY20 directly regulates the expression of the floral pathway integrators FT and SOC1 and the floral meristem identity genes AP1, SEP3, AP3, PI and AG? Further research to answer these questions will shed new light on GsWRKY20-mediated flowering signal transduction.
Supporting Information
Included are three sections, that is, Gene-specific primers used for RT-PCR assays, Locus or accession number of the genes, and Average rosette leaf numbers of WT and GsWRKY20ox plants at the time of flowering after GA3 treatment.
(PDF)
Up or down-regulated genes in the GsWRKY20ox line28 (≥2 Fold Change, compared with the WT).
(XLS)
Acknowledgments
We thank members of the lab for discussions and comments on the manuscript. And we thank professor Zhengjun Xia and Qingyun Bu (Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences) for critical comments and suggestions to the manuscript. We are also grateful to Dr. Erik Bergen (University of Alberta, Canada) for the language editing assistance.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31201223, 31171578); the Research Fund for the Doctoral Program of Higher Education of China (20102325120002); the Key Project of the Ministry of National Education (212049). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Razafimandimbison SG, Ekman S, McDowell TD, Bremer B (2012) Evolution of growth habit, inflorescence architecture, flower size, and fruit type in rubiaceae: its ecological and evolutionary implications. PLOS ONE 7: e40851. doi:10.1371/journal.pone.0040851. PubMed: 22815842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Turnbull C (2011) Long-distance regulation of flowering time. J Exp Bot 62: 4399-4413. doi:10.1093/jxb/err191. PubMed: 21778182. [DOI] [PubMed] [Google Scholar]
- 3. Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci, 68: 1-25. PubMed: 21611891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng Z, Guan H, Leal F, Grey PH, Oppenheimer DG (2013) Mediator subunit18 controls flowering time and floral organ identity in Arabidopsis. PLOS ONE 8: e53924. doi:10.1371/journal.pone.0053924. PubMed: 23326539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corbesier L, Coupland G (2006) The quest for florigen: a review of recent progress. J Exp Bot 57: 3395-3403. doi:10.1093/jxb/erl095. PubMed: 17030536. [DOI] [PubMed] [Google Scholar]
- 6. Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738-749. doi:10.1016/j.cell.2009.06.014. PubMed: 19703399. [DOI] [PubMed] [Google Scholar]
- 7. Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK et al. (1999) Activation tagging of the floral inducer FT. Science 286: 1962-1965. doi:10.1126/science.286.5446.1962. PubMed: 10583961. [DOI] [PubMed] [Google Scholar]
- 8. Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z et al. (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613-1616. doi:10.1126/science.288.5471.1613. PubMed: 10834834. [DOI] [PubMed] [Google Scholar]
- 9. Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949-956. doi:10.1105/tpc.11.5.949. PubMed: 10330478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun H, Jia Z, Cao D, Jiang B, Wu C et al. (2011) GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLOS ONE 6: e29238. doi:10.1371/journal.pone.0029238. PubMed: 22195028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smaczniak C, Immink RG, Muiño JM, Blanvillain R, Busscher M et al. (2012) Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci U S A 109: 1560-1565. doi:10.1073/pnas.1112871109. PubMed: 22238427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoo SY, Kim Y, Kim SY, Lee JS, Ahn JH (2007) Control of flowering time and cold response by a NAC-domain protein in Arabidopsis. PLOS ONE 2: e642. doi:10.1371/journal.pone.0000642. PubMed: 17653269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shin B, Choi G, Yi H, Yang S, Cho I et al. (2002) AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J 30: 23-32. doi:10.1046/j.1365-313X.2002.01264.x. PubMed: 11967090. [DOI] [PubMed] [Google Scholar]
- 14. Tong Z, Hong B, Yang Y, Li Q, Ma N et al. (2009) Overexpression of two chrysanthemum DgDREB1 group genes causing delayed flowering or dwarfism in Arabidopsis. Plant Mol Biol 71: 115-129. doi:10.1007/s11103-009-9513-y. PubMed: 19544047. [DOI] [PubMed] [Google Scholar]
- 15. Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199-205. doi:10.1016/S1360-1385(00)01600-9. PubMed: 10785665. [DOI] [PubMed] [Google Scholar]
- 16. Chen L, Song Y, Li S, Zhang L, Zou C et al. (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819: 120-128. doi:10.1016/j.bbagrm.2011.09.002. PubMed: 21964328. [DOI] [PubMed] [Google Scholar]
