Abstract
Most EEG‐fMRI studies in epileptic patients are analyzed using the general linear model (GLM), which assumes a known hemodynamic response function (HRF) to epileptic spikes. In contrast, independent component analysis (ICA) can extract blood‐oxygenation level dependent (BOLD) responses without imposing constraints on the HRF. ICA might therefore detect responses that vary in time and shape, and that are not detected in the GLM analysis. In this study, we compared the findings of ICA and GLM analyses in 12 patients with idiopathic generalized epilepsy. Spatial ICA was used to extract independent components from the functional magnetic resonance imaging (fMRI) data. A deconvolution method identified component time courses significantly related to the generalized EEG discharges, without constraining the shape of the HRF. The results from the ICA analysis were compared to those from the GLM analysis. GLM maps and ICA maps showed significant correlation and revealed BOLD responses in the thalamus, caudate nucleus, and default mode areas. In patients with a low rate of discharges per minute, the GLM analysis detected BOLD signal changes within the thalamus and the caudate nucleus that were not revealed by the ICA. In conclusion, ICA is a viable alternative technique to GLM analyses in EEG‐fMRI studies related to generalized discharges. This study demonstrated that the BOLD response largely resembles the standard HRF and that GLM analysis is adequate. However, ICA is more dependent on a sufficient number of events than GLM analysis. Hum Brain Mapp, 2011. © 2010 Wiley‐Liss, Inc.
Keywords: independent component analysis, general linear model, EEG‐fMRI, epilepsy
INTRODUCTION
Combined functional magnetic resonance imaging (fMRI) and EEG recording is a noninvasive method that allows to investigate blood‐oxygenation level dependent (BOLD) signal changes related to interictal or ictal discharges seen on the scalp EEG [Gotman et al., 2006; Laufs and Duncan, 2007]. Over the last years, EEG‐fMRI studies have largely contributed to the understanding of pathophysiological mechanisms of both focal and generalized epilepsy. Most of these studies were based on the general linear model (GLM) [Worsley and Friston, 1995; Worsley et al., 2002] in which identified interictal epileptiform discharges (IED) are convolved with the hemodynamic response function (HRF) to detect voxels significantly correlated with the model time course. The HRF, however, may vary between subjects, for different brain regions or for different types of IED [Aguirre et al., 1998; Benar et al., 2003; Handwerker et al., 2004; Menz et al., 2006]. Furthermore, for very frequent IEDs, the GLM analysis may no longer be valid [Jacobs et al., 2008] and BOLD signal responses may be undetected in a large proportion of investigated patients. These limitations of GLM‐based analyses demand alternative analysis techniques.
In contrast to model‐based GLM analyses, data‐driven fMRI analyses such as Independent Component Analysis (ICA) are not constrained by a fixed model‐based hypothesis [McKeown et al., 1998]. ICA divides the data into a large set of independent components, each showing brain activation patterns with a common time course. ICA methods do not impose prior constraints on the shape of the HRF and therefore might detect responses that would not have been revealed by a GLM analysis. However, the challenging step is to identify the BOLD components that are relevant to the investigated event (e.g., the spike‐related BOLD response). In patients with epilepsy, ICA was applied for the first time by Rodionov et al. [2007]. Components were selected if they showed a spatial overlap with positive BOLD signal changes in the GLM analysis and a correlation of the components time course with the model used in the GLM analysis [Rodionov et al., 2007]. Therefore, in this approach, only some of the potential spike‐related components were evaluated (namely, those with corresponding activation in the GLM analysis) and the selection of components depended on the canonical HRF used in the GLM. An ICA method more independent from the canonical HRF was described by LeVan and Gotman [2009], in which a deconvolution method identified component time courses significantly related to simulated focal spikes without constraining the shape of the HRF. When compared to results from the GLM analysis, the ICA was superior to detect BOLD responses with variable HRFs [LeVan and Gotman, 2009]. Applied to ictal EEG‐fMRI data, the method could detect dynamic ictal BOLD responses in focal seizures [LeVan et al., 2010]. In the current study, we applied this ICA approach to patients with idiopathic generalized epilepsy (IGE) and generalized spike wave discharges (GSW) to test whether ICA could be an alternative technique to the GLM‐based analyses most commonly used in spike‐related EEG‐fMRI studies. We decided to test the method on patients with IGE for the following reasons: In comparison to focal discharges, GSW are often associated with more robust results in consistent areas, thus allowing a good comparison between the two methods. Common findings are thalamic activation and deactivation in default mode areas and the caudate nucleus [Aghakhani et al., 2004; Gotman et al., 2005; Hamandi et al., 2006; Moeller et al., 2008a]. Furthermore, early BOLD signal changes not detected by analysis with a standard HRF have been described in some patients with IGE [Hawco et al., 2007; Moeller et al., 2008b]. One might therefore assume that these BOLD signal changes could be better detected by ICA.
