Abstract
Secondary metabolites are known to serve a wide range of specialized functions including communication, developmental control and defense. Genome sequencing of several fungal model species revealed that the majority of predicted secondary metabolite related genes are silent in laboratory strains, indicating that fungal secondary metabolites remain an underexplored resource of bioactive molecules. In this study, we combine heterologous expression of regulatory proteins in Aspergillus nidulans with systematic variation of growth conditions and observe induced synthesis of insect juvenile hormone-III and methyl farnesoate. Both compounds are sesquiterpenes belonging to the juvenile hormone class. Juvenile hormones regulate developmental and metabolic processes in insects and crustaceans, but have not previously been reported as fungal metabolites. We found that feeding by Drosophila melanogaster larvae induced synthesis of juvenile hormone in A. nidulans indicating a possible role of juvenile hormone biosynthesis in affecting fungal-insect antagonisms.
Introduction
Filamentous fungi are capable of synthesizing a wide range of bioactive molecules important for growth and survival in complex and competitive ecological niches [1]–[3]. A substantial number of these metabolites have been found to have beneficial as well as detrimental impact upon human health. Notable examples of both categories include the pharmaceutically important lovastatin and penicillin [4]; and the mycotoxins fumonisin and aflatoxin that cause health hazards and economical losses when they are present in infected crops [5]. With the release of the full genome sequences of several filamentous fungi it has become apparent that the number of predicted secondary metabolite synthases by far exceeds the number of known metabolites [6], [7]. These observations suggest that specific environmental stimuli are required for induction of the majority of secondary metabolites [8]. Despite attempts to identify or mimic these stimuli in order to unravel the secondary metabolism of the model organism Aspergillus nidulans, the product of the majority of predicted synthases are still not known [9], [10]. Genetic approaches have been somewhat successful through manipulation of histone methylation [11] or controlled expression of regulatory proteins [12]. As biosynthetic pathways towards secondary metabolites tend to be clustered in the genome [6], [7] regulatory proteins likely to be involved in secondary metabolism may be identified by genomic co-localization. However, the number of successful applications of this approach is limited, possibly because far from all predicted gene clusters contain regulatory proteins. We decided to investigate whether induction of secondary metabolites could be achieved through heterologous expression of regulatory genes from other filamentous fungi using the expression of A. niger proteins in A. nidulans as a test case. A selection of putative pathway specific regulators was tested for this purpose by expressing the corresponding genes individually from a defined locus using a constitutive promoter [13]. This genetic approach was combined with a screen of several complex media recently demonstrated to influence A. nidulans secondary metabolism [14]. This combinatorial approach resulted in the identification of one regulatory protein that strongly induced metabolites not previously reported from A. nidulans. Among the induced metabolites were the sesquiterpene hormones methyl farnesoate and insect juvenile hormone-III. Juvenile hormones are required in exact concentrations for correct development of insects and crustaceans [15]–[17] and therefore hold a strong potential as insecticides [18], [19]. To the best of our knowledge, this is the first observation of a fungus with the capacity of synthesizing juvenile hormones. In this manuscript, the biological function of juvenile hormones in A. nidulans was addressed through interaction with the saprophagous insect, Drosophila melanogaster. We found that when A. nidulans was challenged by grazing insects, synthesis of juvenile hormones was induced suggesting that juvenile hormones are part of the fungal defense against invertebrates.
Results and Discussion
Procedure for selection of candidate genes
Selection of candidate regulatory proteins associated with secondary metabolism was based on genomic co-localization of gene clusters. We utilized a collection of previously published microarray experiments from A. niger grown under diverse conditions [20]–[22] to identify regulatory genes associated with predicted secondary metabolite gene clusters using a recently described co-expression based algorithm [23]. Seven candidate genes associated with predicted gene clusters containing either polyketide synthases or non-ribosomal peptide synthases, were identified (Table 1). All seven putative transcription factors belong to the binuclear zinc finger class of proteins, a class often associated with secondary metabolism in fungi [24]. BLAST analysis [25] using the predicted protein sequences against the annotated A. nidulans genome (Aspergillus Comparative Database, BROAD Institute) revealed that only one candidate (fge1_pg_C_4000037) had a potential ortholog (ANID_06396, 62% amino acid identity, Table 1). Genes encoding all seven putative regulators were expressed individually in A. nidulans under control of the strong constitutive PgpdA-promoter from the defined locus IS1 [13].
Table 1. Candidate genes from A. niger.
| Strain # | Broad annotation | Transcript ID | Candiate A. nidulans homologues | Identity percentage |
| NID357 | fge1_pg_C_4000037 | 38716 | ANID_06396, ANID_03269 | 62%, 27% |
| NID358 | e_gw1_4.316 | 178503 | ANID_07346 | 26% |
| NID360 | e_gw1_11.945 | 188323 | ANID_08894 | 25% |
| NID366 | gw1_10.247 | 123782 | None | – |
| NID367 | fge1_pg_C_19000192 | 45823 | ANID_11683, ANID_07921 | 43%, 22% |
| NID476 | e_gw1_8.296 | 184613 | ANID_04485 | 30% |
| NID477 | est_fge1_pg_C_150220 | 54836 | None | – |
Candidate genes were selected based on co-localization with predicted gene clusters in A. niger containing either a polyketide synthase, a non-ribosomal peptide synthase or both. Transcript ID = Annotion from the DOE Joint Genome Institute (genome.jgi-psf.org), candidate A. nidulans homologues = Highest scoring potential homologs in A. nidulans, Identity percentage = amino acid identity percentage.