- 17. Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P et al. (2011) WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol J, 10: 2–11. PubMed: 21696534. [DOI] [PubMed] [Google Scholar]
- 18. Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648-1655. doi:10.1104/pp.109.138990. PubMed: 19420325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun C, Palmqvist S, Olsson H, Borén M, Ahlandsberg S et al. (2003) A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15: 2076-2092. doi:10.1105/tpc.014597. PubMed: 12953112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A (2005) MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci U S A 102: 17531-17536. doi:10.1073/pnas.0508418102. PubMed: 16293693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson CS, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359-1375. doi:10.1105/tpc.001404. PubMed: 12084832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang H, Hao J, Chen X, Hao Z, Wang X et al. (2007) Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol 65: 799-815. doi:10.1007/s11103-007-9244-x. PubMed: 17960484. [DOI] [PubMed] [Google Scholar]
- 23. Luo X, Bai X, Sun X, Zhu D, Liu B et al. (2013) Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J Exp Bot 64: 2155-2169. doi:10.1093/jxb/ert073. PubMed: 23606412. [DOI] [PubMed] [Google Scholar]
- 24. Zhu D, Bai X, Chen C, Chen Q, Cai H et al. (2011) GsTIFY10, a novel positive regulator of plant tolerance to bicarbonate stress and a repressor of jasmonate signaling. Plant Mol Biol, 77: 1-13. PubMed: 21805375. [DOI] [PubMed] [Google Scholar]
- 25. Pernas M, García-Casado G, Rojo E, Solano R, Sánchez-Serrano JJ (2007) A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling. Plant J 51: 763-778. doi:10.1111/j.1365-313X.2007.03179.x. PubMed: 17617176. [DOI] [PubMed] [Google Scholar]
- 26. Willems E, Leyns L, Vandesompele J (2008) Standardization of real-time PCR gene expression data from independent biological replicates. Anal Biochem 379: 127-129. doi:10.1016/j.ab.2008.04.036. PubMed: 18485881. [DOI] [PubMed] [Google Scholar]
- 27. Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ et al. (2012) Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24: 3320-3332. doi:10.1105/tpc.112.101014. PubMed: 22942378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moon J, Suh SS, Lee H, Choi KR, Hong CB et al. (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613-623. doi:10.1046/j.1365-313X.2003.01833.x. PubMed: 12940954. [DOI] [PubMed] [Google Scholar]
- 29. Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES et al. (2006) The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci U S A 103: 14631-14636. doi:10.1073/pnas.0606385103. PubMed: 16983073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA et al. (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445-458. doi:10.2307/3870872. PubMed: 10072403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tao Z, Kou Y, Liu H, Li X, Xiao J et al. (2011) OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J Exp Bot 62: 4863-4874. doi:10.1093/jxb/err144. PubMed: 21725029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wei W, Zhang Y, Han L, Guan Z, Chai T (2008) A novel WRKY transcriptional factor from Thlaspi caerulescens negatively regulates the osmotic stress tolerance of transgenic tobacco. Plant Cell Rep 27: 795-803. doi:10.1007/s00299-007-0499-0. PubMed: 18183400. [DOI] [PubMed] [Google Scholar]
- 33. Hinderhofer K, Zentgraf U (2001) Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213: 469-473. doi:10.1007/s004250000512. PubMed: 11506370. [DOI] [PubMed] [Google Scholar]
- 34. Ulker B, Shahid Mukhtar M, Somssich IE (2007) The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226: 125-137. doi:10.1007/s00425-006-0474-y. PubMed: 17310369. [DOI] [PubMed] [Google Scholar]
- 35. Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF (2001) APETALA1 and SEPALLATA3 interact to promote flower development. Plant J 26: 385-394. PubMed: 11439126. [DOI] [PubMed] [Google Scholar]
- 36. Riechmann JL, Krizek BA, Meyerowitz EM (1996) Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci U S A 93: 4793-4798. doi:10.1073/pnas.93.10.4793. PubMed: 8643482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ali GS, Palusa SG, Golovkin M, Prasad J, Manley JL et al. (2007) Regulation of plant developmental processes by a novel splicing factor. PLOS ONE 2: e471. doi:10.1371/journal.pone.0000471. PubMed: 17534421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ebine K, Uemura T, Nakano A, Ueda T (2012) Flowering time modulation by a vacuolar SNARE via FLOWERING LOCUS C in Arabidopsis thaliana. PLOS ONE 7: e42239. doi:10.1371/journal.pone.0042239. PubMed: 22848750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mockler TC, Yu X, Shalitin