SUBJECTS AND METHODS
Twelve patients with IGE and GSW were selected sequentially from our EEG‐fMRI database of patients scanned at 3 Tesla. Patients participated in the research after giving written informed consent, in accordance with the local ethics committee.
EEG Acquisition
The EEG acquisition was performed with 25 MR compatible electrodes (Ag/AgCl) placed on the scalp using the 10–20 (21 usual electrodes without Fpz and Oz, reference at FCz) and 10–10 (F9, T9, P9, F10, T10, and P10) electrode placement systems. Two electrodes located on the back recorded the electrocardiogram. To minimize movement artifacts and for the patient's comfort, the head was immobilized with a pillow filled with foam microspheres (Siemens, Germany). Data were transmitted from a BrainAmp amplifier (Brain Products, Munich, Germany, 5‐kHz sampling rate) via an optic fiber cable to the EEG monitoring computer located outside the scanner room.
fMRI Acquisition
Functional images were continuously acquired using a 3‐T MR scanner (Siemens, Trio, Germany). A T1 weighted anatomical acquisition was first done (1‐mm slice thickness, 256 × 256 matrix; TE = 7.4 ms and TR = 23 ms; flip angle 30°) and used for superposition with the functional images. The functional data were acquired in series of 7–14 runs of 6 min each using a T2* weighted EPI sequence (5 × 5 × 5 mm voxels, 25 slices, 64 × 64 matrix; TE = 30 ms and TR = 1750 ms; flip angle 90°). No sedation was given.
EEG Processing
The Brain Vision Analyser software (Brain Products) was used for off‐line correction of the gradient artifact and filtering of the EEG signal. This software uses the method described by Allen et al. [2000]. A 50‐Hz low‐pass filter was also applied to remove remaining high‐frequency artifact. The ballistocardiogram artifact was removed by ICA [Bénar et al., 2003; Srivastava et al., 2005]. A neurologist reviewed the EEG recording and selected the GSW.
Data Processing
The EPI images were motion corrected and smoothed (6‐mm full width at half maximum) using the software package from the Brain Imaging Center of the Montreal Neurological Institute (http://www.bic.mni.mcgill.ca/). Following these preprocessing steps, the 6‐min runs containing GSW were concatenated and analyzed first using a standard GLM method, and then using ICA.
GLM Analysis
The GLM analysis was performed using the FMRISTAT software package [Worsley et al., 2002]. Temporal autocorrelations were accounted for by fitting an autoregressive (AR) model of order 1 and low frequency drifts in the signal were modeled with a third‐order polynomial fitted to each run. A regressor for the identified GSW was built using the timing and duration of each event convolved with four HRFs with peaks at 3, 5, 7, and 9 s [Bagshaw et al., 2004]. A statistical t‐map was obtained for each regressor in the fMRI analysis (fMRIstat) [Worsley et al., 2002]. At each voxel, the maximum t‐value was taken from the four individual t‐maps created with the four HRFs.
The EPI frames were realigned together using a linear six‐parameter rigid‐body transformation (three translations and three rotations) to correct for movement effects. The six parameters used for the realignment were also integrated in the analysis as confound regressors in the GLM to account for residual movement artifacts. To be significant, a response needed to have a minimum of five contiguous voxels with a |t| > 3.1, corresponding to P < 0.05 corrected for the multiple comparisons resulting from the number of voxels in the brain (correction based on random field theory and spatial extent [Friston et al., 1994]) and the use of four HRFs (Bonferroni correction). The t‐map results were represented using red‐yellow scale corresponding to positive BOLD changes (activation) and blue‐white scale for negative BOLD changes (deactivation).
ICA Analysis
The method requires several steps: the preprocessed images are first decomposed into spatially independent components. To identify which components are related to the GSW, arbitrary HRF shapes time‐locked to the GSW are fitted to the time courses of each component. Finally, activation maps are generated based on the amplitude and sign of the fitted HRFs. The following will describe the analysis steps in detail.