Chemical analysis of mutant strains identifies juvenile hormones as metabolites of A. nidulans
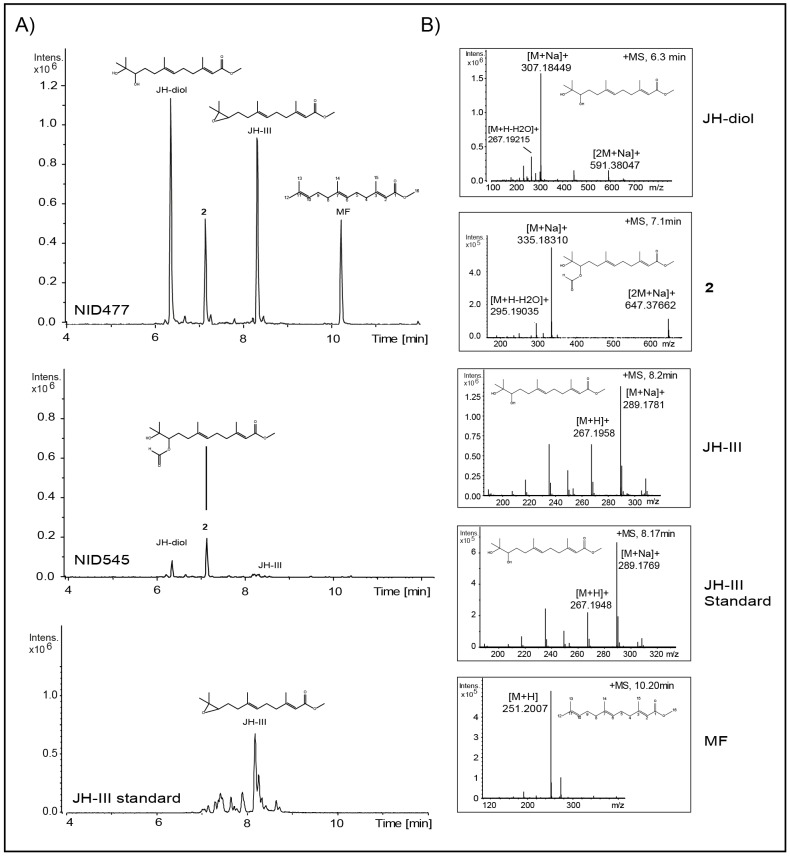
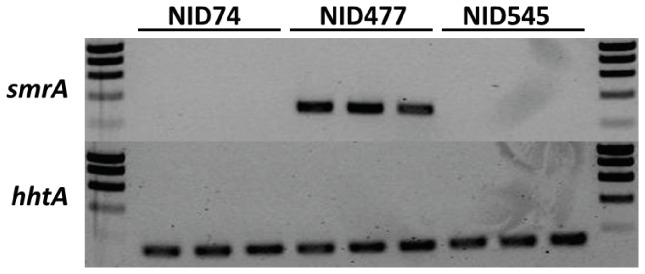
The resulting mutant strains were grown on minimal glucose media as well as four complex media representing diverse physiological conditions. Metabolite profiles of mycelia extracts were analyzed with liquid chromatography-high resolution mass spectroscopy (LC-HRMS) as well as ultra-high pressure liquid chromatography diode array detection (UHPLC-DAD) and compared to a reference strain that constitutively transcribes the E. coli β-galactosidase-gene (lacZ) from IS1 (NID545). Of all combinations of candidate genes and growth conditions, only est_fge1_pg_C_150220 (annotation from Aspergillus Comparative Database, BROAD institute of Harvard and MIT) propagated under high salt conditions had an immediately appreciable impact on secondary metabolism resulting in increased accumulation of several metabolites not previously reported to be produced by A. nidulans (Figure 1A). Hence, we renamed est_fge1_pg_C_150220 Secondary Metabolism associated Regulatory protein A (smrA). The strain that constitutively transcribe smrA was denoted NID477, see Table 2. Correct integration of smrA into IS1, as well as presence of smrA mRNA, was confirmed by Southern blot (Figure 2) and quantitative RT-PCR (Figure 3), respectively. Two induced metabolites displaying very similar UV-spectra were isolated from extracts of NID477 and identified by NMR analysis as the sesquiterpenes: methyl (2E,6E)-10,11-dihydroxy-3,7,11-trimethyl-2,6-dodecadienoate (1) (JH-diol) [26] and its formylated analogue (2). The formylation, however, was subsequently demonstrated to occur during the extraction procedure and 2 is therefore an artificial derivative of 1. The sesquiterpene 1 (JH-diol) represents the hydrated form of insect juvenile hormone-III (JH-III). This observation prompted us to search for JH-III and related metabolites using targeted LC-HRMS analysis. Indeed, metabolites with accurate masses corresponding to JH-III and the related crustacean hormone methyl farnesoate (MF) [15] were strongly induced in NID477 compared to the reference, NID545 (Figure 1A). Final identification of these metabolites as JH-III and MF was established by comparison of retention time and mass spectra with an authentic standard (Figure 1B), or with a reference spectra (Xcalibur software package, Thermo Scientific), respectively. The discovery of JH-III and MF as metabolites of A. nidulans represents to our knowledge the first report of production of invertebrate juvenile hormones in fungi.
Figure 1. Induction of metabolites by SmrA.
A) UHPLC-QTOFMS extracted ion chromatogram of m/z 251 (MF, [M+H]+), 289 (JH-III, [M+Na]+), 307 (JH-diol, [M+H]+) and 335 (X2, [M+H]+) recorded in positive mode of extracts from the strain constitutively expressing smrA (top) and reference (middle) grown under high salt conditions. Chromatograms are normalized by intensity. Chemical structures of JH-diol, compound 2, JH-III and MF are embedded above the corresponding signal peaks. Bottom panel depicts extracted ion chromatogram of m/z 289 (JH-III, [M+Na]+) of an authentic JH-III standard (65% pure) purchased from Sigma Aldrich. Note that the standard contains several impurities. Panel B): Corresponding mass spectra of JH-diol, compound 2, JH-III and MF in the mutant strain constitutively expressing smrA as well as the authentic JH-III standard. Chemical structure of the corresponding molecule is embedded in each panel.
Table 2. Name and description of fungal strains used in this work.
| Strain # | Genotype | Description | Reference |
| NID74 | argBΔ, pyrG89, veA1, nkuAΔ | Parental strain with permanent deletion of nkuA and argB to facilitate gene targeting | This study |
| NID545 | argBΔ, pyrG89, veA1, nkuAΔ IS1::PgpdA-lacZ::argB | Reference strain with E.coli lacZ integrated in IS1. | This study |
| NID357 | argBΔ, pyrG89, veA1, nkuAΔ, IS1:PgpdA:fge1_pg_C_4000037::argB | Constitutive expression of putative binuclear zinc finger transcrption factor fge1_pg_C_4000037 integrated in IS1 | This study |
| NID358 | argBΔ, pyrG89, veA1, nkuAΔ, IS1:PgpdA:e_gw1_4.316::argB | Constitutive expression of putative binuclear zinc finger transcrption factor e_gw1_4.316 integrated in IS1 | This study |
| NID360 | argBΔ, pyrG89, veA1, nkuAΔ, IS1:PgpdA::e_gw1_11.945::argB | Constitutive expression of putative binuclear zinc finger transcrption factor e_gw1_11.945 integrated in IS1 | This study |
| NID366 | argBΔ, pyrG89, veA1, nkuAΔ, IS1:PgpdA:gw1_10.247::argB | Constitutive expression of putative binuclear zinc finger transcrption factor gw1_10.247 integrated in IS1 | This study |
| NID367 | argBΔ, pyrG89, veA1, nkuAΔ, IS1:PgpdA:fge1_pg_C_19000192::argB | Constitutive expression of putative binuclear zinc finger transcrption factor fge1_pg_C_19000192 integrated in IS1 | This study |
| NID476 | argBΔ, pyrG89, veA1, nkuAΔ, IS1:PgpdA::e_gw1_8.296::argB | Constitutive expression of putative binuclear zinc finger transcrption factor e_gw1_8.296 integrated in IS1 | This study |
| NID477 | argBΔ, pyrG89, veA1, nkuAΔ, IS1:PgpdA::smrA::argB | Constitutive expression of smrA (est_fge1_pg_C_150220) integrated in IS1 | This study |
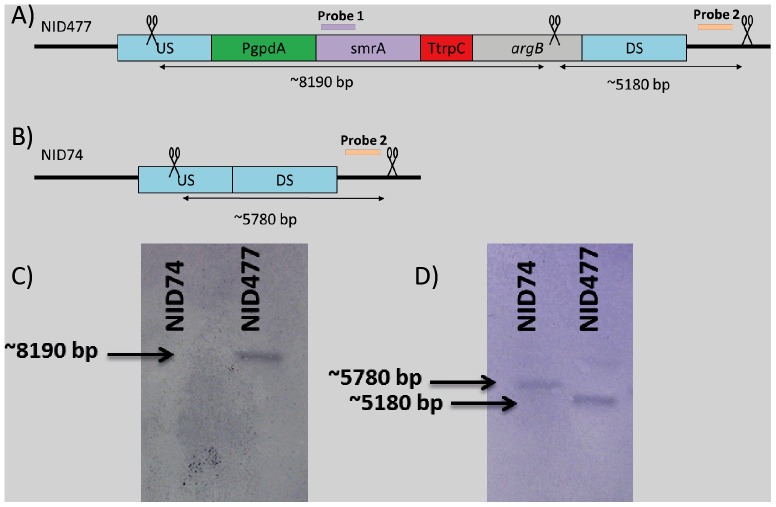
Figure 2. Confirmation of correct integration of smrA in IS1.