D, Parikh D, Michael TP et al. (2004) Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci U S A 101: 12759–12764. doi:10.1073/pnas.0404552101. PubMed: 15310842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zheng BB, Wu XM, Ge XX, Deng XX, Grosser JW et al. (2012) Comparative transcript profiling of a male sterile cybrid pummelo and its fertile type revealed altered gene expression related to flower development. PLOS ONE 7: e43758. doi:10.1371/journal.pone.0043758. PubMed: 22952758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Urbanus SL, Martinelli AP, Dinh QD, Aizza LC, Dornelas MC et al. (2010) Intercellular transport of epidermis-expressed MADS domain transcription factors and their effect on plant morphology and floral transition. Plant J 63: 60-72. PubMed: 20374529. [DOI] [PubMed] [Google Scholar]
- 42. Liu Z, Mara C (2010) Regulatory mechanisms for floral homeotic gene expression. Semin Cell Dev Biol 21: 80-86. doi:10.1016/j.semcdb.2009.11.012. PubMed: 19922812. [DOI] [PubMed] [Google Scholar]
- 43. Wuest SE, O’Maoileidigh DS, Rae L, Kwasniewska K, Raganelli A et al. (2012) Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc Natl Acad Sci U S A 109: 13452-13457. doi:10.1073/pnas.1207075109. PubMed: 22847437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Luo X, Cui N, Zhu Y, Cao L, Zhai H et al. (2012) Over-expression of GsZFP1, an ABA-responsive C2H2-type zinc finger protein lacking a QALGGH motif, reduces ABA sensitivity and decreases stomata size. J Plant Physiol 169: 1192-1202. doi:10.1016/j.jplph.2012.03.019. PubMed: 22705253. [DOI] [PubMed] [Google Scholar]
- 45. Li W, Wang Z, Li J, Yang H, Cui S et al. (2011) Overexpression of AtBMI1C, a polycomb group protein gene, accelerates flowering in Arabidopsis. PLOS ONE 6: e21364. doi:10.1371/journal.pone.0021364. PubMed: 21701597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang L, Ji W, Zhu Y, Gao P, Li Y et al. (2010) GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J Exp Bot 61: 2519-2533. doi:10.1093/jxb/erq084. PubMed: 20400529. [DOI] [PubMed] [Google Scholar]
- 47. Xiao C, Chen F, Yu X, Lin C, Fu YF (2009) Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol Biol 71: 39-50. doi:10.1007/s11103-009-9507-9. PubMed: 19517252. [DOI] [PubMed] [Google Scholar]
- 48. Zhao J, Huang X, Ouyang X, Chen W, Du A et al. (2012) OsELF3-1, an ortholog of Arabidopsis early flowering 3, regulates rice circadian rhythm and photoperiodic flowering. PLOS ONE 7: e43705. doi:10.1371/journal.pone.0043705. PubMed: 22912900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu B, Sathitsuksanoh N, Tang Y, Udvardi MK, Zhang JY et al. (2012) Overexpression of AtLOV1 in Switchgrass Alters Plant Architecture, Lignin Content, and Flowering Time. PLOS ONE 7: e47399. doi:10.1371/journal.pone.0047399. PubMed: 23300513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ren X, Chen Z, Liu Y, Zhang H, Zhang M et al. (2010) ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J 63: 417-429. doi:10.1111/j.1365-313X.2010.04248.x. PubMed: 20487379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247-258. doi:10.1016/j.tplants.2010.02.006. PubMed: 20304701. [DOI] [PubMed] [Google Scholar]
- 52. Van Verk MC, Pappaioannou D, Neeleman L, Bol JF, Linthorst HJ (2008) A Novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol 146: 1983-1995. doi:10.1104/pp.107.112789. PubMed: 18263781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mangelsen E, Kilian J, Berendzen KW, Kolukisaoglu UH, Harter K et al. (2008) Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 9: 194. doi:10.1186/1471-2164-9-194. PubMed: 18442363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cai M, Qiu D, Yuan T, Ding X, Li H et al. (2008) Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ 31: 86-96. PubMed: 17986178. [DOI] [PubMed] [Google Scholar]
- 55. Grierson C, Du JS, de Torres Zabala M, Beggs K, Smith C et al. (1994) Separate cis sequences and trans factors direct metabolic and developmental regulation of a potato tuber storage protein gene. Plant J 5: 815-826. doi:10.1046/j.1365-313X.1994.5060815.x. PubMed: 8054988. [DOI] [PubMed] [Google Scholar]
- 56. Kang X, Li W, Zhou Y, Ni M (2013) A WRKY transcription factor recruits the SYG1-like protein SHB1 to activate gene expression and seed cavity enlargement. PLOS Genet 9: e1003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Included are three sections, that is, Gene-specific primers used for RT-PCR assays, Locus or accession number of the genes, and Average rosette leaf numbers of WT and GsWRKY20ox plants at the time of flowering after GA3 treatment.
(PDF)
Up or down-regulated genes in the GsWRKY20ox line28 (≥2 Fold Change, compared with the WT).
(XLS)