Preprocessing and ICA decomposition
The ICA was performed using the MELODIC toolbox [Beckmann and Smith, 2004] implemented in FSL [Smith et al., 2004]. The fMRI slices within each volume were temporally realigned using sinc interpolation with Hanning windowing, and high‐pass filtered to remove drift effects, separately for each concatenated 6‐min run. Voxels outside the brain were removed using a mask created from the averaged EPI volume [Smith, 2002]. The independent components were then computed by an iterative fixed‐point method maximizing the non‐Gaussianity of the sources, which has been shown to be equivalent to maximizing statistical independence [Hyvarinen and Oja, 2000]. The number of extracted components sources was derived from previous studies that showed reproducible components from 37 to 353 for each patient [LeVan and Gotman, 2009; LeVan et al., 2010] depending on the number of runs. For patients with 3 to 6 runs we chose to extract 100 components, whereas for patients with 9 to 14 runs 200 components were extracted.
Identification of GSW components
The application of ICA on fMRI data typically yields a large number of components, which include any sources of fluctuation such as physiological artifacts, residual motion, and cerebral BOLD activity. Although the ICA decomposition is purely data‐driven, modeling assumptions are required to detect components relevant to the current study, namely the hemodynamic changes related to the GSW events. For this purpose, the ICA time courses were modeled as blocks with the same timings and durations as the GSWs, and convolved with a Fourier basis set. It was assumed that the BOLD response to the GSWs could be contained within an interval from 10 s before to 20 s after the marked events. The Fourier basis set then consisted of five sine and cosine functions of periods equal to submultiples of this 30‐s time interval, and could thus represent arbitrary HRF shapes [Josephs et al., 1997]. The time interval was chosen to accommodate the HRF variability that has been reported in the literature. For example, for the canonical HRF, the peak is at 5.4 s, with the undershoot a further 5.4 s after the peak [Glover, 1999]. However, HRFs have also been observed peaking as early as 3 s before or as late as 9 s after EEG events [Bagshaw et al., 2004; Hawco et al., 2007; Moeller et al., 2008b], so the selected time interval allows for a reasonable margin around these reported values.
The fitting of the Fourier basis set then produced a deconvolved HRF from the time course of each component extracted by ICA. Components significantly related to the GSW were then identified by an F‐test (P < 0.05, corrected for the number of components), with the motion parameters used as confounds as in the GLM analysis. Noise was modeled as an AR process estimated from the residuals using the Yule‐Walker equations. The AR order was determined by performing the estimation using gradually increasing AR orders, until the residuals no longer contained significant autocorrelations (Ljung‐Box test, P < 0.01). The AR parameters were used to compute an adjusted F‐statistic with appropriate effective degrees of freedom [Kruggel et al., 2002]. Statistically significant F‐values were then indicative of component time courses showing significant signal changes time‐locked to the GSW.
Generation of activation maps
The identified time courses had unconstrained HRF shapes, and were each associated with their own spatial distribution. It was then necessary to threshold these spatial topographies to identify the voxels that were associated with the identified ICA time courses. As implemented in the MELODIC toolbox [Beckmann and Smith, 2004], this thresholding was accomplished by normalizing the voxel intensities by a noise amplitude estimated from the ICA residuals, and then modeling these intensities by a mixture of Gaussian and two Gamma distributions, representing nonactivated voxels, positive voxel intensities, and negative voxel intensities, respectively [Beckmann and Smith, 2004]. A voxel was considered as activated if it had a 99.9% or greater probability of not belonging to the nonactivated voxel distribution.
HRFs fitted to the GSW components were investigated to determine the sign of the HRF peaks. The peak was defined as the maximum absolute value of the fitted HRF. Any data outside a window of ±5 s around the peak were then considered as baseline. This window should accommodate the peak width; for example, the canonical HRF has a full‐width at half‐maximum of 5.2 s [Glover, 1999]. The prominence of the peak was then quantified by dividing its amplitude by the standard deviation of the baseline. HRFs with a ratio inferior to 3 were excluded from further analysis. The value of 3 was determined from Monte‐Carlo simulations using false, randomly determined GSW timings, and computing the threshold that would result in the rejection of 95% of the simulated HRFs. This criterion was used to exclude prolonged oscillatory HRFs without a clear peak, as these HRFs are most likely artifactual [Lemieux et al., 2008]. The shape of the HRF within the ±5‐s window was still arbitrary, and the position of the peak with respect to the EEG event was also unconstrained. The identification of the HRF peak then allowed to differentiate between BOLD activations (positive peak) and deactivations (negative peak). An amplitude map was constructed from the signed peak amplitudes and compared with the GLM t‐statistic map using Spearman's correlation coefficient.