A) and B): Schematic overview of the HindIII cut sites (indicated with scissors) and the size of the resulting fragments. Purple and orange bars indicate hybridization site for smrA probe and locus probe respectively. C): Illustration showing placement of the bands relative to each other. D): Southern blot of NID74 and NID477 digested with HindIII and hybridized with smrA probe. E): Southern blot of NID74 and NID477 digested with HindIII and hybridized with locus probe. The illustration is not drawn to scale.
Figure 3. Expression of smrA in NID477 and two control strains.
Measuring the expression of smrA and the control hhtA after cultivating the strains on both MM and CYAs gave the same result. No expression of smrA could be detected in the controls (top panel), but only in the NID477 strain.
Biosynthesis of JH-III, JH-diol and MF in A. nidulans
Biosynthesis of juvenile hormones is well characterized in insects [27]. Since further elucidation of the potential role of juvenile hormones in fungal-insect antagonisms would benefit substantially from the generation of null mutants, we attempted several homology based strategies for identification of the biosynthetic pathway for juvenile hormones in A. nidulans. Initially, BLAST analysis was performed using previously characterized insect enzymes as input, however, no obvious candidates were identified (data not shown). We speculate that the long evolutionary distance between insects and A. nidulans may have obscured a common origin, but it cannot be excluded that an alternative biosynthetic mechanism has evolved in fungi. We tested whether the mixed PKS/NRPS gene cluster in which smrA is located (A. niger transcript ID: 192362, 128601, 191998, 44877, 44878, 44880 and 54837) is conserved in A. nidulans and could provide an alternative biosynthetic route, however, the cluster is not present in A. nidulans as evidenced by BLAST analysis of individual genes (data not shown). Moreover, SmrA does not have any homologs in A. nidulans (Table 1). Thus homology based methods seems to be challenging. We expect that microarray based analysis of the growth condition dependent synthesis of juvenile hormones in NID477 may serve as a more fruitful route for identification of the juvenile hormone synthesis pathway in A. nidulans.
Biological function of Juvenile hormones in A. nidulans
Fungal secondary metabolites are known to play an important role in fungal-insect interactions [2], [3]. Moreover, the role of juvenile hormones in regulating processes of insect metamorphosis, reproduction and metabolism are well described [16], [17]. We therefore hypothesized that the biological function of juvenile hormones in A. nidulans is related to interaction with insects. It is known, that timing and dosage of insect exposure to juvenile hormones is crucial for correct development, with fatal consequences of both under- and overexposure [16]. Consequently, synthesis of JH and MF could be employed as a defense mechanism in A. nidulans and such a strategy has been demonstrated for the plant Cyprus iria [28]. We pursued two experimental lines of evidence in order to test our hypothesis; 1) analysis of the spatial distribution of JH and MF and 2) conducted confrontation experiments between A. nidulans and larvae of the saprophagous insect Drosophila melanogaster.
Distribution of JH-III, JH-diol and MF
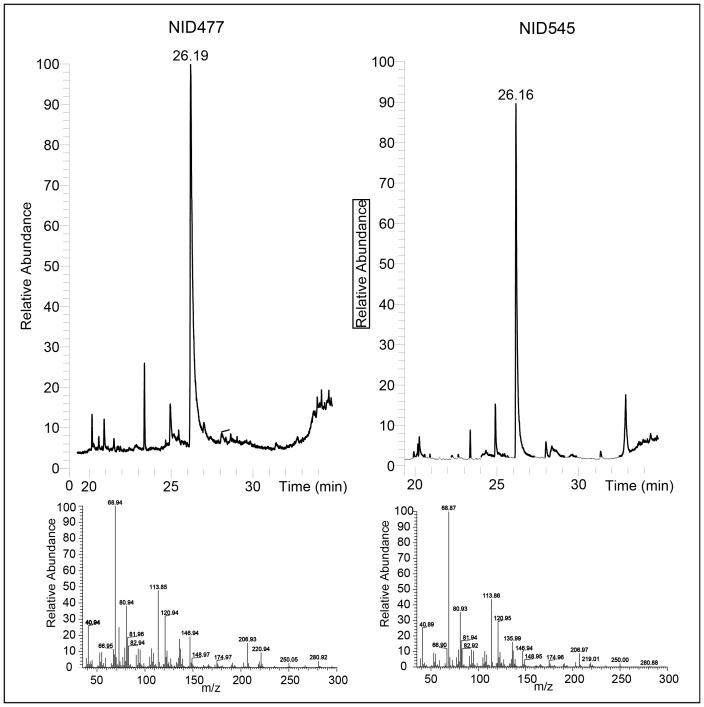
The metabolite composition of growth media extracts and collected volatiles of NID477 and the reference, NID545, grown under juvenile hormone stimulating conditions was analyzed by LC-HRMS and gas chromatography mass spectroscopy (GC-MS), respectively. None of the three terpenes were detectable as extracellular metabolites in the growth media. JH-III and JH-diol were also undetectable among the volatiles whereas MF constituted a major metabolite in the volatile fraction of both strains (Figure 4). Taken together with the presence of JH-III and JH-diol in mycelia extracts (see above), we conclude that JH-III and JH-diol are maintained intracellularly in the mycelium. Therefore, insects will ingest juvenile hormones upon foraging on A. nidulans which may disturb the careful balance of juvenile hormone dosage.
Figure 4. Excretion of MF by A. nidulans.
Top panel: Total MS chromatogram of the collected volatiles from the smrA expressing strain and the reference. Bottom panel: Mass spectrum of the compound eluting at 26.19 minutes. The compound was identified as MF by comparison to the metabolite library of the Xcalibur software package (Thermo Scientific).
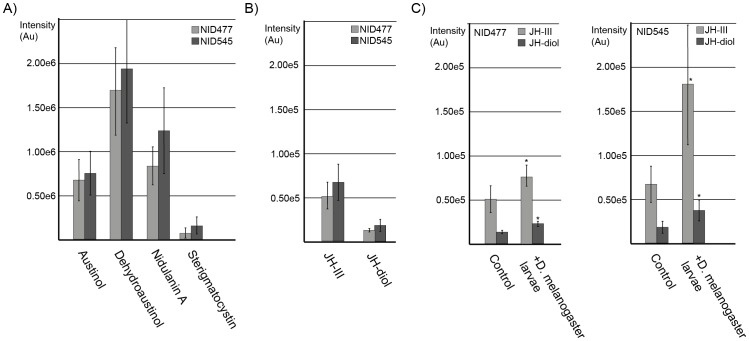
D. melanogaster larvae induce JH-III synthesis upon grazing
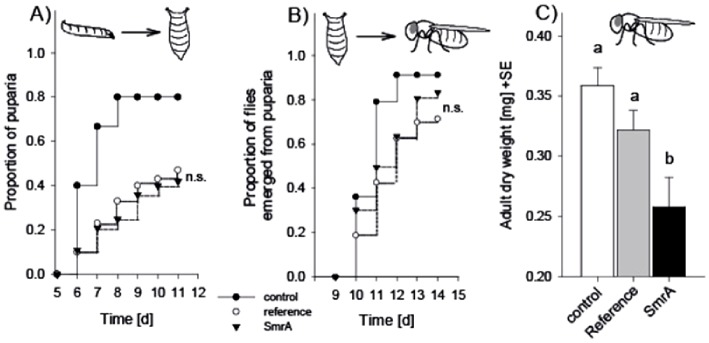
D. melanogaster was chosen for the confrontation experiments since the versatile role of juvenile hormones in D. melanogaster development is well documented [29] and since patterns of interaction between A. nidulans and D. melanogaster larvae have been described previously [30]. The confrontation experiments were initially performed under the conditions where SmrA stimulated JH-III and MF synthesis. However, the high salt content in the media (5% NaCl) caused severe larval mortality even in mock free controls (data not shown). We therefore decided to perform the experiments under less stressful conditions (standard Drosophila medium, [30]). In this experiment, the fitness of grazing D. melanogaster larvae was not significantly different between NID545 and NID477 on two of three parameters evaluated (Figure 5). However, flies emerging from the NID477 treatment displayed a significant decreased dry weight, indicating a negative impact of NID477 on D. melanogaster fitness compared to NID545. We therefore performed a metabolite analysis of fungal extracts produced from the two strains in the presence or absence of larvae in order to correlate the observed effect with differences in the metabolite profile. When NID545 and NID477 were grown on standard Drosophila medium most of the detectable secondary metabolites (austinol, dehydroaustinol, nidulanin A and sterigmatocystin) did not differ significantly between the two strains (Figure 6A). Importantly, JH-III and the JH-diol were detected in both strains, but was not significantly induced in NID477 on this medium (Figure 6B). These observations are in agreement with the results obtained in the initial screening of the NID477 strain, which revealed that the induction of secondary metabolites was highly condition dependent. Interestingly, comparison of the metabolite profiles obtained from NID477 and NID545 strains grown in the presence or in the absence of grazing D. melanogaster larvae demonstrated that the presence of the insects significantly increased the level of both JH-III and JH- diol irrespective of the strain background (p-values JH-III; 0,0288 and 0,00723 and JH-diol; 0,0006 and 0,02415 for NID477 and NID545, respectively, Figure 6C). Curiously, JH-III accumulated to higher levels in NID545 than in NID477 (p-value <0,025). Perhaps, this reflects that when the natural induction of JH-III takes place, the contribution from the presence of the heterologous transcription factor SmrA is detrimental to JH-III biosynthesis. A simple model could be that the natural A. nidulans transcription factor and SmrA bind in a competitive manner to the promoters of the genes involved in JH-III biosynthesis and that activation is less efficient when SmrA is present. We consider it likely that constitutive expression of the SmrA transcription factor has numerous other effects on A. nidulans that is not reflected in our metabolite analysis, and that collectively these effects cause the observed decrease in D. melanogaster fitness. However, the induction of juvenile hormones upon insect feeding, taken together with the well-established involvement of juvenile hormones in insect development and physiology, strongly suggest that JH-III do impact the relation between insects and A. nidulans.
Figure 5. Influence on D. melanogaster larva-to-adult development.
Panel A): Proportion of D. melanogaster larvae that reached the pupal stage as a function of fungal treatment (mold-free control, NID545 or NID477) and time. Panel B): Proportion of flies that emerged from puparia as a function of fungal treatment and time. Panel C): Dry weight of emerged flies as a function of fungal treatment. Different letters indicate statistically significant differences between treatment following a one-way Analysis of Variance (F2,38 = 6.652, p = 0.003) and Holm-Sidak pair-wise comparison. n.s. not significant.
Figure 6. Insect grazing induced alterations of secondary metabolites in A. nidulans.
Quantification of secondary metabolites: JH-III, JH-diol, austinol, dehydroaustinol, nidulanin A and sterigmatocystin from LC-HRMS analysis. For each metabolite, columns display the average and error bars the standard deviation. Statistical analysis was performed with pair-wise comparisons using the student's t-test. Panel A) comparison of austinol, dehydroaustinol, nidulanin A and sterigmatocystin levels in NID477 and NID545. Panel B) comparison of JH-III and JH-diol levels in NID545 and NID477. Panel C) D. melanogaster feeding significantly increases accumulation of JH-III (p-values; 0,0288 and 0,00723) and JH- diol (p-values; 0,0006 and 0,02415) in NID477 and NID545, respectively.
Perspectives
The findings of this manuscript indicate that juvenile hormones represent previously overlooked compounds in chemical interactions between A. nidulans and insects. In addition, the ability of A. nidulans to synthesize juvenile hormones provides the potential for a bio-based source for juvenile hormone production in cell factories. Juvenile hormones are considered to be among the most potent and promising insecticides due to their high specificity and efficiency [18], [19]. Moreover, as A. nidulans releases the juvenile hormone MF to the environment, downstream purification of MF would be simple, as MF could be collected from the volatiles as described previously for other sesquiterpenes [31]. Finally, the findings in this manuscript underline how manipulation of regulatory proteins, systematic variation of physical parameters as well as insect-fungus confrontation systems may be valuable tools for modifying fungal secondary metabolite profiles. The latter approach has the advantage of providing clues to biological function of metabolites. A similar approach simulating bacterial-fungal interactions has previously been successful in identifying novel metabolites in A. nidulans [32] indicating that this more biological approach may constitute a promising route for future studies.
Materials and Methods
Strains and media
Escherichia coli strain DH5α was used to propagate all plasmids. All A. niger genes were amplified from strain ATCC1015. The A. nidulans strain NID74 (argBΔ, veA1, pyrG89, nkuAΔ) was used as background strain for all transformations as it allows gene targeting with the argB marker due to a complete deletion of the A. nidulans argB-open reading frame. NID74 was generated from NID1 (argB2, veA1, pyrG89, nkuAΔ) using the fusion PCR technique essentially as described previously [33]. NID545 (argBΔ, pyrG89, veA1, nkuAΔ, IS1::PgpdA-lacZ-TtrpC::argB) was used as reference strain for metabolite analysis. Genotypes of all strains are summarized in Table 2. All A. nidulans strains were propagated on solid glucose minimal medium (MM) prepared as described by Cove [34], but with 1% glucose, 10 mM NaNO3 and 2% agar. MM was supplemented with 10 mM uridine (Uri), 10 mM uracil (Ura), where required. Complex media used for chemical analysis were prepared as described by Frisvad and Samson [35] and supplemented with 10 mM uridine and 10 mM uracil.