Exploration of ICA specific components
GLM maps and ICA were compared visually to detect areas of BOLD response that were only detected by the ICA analysis. In these components, the shape of the HRF was compared to the shape of HRFs in components in the thalamus, caudate nucleus and default mode areas, regions that are usually detected by GLM analysis.
RESULTS
On average, 0.4 ± 0.3 (mean ± SD) spontaneous brief GSW per minute were recorded with a mean duration of 2.3 ± 1.0 s. The duration per minute (product of events per minute by mean duration) was used as a measure including both duration and occurrence rate of events, thus allowing the comparison of patients. It averaged 1.1 ± 0.9 s across patients. The results for each patient are listed in Figure 1 and Table I.
Figure 1.
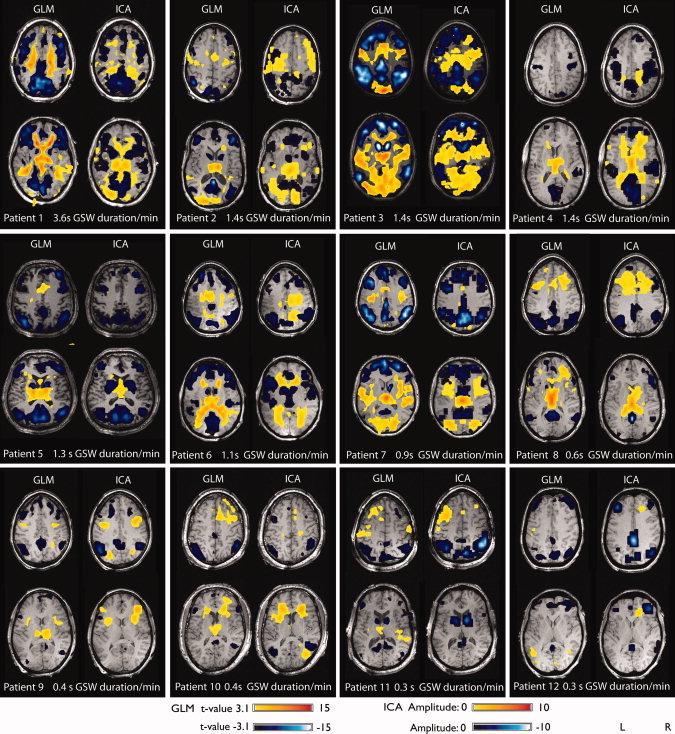
Results of GLM analysis and ICA for every patient. Patients are arranged according to GSW duration per minute.
Table I.
Clinical data and results summary
| Patients | Sex | Diagnosis | Age/onset (yrs) | GSW/mina | Thalamus | Caudate nucleus | Default mode | IC | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GLM | ICA | GLM | ICA | GLM | ICA | ||||||
| 1 | F | CAE | 22/6 | 3.6 | ↑ | ↑ | ↓ | ↓ | ↓ | ↓ | 97 (10) |
| 2 | F | JAE | 51/3 | 1.4 | ↑ | ↑ | ↓ | ↓ | ↓ | ↓ | 33 (4) |
| 3 | F | JAE | 26/10 | 1.4 | ↑ | ↑ | ↓ | ↓ | ↓ | ↓ | 89 (11) |
| 4 | M | JME | 27/13 | 1.4 | ↑ | ↑ | — | — | ↓ | ↓ | 27 (6) |
| 5 | M | JME | 46/15 | 1.3 | ↑ | ↑ | ↓ | ↓ | ↓ | ↓ | 9 (2) |
| 6 | M | JME | 30/15 | 1.1 | ↑ | ↓ | ↓ | ↓ | ↓ | ↓ | 39 (9) |
| 7 | F | CAE | 36/? | 0.9 | ↑ | ↑ | ↓ | ↓ | ↓ | ↓ | 23 (6) |
| 8 | F | JAE | 19/8 | 0.6 | ↑ | ↑ | ↓ | — | ↓ | ↓ | 19 (3) |
| 9 | F | EMA | 27/4 | 0.4 | ↑ | — | — | — | ↓ | ↓ | 12 (4) |
| 10 | M | JAE | 41/10 | 0.4 | ↑ | — | — | — | ↓ | ↓ | 12 (4) |
| 11 | M | JME | 25/14 | 0.3 | ↑ | — | ↓ | ↓ | ↓ | ↓ | 10 (5) |
| 12 | F | JME | 38/19 | 0.3 | — | — | — | — | ↓ | ↓ | 6 (3) |
CAE, childhood absence epilepsy; JAE, juvenile absence epilepsy; EMA, eyelid myoclonia with absences; JME, juvenile myoclonic epilepsy.
GSW duration in seconds per minute (product of events per minute and the mean duration), ↑ positive BOLD signal change, ↓ negative BOLD signal change, IC: number of components significantly related to GSW and having a signal‐to‐noise ratio >3; number of components in default mode area are mentioned in brackets, for thalamus and caudate nucleus not more than 1 component was detected.
GLM Results
All 12 patients showed GSW‐related BOLD signal changes. The volume of significant BOLD responses was strongly correlated with the GSW duration per minute (Pearson's r = 0.68, P = 0.01). Thalamic activation was detected in 11 patients (92%, mean t‐value: 7.9 range 3.5–14.4). The only patient who did not show a thalamic activation had the lowest GSW duration per minute among the investigated patients (0.3 s/min). Although deactivation in default mode areas was detected in all 12 patients (100%, mean t‐value: −10.3 range −5.5 to −18.8), deactivation of the caudate nucleus was found only in 8 patients (67%, mean t‐value: −9.1 range −5.6 to −13.6). Three patients in whom no BOLD signal changes in the caudate nucleus were found had low GSW duration per minute (0.3–0.4 s/min), while one patient had a high rate of GSW duration per minute (1.4/min).
ICA Results
For all 12 patients, components related to GSW could be identified: when fitting an HRF to the time course of the components, there were between 8 and 133 components per patient in which the fitted HRF was significantly related to the GSW. Among these components, those in which the peak of the fitted HRF showed a ratio of peak amplitude to baseline standard deviation (signal‐to‐noise ratio, SNR) of less than 3 were rejected. All patients showed significantly correlated components with SNR greater than 3, ranging from 6 to 97 components per patient. Although not more than one component per patient was detected in the thalamus and the caudate nucleus, the number of components in default mode areas varied from 2 to 11 (Table I). All components of one patient were combined in one map, using the sign of the fitted HRF peak to distinguish between BOLD activation (positive peak) and BOLD deactivation (negative peak). The volume of significant BOLD responses was strongly correlated with the GSW duration per minute (Pearson's r = 0.80, P = 0.002). Thalamic BOLD signal changes were detected in eight patients (67%), in seven patients these BOLD signal changes were classified as activations, whereas in one patient (Patient 6) the BOLD signal change was classified as deactivation. However, when looking at the associated HRF in this patient a clear positive peak at 4 s preceded a negative peak at 10 s (Fig. 2). The negative peak was of slightly higher amplitude than the positive peak. For the generation of the map, the sign of the highest peak was used to distinguish BOLD activations (positive peak) from deactivations (negative peak), so the BOLD signal was classified as a deactivation in this patient. The negative peak can be interpreted as an undershoot of the preceding activation and was also seen in Patients 1, and 3–8 (Fig. 2). The four patients who did not show a thalamic activation had the lowest GSW duration per minute among the investigated patients (0.3–0.4 s/min). Deactivation in default mode areas was detected in all 12 patients (100%), while deactivation of the caudate nucleus was found only in seven patients (58%, mean). Four patients in whom no BOLD signal changes in the caudate nucleus were found had a low GSW duration per minute (0.3–0.6 s/min), while one patient had a higher GSW duration per minute (1.4/min).
Figure 2.
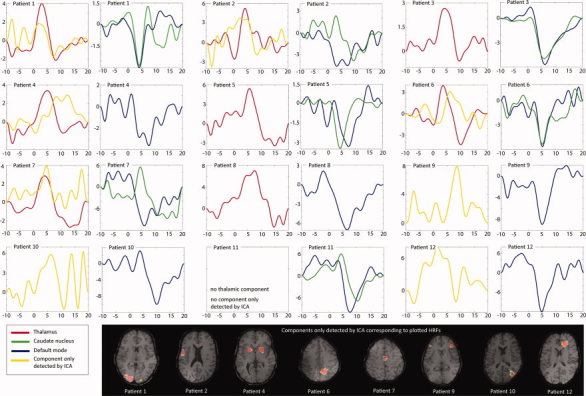
HRFs for components in the thalamus, caudate nucleus, and default mode areas. Additionally, components that were only detected by ICA are shown with their corresponding HRF. Please note that the amplitude scales of BOLD signal changes were adjusted to the HRF and vary from patient to patient.