PCR, USER cloning and A. nidulans strain construction
USER cloning compatible PCR products were amplified with 30 PCR cycles in 50 µl reaction mixtures using proof-reading PfuX7 polymerase [36]. USER vectors were denoted according to the nomenclature introduced by Hansen et al [13]. Putative A. niger genes were amplified from A. niger genomic DNA, USER cloned into pU1111-IS1, and transformed into A. nidulans as described previously [13]. In order to generate the NID545 reference strain, the E. coli lacZ gene was cloned into a pU1014-IS1 vector generating pU1011-IS1:lacZ which was transformed to a pU1110-IS1-lacZ vector by insertion of A. nidulans gpdA promoter in the AsiSI/Nb.BtsI cassette. All expression plasmids were verified by sequencing. Gene targeting events were verified in all A. nidulans transformants by analytical PCR as described previously [13]. Table 3 summarizes the PCR primers used in this study. In addition, NID477 was confirmed by Southern blotting as described in [37]. For each Southern blot 2 µg genomic DNA was digested with HindIII. Two probes for detecting insertion of the smrA gene into IS1 were generated by PCR. Specifically, primers JBN X66 and JBN X67 were used to generate Probe 1, a 896 bp fragment of smrA using genomic DNA from A. niger as template, and primers JBN X64 and JBN X65 were used to generate Probe 2, a 948 bp fragment at the IS1 locus using genomic DNA from A. nidulans as template, see Figure 2. The probes were labeled with Biotin-11-dUTP using the Biotin DecaLabelTM DNA Labeling kit (Fermentas). Detection was performed with the Biotin Chromogenic detection kit (Thermo scientific).
Table 3. PCR primers used in this study.
| Primer pair (fw/rv) | Forward primer | Reverse Primer | Description |
| JBN 2QQ/3QQ | GCCAAGTGGTGGAATGCG | gatccccgggaattgccatgCAACACTATCGCATACTCTCC | Amplifies 2 kb upstream region from argB (ANID_04409) for fusion PCR |
| JBN 4QQ/5QQ | aattccagctgaccaccatgCCGATCACGTAAAAGCCTGTTAG | CAGGGCGTGGGAGATAGC | Amplifies 2 kb downstream region from argB (ANID_04409) for fusion PCR |
| JBN 5A/2K | catggcaattcccggggatcTGGATAACCGTATTACCGCC | GGAAGAGAGGTTCACACC | Amplifies 5′ A. fumigatus pyrG sequence including 300 bp direct repeat and native promoter for fusion PCR |
| JBN 4Q/2B | TGATACAGGTCTCGGTCCC | catggtggtcagctggaattTGCCAAGCTTAACGCGTACC | Amplifies 3′ A. fumigatus pyrG sequence including 300 bp direct repeat and native terminator for fusion PCR |
| JBN 2QQ/2K | GCCAAGTGGTGGAATGCG | GGAAGAGAGGTTCACACC | Amplifies argB (ANID_04409)::A. fumigatus pyrG upstream gene targeting fragment |
| JBN 4Q/5QQ | TGATACAGGTCTCGGTCCC | CAGGGCGTGGGAGATAGC | Amplifies argB (ANID_04409)::A. fumigatus pyrG downstream gene targeting fragment |
| Motni 165/185 | cgtgcgauGCAGTGAGAGCGATCGCAGACACTGCATGACCATGATTACGGATTC | cacgcgauTTATTTTTGACACCAGACCA | Amplifies the E. coli lacZ ORF for cloning into an AsiSI/Nb.BsmI casette. Forward primer introduces an upstream AsiSI/Nb.BtsI USER cloning casette |
| Motni 355/354 | agagcgauTAAGCTCCCTAATTGGCCC | tctgcgauGCGGTAGTGATGTCTGCTCA | Amplifies 0,5 kb of the gpdA promoter for cloning into an AsiSI/Nb.BtsI |
| 287/288 | agagcgauATGTTCGTCGCTCCGACGCTT | tctgcgauTCAGAATAAATTGCTTGGAAC | Amplifies the putative ORF of A. niger gene fge1_pg_C_4000037 |
| 289/290 | agagcgauATGCGCCATCGACTCACAAA | tctgcgauTTAATAGAGAGCCCATCGC | Amplifies the putative ORF of A. niger gene e_gw1_4.316 |
| 297/298 | agagcgauATGAGATCCTCCCAGTCCAA | tctgcgauTCAGCATCCATGCACATGAG | Amplifies the putative ORF of A. niger gene e_gw1_8.296 |
| 299/300 | agagcgauATGAGTCCCGTGTCTGGCCA | tctgcgauTTACATATTCCATGCAAACGA | Amplifies the putative ORF of A. niger gene e_gw1_11.945 |
| 303/304 | agagcgauATGGTCTACTGCGGTCGCC | tctgcgauCTATCCACTTAATGGTGGAC | Amplifies the putative ORF of A. niger gene est_fge1_pg_C_150220 |
| 305/306 | agagcgauATGAACAGTGAACGAAAGCT | tctgcgauCTATGGATTGGCCATAACCT | Amplifies the putative ORF of A. niger gene gw1_10.247 |
| 307/308 | agagcgauATGGATTTGAAACAGAAAGTG | tctgcgauCTATCCTTCGAGAACCTCTT | Amplifies the putative ORF of A. niger gene fge1_pg_C_19000192 |
| BGHA163/502 | GGTCTACTCCCCTGCGTCTA | AGGGAGGCCTTGTCGGTCTT | Check primers for integration in IS1. Amplifies the junction between A. nidulans chromosome 1 and the gpdA promoter |
| BGHA98/162 | GGTTTCGTTGTCAATAAGGGAA | GTTCAGGGTGACGGTGAGAG | Check primers for integration in IS1. Amplifies the junction between A. nidulans chromosome 1 and the argB marker gene |
| JBN X64/X65 | GAACGACGGAACTGTGCTC | CTGCACAATAAGCCACGC | Detection of smrA by Southern blot |
| JBN X66/X67 | ATGGTCTACTGCGGTCGC | CGAGACCGATGACGACGAG | Detection of insertion into IS1 by Southern blot |
| JBN X28/X29 | CACCCAAACCATCGTCCGC | CTGCGCAAGCCCTGCTTC | Check primers for transcription of smrA |
| JBN L39/L52 | GAGGCGACGAGCAAGCTG | GTGCTCTCCAGGAGTCCG | Check primers for transcription of hhtA (ANID_00733) |
Upper case letters indicate annealing nucleotides, lower case indicate tails for user cloning.