Comparison Between GLM and ICA Results
In all patients, both methods detected GSW‐related BOLD signal changes. All 12 patients showed significant correlations (P < 0.001) between the GLM and ICA maps, with an average correlation coefficient of 0.46. The correlation between duration of GSW per minute and the volume of significant BOLD responses was higher for ICA (Pearson's r = 0.80, P = 0.002) than for the GLM analysis (Pearson's r = 0.68, P = 0.01). All patients in whom no BOLD signal changes in the thalamus were detected by ICA had a low GSW duration per minute (0.3–0.4 s/min). In three of these patients, the GLM analysis was able to detect BOLD signal changes. However, two of these patients (pts 10 and 11) showed t‐values just above the threshold of 3.1 (3.5 and 3.6), while the third patient (pt 9) showed a higher t‐value (t = 6.5). In one patient, unilateral deactivation in the caudate nucleus was only found in the GLM analysis (pt 8, GSW duration: 0.6 s/min) but not with the ICA. In all other patients, both methods showed a deactivation in the caudate nucleus in the same patients.
The comparison between GLM maps and ICA maps revealed a few areas of BOLD signal changes that were only detected by ICA in eight patients (Fig. 2). The localization of these components did not follow a common pattern, but were found in the frontal, parietal and occipital cortex and in the putamen. Although components in areas detected by both analyses showed shapes similar to the canonical HRF, the HRFs in components only detected by ICA showed variable shapes, different from the canonical HRF in most cases. However, one component that was only detected by ICA showed an HRF resembling the canonical HRF (Patient 9). When lowering the statistical threshold from 3.1 to 2.8, the GLM analysis also revealed BOLD signal changes in this area.
DISCUSSION
This study evaluated the application of ICA in EEG‐fMRI studies in IGE patients by comparing it to the results revealed by the GLM analysis. In a previous study, ICA was applied in patients with focal epilepsy: in that study, components were selected if they showed a spatial overlap with positive BOLD signal changes in the GLM analysis and a correlation of the component with the model used in the GLM analysis [Rodionov et al., 2007]. In each patient, one component matching the selection criteria was found. However, this approach does not allow a direct comparison between both methods since the selection of the components depended on the GLM analysis. The current study uses an ICA‐based deconvolution method that is independent of the results of the GLM analysis and thus allows a comparison between the methods. The study shows that GLM analysis and ICA yield similar results in GSW‐related EEG‐fMRI studies with highly significant correlations (P < 0.001) between the GLM and ICA maps, with an average correlation coefficient of 0.46. In agreement with previous EEG‐fMRI studies in IGE patients, all using GLM analysis, structures consistently involved were the thalamus, caudate nucleus, and default mode areas. Although a deactivation in default mode areas was detected in all patients, thalamic BOLD signal changes were only detected in 92% (GLM) and 67% (ICA) and deactivation in the caudate nucleus in 67% (GLM) and 58% (ICA) of the patients. The high concordance between the analyses by both methods indicates that BOLD responses with an HRF very different from the canonical response do not appear present at the time of GSW. This is confirmed by the analysis of the HRF, which showed a canonical shape in components of the thalamus, caudate nucleus, and default mode areas. In a few components that were only detected by ICA, noncanonical HRFs occurred. However, these components were rare and did not show any consistent pattern in shape of the HRF or in location. Therefore, one can infer that GSW‐related BOLD signal changes have a canonical response in almost all areas. Such a conclusion can only be reached as a result of the ICA analysis. The advantage of ICA over GLM to detect BOLD responses with variable shapes of HRF different from the canonical HRF did not result in a higher yield of BOLD signal in the patients studied. Although patients with IGE in whom BOLD signal changes preceding the GSW were not detected by the standard GLM analysis have been described [Hawco et al., 2007, Moeller et al., 2008b], this phenomenon seems to apply only to a small subset of IGE patients and might be more frequent in specific EEG patterns such as polyspike wave discharges [Moeller et al., 2008b]. The majority of IGE patients, however, seem to show BOLD responses with an HRF similar to the canonical shape. In this case, the GLM, which is specifically tuned to detect such HRFs, is thus optimal.
Although yielding similar results in patients with frequent GSW, ICA seemed to be less sensitive for patients with infrequent GSW: Most of the patients who did not show BOLD signal changes either in the thalamus or the caudate nucleus had a low rate of GSW per minute (0.3–0.6 s GSW/min). However, the GLM analysis detected BOLD signal changes despite low GSW duration per minute in three patients in the thalamus and in one patient in the caudate nucleus that were not detected in the ICA approach. This confirms that ICA greatly depends on a sufficient number of events [LeVan and Gotman, 2009]. This mirrored the correlation between the rate of GSW per minute and the volume of significant BOLD responses that was higher for ICA (Pearson's r = 0.80, P = 0.002) than for the GLM analysis (Pearson's r = 0.68, P = 0.01).
In conclusion, this study showed that ICA is a viable alternative technique to GLM analyses in EEG‐fMRI studies in IGE patients. The promise of ICA lies in the ability to detect HRF with variable time courses, but such HRFs were not consistently observed in the current study, a result that can only be stated after the analysis with ICA. Also, it appears that the GLM analysis tends to be more sensitive for infrequent GSW events.
REFERENCES
- Aghakhani Y, Bagshaw AP, Benar CG, Hawco C, Andermann F, Dubeau F, Gotman J ( 2004): fMRI activation during spike and wave discharges in idiopathic generalized epilepsy. Brain 127: 1127–1144. [DOI] [PubMed] [Google Scholar]
- Aguirre G, Zarahn E, D'Esposito M ( 1998): The variability of human BOLD hemodynamic responses. Neuroimage 8: 360–369. [DOI] [PubMed] [Google Scholar]
- Allen PJ, Josephs O, Turner R ( 2000): A method for removing imaging artifact from continuous EEG recorded during functional MRI. Neuroimage 12: 230–239. [DOI] [PubMed] [Google Scholar]
- Bagshaw AP, Aghakhani Y, Bénar CG, Kobayashi E, Hawco C, Dubeau F, Pike GB, Gotman J ( 2004): EEG‐fMRI of focal epileptic spikes: analysis with multiple haemodynamic functions and comparison with gadolinium‐enhanced MR angiograms. Hum Brain Mapp 22: 179–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann C, Smith S ( 2004): Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging 23: 137–152. [DOI] [PubMed] [Google Scholar]
- Bénar C, Aghakhani Y, Wang Y, Izenberg A, Al‐Asmi A, Dubeau F, Gotman J ( 2003): Quality of EEG in simultaneous EEG‐fMRI for epilepsy. Clin Neurophysiol 114: 569–580. [DOI] [PubMed] [Google Scholar]
- Friston K, Worsley K, Frackowiak R, Mazziotta J, Evans A ( 1994): Assessing the significance of focal activations using their spatial extent. Hum Brain Mapp 1: 210–220. [DOI] [PubMed] [Google Scholar]
- Glover G ( 1999): Deconvolution of impulse response in event‐related BOLD fMRI. Neuroimage 9: 416–429. [DOI] [PubMed] [Google Scholar]
- Gotman J, Grova C, Bagshaw A, Kobayashi E, Aghakhani Y, Dubeau F ( 2005): Generalized epileptic discharges show thalamocortical activation and suspension of the default state of the brain. Proc Natl Acad Sci USA 102: 15236–15240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotman J, Kobayashi E, Bagshaw AP, Bénar CG, Dubeau F ( 2006): Combining EEG and fMRI: A multimodal tool for epilepsy research. J Magn Reson Imaging 23: 906–920. [DOI] [PubMed] [Google Scholar]
- Hamandi K, Salek‐Haddadi A, Laufs H, Liston A, Friston K, Fish DR, Duncan JS, Lemieux L ( 2006): EEG‐fMRI of idiopathic and secondarily generalized epilepsies Neuroimage 31: 1700–1710. [DOI] [PubMed] [Google Scholar]
- Handwerker DA, Ollinger JM, D'Esposito M ( 2004): Variation of BOLD hemodynamic responses across subjects and brain regions and their effects on statistical analyses. Neuroimage 21: 1639–1651. [DOI] [PubMed] [Google Scholar]
- Hawco C, Bagshaw A, Lu Y, Dubeau F, Gotman J ( 2007): BOLD changes occur prior to epileptic spikes seen on scalp EEG. Neuroimage 35: 1450–1458. [DOI] [PubMed] [Google Scholar]