RNA isolation and quantitative RT-PCR
RNA isolation from the A. nidulans strains and quantitative RT-PCR reactions were done as previously described in [13], except that disruption of biomass for RNA isolation was prepared with a Tissue-Lyser LT (Qiagen) by treating samples for 1 min at 45 mHz. The A. nidulans histone 3 encoding gene, hhtA (AN0733) was used as an internal standard for normalization of expression levels. All primers used for quantitative RT-PCR are shown in Table 3.
Chemical characterization of mutant strains by UHPLC-DAD and LC-HRMS
All strains were grown as three point inoculations for 7 days at 37°C in the dark on solid glucose minimal, CYAs, RTO and YES media [35]. Extraction of metabolites was performed by the agar plug extraction method [38] using three 6 mm agar plugs/extract. Extracts were analyzed by UHPLC-DAD and LC-HRMS. UHPLC-DAD analysis was performed on a Dionex RSLC Ultimate 3000 (Dionex, Sunnyvale, CA) equipped with a diode-array detector. Separation was performed at 60°C on a 150 mm×2.1 mm ID, 2.6 µm Kinetex C18 column (Phenomenex, Torrence, CA) using a linear water/MeCN (both buffered with 50 ppm tri-fluoroacetic acid (TFA)) gradient starting from 15% MeCN to 100% over 7 min at a flow rate of 0.8 mL min−1. LC-HRMS analysis was performed on a MaXis 3G QTOF (Bruker Daltronics) coupled to a Dionex Ultimate 3000 UHPLC system equipped with a 100×2.0 mm, 2.6 µm, Kinetex C-18 column. The separation column was held at a temperature of 40°C and a gradient system composed of A: 20 mM formic acid in water, and B: 20 mM formic acid in acetonitrile was used. The flow was 0.4 ml/min, 85% A graduating to 100% B in 0–10 min, 100% B 10–13 min, 85% A 13.1–15 min. For calibration, a mass spectrum of sodium formate was recorded at the beginning of each chromatogram using a divert valve (0.3–0.4 min). Samples were analyzed both in positive and negative ionization mode. De-replication of induced compounds were performed by comparison of accurate mass to the metabolite database Antibase2009 [39], comparison of UV spectra to published data as well as authentic standards (JH-III, Sigma Aldrich).
Chemical characterization of mutant strains by GC-MS
Volatile metabolites were collected during days 5–7 for the strains inoculated in CYAs. To collect the volatiles, a stainless steel Petri dish lid with a standard 1/4 Swagelock™ replaced the usual lid [40]. This lid possessed a standard 1/4 Swagelok fitting with PTFE insert in the centre that is used to hold a charcoal tube (SKC, 226-01). The collected volatiles were extracted from the charcoal tube with 0.3 mL of ether (Sigma Aldrich). The samples were concentrated to approximately 0.1 mL using a nitrogen flow in a GC vial and analysed using a Finnigan Focus GC coupled to a Finnigan Focus DSQ mass selective detector. The separation of the volatiles was done on a Supelco SLB™-5 MS capillary column, using He as carrier gas, at 1.2 mL/min. The injection and detection temperature was set to 220°C. One microlitre of each sample was injected into the GC–MS system. Chromatographic conditions were set to an initial temperature of 35°C for 1 min, raised at 6°C/min to 220°C and then 20°C/min to 260°C for 1 min. The separated compounds were characterized by their mass spectra generated by electron ionization (EI) at 70 eV at a scan range from m/z 35–300.
Isolation of methyl (2E,6E)-10,11-dihydroxy-3,7,11-trimethyl-2,6-dodecadienoate (JH-diol)
NID477 was cultured on 100 CYAs plates for 7 days at 37°C in the dark. The plates were homogenized using a Stomacher homogenizer and 100 mL ethyl acetate (EtOAc) +1% formic acid (FA) pr. 10 plates. The extract was filtered after 1 hour and the remaining broth was extracted with EtOAc +1% FA for 24 hours. The extract was filtered and the two fractions pooled and dried down on a freeze drier. The crude extract was separated into three phases by dissolving it in 9∶1 MeOH:H2O – Milli-Q and extracted into a heptane phase followed by a dichloromethane (DCM) phase. The DCM phase was fractionated with a 10 g ISOL Diol column, using 13 steps of stepwise Hexane-dichloromethane-EtOAc-MeOH. JH-diol was present in the DCM fraction (9.5 mg) and was purified on a Waters HPLC W600/996PDA (Milford, MA, USA) using a RP column (Phenomenex Luna C18 (2), 250×10 mm, 5 µm, Torrance, CA, USA) using a gradient of 40% MeCN (H2O – Milli-Q (Millipore, MA, USA)) to 100% over 20 min with 50 ppm TFA and a flow of 4 mL/min. The fractions were concentrated on a rotarvap (Büchi V-855/R-215) and dried down under N2(g) to yield 2.0 mg of JH-diol. 1 and 2D NMR characterization (1H, DQF-COSY, H2BC, HMBC and HSQC) of the compound showed that the compound was a racemic mixture with a 2∶3 ratio of JH-diol a: JH-diol b. This is in agreement with an optical rotation of 0.0. The chemical shifts differed most in the reduced end of JH-diol, where the stereocenter is present, whereas the chemical shifts from C5 to C1 were overlaying. The difference of chemical shifts of the two methyl groups (H12/C12 and H13/C13) and the two CH2 groups next to the stereocenter are due to the presence of the chiral center. The two diastereomers present in the JH-diol solution must be due to the presence of JH-diol in both the E- and Z-conformation at the C6 and C7 double bond. The carbon shifts are in good agreement with published data [26].
Isolation of methyl (2E,6E)-10-hydroxy-11-formyl-3,7,11-trimethyl-2,6-dodecadienoate (compound 2)
Compound 2 was present in the 60∶40 DCM:EtOAc fraction (13.1 mg) of the Diol fractionation as described above and was purified on a Waters HPLC W600/996PDA (Milford, MA, USA) using a RP column (Phenomenex Luna C18(2), 250×10 mm, 5 µm, Torrance, CA, USA) using a gradient of 40% MeCN (H2O – Milli-Q (Millipore, MA, USA)) to 100% over 20 min. with 50 ppm TFA and a flow of 4 mL/min. The collections were concentrated on a rotarvap (Büchi V-855/R-215) and dried under N2(g) to yield 2.6 mg of compound 2.
Isolation of methyl (2E,6E)-10,11-epoxid-3,7,11-trimethyl-2,6-dodecadienoate (JH III)
JH-III was present in the 46∶60 DCM:EtOAc fraction (26.2 mg) of the Diol fractionation as described for JH-diol and was purified on a Waters HPLC W600/996PDA (Milford, MA, USA) using a RP column (Phenomenex Luna C18(2), 250×10 mm, 5 µm, Torrance, CA, USA) using a gradient of 55% MeCN (H2O – Milli-Q (Millipore, MA, USA)) to 65% over 20 min. with 50 ppm TFA and a flow of 4 mL/min. The fractions were concentrated on a rotarvap (Büchi V-855/R-215) and dried under N2(g) to yield 1.4 mg of JH-III. However, the purified JH-III degraded before NMR experiments could be conducted. Instead, JH-III was identified based on comparison of accurate mass and retention time with authentic standard. HRMS (m/z): [M+H]+ calcd. for C16H27O3, 267.1955; found, 267.1957.; [M+Na]+ calcd. For C16H26O3Na, 289.1780; found, 289.1774.
NMR studies and structure elucidation
NMR spectra were acquired in DMSO-d6 on a Varian Unity Inova 500 MHz spectrometer for JH-diol and JH-III and on a Bruker Avance 800 MHz spectrometer at the Danish Instrument Center for NMR Spectroscopy of Biological Macromolecules for compound 2 using standard pulse sequences. The spectra were referenced to this solvent with resonances δH = 2.49 and δC = 39.5.
Characterization data of methyl (2E,6E)-10,11-dihydroxy-3,7,11-trimethyl-2,6-dodecadienoate (JH-diol)
NMR data for JH-diol-a: 1H NMR (500 MHz, DMSO-d6): δ 5.65 (s, 1 H), 5.07 (m, 1 H), 4.25 (d, J = 5.6 Hz, 1 H), 4.01 (s, 1 H), 3.57 (s, 3 H), 3.02 (ddd, J = 10.0, 5.6, 2.5 Hz, 1 H), 2.15 (m, 2 H), 2.15-2.11 (m, 2 H), 2.09 (s, 3 H), 1.87 (m, 2 H), 1.60 (m, 1 H), 1.56 (s, 3 H), 1.15 (m, 1 H), 1.02 (s, 3 H), 0.97 (s, 3 H); 13C NMR (125 MHz): δ 166.1, 159.7, 135.9, 122.2, 114.7, 76.6, 71.5, 50.4, 39.7, 36.2, 29.4, 26.1, 25.1, 24.2, 18.2, 15.7.
NMR data for JH-diol-b: 1H NMR (500 MHz, DMSO-d6): δ 5.65 (s, 1 H), 5.08 (m, 1 H), 3.74 (dd, J = 10.0, 3.0 Hz, 1 H), 3.57 (s, 3 H), 2.15 (m, 2 H), 2.15-2.11 (m, 2 H), 2.10 (m, 1 H), 2.09 (s, 3 H), 1.96 (m, 1 H), 1.57 (s, 3 H), 1.48 (m, 1 H), 1.41 (m, 1 H), 1.21 (s, 3 H), 1.08 (s, 3 H); 13C NMR (125 MHz): 166.1, 159.7, 134.9, 123.0, 114.7, 82.6, 79.4, 50.4, 39.7, 35.7, 29.2, 27.7, 25.1, 22.8, 18.2, 15.6. HRMS (m/z): [M+H]+ calcd. For C16H29O4, 285.2060; found, 285.2025; [M+Na]+ calcd. For C16H28O4Na, 307.1885; found, 307.1887; [α]D = 0.0 (MeOH).
Characterization data of methyl (2E,6E)-10-hydroxy-11-formyl-3,7,11-trimethyl-2,6-dodecadienoate (compound 2)
1H NMR (800 MHz, DMSO-d6): δ 8.23 (s, 1 H), 5.65 (q, J = 1.0 Hz, 1H), 5.05 (t, J = 6.9, 1 H), 4.62 (s, 1 H), 4.52 (d, J = 10.2 Hz, 1 H), 3.57 (s, 3 H), 2.15 (m, 2 H), 2.11 (m, 2 H), 2.09 (d, J = 1 Hz, 3 H), 1.93 (m, 1 H), 1.84 (m, 1 H), 1.75 (m, 1 H), 1.55 (s, 3 H), 1.46 (m, 1 H), 1.04 (s, 3 H), 1.03 (s, 3 H); 13C NMR (200 MHz): δ 166.2, 162.3, 159.9, 134.6, 123.4, 114.7, 79.7, 70.2, 50.4, 39.6, 35.7, 26.9, 25.2, 25.1, 25.1, 18.3, 15.6; HRMS (m/z): [M+H]+ calcd. for C17H29O5, 313.2010; found, 313.2010. [M+Na]+ calcd. for C17H28O5Na, 335.1828; found, 335.1831.
Characterization data of methyl (2, 6, 10) -3,7,11-trimethyl-2,6-dodecadienoate (MF)
HRMS (m/z): [M+H]+ calcd. for C16H27O2, 251.2006; found, 251.2007.
Confrontation with D. melanogaster larvae
Fungal strains were point-inoculated (1000 conidia in 1 µl Ringer solution) on 3 ml standard Drosophila medium [30] filled in 3.5 cm diameter Petri dishes. Prior to the transfer of ten sterile D. melanogaster larvae per plate, colonies were pre-incubated for two days at 25°C and constant darkness. Colonies were exposed to insects for four days. Subsequently, insects were removed and the plates snap frozen for metabolite profile analysis. Quantitative metabolite profile analyses were performed on groups of five biological replicates. Statistical analysis was performed with pair-wise comparisons using the student's t-test procedure. Evaluation of insect fitness followed the procedure described in Trienens et al. [30]. We confronted the larval stage of the fruit fly Drosophila melanogaster (wild type Oregon R strain) with the reference NID545 or the JH-producer strain NID477. Sterile two-day first-instar larvae were exposed to A. nidulans colonies growing on autoclaved standard Drosophila culture medium in 2 ml micro-tubes. There were N = 20 experimental units per fungal treatment and N = 10 mold-free control units. Insect developmental success was monitored in terms of (1) larva-to-pupa survival and development time, (2) emergence of flies, and (3) fly dry weight. Short development time and high body mass are considered to be positively correlated with fitness in Drosophila [41]. Experimental tubes were checked for pupae and emerged flies at about 2 p.m. each day for a total of 14 days after larval transfer. Emerged flies were removed from the tubes and stored deep-frozen. Subsequently, flies were lyophilized for 24 hours and the dry weight of all flies within each experimental unit was determined as a single value using a micro-balance.
Acknowledgments
The authors would like to acknowledge the Danish Instrument Center for NMR Spectroscopy of Biological Macromolecules for the use of NMR facilities and Jesper Mogensen for technical assistance.