- Hyvarinen A, Oja E ( 2000): Independent component analysis: Algorithms and applications. Neural Netw 13: 411–430. [DOI] [PubMed] [Google Scholar]
- Jacobs J, Hawco C, Kobayashi E, Boor R, LeVan P, Stephani U, Siniatchkin M, Gotman J ( 2008): Variability of the hemodynamic response as a function of age and frequency of epileptic discharge in children with epilepsy. Neuroimage 40: 601–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs O, Tuner R, Friston KJ ( 1997): Event‐related fMRI. Hum Brain Mapp 5: 243–248. [DOI] [PubMed] [Google Scholar]
- Kruggel F, Pelegrini‐Issac M, Benali H ( 2002): Estimating the effective degrees of freedom in univariate multiple regression analysis. Med Image Anal 6: 63–75. [DOI] [PubMed] [Google Scholar]
- Laufs H, Duncan JS ( 2007): Electroencephalography/functional MRI in human epilepsy: What it currently can and cannot do. Curr Opin Neurol 4: 417–423. [DOI] [PubMed] [Google Scholar]
- Lemieux L, Laufs H, Carmichael D, Paul J, Walker M, Duncan J ( 2008): Noncanonical spike‐related BOLD responses in focal epilepsy. Hum Brain Mapp 29: 329–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVan P, Gotman J ( 2009): Independent component analysis as a model‐free approach for the detection of BOLD changes related to epileptic spikes: A simulation study. Hum Brain Mapp 30: 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVan P, Tyvaert L, Moeller F, Gotman J ( 2010): Independent component analysis reveals dynamic ictal BOLD responses in EEG‐fMRI data from focal epilepsy patients. Neuroimage 49: 366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown M, Makeig S, Brown G, Jung T, Kindermann S, Bell A, Sejnowski T ( 1998): Analysis of fMRI data by blind separation into independent spatial components. Hum Brain Mapp 6: 160–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz M, Neumann J, Muller K, Zysset S ( 2006): Variability of the BOLD response over time: An examination of within‐session differences. Neuroimage 32: 1185–1194. [DOI] [PubMed] [Google Scholar]
- Moeller F, Siebner H, Wolff S, Muhle H, Granert O, Jansen O, Stephani U, Siniatchkin M ( 2008a): Simultaneous EEG‐fMRI in drug naïve children with newly diagnosed absence epilepsy. Epilepsia 49: 1510–1519. [DOI] [PubMed] [Google Scholar]
- Moeller F, Siebner H, Wolff S, Muhle H, Boor R, Granert O, Jansen O, Stephani U, Siniatchkin M ( 2008b): Changes in activity of striato‐thalamo‐cortical network precede generalized spike wave discharges. Neuroimage 39: 1839–1849. [DOI] [PubMed] [Google Scholar]
- Rodionov R, De Martino F, Laufs H, Carmichael D, Formisano E, Walker M, Duncan J. Lemieux L ( 2007): Independent component analysis of interictal fMRI in focal epilepsy: Comparison with general linear model‐based EEG correlated fMRI. Neuroimage 38: 488–500. [DOI] [PubMed] [Google Scholar]
- Smith S ( 2002): Fast robust automated brain extraction. Hum Brain Mapp 17: 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Jenkinson M, Woolrich M, Beckmann C, Behrens T, Johansen‐Berg H, Bannister P, De Luca M, Drobnjak I, Flitney D, Niazy R, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady J, Matthews P ( 2004): Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23 ( Suppl 1): S208–S219. [DOI] [PubMed] [Google Scholar]
- Srivastava G, Crottaz‐Herbette S, Lau KM, Glover GH, Menon V ( 2005): ICA‐based procedures for removing ballistocardiogram artifacts from EEG data acquired in the MRI scanner. Neuroimage 24: 50–60. [DOI] [PubMed] [Google Scholar]
- Worsley K, Friston K ( 1995): Analysis of fMRI time‐series revisited‐again. Neuroimage 2: 173–181. [DOI] [PubMed] [Google Scholar]
- Worsley K, Liao C, Aston J, Petre V, Duncan G, Morales F, Evans A ( 2002): A general statistical analysis for fMRI data. Neuroimage 15: 1–15. [DOI] [PubMed] [Google Scholar]