Funding Statement
This work was supported by the Danish Research Agency for Technology and Production, grant # 09-064967, and the Research School for Biotechnology at the faculty of Life Sciences, University of Copenhagen. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Aminov RI (2009) The role of antibiotics and antibiotic resistance in nature. Environmental Microbiology 11: 2970–2988. [DOI] [PubMed] [Google Scholar]
- 2. Kempken F, Rohlfs M (2010) Fungal secondary metabolite biosynthesis - a chemical defence strategy against antagonistic animals? Fungal Ecology 3: 107–114. [Google Scholar]
- 3. Rohlfs M, Churchill ACL (2011) Fungal secondary metabolites as modulators of interactions with insects and other arthropods. Fungal Genetics and Biology 48: 23–34. [DOI] [PubMed] [Google Scholar]
- 4. Newman DJ, Cragg GM (2007) Natural products as sources of new drugs over the last 25 years. Journal of Natural Products 70: 461–477. [DOI] [PubMed] [Google Scholar]
- 5. Reddy K, Salleh B, Saad B, Abbas H, Abel C, et al. (2010) An overview of mycotoxin contamination in foods and its implications for human health. Toxin Reviews 29: 3–26. [Google Scholar]
- 6. Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, et al. (2005) Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae . Nature 438: 1105–1115. [DOI] [PubMed] [Google Scholar]
- 7. Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, et al. (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotech 25: 221–231 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 8. Gross H (2007) Strategies to unravel the function of orphan biosynthesis pathways: recent examples and future prospects. Applied Microbiology and Biotechnology 75: 267–277. [DOI] [PubMed] [Google Scholar]
- 9. Brakhage AA, Schroeckh V (2011) Fungal secondary metabolites - Strategies to activate silent gene clusters. Fungal Genetics and Biology 48: 15–22. [DOI] [PubMed] [Google Scholar]
- 10. Chiang YM, Chang SL, Oakley BR, Wang CCC (2011) Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Current Opinion in Chemical Biology 15: 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bok JW, Chiang YM, Szewczyk E, Reyes-Domingez Y, Davidson AD, et al. (2009) Chromatin-level regulation of biosynthetic gene clusters. Nature Chemical Biology 5: 462–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bergmann S, Funk AN, Scherlach K, Schroeckh V, Shelest E, et al. (2010) Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic non-ribosomal peptide synthetase gene cluster. Applied and Environmental Microbiology 76: 8143–8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hansen BG, Salomonsen B, Nielsen MT, Nielsen JB, Hansen NB, et al. (2011) Versatile enzyme expression and characterization system for Aspergillus nidulans, with the Penicillium brevicompactum polyketide synthase gene from the mycophenolic acid gene cluster as a test case. Applied and Environmental Microbiology 77: 3044–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nielsen ML, Nielsen JB, Rank C, Klejnstrup ML, Holm DK, et al. (2011) A genome-wide polyketide synthase deletion library uncovers novel genetic links to polyketides and meroterpenoids in Aspergillus nidulans . Fems Microbiology Letters 321: 157–166. [DOI] [PubMed] [Google Scholar]
- 15. Nagaraju GPC (2007) Is methyl farnesoate a crustacean hormone? Aquaculture 272: 39–54. [Google Scholar]
- 16. Wilson TG (2004) The molecular site of action of juvenile hormone and juvenile hormone insecticides during metamorphosis: how these compounds kill insects. Journal of Insect Physiology 50: 111–121. [DOI] [PubMed] [Google Scholar]
- 17. Gilbert LI, Granger NA, Roe RM (2000) The juvenile hormones: historical facts and speculations on future research directions. Insect Biochemistry and Molecular Biology 30: 617–644. [DOI] [PubMed] [Google Scholar]
- 18. Marrs TC (2012) Toxicology of insecticides to mammals. Pest Management Science 68: 1332–1336. [DOI] [PubMed] [Google Scholar]
- 19. Minakuchi C, Riddiford LM (2006) Insect juvenile hormone action as a potential target of pest management. Journal of Pesticide Science 31: 77–84. [Google Scholar]
- 20. Andersen MR, Vongsangnak W, Panagiotou G, Salazar MP, Lehmann L, et al. (2008) A trispecies Aspergillus microarray: Comparative transcriptomics of three Aspergillus species. Proceedings of the National Academy of Sciences of the United States of America 105: 4387–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panagiotou G, Andersen MR, Grotkjaer T, Regueira TB, Hofmann G, et al. (2008) Systems analysis unfolds the relationship between the phosphoketolase pathway and growth in Aspergillus nidulans Plos One 3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Panagiotou G, Andersen MR, Grotkjaer T, Regueira TB, et al. (2009) Studies of the production of fungal polyketides in Aspergillus nidulans by using systems biology tools. Applied and Environmental Microbiology 75: 2212–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Andersen MR, Nielsen JB, Klitgaard A, Petersen LM, Zachariasen M, et al. (2013) Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proceedings of the National Academy of Sciences of the United States of America 110: E99–E107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. MacPherson S, Larochelle M, Turcotte B (2006) A fungal family of transcriptional regulators: The zinc cluster proteins. Microbiology and Molecular Biology Reviews 70: 583–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic Local Alignment Search Tool. Journal of Molecular Biology 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 26. Kuhnz W, Rembold H (1981) C-13 nuclear magnetic-resonance spectra of juvenile hormone-III, some of Its derivatives, and of analogous compounds. Organic Magnetic Resonance 16: 138–140. [Google Scholar]
- 27.Belles X, Martin D, Piulachs MD (2005) The mevalonate pathway and the synthesis of juvenile hormone in insects. PALO ALTO: ANNUAL REVIEWS. 199. [DOI] [PubMed]
- 28. Toong YC (1988) Schooley DA, Baker FC (1988) Isolation of insect juvenile Hormone-III from a plant. Nature 333: 170–171. [Google Scholar]
- 29. Flatt T, Tu MP, Tatar M (2005) Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. Bioessays 27: 999–1010. [DOI] [PubMed] [Google Scholar]
- 30. Trienens M, Keller NP, Rohlfs M (2010) Fruit, flies and filamentous fungi - experimental analysis of animal-microbe competition using Drosophila melanogaster and Aspergillus mould as a model system. Oikos 119: 1765–1775. [Google Scholar]
- 31. Asadollahi MA, Maury J, Patil KR, Schalk M, Clark A, et al. (2009) Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metabolic Engineering 11: 328–334. [DOI] [PubMed] [Google Scholar]
- 32. Schroeckh V, Scherlach K, Nutzmann HW, Shelest E, Schmidt-Heck W, et al. (2009) Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans . Proceedings of the National Academy of Sciences of the United States of America 106: 14558–14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nielsen ML, Albertsen L, Lettier G, Nielsen JB, Mortensen UH (2006) Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans . Fungal Genetics and Biology 43: 54–64. [DOI] [PubMed] [Google Scholar]
- 34.Cove DJ (1966) Induction and Repression of Nitrate Reductase in Fungus Aspergillus Nidulans. Biochimica et Biophysica Acta 113: : 51–&. [DOI] [PubMed] [Google Scholar]
- 35. Frisvad JC, Samson RA (2004) Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicilia and their mycotoxons. Studies in Mycology 49: 1–173. [Google Scholar]
- 36. Norholm MHH (2010) A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. Bmc Biotechnology 10 10.1186/1472-6750-10-21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Southern E (2006) Southern blotting. Nature Protocols 1: 518–525. [DOI] [PubMed] [Google Scholar]
- 38. Smedsgaard J (1997) Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. Journal of Chromatography A 760: 264–270. [DOI] [PubMed] [Google Scholar]
- 39.Laatsch H (2009) AntiBase 2009. The natural compound identifier. Wiley-VCH GmpH & Co, Weinheim, Germany.
- 40. Larsen TO, Frisvad JC (1994) A simple method for collection of volatile metabolites from fungi based on diffusive sampling from petri dishes. Journal of Microbiological Methods 19: 297–305. [Google Scholar]
- 41. Rohlfs M (2006) Genetic variation and the role of insect life history traits in the ability of Drosophila larvae to develop in the presence of a competing filamentous fungus. Evolutionary Ecology 20: 271–289. [Google Scholar]